Abstract
G-quadruplexes (G4s) are four-stranded DNA structures formed by Hoogsteen base pairing between stacked sets of four guanines. Pif1 helicase plays critical roles in suppressing genomic instability in the yeast Saccharomyces cerevisiae by resolving G4s. However, the structural properties of G4s in S. cerevisiae and the substrate preference of Pif1 for different G4s remain unknown. Here, using CD spectroscopy and 83 G4 motifs from S. cerevisiae ranging in length from 30 to 60 nucleotides, we first show that G4 structures can be formed with a broad range of loop sizes in vitro and that a parallel conformation is favored. Using single-molecule FRET analysis, we then systematically addressed Pif1-mediated unwinding of various G4s and found that Pif1 is sensitive to G4 stability. Moreover, Pif1 preferentially unfolded antiparallel G4s rather than parallel G4s having similar stability. Furthermore, our results indicate that most G4 structures in S. cerevisiae sequences have long loops and can be efficiently unfolded by Pif1 because of their low stability. However, we also found that G4 structures with short loops can be barely unfolded. This study highlights the formidable capability of Pif1 to resolve the majority of G4s in S. cerevisiae sequences, narrows the fractions of G4s that may be challenging for genomic stability, and provides a framework for understanding the influence of different G4s on genomic stability via their processing by Pif1.
Keywords: DNA helicase, G-quadruplex, Saccharomyces cerevisiae, genomic instability, single-molecule biophysics, DNA-protein interaction, enzyme mechanism, DNA structure, fluorescence resonance energy transfer (FRET), DNA unwinding
Introduction
G-quadruplex (G4)2 structures function in multiple cellular processes, including DNA replication, transcription, and telomere maintenance (1, 2). A high-resolution sequencing-based method has identified 716,310 G4 structures in human genome, highlighting the importance of G4 structure for genome stability (3). In Saccharomyces cerevisiae genome, bioinformatics studies have found enrichment of G4 motifs in upstream promoter regions, open reading frames (ORFs), ribosomal DNA, mitochondrial DNA, and meiotic double-strand breaks (DSBs) (4, 5). The number of intramolecular G4-forming motifs increases significantly with the lengthening of the window size from 30 to 50 nt (143–854 G4s) (4). In a similar study, the overall number of G4 DNA motifs was estimated to be 668 with loop threshold in the range of 5–50 nt (5). However, it is still unknown how many G4 motifs in those predicted sequences from S. cerevisiae genome may actually fold into G4 structures. In addition, their structural properties, including thermal stability and folding conformation, are also to be determined.
Pif1 helicase plays critical roles in preventing replication pausing and DSBs at G4 structures in S. cerevisiae (6–8). With its strong unwinding activity, Pif1 stands out among the helicases that can resolve G4 structures in vivo and in vitro (8–14). The observation that, in Pif1-deficient strain, failure to unravel G4 structures leads to replication-fork impairment, unusual epigenetic silencing, and gross chromosomal rearrangement highlights the essential function of Pif1 in rescuing the genome from the negative effects of G4 (9, 15, 16). By introducing human G4-forming minisatellite CEB1 into S. cerevisiae genome, Piazza et al. (12) discovered that phenanthroline DC (Phen-DC) compounds specifically induce recombination-dependent DNA rearrangement, suggesting that Pif1 may not be able to resolve those stabilized G4 structures. Minisatellites of CEB25 variants with short loop length and high thermal stability have also been shown to trigger genomic instability in S. cerevisiae (17). In those studies, exogenous G4 structures were used to probe the functions of G4s in genomic stability. However, whether Pif1 is capable of resolving all kinds of endogenous G4 structures in S. cerevisiae or whether Pif1 just preferentially unfolds specific G4s is still unknown. Paeschke et al. (16) reported that only 25% of G4 motifs were associated with Pif1 in the S. cerevisiae genome. In addition, several other helicases in S. cerevisiae, including Sgs1, Srs2, Dna2, and Rrm3, have also been discovered to unfold G4 structures (9). This evidence suggests that different helicases may be responsible for processing different G4 structures in S. cerevisiae. Therefore, a systematic study of Pif1 unwinding activity toward various G4 structures in vitro may provide insight into its in vivo G4 substrates in S. cerevisiae.
Besides resolving G4s, Pif1 is also involved in inhibiting telomerase activity at telomeres and DSBs, processing Okazaki fragments, and ribosomal DNA replication in nucleus. Moreover, Pif1 participates in mitochondrial DNA recombination, replication, and maintenance (18), and Pif1 has been indicated to be part of the mitochondrial replisome (19). Some of those functions require Pif1 to translocate on ssDNA and unwind duplex DNA into single strands (20). However, formation of G4 structures may directly affect the activity of Pif1 to unwind the downstream duplex. Therefore, studying the impact of G4 structures on Pif1 activity will be important to understand how G4 structures can modulate the rate of DNA transactions such as mitochondrial DNA replication in S. cerevisiae. We have shown that for a G4-duplex conjugate, at low protein concentration, monomeric Pif1 unfolds G4 structures repetitively in successive runs but without further unwinding the downstream duplex as Pif1 dimerization is required for optimal duplex DNA unwinding (14). At high protein concentration, G4 structure is able to stimulate Pif1-catalyzed downstream duplex unwinding (21). Of note, those experiments were performed in 50 mm NaCl with human telomeric G4 sequence, resulting in a less stabilized G4 structure. In contrast, Pif1 was reported to unfold the parallel c-Myc G4 more slowly relative to unwinding of duplex in 50 mm KCl (22). Mendoza et al. (23) also demonstrated that a stable intramolecular G4 can resist unfolding by Pif1 helicase in 100 mm KCl. Therefore, whether G4 structures with different thermal stabilities or folding conformations can stimulate or impair Pif1 activity is still controversial.
In this study, we first showed that sequences from S. cerevisiae can form G4 structures with a broad range of loop sizes in vitro. They may adopt different conformations, but parallel form is favored. In addition, their thermal stability gradually decreases with increasing G4 motif length. Afterward, we characterized Pif1 unwinding activity toward various G4 structures systematically. We showed that Pif1 is sensitive to the stability of G4, and downstream duplex unwinding is limited by how fast G4 structure is unfolded. In addition, Pif1 preferentially unwinds antiparallel G4s over parallel G4s with similar stability, and conformation is most influential for G4s with very high stability. More importantly, we discovered that Pif1 should be able to efficiently resolve the majority of G4 structures in S. cerevisiae because of their low thermal stability. However, Pif1 demonstrates very poor activity toward G4 structures with short loops as a result of their high stability and parallel conformation, suggesting this kind of G4 may be resolved by other helicases, or additional in vivo factors may assist Pif1 to remove them. The present work sheds new light on the formation and structural properties of G4s in S. cerevisiae and reveals the factors that may influence their removal by Pif1.
Results
Sequences from S. cerevisiae can form G4 structures with a broad range of loop sizes in vitro
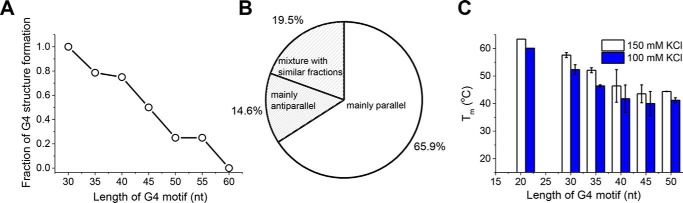
Hershman et al. (4) have predicted the DNA sequences with G4-forming potential in S. cerevisiae genome using a bioinformatics method. Based on their results, we analyzed the features of G4 motifs from 15 to 60 nt. Fig. S1A demonstrates that the number of G4 motifs increases significantly with increasing sequence length. In particular, there are a large number of G4 motifs with a sequence longer than 30 nt. However, it is still unknown how many of those predicted G4 motifs actually fold into G4 structures. To this end, 83 G4 motifs were randomly selected from S. cerevisiae (Table S2) (4), and their folding properties were examined by circular dichroism (CD). It is generally accepted that parallel G4 will generate a characteristic maximum near 265 nm and a minimum near 240 nm. Antiparallel G4 has a distinct spectrum with a maximum near 290 nm accompanied by a minimum near 260 nm (24). In-depth studies further demonstrated that the CD profile depends mainly on the type of stacking, including anti-anti, syn-syn, anti-syn, and syn-anti, between G-tetrads rather than the strand orientations, and quantitative secondary structural information may be obtained by analyzing the CD profile to resolve the components in a mixture of topologies (25, 26). According to previous reports (27–29), besides the parallel conformation with double chain reversal loops, antiparallel with all lateral loops, antiparallel with lateral-diagonal-lateral loops, hybrid with double chain reversal-lateral-lateral loops, and hybrid with lateral-lateral-double chain reversal loops have also been found experimentally.
The CD spectra in Fig. S2 demonstrate that G4 structures can indeed be formed in 41 of those G4 motifs with a broad range of loop sizes. For instance, one single loop can be as long as 20, 27, or more than 30 nt, updating our knowledge about the loop length of G4 structures. We then chose 10 sequences from Fig. S2 and further confirmed the formation of G4 structures with an engineered, structure-specific antibody that binds with high selectivity to G4 structure (30). Fig. S3 shows that although ssDNA fails to bind to the antibody, those G4s with 30–55-nt sequences can bind well to the antibody, indicating G4 structures can indeed be formed, consistent with the results of CD spectra. Next, we determined the fractions of G4 motifs that can fold into G4 structures based on the CD spectra. The number of sequences with G4 formation was divided by the total number of sequences with a specific G4 length in Table S2, generating the folding fraction. Fig. 1A shows that G4 structures are formed in all of the 30-nt G4 motifs; however, the probability of G4 formation decreases gradually as the sequence length increases. For the 45-nt G4 motifs, 50% of the sequences can be folded, whereas none of the sequences are folded once the length reaches 60 nt, indicating that G4 structures with sequences longer than 60 nt may not exist in S. cerevisiae. Fig. S2 also demonstrates that, based on the folding conformation, those G4 structures can be classified as mainly parallel, a mixture of parallel and antiparallel with similar fractions, and mainly antiparallel. Fig. 1B shows that the majority of the sequences adopt a mainly parallel conformation, suggesting that parallel G4 topology is favored in S. cerevisiae. Previous studies reported that parallel G4 structures are only present when the loops are too short to accommodate the antiparallel conformation (24, 31–33). In those studies, the loops were usually pure thymine. Fig. S4 demonstrates that G4 motifs with the same loop length but different loop sequence can indeed fold into different conformational G4 structures, and G4 motifs with poly(T) loops mainly adopt an antiparallel conformation, consistent with previous results (24, 31–33). Therefore, the preference of parallel conformation in S. cerevisiae may be related to loop sequences that are particularly rich in thymine and guanine (Table S2).
Figure 1.
DNA sequences from S. cerevisiae can form G4 structures with a broad range of loop sizes in vitro. A, the fractions of G4 motifs that can form G4 structures decrease with increasing sequence length. B, G4 structures in S. cerevisiae sequences can adopt different conformations, and parallel conformation is favored. C, the stability of G4 structure decreases with increasing sequence length. For each specific length from 30 to 50 nt, two to three sequences in Table S1 were measured by FRET melting. The average Tm and standard deviation values are plotted. Error bars represent S.D. Their stabilities are lower than that of 3G4 with three 3-nt loops (21 nt in G4 motif length).
The thermal stability of those G4 structures has also been characterized by fluorescence resonance energy transfer (FRET) melting assay (34). G4 sequences were modified by donor and acceptor pairs (FAM and TAMRA) at both ends. The unfolding of G4 with increasing temperature will lead to an increased emission intensity in FAM. For each specific G4 length, two or three sequences (Table S1) were selected from the motifs that can fold into G4 structures. Fig. 1C shows that G4 stability decreases as the sequence becomes longer. For 32-3-3 with a 50-nt length, the melting temperature (Tm) is 40.6 °C in 100 mm KCl, which is much lower than the 60.1 °C value for three-layered G4 with 3-3-3 loops (named 3G4) in the same buffer (Table S3). The above results reflect that G4 structures with long DNA sequences in S. cerevisiae may have lower thermal stability.
Pif1 unfolds different G4 structures with different efficiencies
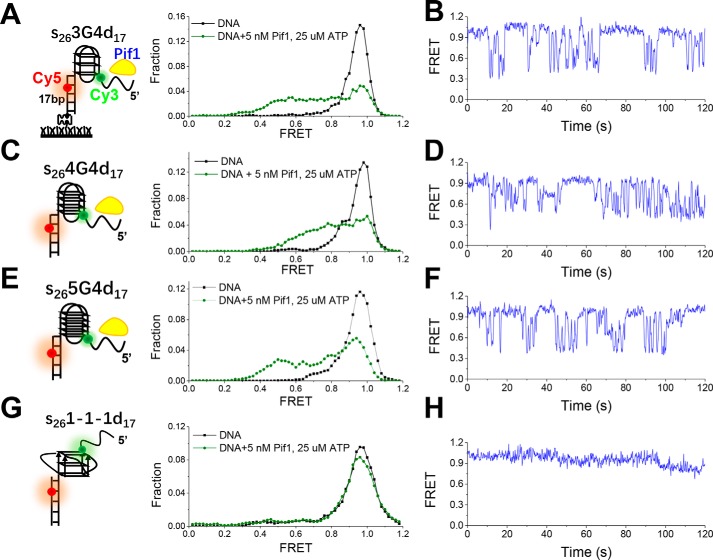
After characterizing the formation and structural properties of G4s in S. cerevisiae sequences, we wanted to address how these structures with different thermal stabilities and conformational diversity are resolved by Pif1. As Paeschke et al. (16) reported that only a portion of G4 motifs were associated with Pif1 in S. cerevisiae, we hypothesized that Pif1 may be sensitive to the structural properties of G4s and preferentially act on specific G4 structures. We first determined whether Pif1 indeed reacts differently toward different G4 structures with varied G-tetrad layers or loop lengths using a fluorescently labeled substrate as described under “Experimental procedures.” The substrate contains a G4 structure with a 5′ tail of 26-nt ssDNA and a 3′ tail of 17-bp dsDNA, mimicking the ongoing synthesis of a leading or lagging strand stalled by G4 structure. The substrate was labeled with Cy3 at the 26th nucleotide from the 5′ end in the G4 strand and with Cy5 at the fifth nucleotide from the 3′ end in the complementary strand (Fig. 2A, C, E, and G, left panels). This spacing of the fluorophores ensured that the FRET signal may sensitively report the conformational change of G4 structure, and the simultaneous disappearance of Cy3 and Cy5 signals would reflect the escape of the unwound DNA from the coverslip.
Figure 2.
Unfolding of different G4 structures by Pif1 in 50 mm NaCl. A, C, E, and G, schematic representations of experimental setup (left panels) and FRET histograms of DNA substrates alone and in the presence of 5 nm Pif1 and 25 μm ATP (right panels). Sequences are listed in Table S1. Each FRET histogram was constructed from more than 300 traces. B, D, F, and H, representative FRET traces for Pif1-catalyzed unfolding of different DNA substrates.
G4 structure in s263G4d17 can be well-formed in 25 mm Tris-HCl, pH 7.5, containing 50 mm NaCl as shown in our previous study (14). Therefore, this buffer condition was adopted in the unfolding study in the beginning. The FRET distribution of s263G4d17 is composed of a major Gaussian peak around 0.95, corresponding to the well-folded G4 structure (Fig. 2A, right panel). In the presence of 5 nm Pif1 and 25 μm ATP, the high EFRET population decreases accompanied by an increase of low EFRET populations, suggesting the disruption of G4 structure by Pif1. The individual FRET trajectory shows several cycles of repetitive G4 unfolding without duplex unwinding, consistent with our previous report (14), as Pif1 monomer is able to resolve the G4 structure, but dimer would be required for optimal duplex unwinding (35, 36). Fig. 2, C–F, demonstrate that a four-layered G4 and a five-layered G4 (named 4G4 and 5G4) can both be disrupted as efficiently as 3G4. It is worth noting that 89.6% of 3G4, 90.8% of 4G4, and 90.6% of 5G4 show the dynamic behavior in 2 min; however, a three-layered G4 named 1-1-1, which has been reported to have a parallel conformation with strong thermal stability (17), was barely disrupted (Fig. 2, G–H). Altogether, the above results clearly indicate that Pif1 responds differently toward different G4 structures.
Unwinding of the downstream duplex is limited by how fast G4 structure is unfolded
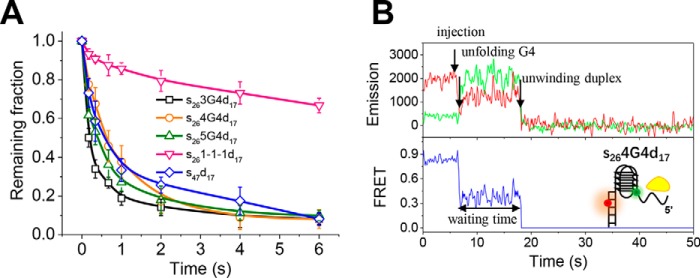
We have shown previously that, in 50 mm NaCl, 3G4 with 3-3-3 loops stimulates Pif1-catalyzed downstream duplex DNA unwinding through reducing the waiting time for Pif1 dimerization at the ss/dsDNA junction (21). Here, we further probed whether the unwinding of downstream duplex DNA is stimulated differently by different G4 structures. To observe the unwinding of duplex DNA, 80 nm Pif1 and 2 mm ATP were introduced to DNA substrates. We first determined the unwinding fractions by counting the number of Cy5 spots over time with various DNA substrates. Fig. 3A demonstrates that the remaining fractions decrease with time for all DNA substrates to a different degree. The differences between the remaining fractions of s263G4d17, s264G4d17, s265G4d17, and the partial duplex s47d17 are not significant within the error of the measurement. However, the unwinding of s261-1-1d17 is much worse than the unwinding of s47d17.
Figure 3.
Pif1-catalyzed downstream duplex DNA unwinding at 80 nm Pif1 and 2 mm ATP. A, fractions of remaining DNA molecules on the coverslip versus time after addition of 80 nm Pif1 and 2 mm ATP in 50 mm NaCl. All lines are the simple connections of the individual data points by Origin 8.0. Error bars represent S.D. B, representative fluorescence emission and FRET traces for Pif1-catalyzed unwinding of s264G4d17. Upon Pif1 and ATP addition as indicated by the arrow, FRET signal decreases and then remains at a constant value with a waiting time for Pif1 dimerization until both Cy3 and Cy5 signals disappear abruptly.
By inspecting individual FRET traces, we found that upon addition of Pif1, the FRET values for s263G4d17, s264G4d17, and s265G4d17 decrease from ∼0.9 to ∼0.3 and then remain at a stable level until the Cy3 and Cy5 signals suddenly disappear simultaneously, reflecting the separation of two DNA strands (Ref. 21, Fig. 3B, and Fig. S5A). In this condition, repetitive G4 unfolding/refolding cannot be observed as in Fig. 2; instead, G4 is kept at linear status until duplex unwinding because once G4 is refolded it will be unfolded rapidly due to the high Pif1 concentration. The time from G4 disruption to final duplex unwinding was designated as the waiting time. During this time, Pif1 undergoes dimerization at the ss/ds DNA junction as a monomeric Pif1 can unfold the G4 structure (13, 14), but a dimer would be required to unwind the duplex optimally (21, 35, 36). For s261-1-1d17, the FRET values of most molecules did not change during the 2-min recording time. The above results suggest that unwinding of the downstream duplex is limited by how fast the G4 structure is unfolded. As the unwinding fractions of duplex DNA with time are convenient to record and quantify, this parameter provides a reliable assessment of how efficiently the upstream G4 structure is unfolded.
Pif1 is sensitive to the thermal stability of G4 structures
As Pif1 unfolds different G4 structures with different efficiencies, we further explored the factors that may influence the unfolding activity of Pif1, including the substrate binding affinity and G4 structural properties such as thermal stability, folding conformation, and loop sizes. First, the DNA binding properties of Pif1 were characterized under equilibrium conditions using fluorescence anisotropy measurements (37). The DNA constructs were the same as those used in the single-molecule FRET experiments but had a FAM fluorophore attached to the 5′ end of the stem strand instead of Cy3 and Cy5 (Fig. S6A, inset). The apparent dissociation constant KD was obtained by fitting the binding curve to the Hill equation. Fig. S6, A–D, and Table S4 show that no major difference can be detected for the binding of Pif1 to the above substrates in all buffer conditions, suggesting that binding affinity may not be responsible for the discrepancy in Pif1 unfolding efficiency.
Afterward, we probed whether thermal stability may lead to the different unfolding efficiencies of Pif1. The Tm values of various G4 structures in different buffer conditions were measured by the FRET melting assay (Fig. S6, E–H) and are summarized in Table S3. These results show that the thermal stability of 1-1-1 is slightly stronger than that of 5G4 but much stronger than those of 4G4 and 3G4, possibly leading to the differences in unfolding.
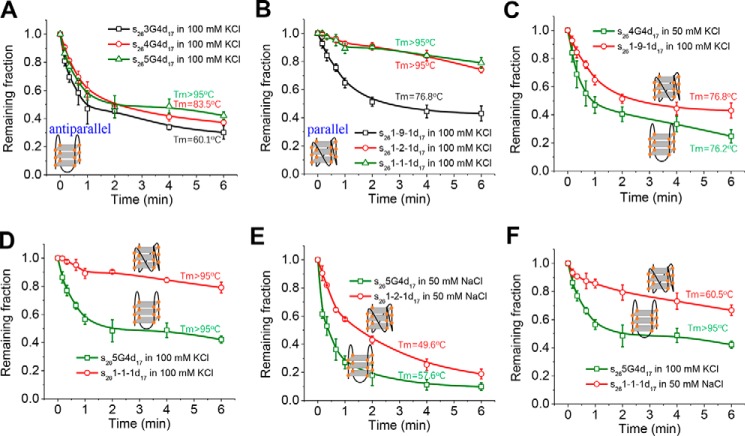
Thereafter, we systematically evaluated how G4 stability may affect Pif1-catalyzed G4 and downstream duplex unwinding. Because the G4 structure is known to be more stabilized by K+ than by Na+ (38), we compared the unwinding in buffers with different ion types and concentrations. Fig. S6, E–H, demonstrates the melting curves of the above mentioned G4 structures in different buffers. In general, their stabilities all increase with the same trend from 50 mm NaCl to 100 mm KCl (Table S3). Next, we prepared all DNA substrates in 50–100 mm KCl and characterized their unwinding at 80 nm Pif1 and 2 mm ATP, which should reflect the unfolding efficiency of the upstream G4. The control experiment in Fig. S7 shows that Pif1 unwinds the partial duplex DNA with similar efficiencies in 50 mm NaCl, 50 mm KCl, and 100 mm KCl, indicating that those buffer conditions should not influence the Pif1 activity toward duplex DNA. However, with the increase in G4 thermal stability, the unwinding of s263G4d17, s264G4d17, and s265G4d17 becomes worse in 50 mm KCl and 100 mm KCl (Fig. 4, A–C), suggesting that Pif1 is sensitive to the stability of G4 structure. For s261-1-1d17, there is a slight decrease in the unwinding fractions in 100 mm KCl, although its unwinding is always poor in all buffers (Fig. 4D).
Figure 4.
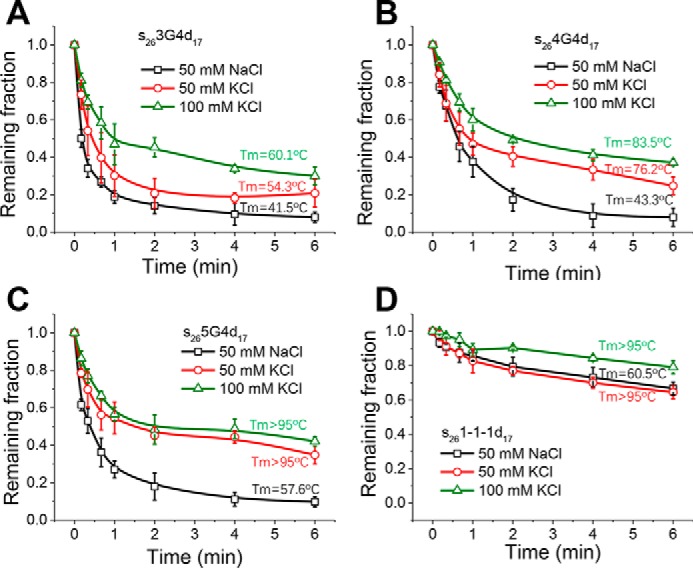
Pif1-catalyzed unwinding of downstream duplex DNA in different buffer conditions at 80 nm Pif1 and 2 mm ATP. A–D, fractions of remaining DNA molecules on the coverslip versus time for s263G4d17 (A), s264G4d17 (B), s265G4d17 (C), and s261-1-1d17 (D). All lines are the simple connections of the individual data points by Origin 8.0. Error bars represent S.D.
Pif1 unfolds antiparallel G4s more efficiently than parallel G4s with similar stability
After characterizing the effects of G4 stability on Pif1 unfolding, we next examined whether Pif1 preferentially unfolds G4s with specific conformations. We measured the CD spectra of s263G4d17, s264G4d17, s265G4d17, and s261-1-1d17 and obtained the CD signal of pure G4s by separating the contribution of the proximal ssDNA and the duplex from the entire DNA substrates (Fig. S8A) as the presence of proximal DNA may affect the folding conformation of G4 (38). Fig. S8, B and C, demonstrate that in either NaCl or KCl, 1-1-1 adopts the parallel conformation; however, 3G4, 4G4, and 5G4 all adopt conformations different from 1-1-1. The CD profiles of 3G4 in NaCl or KCl and 4G4 and 5G4 in NaCl are similar to those of other antiparallel G4s in which each strand is adjacent to two strands oriented in opposite directions (31). The CD spectra of both 4G4 and 5G4 in KCl display a dominant peak at ∼290 nm. However, these spectra differ from the typical antiparallel G4 spectra with a small peak near 270 nm or a diminished valley at 260 nm. These spectra are also different from the hybrid G4 spectrum, which displays a maximum near 290 nm accompanied by a shoulder near 260 nm and a minimum near 245 nm (28). Therefore, besides the antiparallel form, other topologies such as hybrid may also exist, resulting in the complex CD profile. For simplification, here we categorized 3G4, 4G4, and 5G4 as antiparallel G4. However, it should be noted that other conformational forms may also coexist.
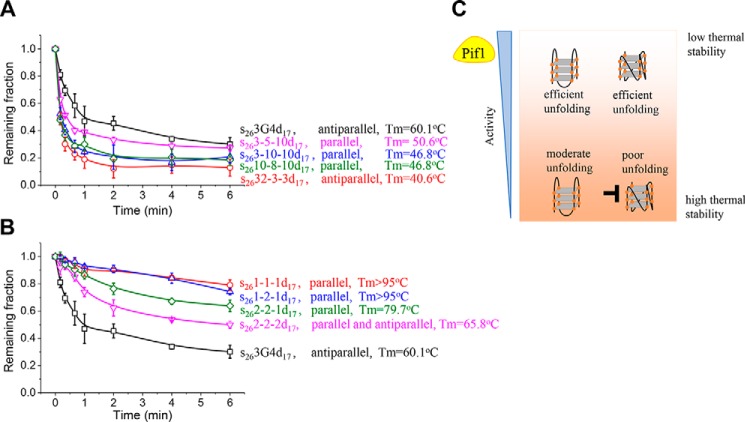
Fig. 5A demonstrates that the unwinding fractions of antiparallel G4s in 100 mm KCl decrease slightly as the G4 stability increases (Tm from 60.1 to above 95 °C). For 1-1-1 and two other three-layered parallel G4s, 1-9-1 and 1-2-1, which were named by their loop sizes (Fig. S8D), with the increase in the Tm value from 76.8 to above 95 °C, there is a remarkable reduction in the unwinding fractions (Fig. 5B).
Figure 5.
Comparison of Pif1-catalyzed duplex DNA unwinding with upstream antiparallel or parallel G4s at 80 nm Pif1 and 2 mm ATP. A, fractions of remaining DNA molecules on the coverslip versus time for substrates with antiparallel G4s in 100 mm KCl. B, fractions of remaining DNA molecules on the coverslip versus time for substrates with parallel G4s in 100 mm KCl. C and D, fractions of remaining DNA molecules on the coverslip versus time with antiparallel or parallel G4s of similar stability. E and F, fractions of remaining DNA molecules on the coverslip versus time with antiparallel G4s more stabilized than parallel G4s. All lines are the simple connections of the individual data points by Origin 8.0. Error bars represent S.D.
To determine whether Pif1 indeed exhibits different abilities to unfold antiparallel G4s and parallel G4s, we made two types of comparisons. The first comparison was the unwinding fractions versus time for antiparallel and parallel G4s with approximate Tm values. Fig. 5C shows that although the stability of 4G4 in 50 mm KCl is fairly close to that of 1-9-1 in 100 mm KCl, Pif1 apparently unwinds s264G4d17 better than s261-9-1d17, suggesting that Pif1 has stronger unwinding activity for antiparallel G4s. In addition, Fig. 5D demonstrates that although their Tm values are both higher than 95 °C, the unwinding of s265G4d17 is much better than that of s261-1-1d17 in 100 mm KCl. The second comparison was the unwinding fractions versus time of antiparallel G4s with higher Tm values than parallel G4s. Fig. 5, E and F, demonstrate that in some circumstances, even if antiparallel G4 is more stabilized, it can still be better unwound by Pif1, strongly indicating that Pif1 unfolds antiparallel G4s more efficiently than parallel G4s.
Pif1 may efficiently unfold majority of G4 structures in S. cerevisiae
Because the G4 motifs in S. cerevisiae genome display a wide range of loop sizes, next we characterized the effects of different G4 loop lengths on Pif1 unfolding efficiency. Therefore, the loop properties of G4 motifs were analyzed based on the prediction (4). The upper limit for the length of G4 motifs was set as 35, 50, and 60 nt. Fig. S1B demonstrates that as the threshold increases, 85–87% of the G4 motifs possess three loops longer than 2-2-2 G4, and 68–76%, i.e. majority of the G4 motifs in S. cerevisiae, possess three loops longer than 3G4. The CD spectra in Fig. S9 show that G4 structures with long loops can also be formed in 100 mm KCl in the presence of proximal duplex and ssDNA (Table S1). In addition, the parallel conformation is favored, consistent with the results for bare G4 motifs.
Afterward, the unwinding of four randomly selected substrates with the G4 motif lengths from 30 to 50 nt was measured at 80 nm Pif1 in 100 mm KCl. Fig. 6A demonstrates that Pif1 unfolds all of these G4 substrates with higher efficiency than 3G4. In addition, the unfolding efficiency increases obviously with decreased G4 stability. For instance, ∼70 and ∼ 80% of the molecules can be unwound in 1 min for the 3-10-10 and 32-3-3 G4 substrates, respectively, highlighting the strong activity of Pif1 on G4 structures with long loops. Therefore, we speculate that Pif1 should be able to unfold the majority of the G4 structures in S. cerevisiae as 76% of the G4 motifs from 15 to 60 nt possess three loops longer than 3 nt. As a result, the folded G4 structures usually have lower thermal stability than 3G4 (Fig. 1C). In addition, these results suggest that conformation may not be a key determinant for the less stabilized G4 structures as those G4s with parallel form can still be removed efficiently by Pif1. However, conformation is most influential for G4 structures with high stability (Fig. 5).
Figure 6.
Effects of G4 loop lengths on Pif1 activity at 80 nm Pif1 and 2 mm ATP in 100 mm KCl. A, fractions of remaining DNA molecules on the coverslip versus time for G4 substrates with three loops no shorter than 3 nt. B, fractions of remaining DNA molecules on the coverslip versus time for G4 substrates with three loops no longer than 3 nt. All lines are the simple connections of the individual data points by Origin 8.0. Error bars represent S.D. For the remaining fractions for s263G4d17, s261-1-1d17, and s261-2-1d17, we reused the results in Fig. 5, A and B, to display the effects of G4 loop lengths on Pif1 unwinding activity. C, summary of the impacts of G4 stability and conformation on Pif1 activity.
Pif1 demonstrates poor activity toward G4 structures with short loops
After characterizing the unfolding of G4 structures with long loops, next we evaluated how Pif1 resolves G4 structures with short loops. Three-layered G4 structures with decreased loop size, including the 3-3-3, 2-2-2, 2-2-1, 1-2-1, and 1-1-1 forms, were chosen. On the one hand, G4 stability becomes stronger with decreasing loop sizes (Table S3 and Fig. 6B), consistent with a previous report (17). On the other hand, after the loops are shortened from 3-3-3 to 2-2-2, the three-layered G4 adopts a mixture of antiparallel and parallel conformations instead of mainly antiparallel (Fig. S8D). Then, with further shortening of the loops into 2-2-1, 1-2-1, and 1-1-1, a single population of parallel G4s is formed. Therefore, loop sizes also have essential effects on the G4 folding conformation. Fig. 6B demonstrates that the unwinding efficiency decreases dramatically with the shortening of G4 loops, most likely due to contributions from both the increased thermal stability and the altered folding conformation.
One may wonder whether the Pif1 unfolding efficiency might be directly influenced by the G4 loop length rather than the folding conformation. Therefore, we compared three-, four-, and five-layered G4 structures with the same loop TA. Notably, although all of these G4 motifs have the same loops, 2-2-2 folds into both antiparallel and parallel conformations (Fig. S8D), whereas 4G4 and 5G4 mainly adopt the antiparallel form (Fig. S8C). Fig. S10 shows that despite their much stronger thermal stabilities, s264G4d17 and s265G4d17 both have better unwinding than s262-2-2d17. Therefore, it is not the loop sizes but the folding conformation that has a direct impact on Pif1 unfolding ability.
Discussion
Pif1 is an important helicase in S. cerevisiae for promoting DNA replication through G4 obstacles. However, it is still unknown what kinds of G4 structures Pif1 may act on. To answer this question, we first investigated the formation and structural properties of G4 structures in S. cerevisiae and then examined the effects of G4 thermal stability, folding conformation, and loop sizes on Pif1 unfolding activity. We discovered that DNA sequences from S. cerevisiae can form G4 structures with a broad range of loop sizes in vitro, and the parallel conformation is favored. Pif1 is sensitive to both the thermal stability and folding conformation of G4 structures. Importantly, our results suggest that Pif1 may be capable of resolving the majority of G4 structures in S. cerevisiae because of their low thermal stability; however, Pif1 is incapable of handling G4 structures with very short loops, which may pose challenges for genomic stability.
Pif1 responds differently to the stability changes of antiparallel and parallel G4 structures in vitro
In this study, we discovered that as the G4 stability increases for each DNA substrate, the unfolding efficiency of Pif1 decreases as shown in Fig. 4. Here, we further determined the relationship between G4 stability and unfolding efficiency via the evaluation of various G4 structures with different sequences for antiparallel G4s and parallel G4s separately as the G4 conformation also impacts its unfolding. Fig. S11 demonstrates that for antiparallel G4s, Pif1 is not very sensitive to stability changes in the low Tm range. Then, the unfolding efficiency of Pif1 decreases abruptly with a further increase in the G4 stability. Afterward, the unfolding efficiency decreases slightly within a broad Tm range. Based on this observation, we speculate that Pif1-catalyzed G4 unfolding can be regulated by subtle changes in G4 stability, either smoothly when acting on the structural stability below a certain threshold or more sharply when the threshold is exceeded.
For parallel G4s, previous studies by extensive mutagenesis of the CEB25 minisatellite motifs revealed that only the variants with very short G4 loops trigger genomic instability in S. cerevisiae (17). The shortening of loops did not change the monomorphic G4 structure of CEB25 variants but drastically increased its thermal stability in correlation with the in vivo instability (17). We clearly demonstrated that for CEB25 G4 (1-9-1 in the current study), shortening of the middle loop (1-2-1 and 1-1-1) will increase the thermal stability of G4 without changing the folding conformation and will finally lead to a significant decrease in the unfolding fractions (Fig. 5B). Therefore, our results provide a direct link between G4 thermal stability and the unfolding efficiency by Pif1 in vitro and may explain the phenotype observed by Piazza et al. (17) that G4 thermal stability is a key determinant for triggering genomic instability by acting as a stable roadblock for the replicative polymerases. In addition to the phenomenon in which increased thermal stability may lead to lower unwinding fractions as shown in Fig. 5B, Fig. S11 demonstrates that Pif1 unfolds 1-1-1 in 50 mm NaCl with an apparently lower efficiency than other parallel G4 structures with even higher stability, suggesting that Pif1 is especially poor at unfolding G4 with three extremely short loops as well as being affected by the thermal stability and conformation.
Pif1 may act on different G4 structures with the same oligomeric state
Pif1 exists as free monomer in the solution, and ssDNA binding can induce dimerization of Pif1 (35). For a G4-duplex conjugate, at low protein concentration, the monomeric Pif1 unfolds G4 structures repetitively in successive cycles but without further unwinding the downstream duplex (Fig. 2). At high protein concentration, Pif1 first unfolds the G4 structure as a monomer and then dwells at the ss/dsDNA junction with a “waiting time.” During this step, Pif1 undergoes dimerization, which is required for optimal duplex DNA unwinding (Fig. 3). We have previously discovered that the waiting time decreases with the increase of Pif1 concentration from 80 to 200 nm (12.3–3.0 s) (21). Therefore, even at very high concentration, Pif1 may also unfold G4 structure as a monomer; otherwise, there would not be the waiting step for protein dimerization.
In the current study, we further discovered that the unwinding behavior of Pif1 is similar on different G4 substrates. For instance, at low protein concentration, Pif1 unfolds the G4 structure with long loops similarly as s263G4d17 (Fig. S12A). In addition, at high Pif1 concentration, G4 substrates with different stability or different conformations all display the similar unwinding phenomenon, characterized by a waiting step before duplex unwinding (Ref. 21, Fig. 3B, Fig. S5A, and Fig. S12, B–D). Therefore, we speculate that Pif1 acts on the different G4 structures with the same oligomeric state.
G4 structure can either stimulate or impede the unwinding activity of Pif1
The waiting time of Pif1 varies from one G4 substrate to another. For instance, at 80 nm Pif1 and 2 mm ATP, the waiting periods of s263G4d17, s264G4d17, and s265G4d17 are 12.3, 10.6, and 10.0 s, respectively (Ref. 21 and Fig. S5, B and C). During this step, G4 structure is linearized into the ssDNA strand, and Pif1 undergoes dimerization at the ss/dsDNA junction. We have previously shown that the G-rich DNA sequence without a G4 motif is also able to accelerate Pif1 dimerization, displaying a shorter waiting time than the poly(T) sequence (21). Therefore, it is likely that the waiting time will gradually decrease with the increase of guanine richness in the DNA sequence (57.1% for 3G4, 72.7% for 4G4, and 76.9% for 5G4).
We have shown previously that 3G4 in 50 mm NaCl can stimulate the downstream duplex unwinding by accelerating Pif1 dimerization on the linearized G4 sequence with the waiting time reduced from 23.3 (poly(T) sequence) to 12.3 s (21). Here, we further demonstrated that the stimulation effect of G4s on Pif1 occurs just under certain conditions, and G4 structures exist as a roadblock for Pif1 in most cases (Figs. 4–6). When the G4 structure is immediately disrupted by Pif1, the reduced waiting time will lead to better unwinding of downstream duplex (21). However, once the G4 structure becomes more stabilized or folds into parallel conformation (Fig. S12, B and C), it cannot be unfolded by Pif1 easily (there is a long period over 10 s before G4 unfolding). As a result, it may exist as an obstacle for Pif1 even though the following waiting step for Pif1 dimerization might be short. Taken together, whether a G4 structure can stimulate or impede the unwinding activity of Pif1 not only depends on the waiting time of Pif1 dimerization but also depends on how long it takes for Pif1 to unfold the G4 structure in the beginning (Fig. S12, E and F).
Potential biological significance of the sensitivity of Pif1 to the thermal stability and conformation of G4 structures
In S. cerevisiae, G4 motifs may fold into G4 structures with diverse conformations and different thermal stabilities. We discovered that Pif1 can unfold G4 structures with low stability very efficiently for both the parallel and antiparallel conformations (Fig. S11), indicating that Pif1 is not very sensitive to the conformation of G4s with low stability (Fig. 6C). However, the contrast between antiparallel and parallel G4s with high stability is significant (Fig. S11), suggesting that conformation is most influential for highly stable G4 structures (Fig. 6C).
Fig. 6A demonstrates that Pif1 efficiently unfolds the four G4 structures with long loops, suggesting that Pif1 may be able to process the majority of G4 structures in S. cerevisiae as 76% of G4 motifs from 15 to 60 nt have three loops longer than 3G4. Therefore, our results highlight the formidable function of Pif1 in unfolding G4 structures with conformational diversity and different thermal stabilities. According to Paeschke et al. (16), only 25% of the G4 motifs in S. cerevisiae were associated with Pif1. Our results suggest that this value might be an underestimate for the binding fractions of G4 structures by Pif1, which has also been suggested by Paeschke et al. (16), as not all G4 motifs in S. cerevisiae can fold into G4 structures (Fig. 1A).
Based on the bioinformatics prediction, G4 motifs with short loops are rare in S. cerevisiae (4). For example, there are only four G4 motifs with three 1-nt loops and one G4 motif with two 1-nt loops and one 2-nt loop (17). As it is very difficult for Pif1 to remove those structures, they may cause a high risk for genomic stability in S. cerevisiae. However, three-layered G4 structures with 1-1-1 and 1-2-1 loops might be efficiently resolved by other helicases such as human RNA helicase associated with AU-rich element (RHAU) (39). Therefore, it is likely that other helicases in S. cerevisiae, including Sgs1, Srs2, Dna2, and Rrm3 (9), might be responsible for resolving those highly stable parallel G4 structures. Alternatively, other in vivo factors may assist Pif1 to remove these structures. Otherwise, they may cause genomic instability by acting as a stable roadblock for the replicative polymerases. According to Piazza et al. (17), there are 1172 G4 motifs with three 1-nt loops among the 2,226 total G4 motifs in Caenorhabditis elegans. In the human genome, 18,153 motifs have three 1-nt loops among the 376,000 predicted motifs. These comparisons suggest that the rarity of short-loop G4 motifs in S. cerevisiae could be related to evolutionary selection as their folded structures may be difficult to resolve by helicases in S. cerevisiae.
Experimental procedures
Buffers
The Pif1 reaction buffer contains 5 mm MgCl2 and 50–100 mm NaCl or KCl in 25 mm Tris-HCl, pH 7.5. For single-molecule measurements, 0.8% d-glucose, 1 mg/ml glucose oxidase (266,600 units/g; Sigma), 0.4 mg/ml catalase (2000–5000 units/mg; Sigma), and 4 mm Trolox were added to the reaction buffer (40).
DNA constructs
All oligonucleotides were purchased from Sangon Biotech (Shanghai, China). The sequences and labeling positions of all the oligonucleotides are listed in Table S1. For the DNA constructs used in single-molecule measurements, DNA was annealed with a 1:2 mixture of the stem and G4 or ssDNA strands by incubating the mixture at 95 °C for 5 min and then slowly cooling down to room temperature in about 7 h. The strand without biotin was used in excess to reduce the possibility of having nonannealed strand anchored at the coverslip surface. The concentration of the stem strand was 5 nm, and all annealing procedures were carried out in annealing buffer containing 50–100 mm NaCl or KCl and 25 mm Tris-HCl, pH 7.5.
Pif1 purification
All of our experiments were performed with S. cerevisiae Pif1, which can be easily expressed and purified in Escherichia coli. The plasmid was a gift from Dr. Zakian. Protein expression and purification were performed essentially according to Boulé and Zakian (41) with minor modifications. Protein expression was induced at 16 instead of 23 °C by addition of 0.3 mm isopropyl 1-thio-β-d-galactopyranoside to a mid-log phase culture.
CD spectropolarimetry
CD experiments were performed with a Bio-Logic MOS450/AF-CD optical system (Bio-Logic Science Instruments, France). A 2 μm solution of G4 sequences from the S. cerevisiae genome in Table S2 was incubated at 95 °C for 5 min and then slowly cooled down to room temperature in about 7 h. CD spectra were recorded in the UV (220–320 nm) regions in 0.75-nm increments with an averaging time of 2 s at 25 °C. A 2.5 μm solution of DNA in Table S1 was annealed with a 1:1 mixture of the stem and G4 strands by incubating the mixture at 95 °C for 5 min followed by slow cooling to room temperature in about 7 h. The CD profile of G4 structure was obtained by subtracting the spectra of the duplex and ssDNA overhangs to separate their contributions as described in a previous study (42).
FRET melting
FRET melting experiments were conducted with the FAM-TAMRA dual-labeled oligomers listed in Table S1 using a Rotor-Gene Q real-time PCR machine (Qiagen) as described in a previous study (34). The oligonucleotides were generally measured at 0.5 μm strand concentration in 25 mm Tris-HCl, pH 7.5, containing different concentrations of NaCl or KCl. The emission of the FAM fluorophore was normalized between 0 and 1, and the Tm was determined as the temperature at which the normalized emission equaled 0.5.
Single-molecule fluorescence data acquisition
Single-molecule fluorescence experiments were performed as described previously (14). Imaging was initiated before Pif1 and ATP were flowed into the chamber. We used an exposure time of 100 ms for all single-molecule measurements at a constant temperature of 22 °C. To determine the fractions of unwound DNA with time, a series of movies were recorded with 1-s duration at different times, and the acceptor Cy5 spots were counted to represent the number of remaining DNA molecules.
FRET data analysis
The FRET efficiency was calculated using IA/(ID + IA) where ID and IA represent the intensity of donor and acceptor, respectively. Basic data analysis was carried out by scripts written in MATLAB, and all data fitting was performed by Origin 8.0. All fluorescent spots in each movie were selected unless the trace showed a poor signal/noise ratio or the intensity changes of the donor and acceptor did not match well. Each FRET histogram was constructed from more than 300 traces.
Equilibrium DNA-binding assay with G4 antibody and Pif1
The plasmid encoding the gene of G4 antibody was a gift from Dr. Balasubramanian. Protein expression and purification were performed essentially according to Biffi et al. (30). The binding of protein to DNA was analyzed by a fluorescence polarization assay using Infinite F200 PRO (Tecan group, Switzerland) at 25 °C. Various amounts of protein were added to a 150-μl aliquot of binding buffer containing 5 nm FAM-labeled DNA. Each sample was allowed to equilibrate in the solution for 5 min after which fluorescence polarization was measured.
Author contributions
L. W. and X.-M. H. data curation; L. W. and X.-M. H. formal analysis; L. W., Q.-M. W., Y.-R. W., X.-G. X., and X.-M. H. investigation; X.-M. H. conceptualization; X.-M. H. supervision; X.-M. H. funding acquisition; X.-M. H. writing-original draft.
Supplementary Material
Acknowledgments
We thank Dr. Ke-Yu Lu and Dr. Fang-Yuan Teng for advice and suggestions on this work and Dr. Wen-Qiang Wu for critical reading of the manuscript.
This work was supported by Chinese Universities Scientific Fund Grant Z109021718 and National Natural Science Foundation of China Grant 11574252. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S12 and Tables S1–S4.
- G4
- G-quadruplex
- DSB
- double-strand break
- ssDNA
- single-stranded DNA
- nt
- nucleotide(s)
- FAM
- fluorescein amidite
- TAMRA
- tetramethylrhodamine
- Tm
- melting temperature.
References
- 1. Murat P., and Balasubramanian S. (2014) Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 25, 22–29 10.1016/j.gde.2013.10.012 [DOI] [PubMed] [Google Scholar]
- 2. Bochman M. L., Paeschke K., and Zakian V. A. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 10.1038/nrg3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chambers V. S., Marsico G., Boutell J. M., Di Antonio M., Smith G. P., and Balasubramanian S. (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 33, 877–881 10.1038/nbt.3295 [DOI] [PubMed] [Google Scholar]
- 4. Hershman S. G., Chen Q., Lee J. Y., Kozak M. L., Yue P., Wang L. S., and Johnson F. B. (2008) Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 36, 144–156 10.1093/nar/gkm986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capra J. A., Paeschke K., Singh M., and Zakian V. A. (2010) G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 6, e1000861 10.1371/journal.pcbi.1000861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Byrd A. K., and Raney K. D. (2017) Structure and function of Pif1 helicase. Biochem. Soc. Trans. 45, 1159–1171 10.1042/BST20170096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chung W. H. (2014) To peep into Pif1 helicase: multifaceted all the way from genome stability to repair-associated DNA synthesis. J. Microbiol. 52, 89–98 10.1007/s12275-014-3524-3 [DOI] [PubMed] [Google Scholar]
- 8. Mendoza O., Bourdoncle A., Boulé J. B., Brosh R. M. Jr., and Mergny J. L. (2016) G-quadruplexes and helicases. Nucleic Acids Res. 44, 1989–2006 10.1093/nar/gkw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paeschke K., Bochman M. L., Garcia P. D., Cejka P., Friedman K. L., Kowalczykowski S. C., and Zakian V. A. (2013) Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 497, 458–462 10.1038/nature12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sauer M., and Paeschke K. (2017) G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 45, 1173–1182 10.1042/BST20170097 [DOI] [PubMed] [Google Scholar]
- 11. Ribeyre C., Lopes J., Boulé J. B., Piazza A., Guédin A., Zakian V. A., Mergny J. L., and Nicolas A. (2009) The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 5, e1000475 10.1371/journal.pgen.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Piazza A., Boulé J. B., Lopes J., Mingo K., Largy E., Teulade-Fichou M. P., and Nicolas A. (2010) Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 38, 4337–4348 10.1093/nar/gkq136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou R., Zhang J., Bochman M. L., Zakian V. A., and Ha T. (2014) Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife 3, e02190 10.7554/eLife.02190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hou X. M., Wu W. Q., Duan X. L., Liu N. N., Li H. H., Fu J., Dou S. X., Li M., and Xi X. G. (2015) Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 466, 189–199 10.1042/BJ20140997 [DOI] [PubMed] [Google Scholar]
- 15. Boulé J. B., and Zakian V. A. (2006) Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 34, 4147–4153 10.1093/nar/gkl561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paeschke K., Capra J. A., and Zakian V. A. (2011) DNA Replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145, 678–691 10.1016/j.cell.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Piazza A., Adrian M., Samazan F., Heddi B., Hamon F., Serero A., Lopes J., Teulade-Fichou M. P., Phan A. T., and Nicolas A. (2015) Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 34, 1718–1734 10.15252/embj.201490702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bochman M. L., Sabouri N., and Zakian V. A. (2010) Unwinding the functions of the Pif1 family helicases. DNA Repair 9, 237–249 10.1016/j.dnarep.2010.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng X., Dunaway S., and Ivessa A. S. (2007) The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion 7, 211–222 10.1016/j.mito.2006.11.023 [DOI] [PubMed] [Google Scholar]
- 20. Ramanagoudr-Bhojappa R., Chib S., Byrd A. K., Aarattuthodiyil S., Pandey M., Patel S. S., and Raney K. D. (2013) Yeast Pif1 helicase exhibits a one-base-pair stepping mechanism for unwinding duplex DNA. J. Biol. Chem. 288, 16185–16195 10.1074/jbc.M113.470013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang B., Wu W. Q., Liu N. N., Duan X. L., Li M., Dou S. X., Hou X. M., and Xi X. G. (2016) G-quadruplex and G-rich sequence stimulate Pif1p-catalyzed downstream duplex DNA unwinding through reducing waiting time at ss/dsDNA junction. Nucleic Acids Res. 44, 8385–8394 10.1093/nar/gkw669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Byrd A. K., and Raney K. D. (2015) A parallel quadruplex DNA is bound tightly but unfolded slowly by Pif1 helicase. J. Biol. Chem. 290, 6482–6494 10.1074/jbc.M114.630749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mendoza O., Gueddouda N. M., Boulé J. B., Bourdoncle A., and Mergny J. L. (2015) A fluorescence-based helicase assay: application to the screening of G-quadruplex ligands. Nucleic Acids Res. 43, e71 10.1093/nar/gkv193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bugaut A., and Balasubramanian S. (2008) A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry 47, 689–697 10.1021/bi701873c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Masiero S., Trotta R., Pieraccini S., De Tito S., Perone R., Randazzo A., and Spada G. P. (2010) A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 8, 2683–2692 10.1039/c003428b [DOI] [PubMed] [Google Scholar]
- 26. Del Villar-Guerra R., Trent J. O., and Chaires J. B. (2018) G-quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew. Chem. Int. Ed. Engl. 57, 7171–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lane A. N., Chaires J. B., Gray R. D., and Trent J. O. (2008) Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 36, 5482–5515 10.1093/nar/gkn517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ambrus A., Chen D., Dai J., Bialis T., Jones R. A., and Yang D. (2006) Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 34, 2723–2735 10.1093/nar/gkl348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lim K. W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D. J., Luu K. N., and Phan A. T. (2009) Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 131, 4301–4309 10.1021/ja807503g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Biffi G., Tannahill D., McCafferty J., and Balasubramanian S. (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186 10.1038/nchem.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hazel P., Huppert J., Balasubramanian S., and Neidle S. (2004) Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 126, 16405–16415 10.1021/ja045154j [DOI] [PubMed] [Google Scholar]
- 32. Rachwal P. A., Findlow I. S., Werner J. M., Brown T., and Fox K. R. (2007) Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 35, 4214–4222 10.1093/nar/gkm316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guédin A., Gros J., Alberti P., and Mergny J. L. (2010) How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 38, 7858–7868 10.1093/nar/gkq639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Cian A., Guittat L., Kaiser M., Saccà B., Amrane S., Bourdoncle A., Alberti P., Teulade-Fichou M. P., Lacroix L., and Mergny J. L. (2007) Fluorescence-based melting assays for studying quadruplex ligands. Methods 42, 183–195 10.1016/j.ymeth.2006.10.004 [DOI] [PubMed] [Google Scholar]
- 35. Barranco-Medina S., and Galletto R. (2010) DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry 49, 8445–8454 10.1021/bi100984j [DOI] [PubMed] [Google Scholar]
- 36. Lu K. Y., Chen W. F., Rety S., Liu N. N., Wu W. Q., Dai Y. X., Li D., Ma H. Y., Dou S. X., and Xi X. G. (2018) Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase. Nucleic Acids Res. 46, 1486–1500 10.1093/nar/gkx1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dou S. X., Wang P. Y., Xu H. Q., and Xi X. G. (2004) The DNA binding properties of the Escherichia coli RecQ helicase. J. Biol. Chem. 279, 6354–6363 10.1074/jbc.M311272200 [DOI] [PubMed] [Google Scholar]
- 38. Hou X. M., Fu Y. B., Wu W. Q., Wang L., Teng F. Y., Xie P., Wang P. Y., and Xi X. G. (2017) Involvement of G-triplex and G-hairpin in the multi-pathway folding of human telomeric G-quadruplex. Nucleic Acids Res. 45, 11401–11412 10.1093/nar/gkx766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tippana R., Hwang H., Opresko P. L., Bohr V. A., and Myong S. (2016) Single-molecule imaging reveals a common mechanism shared by G-quadruplex-resolving helicases. Proc. Natl. Acad. Sci. U.S.A. 113, 8448–8453 10.1073/pnas.1603724113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roy R., Hohng S., and Ha T. (2008) A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 10.1038/nmeth.1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boulé J. B., and Zakian V. A. (2007) The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 35, 5809–5818 10.1093/nar/gkm613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ying L., Green J. J., Li H., Klenerman D., and Balasubramanian S. (2003) Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 100, 14629–14634 10.1073/pnas.2433350100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.