Abstract
The animal circadian timing system interprets environmental time cues and internal metabolic status to orchestrate circadian rhythms of physiology, allowing animals to perform necessary tasks in a time-of-day-dependent manner. Normal progression of circadian rhythms is dependent on the daily cycling of core transcriptional factors that make up cell-autonomous molecular oscillators. In Drosophila, PERIOD (PER), TIMELESS (TIM), CLOCK (CLK), and CYCLE (CYC) are core clock proteins that function in a transcriptional–translational feedback mechanism to regulate the circadian transcriptome. Posttranslational modifications of core clock proteins provide precise temporal control over when they are active as regulators of clock-controlled genes. In particular, phosphorylation is a key regulatory mechanism that dictates the subcellular localization, stability, and transcriptional activity of clock proteins. Previously, casein kinase 1α (CK1α) has been identified as a kinase that phosphorylates mammalian PER1 and modulates its stability, but the mechanisms by which it modulates PER protein stability is still unclear. Using Drosophila as a model, we show that CK1α has an overall function of speeding up PER metabolism and is required to maintain the 24 h period of circadian rhythms. Our results indicate that CK1α collaborates with the key clock kinase DOUBLETIME (DBT) in both the cytoplasm and the nucleus to regulate the timing of PER-dependent repression of the circadian transcriptome. Specifically, we observe that CK1α promotes PER nuclear localization by antagonizing the activity of DBT to inhibit PER nuclear translocation. Furthermore, CK1α enhances DBT-dependent PER phosphorylation and degradation once PER moves into the nucleus.
SIGNIFICANCE STATEMENT Circadian clocks are endogenous timers that integrate environmental signals to impose temporal control over organismal physiology over the 24 h day/night cycle. To maintain the 24 h period length of circadian clocks and to ensure that circadian rhythms are in synchrony with the external environment, key proteins that make up the molecular oscillator are extensively regulated by phosphorylation to ensure that they perform proper time-of-day-specific functions. Casein kinase 1α (CK1α) has previously been identified as a kinase that phosphorylates mammalian PERIOD (PER) proteins to promote their degradation, but the mechanism by which it modulates PER stability is unclear. In this study, we characterize the mechanisms by which CK1α interacts with DOUBLETIME (DBT) to achieve the overall function of speeding up PER metabolism and to ensure proper time-keeping.
Keywords: casein kinase 1, circadian clock, CK1alpha, Drosophila, phosphorylation
Introduction
Circadian clocks regulate molecular oscillations that manifest into physiological and behavioral rhythms in all kingdoms of life, from bacteria to mammals (Zhang and Kay, 2010; Hardin and Panda, 2013; Andreani et al., 2015). Although circadian clock genes are not highly conserved across kingdoms, the design principle of circadian oscillators in all organisms studied to date appears to be variations on the same theme. Transcriptional regulators, the daily mRNA and protein levels of which are tightly controlled, serve as key components of the molecular oscillator and operate through transcriptional–translational feedback loops to control cyclical expression of the circadian transcriptome. In Drosophila melanogaster, these core oscillator proteins are PERIOD (PER), TIMELESS (TIM), CLOCK (CLK), and CYCLE (CYC) (Allada and Chung, 2010; Dubowy and Sehgal, 2017). Starting in late day, CLK and CYC heterodimerize to activate the transcription of per, tim, as well as other clock-controlled genes (McDonald and Rosbash, 2001; Yu et al., 2006; Liu et al., 2017). Although per and tim mRNAs accumulate, PER and TIM protein levels remain low as PER requires TIM binding for stabilization, but TIM is degraded in the presence of light (Hunter-Ensor et al., 1996; Peschel et al., 2009; Hara et al., 2011; Jang et al., 2015). During early night, however, TIM and PER accumulate and enter the nucleus, where they repress the activity of the CLK-CYC heterodimer. In the early morning, proteasome-dependent PER degradation relieves this repression to initiate another round of CLK-CYC-mediated transcription (Grima et al., 2002; Ko et al., 2002).
The abundance, subcellular localization, and transcriptional activity of core clock proteins are critical for normal progression of circadian rhythms. To precisely control their phase-specific functions and to allow for environmental and metabolic input, core clock proteins are heavily regulated by posttranslational modifications (Hirano et al., 2016). Most notably, the progressive phosphorylation of PER over the circadian cycle has been shown to be closely linked to the speed of the circadian oscillator and the 24 h period of circadian rhythms. A number of kinases have been shown to modulate the phosphorylation status of PER, including DOUBLETIME (DBT/CK1δ/ε) (Kloss et al., 1998, 2001; Price et al., 1998), casein kinase 2 (CK2) (Smith et al., 2008; Szabó et al., 2013; Top et al., 2016), SHAGGY (SGG/GSK3β) (Martinek et al., 2001; Ko et al., 2010), and NEMO-like kinase (NLK) (Chiu et al., 2011; Yu et al., 2011). Specifically, DBT first phosphorylates PER in the cytoplasm during the day to prevent premature nuclear entry (Cyran et al., 2005; Muskus et al., 2007). Upon PER nuclear entry in the middle of the night, additional DBT-dependent phosphorylation events potentiate the transcriptional repressor activity of PER while progressively enhancing its proteasome-dependent degradation, which peaks at dawn (Nawathean and Rosbash, 2004; Kivimäe et al., 2008; Top et al., 2018). Whereas DBT-dependent PER phosphorylation at the N terminus promotes binding to the F-box protein SLIMB (β-TrCP), resulting in proteasome-dependent PER degradation (Chiu et al., 2008), phosphorylation of the perS region by DBT creates a phospho-delay mechanism that delays N-terminal phosphorylation (Chiu et al., 2011). The phospho-delay circuit is initiated by NLK-dependent phosphorylation of PER(S596) and has the overall function of extending the PER phosphorylation cycle to 24 h. Finally, CK2 and SGG have both been shown to promote PER nuclear translocation (Lin et al., 2002, 2005; Akten et al., 2003; Ko et al., 2010).
CK1α is a member of the CK1 family along with DBT. It has been identified as one of the targets of Longdaysin, a small-molecule inhibitor that lengthens the period of zebrafish and human U2OS cells (Hirota et al., 2010). Biochemically, CK1α phosphorylates mammalian PER1 and accelerates its degradation in HEK293T cells (Hirota et al., 2010). Although the role of CK1α as a clock kinase has been established, the mechanism by which it modulates PER protein stability is still unclear. Here, we confirmed the role of CK1α in regulating the Drosophila clock and characterized the mechanisms by which CK1α collaborates with DBT to regulate PER function and the circadian oscillator.
Materials and Methods
Transgenic flies and locomotor activity assays.
Targeted expression of ck1α RNAi in tim-expressing and pdf-expressing clock neurons was achieved using the UAS/Gal4 system (Brand and Perrimon, 1993). The Gal4 driver lines used here were w; UAS-dicer2; tim-UAS-Gal4 (TUG) (Blau and Young, 1999) and w;UAS-dicer2; pdf-Gal4 (Park et al., 2000). Four independent responder RNAi lines, two from the Bloomington Drosophila Stock Center (BDSC stock nos. 25786 and 41711) and two from the Vienna Drosophila Resource Center (VDRC stock nos. 110768 and 13664), were evaluated. The regions targeted by the respective responder ck1α RNAi lines are as follows: UAS-ck1α RNAi25786 targets ck1α nucleotide numbers 145–590, UAS-ck1α RNAi110768 targets ck1α nucleotide numbers 299–590, UAS-ck1α RNAi13664 targets ck1α nucleotide numbers 317–670, and UAS-ck1α RNAi41711 targets ck1α nucleotide numbers 1101–1121. Nucleotide numbering corresponds to Flybase transcript variant RF of ck1α. Regions targeted by all responder lines are present in all transcript variants of ck1α. Male flies from the driver lines were crossed to female flies of the UAS-RNAi responder lines. Male progenies of the crosses were then assayed for locomotor activity using the Drosophila Activity Monitoring System (DAMS, Trikinetics) as described previously (Chiu et al., 2010). Control flies for these experiments included all parental lines for the crosses and progenies of the cross between w; UAS-dicer2; tim-UAS-Gal4 (TUG) and the genetic background control w1118 in the case of ck1α RNAi knock-down experiments. Flies were entrained for 4 d in light/dark (LD) cycles (12 h light/12 h dark), followed by 7 d of total darkness (DD) to access their free-running rhythms. Experiments were performed at 25°C in a Percival incubator. To ensure that any changes in oscillator function were not the result of impaired neuronal development, we used tub-Gal80ts to repress the activity of tim-UAS-Gal4 in driving the RNAi construct until adulthood as described previously in McGuire et al. (2003) (BDSC stock no. 7108). Flies were reared at 18°C after genetic crosses. Three days before and also during behavioral assays, the flies were shifted from 18°C to 29°C, thus inactivating tub-Gal80ts and allowing the expression of the RNAi construct in clock neurons.
Drosophila S2 cell culture and transfection to characterize CK1α function.
Drosophila S2 cells and DES expression medium were obtained from Life Technologies. For all cell culture experiments except immunofluorescence staining, S2 cells were seeded at 3 × 106 cells/ml. For immunofluorescence staining, S2 cells were seeded at 2 × 105 cells/ml. Transient transfection was performed using Effectene (Qiagen) according to the manufacturer's protocol. The following amounts of plasmids were used for transfection. For coimmunoprecipitation (co-IP) assays: S2 cells were cotransfected with 0.8 μg of pAc-per-V5-His and 0.8 μg pMT-ck1α-6xc-myc for detection of protein–protein interactions or transfected singly with 0.8 μg of pAc-per-V5-His or 0.8 μg of pMT-ck1α-6xc-myc in control IPs to detect nonspecific binding. For ck1α coexpression, lambda phosphatase (λ-PP), and cycloheximide (CHX) chase assays: 0.8 μg of pAc-per-V5-His was transfected with 0.6 μg of pMT-ck1α-3xFLAG-6xHis (herein referred to as pMT-ck1α-FH), 0.6 μg of pMT-ck1α(K49R)-FH, or an empty vector pMT-FH. For GST pull-down assay: 0.8 μg of pAc-per(NLS)-V5-His was transfected with 0.6 μg of pMT-ck1α-FH, pMT-ck1α(K49R)-FH, or an empty vector. pAc-per(NLS)-V5-His is a tissue culture expression construct in which the nuclear localization sequence (NLS) sequence of simian virus 40 large-T antigen is appended to the C terminus of the per coding sequence to promote nuclear localization of PER. This construct was used previously to characterize the DBT kinase binding domain on PER (Kim et al., 2007; Nawathean et al., 2007). For mobility shift assay: 0.8 μg of pAc-per-V5-His was transfected with either 0.6 μg of pMT-FH, 0.6 μg of pMT-ck1α-FH, 0.2 μg of pMT-dbt-FH, or both 0.6 μg of pMT-ck1α-FH and 0.2 μg of pMT-dbt-FH. For immunofluorescence staining: 1.5 μg of pAc-per-V5-His or pAc-per(NLS)-V5-His was used for transfection. Expression of kinases was induced with 500 μm CuSO4.
CK1α mobility shift assay.
Twenty-four hours following transient transfection of Drosophila S2 cells, expression of kinases was induced for 16 h. For mobility shift assay, cells were harvested and lysed with EB2 (20 mm HEPES pH 7.5, 100 mm KCl, 5% glycerol, 5 mm EDTA, 1 mm DTT, 0.5 mm PMSF, 0.1% Triton X-100, 25 mm NaF, 0.01 mg/ml Aprotinin, 0.005 mg/ml Leupeptin, 0.001 mg/ml pepstatin A). Proteins were quantified using Coomassie Plus Reagent (Thermo Fisher Scientific) in combination with Biophotometer (Eppendorf) and were analyzed by Western blotting (see “Western blotting and antibodies” section).
Co-IP.
Expression of ck1α was induced immediately following transfection (see “Drosophila S2 cell culture and transfection to characterize CK1α function” section for amounts of plasmids used). Following 40 h of induction, cells were harvested, washed once with 1× PBS, and lysed with modified RIPA (50 mm Tris HCl pH 7.5, 150 mm NaCl, 1% NP40, 0.25% Na-deoxycholate) supplemented with 1 mm EDTA, 25 mm NaF, 0.5 mm PMSF, and complete EDTA-free protease inhibitor mixture (Roche). Each IP sample was performed using 15 μl of settled beads for 4 h at 4°C. Beads were washed three times in modified RIPA with 300 mm NaCl. Reciprocal IP was performed. Immune complexes were analyzed by Western blotting (see “Western blotting and antibodies” section).
λ-PP treatment, time course mobility assay, and Phos-Tag SDS-PAGE.
Expression of kinases was induced at 36 h following transfection (see “Drosophila S2 cell culture and transfection to characterize CK1α function” section for plasmid amount). For time course mobility assay, cells were harvested at indicated time points post kinase induction and lysed with EB2 (20 mm HEPES pH 7.5, 100 mm KCl, 5% glycerol, 5 mm EDTA, 1 mm DTT, 0.5 mm PMSF, 0.1% Triton X-100, 25 mm NaF, 0.01 mg/ml aprotinin, 0.005 mg/ml leupeptin, 0.001 mg/ml pepstatin A). For λ-PP, after 24 h of inductions, cells were harvested and lysed with EB2 supplemented with PhosStop (Roche). Protein lysates were then subjected to IP to α-V5 beads (15 μl of settled beads per IP) for 4 h at 4°C. Beads were then washed twice in EB2 without any NaF or PhosStop (Roche) and once in λ-PP buffer (New England Biolabs) before being resuspended in 40 μl of λ-PP buffer. Control reactions were mock treated at the same temperature. Immunocomplexes were analyzed by Western blotting (see “Western blotting and antibodies” section). For Phos-Tag SDS-PAGE, after 24 h of kinase induction, CHX and MG132 were added to a final concentration of 10 μg/ml and 25 μg/ml, respectively, and incubated for 4 h. Cells were then harvested and lysed with EB2 without EDTA. Protein extracts were quantified to ensure that equal amounts were used for the analysis. Proteins were resolved using 5% SDS-PAGE supplemented with 10 μm Phos-Tag (Wako). Once resolved, gels were incubated for 10 min with gentle agitation first in transfer buffer containing 1 mm EDTA, followed by transfer buffer without EDTA. Proteins were then transferred onto PVDF membranes and visualized as described in the “Western blotting and antibodies” section.
CHX chase experiment.
Twenty-four hours following transfection, ck1α expression was induced for 16 h. Following kinase induction, CHX was added to a final concentration of 10 μg/ml. Cells were harvested and lysed with EB2 (see section on “CK1α mobility shift assay”) at the indicated times. Proteins were analyzed by Western blotting using the following antibodies: α-V5 (Life Technologies) at 1:5000 for detection of PER with α-mouse IgG-HRP secondary antibody (GE Healthcare) used at 1:2000; α-HSP70 (Sigma-Aldrich) at 1:3000 for detection of HSP 70 with α-mouse IgG-HRP (GE Healthcare) at 1:3000.
GST pull-down assays in Drosophila S2 cells and in flies.
Pull-down assays were performed as described previously (Chiu et al., 2008). For pull-down assays in flies, prey proteins were extracted from 250 μl of fly heads per reaction.
Analysis of PER protein cycling in flies.
Flies were entrained in light/dark (LD) cycles (12 h light/12 h dark) at 25°C for 3 d and collected at 6 time points over a circadian day. Fly head extracts for Western blotting were homogenized using motorized pestle using RBS (20 mm HEPES pH 7.5, 50 mm KCl, 10% glycerol, 2 mm EDTA, 1% Triton X-100, 0.4% NP-40, 1 mm DTT, 0.5 mm PMSF, 0.01 mg/ml aprotinin, 0.005 mg/ml leupeptin, and 0.001 mg/ml pepstatin A). Extracts were sonicated for 5 × 5 s with 10 s pauses. Proteins were resolved using 5% Criterion Tris-HCl SDS-PAGE gels (Bio-Rad) and loading was assessed using α-HSP70 (Sigma-Aldrich). To classify and quantify hypophosphorylated versus hyperphosphorylated PER isoforms by Western blotting, we used PER isoforms present at ZT8-16 in TUG control flies as reference because PER has been shown to be hypophosphorylated at those circadian time points (Edery et al., 1994). PER signals above the hypophosphorylated PER isoforms present at ZT20, ZT24, and ZT4 were therefore designated and quantified as hyperphosphorylated PER isoforms.
Western blotting and antibodies.
Blocking reagent (5%; Bio-Rad) in 1× TBST was used for incubation with all antibodies except in the case for α-pS47, in which 2.5% blocking solution was used. Primary antibodies for Western blotting analysis were used at the following dilution: mouse α-V5 (Life Technologies) at 1:5000, mouse α-c-myc (Sigma-Aldrich) at 1:7000, mouse α-HSP70 (Sigma-Aldrich) at 1:7000, rat α-HA 3F10 (Roche) at 1:1000, α-PER (GP5620; RRID:AB_2747405) (for detection of PER in fly extracts) at 1:3000, rabbit α-pS47 (Chiu et al., 2008; RRID:AB_2747407) at 1:1000. Secondary antibodies were used as follow: α-mouse-IgG-HRP (GE Healthcare) at 1:2000, α-rat-IgG-HRP (GE Healthcare) at 1:1000, α-rabbit-IgG-HRP (GE Healthcare) at 1:2000, α-guinea pig-IgG-HRP (Sigma-Aldrich) at 1:2000.
Real-time PCR to analyze circadian gene expression and level of ck1α knock-down.
Flies expressing dsRNA targeting ck1α and control flies (parental driver line w; UAS-dicer2; tim-UAS-Gal4; Blau and Young, 1999) were entrained for 3 d in LD cycles and collected every 4 h on the fourth day. RNA extraction was performed as described previously (Chiu et al., 2008). cDNA was generated using Thermoscript RT (Life Technologies) and real-time PCR was performed using SsoAdvanced SYBR green supermix (Bio-Rad) in a CFX96 (Bio-Rad). Primers were designed to detect these circadian transcripts (for per: 5′-GACCGAATCCCTGCTCAA-3′ and 5′-GTGTCATTGGCGGACTTC-3′; for tim: 5′-CCCTTATACCCGAGGTGGAT-3′ and 5′-TGATCGAGTTGCAGTGCTTC-3′; for pdp1ε: 5′-GCGGCAACTGGTAATG-3′ and 5′-ATTTCCTGCCTGAGCT-3′ (Cyran et al., 2005); for vrille: 5′-ATGAACAACGTCCGGCTATC-3′ and 5′-CTGCGGACTTATGGATCCTC-3′; for ck1α: 5′-ATTCTGAGCGGCGGCGTTGG-3′ and 5′-ATGGATGAAGCACTTGAGATGG-3′; for dbt: 5′-AGCAGCACAAGGTCAATGC-3′ and 5′-TCGAGTCGTCCATGTTGAGT-3′) and cbp20 transcripts (5′-GTCTGATTCGTGTGGACTGG-3′ and 5′-CAACAGTTTGCCATAACCCC-3′) were used for normalization. Cycling conditions were 95°C for 30 s, 40 cycles of 95°C for 5 s, followed by an annealing/extension phase for 30 s at the primer optimal annealing temperature. The reaction was concluded with a melt curve analysis going from 65°C to 95°C in 0.5°C increments at 5 s per step. Three technical replicates were performed for each biological replicate. Three biological replicates were performed for analysis of each gene. Data were analyzed using the standard ΔΔCt method and target gene mRNA expression levels were normalized to the reference gene mRNA levels (cbp20), which remain unchanged over a circadian day. Finally, Ct values for all time points were divided by the highest Ct value to generate a scale from 0 to 1.
ChIP.
ChIP was performed as described previously (Kwok et al., 2015).
IP to detect phosphorylated PER(S47) in flies.
IP was performed essentially as described in Chiu et al. (2008) with the following modifications. Approximately 300 μl of fly heads were homogenized using liquid-nitrogen-chilled ceramic mortar and pestle, mixed with 1.5 ml of modified RIPA buffer (see “Co-IP” section for formula) supplemented with complete EDTA-free protease inhibitor mixture (Sigma-Aldrich) and PhosStop (Roche), and dounced with a glass homogenizer (Kimble Chase) for 15 strokes using the tight “B” pestle. Homogenate was then incubated with gentle rotation for 20 min at 4°C. Protein extracts were quantified using a biophotometer (Eppendorf) to ensure that an equal amount of protein extracts was used for each IP. Extracts were incubated with α-PER (GP5620) at 4°C for 4 h, followed by a 2 h incubation with gamma-bind Sepharose beads (GE Healthcare), IP complexes were then washed once with modified RIPA buffer, eluted with 2× SDS sample buffer, and resolved using a 5% Criterion Gel (Bio-Rad).
Immunofluorescence staining of Drosophila S2 cells or clocks neurons.
Drosophila S2 cells were seeded onto glass coverslips coated with 1 mg/ml of the lectin concanavalin A (Con A) and allowed to adhere to coverslips for 4 h before transfection. Twenty-four hours following transfection, cells were fixed with 4% paraformaldehyde for 35 min. Fixed cells were then washed with washing solution (0.2% Triton X-100 in PBS) 6 times (3 quick washes followed by 3 washes at 10 min each) and incubated with blocking solution (5% goat serum and 0.2% Triton X-100 in PBS) at 4°C for 40 min. Mouse α-V5 (Life Technologies) was added at 1:1000 and incubated overnight at 4°C. Cells were washed with washing solution for 6 times at 20 min each and incubated overnight with α-mouse IgG Alexa Fluor 594 (Life Technologies) at 1:1000. Cells were then washed with washing solution 6 times at 20 min each, rinsed with PBS, and mounted onto slides using ProLong Diamond Antifade Mountant with DAPI (Life Technologies). Images were collected with an EVOS FL Imaging System at 20× magnification.
Brain dissections and immunofluorescence staining procedures were as described in Fu et al. (2016). α-PDF (C7-C; DHSB) and α-PER (Rb s3968–1; RRID:AB_2747406) were added at 1:1000 and 1:500, respectively, and incubated for 16 h at 4°C. α-mouse IgG Alexa Fluor 647 (Cell Signaling Technology) and α-rabbit IgG Alexa Fluor (Life Technologies) were added at 1:1000 and 1:500, respectively. Images were collected with an inverted Zeiss Observer LSN710 confocal microscope using 40× oil-based objective and analyzed with Fiji software. Cytoplasmic and nuclear regions were manually selected using the cytoplasmic PDF marker and a nearby area was selected as background. For each neuron, the intensity of PER staining within cytoplasmic and nuclear regions was calculated and average background intensity from the PER channel was subtracted from each. Nuclear PER intensity or cytoplasmic PER intensity was divided by total PER intensity to calculate the fraction of PER in the nucleus or cytoplasm, respectively, at each time point. The nuclear/total value from each neuron was averaged and SEM was calculated for each group. The cytoplasmic/total value was computed similarly. Fifteen large ventral lateral clock neurons from 10 brains were analyzed for each group.
Statistical analysis.
All numerical data described were analyzed using Excel 2016 (Microsoft). Statistical significances were determined using unpaired Student's t test.
Results
CK1α regulates clock-controlled locomotor activity rhythm in Drosophila
To characterize the role of CK1α in regulating the Drosophila circadian clock, we first analyzed the locomotor activity rhythms of transgenic flies expressing dsRNA against ck1α under the control of tim-UAS-Gal4 driver (TUG) (Blau and Young, 1999) to restrict dsRNA expression to tim-expressing cells and clock neurons (TUG>UAS-ck1α RNAi). Following entrainment in LD conditions at 25°C, the free-running circadian periods of parental controls and TUG>UAS-ck1α RNAi (ck1α RNAi) flies were assayed in DD conditions. Two independent responder ck1α RNAi lines from the BDSC (25786 and 41711) and two from the VDRC (110768 and 13664) were evaluated (Table 1). Compared with the parental controls, progenies expressing ck1α dsRNA displayed a lengthening of the endogenous period in locomotor activity rhythm by ∼1.5 to 2 h (Fig. 1A, Table 1). The only exception is the genotype TUG>UAS-ck1α RNAi41711, which exhibited arrhythmic locomotor activity (Table 1). However, because we observed low survival rates for these flies, we suspect that nontarget effect to overall health of the flies might have contributed to the observed arrhythmicity. We therefore opted to use TUG>UAS-ck1α RNAi25786 for subsequent functional characterization as described below.
Table 1.
Daily locomotor activity rhythms of flies with altered ck1α expression level
| Genotypea | Period (h) (mean ± SEM) | Powerb | Rhythmicity (%)c | No. of flies tested | No. of flies survivingd |
|---|---|---|---|---|---|
| RNAi, tim driver | |||||
| w1118 | 23.4 ± 0.06 | 71.1 | 90.3 | 32 | 31 |
| w; UAS-dicer2; tim(UAS)-Gal4(=TUG) | 23.6 ± 0.06 | 75 | 82.1 | 32 | 28 |
| UAS-ck1α RNAi25786 | 23.9 ± 0.10 | 54.4 | 60.0 | 32 | 25 |
| UAS-ck1α RNAi110768 | 22.9 ± 0.11 | 44.4 | 43.8 | 32 | 32 |
| UAS-ck1α RNAi13664 | 23.5 ± 0.04 | 124.6 | 96.9 | 32 | 32 |
| UAS-ck1α RNAi41711 | 23.7 ± 0.06 | 107 | 96.6 | 32 | 29 |
| TUG>w1118 | 23.7 ± 0.05 | 108.7 | 96.9 | 32 | 31 |
| TUG>UAS-ck1α RNAi25786 | 25.8 ± 0.19 | 65 | 66.7 | 32 | 30 |
| TUG>UAS-ck1α RNAi110768 | 24.3 ± 0.12 | 90.5 | 100 | 18 | 18 |
| TUG>UAS-ck1α RNAi13664 | 24.9 ± 0.29 | 88.4 | 96.9 | 32 | 32 |
| TUG>UAS-ck1α RNAi41711 | AR | ND | 0 | 32 | 4 |
| RNAi, pdf driver | |||||
| w; UAS-dicer2; pdf-Gal4 (=pdf) | 24.0 ± 0.07 | 83.3 | 89.7 | 32 | 29 |
| UAS-ck1α RNAi25786 | 23.9 ± 0.16 | 35 | 31.3 | 32 | 32 |
| pdf>UAS-ck1α RNAi25786 | 26.4 ± 0.61 | 47.7 | 19.4 | 32 | 31 |
| RNAi, tim driver in adult flies | |||||
| TUG | 23.4 ± 0.06 | 59.1 | 86.4 | 48 | 44 |
| w; tub-Gal80(ts); UAS-ck1αRNAi25786 | 24.0 ± 0.06 | 87.7 | 81.5 | 32 | 27 |
| TUG>w; tub-Gal80(ts); UAS-ck1αRNAi25786 | 25.9 ± 0.35 | 57.2 | 37.5 | 32 | 16 |
| Overexpression, tim driver | |||||
| TUG(II) | 24.3 ± 0.10 | 58.6 | 53.3 | 32 | 30 |
| UAS-ck1α55067 | 23.7 ± 0.09 | 37.1 | 51.6 | 32 | 31 |
| TUG(II)>UAS-ck1α55067 | 23.9 ± 0.29 | 34.8 | 16.7 | 32 | 30 |
ack1α RNAi and overexpressor transgenic lines were identified using the stock numbers from BDSC (25786, 41711) or VDRC (13664, 110768).
bMeasures the strength or amplitude of the locomotor activity rhythm (in arbitrary units).
cPercentage of flies that are rhythmic.
dNumber of flies that survived until the end of the experiment.
AR, Arrhythmic; ND, not determined.
Figure 1.
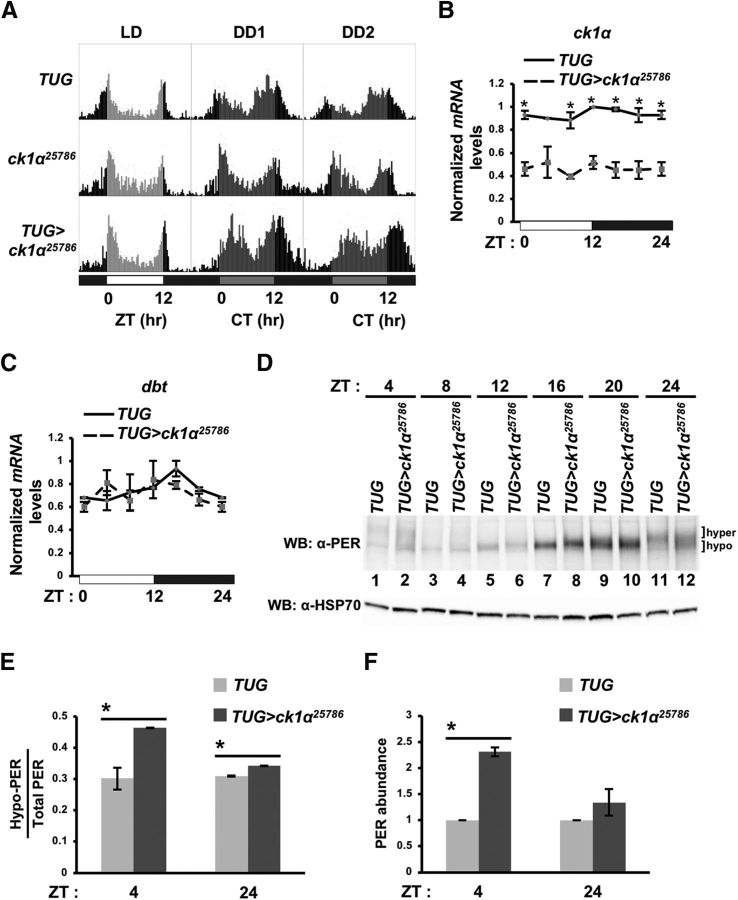
Locomotor activity rhythms and temporal profile of PER phosphorylation are altered in ck1α RNAi flies. A, Flies were entrained for 4 d in LD followed by 7 d in DD (complete darkness). Shown are eduction graphs for LD3, DD1, and DD2 for TUG>ck1α RNAi25786 and parental controls. B, C, Steady-state mRNA levels of ck1α (B) or dbt (C) of TUG control (solid) and TUG>ck1α RNAi25786 (dashed) flies. mRNA level at each time point is normalized to be relative to the maximum value over the time course, which is set to 1. White and black horizontal bars below the x-axis represent periods of lights on and off, respectively. Error bars indicate SEM from two biological replicates. ZT24 data point was plotted twice as ZT0 and 24 to facilitate viewing of daily cycling. *p < 0.05. D, Flies were collected at the indicated times (ZT) on the third day of LD entrainment. Head extracts were analyzed by immunoblotting in the presence of α-PER (GP5620) (top). Equal loading was verified using α-HSP70 (bottom). Hyper and hypo, Hyperphosphorylated and hypophosphorylated PER isoforms, respectively. E, Quantification of hypophosphorylated PER shown in D using ImageLab (Bio-Rad). Following quantification, the amounts of hypophosphorylated PER at ZT4 and ZT24 were presented as a fraction of total PER isoforms. Error bars indicate SEM from two biological replicates. *p < 0.05. F, Total PER abundance at ZT4 and 24 were quantified using ImageLab (Bio-Rad). Error bars indicate SEM from two biological replicates. p < 0.05.
We first validated that the period-lengthening effect observed in TUG>UAS-ck1α RNAi25786 resulted from disrupted oscillators in clock neurons by restricting dsRNA expression to only the pdf-positive neurons (w; UAS-Dicer2; pdf-Gal4>UAS-ck1α RNAi25786), the principle pacemaker cells necessary for sustaining the endogenous rhythm in constant conditions (Helfrich-Förster, 1998). Restricting ck1α dsRNA expression to pdf-positive cells also lengthened the endogenous period (Table 1). The locomotor activity rhythmicities of these flies were also greatly reduced. To exclude the possibility that loss of ck1α function lengthened the endogenous period by altering clock cell development because CK1α is a known component of the Wnt and Hh signaling pathways (Katoh and Katoh, 2007; Chen et al., 2011), we postponed the initiation of ck1α dsRNA expression until animals reached adulthood using the thermogenetic Gal80 system (McGuire et al., 2003). Flies in which ck1α dsRNA was only expressed in adult tim-expressing cells also displayed a period-lengthening of ∼2 h (Table 1). Finally, overexpression of ck1α in tim-expressing cells did not result in period shortening, but substantially reduced the percentage of rhythmic flies (from 52–53% to 17%), suggesting that a high amount of CK1α may also disrupt the circadian oscillator (Table 1). Together, these results not only support CK1α as a regulator of the Drosophila clock, but also suggested that CK1α activity normally functions to accelerate the pace of the endogenous clock in wild-type flies to maintain 24 h circadian rhythms.
Because ck1α shows sequence similarity to dbt (homolog of mammalian ck1δ/ε) and some mutations in dbt, for example, dbtL, exhibit period lengthening in locomotor activity rhythms (Price et al., 1998; Suri et al., 2000), it is important to rule out nontarget effects of ck1α RNAi on dbt. To confirm the specificity of ck1α RNAi knock-down in TUG>UAS-ck1α RNAi25786 flies before using them for further analysis, we assayed both ck1α and dbt transcripts using qPCR. We determined that, whereas ck1α mRNA level was reduced by 40–75% in TUG>UAS-ck1α RNAi25786 flies compared with TUG parental control flies (Fig. 1B; ZT4: t(2) = 2.81, p = 0.106; ZT8: t(2) = 6.76, p = 0.021; ZT12: t(2) = 8.69, p = 0.013; ZT16: t(2) = 7.56, p = 0.017; ZT20: t(2) = 4.88, p = 0.039; ZT24 or ZT0: t(2) = 6.74, p = 0.021), dbt mRNA level was not affected (Fig. 1C; ZT4: t(2) = 1.13, p = 0.377; ZT8: t(2) = 0.38, p = 0.74; ZT12: t(2) = 0.45, p = 0.698; ZT16: t(2) = 1.84, p = 0.207; ZT20: t(2) = 1.61, p = 0.248; ZT24 or ZT0: t(2) = 1.79, p = 0.215).
Temporal profile of PER protein is affected in flies in which ck1α is knocked down
Because CK1α regulates PER1 stability in mammalian cells, we investigated whether the period-lengthening phenotype observed in flies could occur through modulating PER phosphorylation and metabolism over the circadian cycle. We examined the temporal profile of PER extracted from whole heads of TUG (parental control) and TUG>UAS-ck1α RNAi25786 (ck1α RNAi) flies (Fig. 1D). In control flies, PER showed daily oscillations in abundance and phosphorylation that are consistent with published data (Edery et al., 1994), with newly synthesized hypophosphorylated species appearing at ∼ZT8, peaking in abundance at ∼ZT16 to ∼ZT20, attaining maximal phosphorylation at ∼ZT24 to ∼ZT4 and finally by a rapid decrease in protein levels (Fig. 1D, top). In comparison, although PER in ck1α RNAi flies displayed oscillations in abundance and phosphorylation, there are some clear observable differences. First, PER in ck1α RNAi flies exhibited a delay in hyperphosphorylation and degradation that can be clearly observed at ZT24 to ZT4 (Fig. 1D, cf. lane 11 with lane 12 and lane 1 with lane 2). Quantification of PER isoforms at ZT24 and ZT4 showed significantly higher levels of hypophosphorylated PER in ck1α RNAi flies compared with that in control flies (Fig. 1E: ZT4: t(2) = 4.65, p = 0.043; ZT24: t(2) = 18.93, p = 0.0028). Furthermore, quantification of total PER isoforms also showed more abundant PER at ZT4 in ck1α RNAi flies (Fig. 1F: ZT4: t(2) = 14.74, p = 0.0023; ZT24: t(2) = 1.33, p = 0.31). Because DBT is the primary kinase that contributes to PER phosphorylation and mobility shift, as observed in Western blotting (Price et al., 1998; Ko et al., 2010), our results suggest that wild-type CK1α activity normally enhances DBT-dependent PER phosphorylation at these time points when PER is nuclear, thus promoting PER degradation. This is consistent with higher abundance of total PER at ZT24 and ZT4 (Fig. 1D) and the period-lengthening phenotype of ck1α RNAi flies.
Interestingly, upon de novo synthesis of PER in the daytime (ZT12) and progression of the PER protein cycle into the early night (ZT16) when PER is still localized in the cytoplasm, PER isoforms in ck1α RNAi flies exhibited slower mobility (Fig. 1D, cf. lane 5 with lane 6 and lane 7 with lane 8), suggesting that CK1α activity is normally required for inhibiting PER phosphorylation by other kinases in the cytoplasm. As mentioned earlier, because DBT has been identified as the main kinase that phosphorylates PER and leads to an observable mobility shift on Western blots, our results point to the possibility that CK1α may antagonize DBT-dependent PER phosphorylation in the cytoplasm. The loss of ck1α function eliciting opposite effects on PER phosphorylation status that coincides with times in which PER alters its subcellular distribution (Curtin et al., 1995) suggests that CK1α performs distinct regulatory functions on PER depending on its subcellular localization.
CK1α phosphorylates PER in Drosophila S2 cells
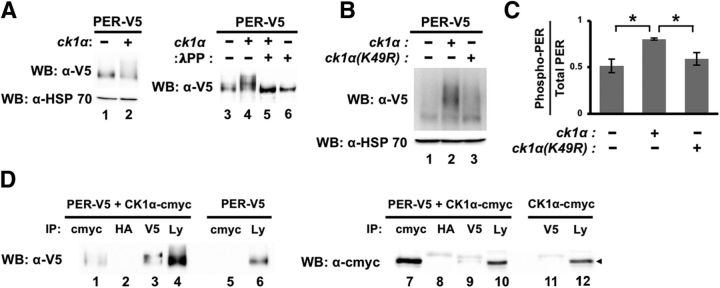
To confirm that PER is a CK1α substrate, we coexpressed PER and CK1α in Drosophila S2 cells and observed slower-migrating PER isoforms that likely represent phosphorylated isoforms (Fig. 2A,B). The slower-migrating isoforms disappeared after treatment with lambda phosphatase, thus confirming that they are indeed phosphorylated isoforms (Fig. 2A, cf. lanes 4–5). To determine whether CK1α kinase activity is required for the observed PER phosphorylation, we analyzed extracts of PER coexpressed with either CK1α or CK1α(K49R), a catalytically “inactive” mutant that is predicted by sequence analysis to have reduced kinase activity. Here, we used Phos-Tag SDS-PAGE to enhance the phosphorylation-dependent mobility shift (Kinoshita et al., 2004, 2006) and, once again, observed prominent slower-migrating PER isoforms in the presence of CK1α (Fig. 2B,C). The amount of slower-migrating PER isoforms was clearly reduced when PER was coexpressed with CK1α(K49R) instead of CK1α(WT) (Fig. 2B,C; PER vs PER+CK1α: t(4) = 3.95, p = 0.0168; PER+CK1α vs PER+ CK1α(K49R): t(4) = 3.16, p = 0.034; PER vs PER+CK1α(K49R): t(4) = 0.795; p = 0.471). Although protein–protein interaction is not a requirement for a kinase to phosphorylate a substrate, we were able to detect interaction of PER and CK1α by reciprocal co-IP when both proteins were coexpressed in S2 cells (Fig. 2D).
Figure 2.
PER is a substrate of CK1α. A, B, Drosophila S2 cells were transfected with pAc-per-V5-6XHis together with an empty plasmid (pMT-FH; FH denotes 3XFLAG-6XHis), pMT-ck1α-FH, or pMT-ck1α(K49R)-FH. Protein extracts were either analyzed directly by Western blotting (A, left), with detection of HSP70 as loading control, or subjected to lambda phosphatase treatment before Western blotting analysis (A, right), or visualized using Phos-Tag SDS-PAGE (B). C, Quantification of phosphorylated PER as shown in B using ImageLab (Bio-Rad). Following quantification, the amount of phosphorylated PER for each condition was presented as a fraction of the total PER isoforms present. Error bars indicate SEM from three biological replicates. *p < 0.05. D, S2 cells were either co-transfected with pAc-per-V5-6XHis and pMT-ck1α-6Xcmyc or singly transfected with pAc-per-V5-6XHis or pMT-ck1α-6Xcmyc. Protein extracts were divided into equal aliquots and each aliquot was independently incubated with α-cmyc beads, α-HA beads, or α-V5 beads (Sigma-Aldrich). Immunocomplexes were analyzed by Western blotting in the presence of the indicated antibody. The inputs (Ly) were also assayed. Arrow on the right panel indicates CK1α. Empty, unloaded lanes are present between lanes 4 and 5 and between lanes 10 and 11.
CK1α regulates PER stability through DBT
Because we observed a delay in PER degradation in ck1α RNAi flies (Fig. 1D), we investigated whether CK1α can directly regulate PER stability. We performed CHX chase assays in S2 cells and examined PER degradation rate in the presence or absence of CK1α coexpression (Fig. 3A). Our results indicate that this does not appear to be the case because PER degradation rate is similar in the absence or presence of CK1α without coexpression of other clock kinases (Fig. 3B).
Figure 3.
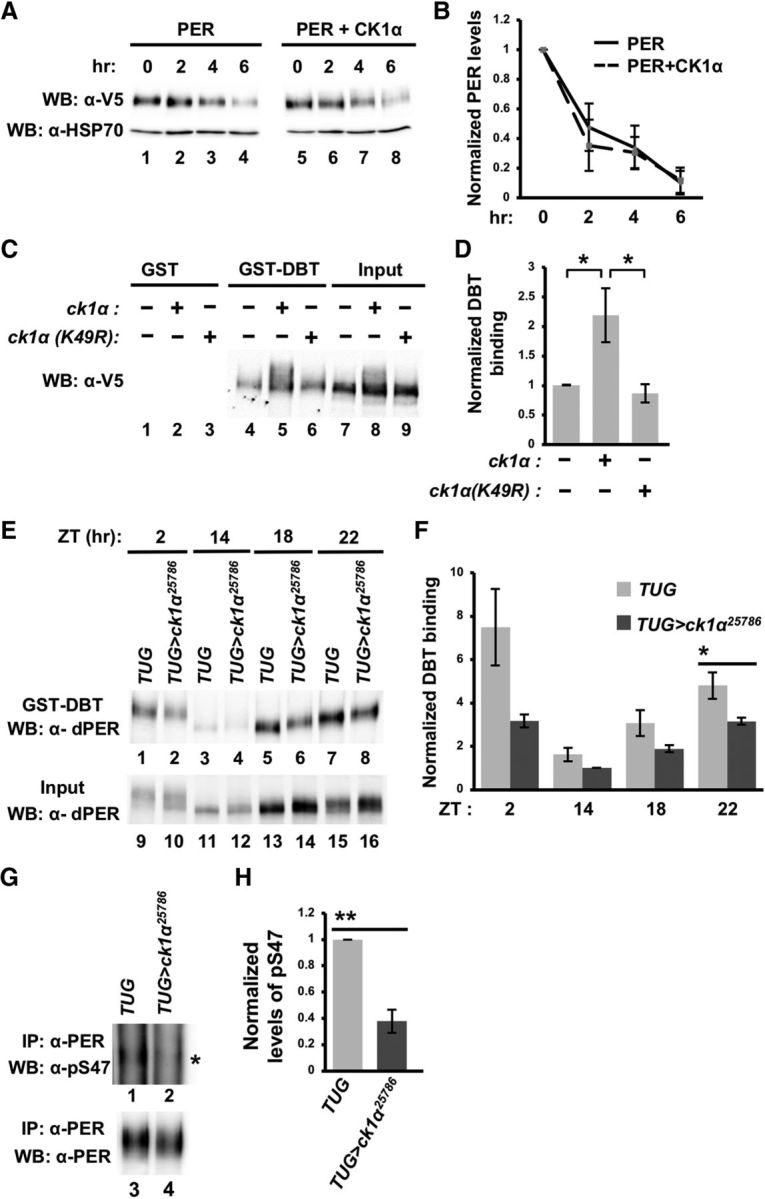
CK1α promotes PER-DBT interactions to regulate PER stability. A, Drosophila S2 cells were cotransfected with pAc-per-V5-6XHis together with either an empty plasmid (pMT-FH; FH denotes 3XFLAG-6XHis) or pMT-ck1α-FH. Following a 24 h incubation period, kinase expression was induced with CuSO4 for 16 h. Following induction, CHX was added and cells were harvested for protein extractions at the indicated times (in hours). Proteins were visualized by Western blotting and detected with α-V5. B, Western blots were quantified using ImageLab (Bio-Rad) and normalized against α-HSP70. Error bars indicate SEM from two biological replicates. C, S2 cells were transfected with pAc-per(NLS)-V5-His together with an empty plasmid (pMT-FH), pMT-ck1α(WT)-FH, or pMT-ck1α(K49R)-FH. Equal amounts of the extracted proteins were then incubated with glutathione resin bound with GST or GST-DBT. Bound PER (lanes 4–6) was eluted with 2× SDS loading buffer and visualized via Western blotting in the presence of α-V5. D, Western blots from C were quantified using ImageLab (Bio-Rad) and normalized against input (lanes 7–9). Values were normalized against the PER-NLS sample (without expression of the kinase), which was set as 1. Error bars indicate SEM from four biological replicates. *p < 0.05. E, Head extracts from TUG control or TUG>ck1α RNAi25786 flies were prepared and aliquots containing equal amount of total protein for each sample were incubated with glutathione beads bound with GST-DBT. The amount of PER in total head extracts (input) and those that are bound to GST-DBT were visualized by immunoblotting in the presence of α-PER (GP5620). F, Quantification of GST-DBT pull-down assays presented in E. Error bars indicate SEM from four biological replicates. *p < 0.05. G, Head extracts from TUG control or TUG>ck1α RNAi flies were immunoprecipitated with α-PER (GP5620) before Western blotting analysis in the presence of α-PER(pS47) (top) and α-PER (GP5620) (bottom). Asterisk indicates the majority of PER(pS47)-containing isoforms (top). H, Western blots from G were quantified using ImageLab (Bio-Rad) and normalized against the amount of immunoprecipitated total PER. Levels of PER(pS47)-containing isoforms in TUG control were set at 1. Error bars indicate SEM from three biological replicates. **p < 0.005.
However, because we observed more hypophosphorylated and abundant PER in ck1α RNAi flies at ZT24 and ZT4, times at which PER is in the nucleus and is targeted for degradation by DBT, we hypothesized that CK1α may promote DBT-dependent PER phosphorylation, leading to PER degradation, even though CK1α does not promote PER degradation on its own. First, we examined DBT-PER interactions by incubating glutathione resin bound with GST-DBT with extracts harvested from S2 cells coexpressing PER-NLS (PER fused to nuclear localization sequence of simian virus 40 large-T antigen) with either CK1α or CK1α(K49R). PER-NLS was used for this experiment to target nuclear-localized PER isoforms. Previous experiments have shown that expression of per in S2 cells without the addition of NLS largely resulted in cytoplasmic-localized PER proteins (Chang and Reppert, 2003; Kim et al., 2007; Nawathean et al., 2007). We observed an enhanced DBT-PER-NLS interaction in the presence of CK1α, suggesting that CK1α-dependent phosphorylation of PER-NLS strengthens the interaction between DBT and PER-NLS (Fig. 3C,D; PER vs PER+CK1α: t(6) = 2.6, p = 0.041; PER+CK1α vs PER+CK1α(K49R): t(6) = 2.73, p = 0.034; PER vs PER+CK1α(K49R): t(6) = 0.85, p = 0.43). We then validated our finding in animals using head extracts of control TUG and ck1α RNAi flies (Fig. 3E,F). Compared with control, ck1α RNAi flies displayed significantly reduced DBT-PER interactions at ZT22 (ZT2: t(6) = 2.41, p = 0.052; ZT14: t(6) = 1.96, p = 0.098; ZT18: t(6) = 1.92, p = 0.10; ZT22: t(6) = 2.64, p = 0.039) even though PER level is higher in ck1α RNAi flies at this time point (Fig. 3E, cf. lanes 15–16).
To determine whether the lower levels of nuclear PER-DBT binding in ck1α RNAi flies correlate to more stable PER, we next compared the phospho-occupancy of serine 47 (pS47) of PER between control and ck1α RNAi flies. DBT-dependent phosphorylation of PER(S47) is a key determinant that regulates its degradation through the proteasome pathway, with phosphorylated S47 acting as a recognition signal for SLIMB, a component of the E3 ubiquitin ligase complex (Chiu et al., 2008). We observed significantly less PER(pS47) signal in ck1α RNAi flies at ZT24 compared with control flies (Fig. 3G,H; t(4) = 7.18, p = 0.002). The increased in PER stability in response to the loss of ck1α function can thereby be attributed to a reduction in nuclear PER-DBT interaction, leading to a decrease in PER(S47) phosphorylation. Together, our data suggest that CK1α phosphorylates PER to promote PER-DBT interactions, leading to DBT-dependent modification of the phosphodegron and subsequent degradation of PER.
Knock-down of ck1α increases PER-dependent repression of clock genes
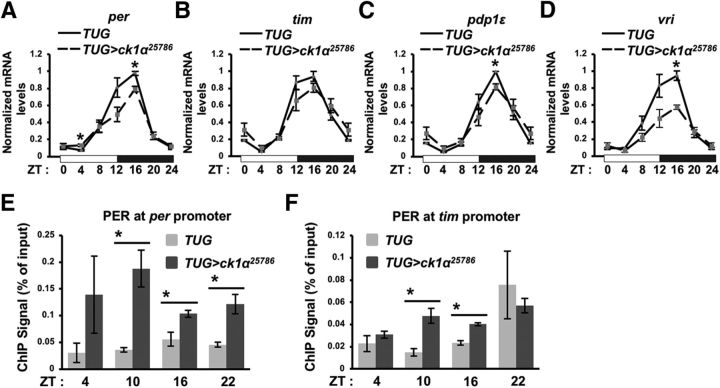
Because CK1α collaborates with DBT to modulate PER stability in the nucleus, we expect that knock-down of ck1α should result in reduced clock gene expression as PER is the key repressor of CLK-CYC activity. A comparison of steady-state mRNA levels of clock genes (per, tim, vri, and pdp1ε) between control and ck1α RNAi flies revealed that ck1α RNAi flies displayed lower levels of circadian transcripts during the transcriptional activation phase of the circadian cycle (Fig. 4A–D) (Fig. 4A; ZT4: t(4) 3.33, p = 0.029; ZT16: t(4) = 4.74, p = 0.009; Fig. 4C; ZT16: t(4) = 5.1, p = 0.007; Fig. 4D; ZT16: t(4) = 5.98, p = 0.0039). This is likely due to the persistence of PER proteins and repression of CLK-CYC activity because of increased PER stability. This conclusion is supported by increased PER occupancy observed at per and tim promoters in ck1α RNAi flies as assayed by ChIP-qPCR (Fig. 4E; ZT4: t(4) = 1.47, p = 0.215; ZT10: t(4) = 4.35, p = 0.012; ZT16: t(4) = 3.2, p = 0.033; ZT22: t(4) = 4.02, p = 0.016) and (Fig. 4F; ZT4: t(2) = 1.03, p = 0.411; ZT10: t(2) = 4.44, p = 0.047; ZT16: t(4) = 7.04, p = 0.02; ZT22: t(2) = 0.602, p = 0.608).
Figure 4.
ck1α knock-down lowers clock gene transcription through increased PER occupancy at clock gene promoters. A–D, Clock gene expression was quantified by real-time PCR and normalized to noncycling cbp20 expression. Values were processed so that the mRNA level at each time point is relative to the maximum value over the time course, which is set at 1. White and black horizontal bars on the x-axis represent lights on and off, respectively. Error bars indicate SEM from three biological triplicates. ZT24 data point was plotted twice as ZT0 and ZT24 to facilitate viewing of daily cycling. Solid and dashed lines represent TUG control and TUG>ck1α RNAi25786 respectively. *p < 0.05. E, F, Chromatin IP analysis of PER occupancy in TUG control and TUG>ck1α RNAi25786. Shown is the amount of ChIP signal on per and tim promoter as measured by qPCR relative to the input at the indicated time points. Error bars indicate SEM from biological replicates (n = 3 for per promoter, n = 2 for tim promoter). *p < 0.05.
CK1α antagonizes DBT-dependent phosphorylation of PER in the cytoplasm
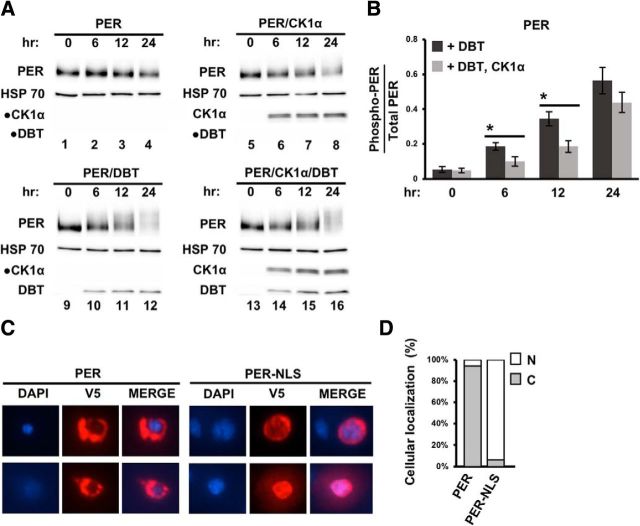
Next, we sought to understand the potential regulatory function of CK1α on cytoplasmic PER because loss of ck1α function in flies appeared to slow down PER mobility at ZT12 and ZT16 (Fig. 1D, cf. lane 5 with lane 6 and lane 7 with lane 8). Because PER mobility shift by SDS-PAGE is predominately influenced by DBT activity, we investigated whether CK1α normally antagonizes DBT activity in the cytoplasm. This could explain the slower-migrating PER isoforms in ck1α RNAi flies that are prominently observed at ZT12 and ZT16. We coexpressed PER with CK1α, DBT, or both kinases in Drosophila S2 cells and monitored PER phosphorylation status following kinase induction (Fig. 5A,B). PER lacking NLS will largely localize to the cytoplasm upon induction in S2 cells, allowing us to examine the effect of CK1α on cytoplasmic PER (Fig. 5C,D) (Saez and Young, 1996; Chang and Reppert, 2003; Nawathean and Rosbash, 2004; Cyran et al., 2005; Kim et al., 2007; Nawathean et al., 2007). In the presence of CK1α, cytoplasmic PER exhibited a delay in DBT-dependent phosphorylation that was significant at 6 and 12 h after kinase induction (Fig. 5A, cf. bottom two panels) (Fig. 5B, 0 h: t(6) = 0.27, p = 0.795; 6 h: t(6) = 2.57, p = 0.042; 12 h: t(6) = 3.07, p = 0.022; 24 h: t(6) = 1.31, p = 0.237). Our results support the hypothesis that CK1α antagonizes DBT kinase activity on PER in the cytoplasm. CK1α inhibition of DBT-dependent PER phosphorylation could be the result of a kinase cascade on PER or a reduction of DBT activity due to CK1α phosphorylation of DBT. Our current data cannot rule out either scenario at this time.
Figure 5.
CK1α antagonizes DBT activity on cytoplasmic PER. A, Drosophila S2 cells were transfected with pAc-per-V5-His together with one of the following: no kinase plasmids; pMT-ck1α-FH only, FH denotes 3XFLAG-6XHis; pMT-dbt-V5–6XHis only; or pMT-ck1α-FH and pMT-dbt-V5–6XHis. Cells were harvested and proteins extracted at the indicated times (in hours) post kinase induction. PER, DBT, and CK1α proteins were visualized by Western blotting in the presence of α-V5 or α-FLAG antibodies. Detection of α-HSP70 represents loading control. Circular dots indicate blots without any signal detected as expected. B, Quantification of phosphorylated PER (phospho-PER) using ImageLab (Bio-Rad). Following quantification, the number of phospho-PER isoforms for each time point was presented as a fraction of total PER isoforms present. Error bars indicate SEM from four biological replicates. *p < 0.05. C, Immunofluorescence of PER in S2 cells. Left, Nuclear staining using DAPI. Middle, PER staining using α-V5; (right) merged image showing localization of PER or PER-NLS. Top and bottom, Representative images of different S2 cells. D, The subcellular localization of PER and PER-NLS were determined and expressed as the percentage of total number of cells analyzed per construct (n = 34). N, Nuclear; C, cytoplasmic localization.
CK1α promotes nuclear entry of PER
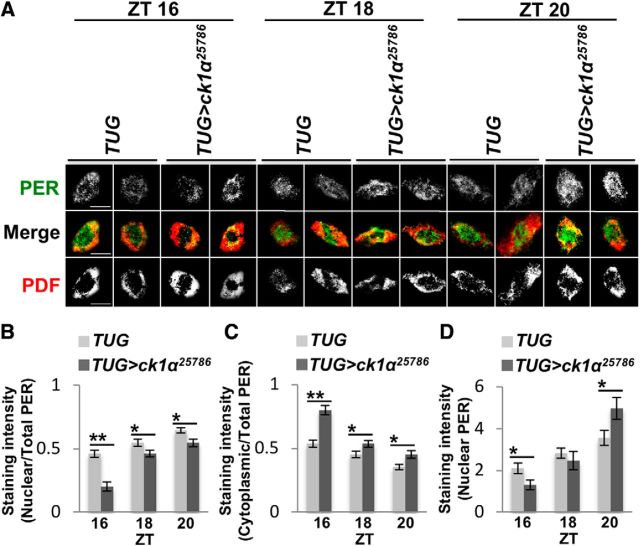
Given that DBT was shown to prevent premature PER nuclear entry in flies (Cyran et al., 2005; Muskus et al., 2007) and our data suggest that CK1α can antagonize DBT-dependent PER phosphorylation in the cytoplasm, we proceeded to determine whether knock-down of ck1α in tim-expressing cells can slow down PER nuclear translocation. This could also contribute to the period-lengthening phenotype of ck1α RNAi flies. Immunostaining of clock neurons (l-LNvs) revealed that nuclear translocation of PER was indeed delayed in ck1α RNAi flies compared with control (TUG) flies (Fig. 6A); we observed a significantly smaller fraction of PER in the nucleus in ck1α RNAi flies at ZT16 (∼20%) compared with control flies at the same time point (∼46%) (Fig. 6B; ZT16: t(28) = 5.65, p = 0.0000046; ZT18: t(28) = 2.32, p = 0.03; ZT20: t(28) = 2.7, p = 0.01). This difference in nuclear PER localization between the two genotypes was diminished at ZT18 and ZT20, suggesting that CK1α kinase is involved in regulating the timing of nuclear PER translocation. The fraction of PER located in clock neuron cytoplasm was also computed and the results supported the same conclusion (Fig. 6C; ZT16: t(28) = 5.65, p = 0.0000046; ZT18: t(28) = 2.32, p = 0.03; ZT20: t(28) = 2.7, p = 0.01). Finally, we investigated the amount of nuclear PER, expressed as pixel intensity, and observed that it was significantly higher in ck1α RNAi flies at ZT20 even though our results indicated that PER nuclear translocation was delayed in these flies compared with control flies (Fig. 6D; ZT16: t(28) = 2.39, p = 0.02; ZT18: t(28) = 0.72, p = 0.48; ZT20: t(28) = 2.2, p = 0.04). This can be explained by our observation that PER in ck1α RNAi flies is more stable than in control flies upon entry into the nucleus. Together, our findings support that CK1α promotes the nuclear entry of cytoplasmic PER and degradation of nuclear PER and therefore has an overall function of speeding up the circadian oscillator.
Figure 6.
CK1α promotes nuclear entry of PER. A, Confocal images of brains from adult TUG control and ck1α RNAi25786 flies stained with α-PER Rb s3968-1 (green) and α-PDF (red) in l-LNv clock neurons. Scale bars are shown on the left-most panels only and indicate 10 μm. Flies were entrained for 3 d in LD cycles and collected at the indicated times on LD3 for fixation and immunostaining. Duplicate panels are shown for each condition. B, Bar graph displaying the fraction of PER in the nucleus presented as nuclear PER intensity divided by total intensity of PER. C, Bar graph displaying the fraction of PER in the cytoplasm presented as cytoplasmic PER intensity divided by total intensity of PER. D, Bar graph showing the amount of nuclear PER expressed as pixel intensity (in thousands). Error bars indicate SEM of 15 neurons from 10 brains. *p < 0.05, **p < 0.005.
Discussion
CK1α was previously identified as a clock kinase in mammalian cells via a high-throughput chemical screen (Hirota et al., 2010) and was shown to regulate the clock through modulation of PER1 stability. However, the mechanisms by which CK1α regulates PER metabolism and in turn the circadian oscillator remain unclear. Using the Drosophila model, we provided further support for a role of CK1α in animal clocks and, more importantly, provided additional insights into the mechanisms by which CK1α acts in coordination with DBT in both the cytoplasm and the nucleus to regulate the function of the molecular oscillator. First, our analysis using Drosophila S2 cell culture demonstrated the ability of CK1α to phosphorylate PER. This was corroborated in whole animals where RNAi knock-down of ck1α in tim-expressing cells affected the phosphorylation program of PER in vivo. Knock-down of ck1α by RNAi led to period-lengthening of locomotor activity rhythms in flies, strongly suggesting that CK1α functions to speed up the clock in wild-type animals to maintain 24 h rhythms.
Similar to DBT (CK1δ/ε), another CK1 isoform, our results suggested that CK1α plays multiple regulatory roles in the molecular oscillator. Due to the period-lengthening phenotype of ck1α RNAi flies, we initially hypothesized that CK1α promotes PER degradation to accelerate the pace of the clock in wild-type animals independently of DBT. Indeed, reducing ck1α expression in flies using RNAi knock-down seemed to produce more stable PER at ZT24 to ZT4, when PER should be degrading rapidly. Further analysis using both tissue culture cells and fly extracts revealed that the increase in PER stability upon reduction of CK1α activity resulted primarily from weakened interactions between PER and DBT. This consequently led to the reduction of PER(S47) phosphorylation, which is known to promote proteasomal-dependent PER degradation. Our data are consistent with previous results demonstrating a negative correlation between the interaction of PER and DBT and the stability of PER (Kim et al., 2007; Nawathean et al., 2007). PER proteins lacking the DBT-binding domain remain hypophosphorylated and at constant high levels over the circadian cycle. Interestingly, CK1α was observed to elicit these effects only during the latter part of the circadian cycle, suggesting that CK1α regulates the turnover of nuclear PER.
In addition to its role on PER degradation, we also demonstrated that CK1α has a role in the cytoplasm to promote PER nuclear localization by antagonizing the activity of DBT. Although DBT has been proposed to play a role in regulating PER nuclear translocation since dbt mutants were first characterized (Price et al., 1998) and the gene that encodes the kinase was first cloned (Kloss et al., 1998), whether its role is stimulatory or inhibitory is not always unequivocal. Evidence that supports the stimulatory role of DBT with regard to PER nuclear translocation mostly come from experiments using Drosophila S2 tissue culture (Nawathean and Rosbash, 2004; Nawathean et al., 2007). Nawathean and Rosbash (2004) knocked down endogenous dbt in S2 cells using dsRNA and observed a significant reduction in the nuclear accumulation of PER and a substantial decrease in transcriptional repressor activity. This prompted them to suggest that DBT may promote PER nuclear localization, although they proposed that DBT likely plays a greater role in activating the repressor function of PER, whereas its regulation on PER nuclear translocation may be an indirect consequence of the association of active PER and DNA. In a follow-up study, Nawathean et al. (2007) characterized a PER variant containing a deletion in what is now termed the DBT binding domain of PER (PERΔ). They observed that this PER variant is resistant to DBT-dependent phosphorylation and is highly stable, but exhibited a low level of nuclear accumulation and repressor activity when expressed in S2 cells. Although they performed rescue experiments in flies and determined that PERΔ was not able to rescue the locomotor activity rhythm of per0 mutants, they did not examine PER subcellular localization in PERΔ fly brains. Nevertheless, they regarded this as further support for their previous conclusion in linking DBT-dependent phosphorylation to promotion of PER repressor activity. Their additional studies on the PERΔ also provided them with more confidence to suggest that DBT actively phosphorylates PER in the cytoplasm to promote nuclear translocation.
Conversely, evidence supporting the inhibitory role of DBT on PER nuclear entry mostly originated from in vivo fly studies in which various dbt mutants were characterized. First, analysis of the strongly hypomorphic dbtP mutant containing a P-element insertion showed that significant reduction in DBT activity resulted in a lack of daily rhythmic PER nucleocytoplasmic shuttling, with PER appearing to be constitutively nuclear in clock neurons (Kloss et al., 1998; Price et al., 1998). Second, characterization of dbtAR, another hypomorphic dbt mutant with a histidine to tyrosine point mutation at amino acid 126 (Rothenfluh et al., 2000), demonstrated that PER can translocate to the nucleus even in the absence of TIM (i.e., in tim0 mutant), but only when DBT activity is severely dampened (Cyran et al., 2005). This suggest that perhaps DBT acts to prevent premature nuclear entry of PER in wild-type flies in the early part of the circadian cycle before the accumulation of TIM proteins. Finally, nuclear PER was observed in mutant flies expressing kinase-inactive DBT (dbtK/R) during periods in which PER would be cytoplasmic (e.g., early nights), thus supporting an inhibitory function of DBT on PER nuclear localization (Muskus et al., 2007). Given that S2 cell culture does not possess a cycling molecular oscillator or fully recapitulate in vivo physiological condition, it is perhaps logical to put more weight on the inhibitory role of DBT supported by fly studies.
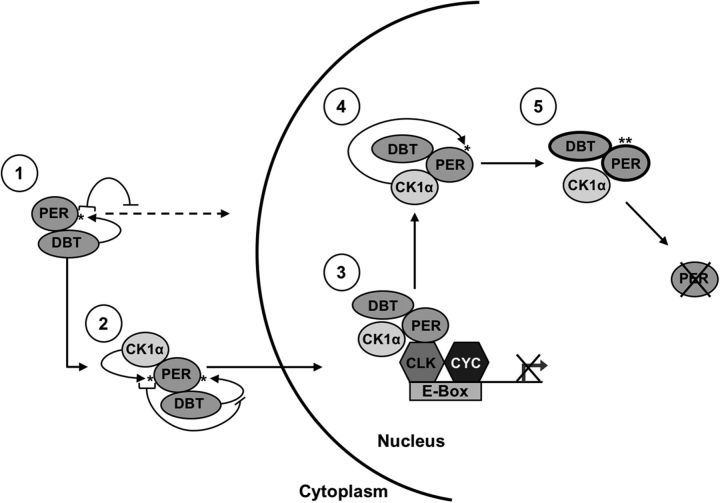
Significantly, our study adds new supporting data to the inhibitory role of DBT activity on PER nuclear translocation. Integrating our results with published data regarding the effects of DBT-dependent PER phosphorylation, we propose the following model (Fig. 7). Starting in late day, PER levels in the cytoplasm begin to accumulate in response to heterodimerization with, and stabilization by TIM (Kloss et al., 1998; Price et al., 1998; Ko et al., 2002). DBT-dependent phosphorylation temporarily retains PER in the cytoplasm to prevent premature entry of PER into the nucleus to initiate transcription repression of circadian genes (Cyran et al., 2005). During early night, CK1α phosphorylates PER to antagonize DBT-dependent PER phosphorylation, thereby promoting PER nuclear entry. At the moment, we cannot rule out the alternative hypothesis in which CK1α phosphorylates DBT to reduce its kinase activity and phosphorylation of PER. Once localized to the nucleus, PER initiates the repression phase of circadian transcription by inhibiting CLK-CYC activity. Toward late night transitioning into early day, CK1α phosphorylates nuclear PER, which promotes or strengthens interactions between PER and DBT. This eventually leads to DBT-dependent phosphorylation of PER(S47), which promotes PER-SLIMB interaction and PER degradation via the proteasome pathway.
Figure 7.
Model describing the proposed function of CK1α in the circadian oscillator. Step 1: In the cytoplasm, DBT phosphorylates PER to prevent premature nuclear entry of PER. Step 2: During times of nuclear translocation, CK1α phosphorylates PER to inhibit its DBT-dependent phosphorylation, therefore favoring nuclear entry of PER. Step 3: Upon entry into the nucleus, PER in complex with multiple kinases and then interacts with and represses the transcriptional activity of CLK-CYC heterodimer on circadian promoters. Not all kinases in the complex are shown. Steps 4 and 5: Finally, in preparation for the new circadian cycle, nuclear PER is phosphorylated by CK1α, which enhances/stabilizes PER-DBT interaction to promote phosphorylation of PER in the phosphodegron and its subsequent degradation. Line width and asterisks denote strength of interactions and phosphorylation events, respectively.
Future experiments to determine whether the various regulatory roles of CK1α are conserved in mammalian clock will be important to understand the full extent of the effects of the CK1α-targeting Longdaysin (Hirota et al., 2010), especially if this chemical compound were to be considered as a therapeutic option for circadian disorders. Our results highlighting the multiple roles of CK1α and its coordination with DBT in regulating PER proteins are in congruence with the observation that small molecules such as Longdaysin that target multiple kinases of the same substrate due to their similarities in active sites often produce the largest period effects and may represent the most effective therapeutic compounds (Hirota et al., 2010).
Footnotes
This work was supported by the National Institutes of Health (Grant R01GM10225 to J.C.C.) and the National Science Foundation (Grant IOS1456297 to J.C.C.). We thank Dr. Michael Paddy (UC Davis) and Dr. Yong Zhang (University of Nevada, Reno) for technical advice in immunofluorescence and confocal imaging; Paul Hardin for providing the pdf-Gal4 fly line; and the Bloomington Drosophila stock center and VDRC for supplying ck1α RNAi responder and overexpressor fly lines.
The authors declare no competing financial interests.
References
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR (2003) A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 6:251–257. 10.1038/nn1007 [DOI] [PubMed] [Google Scholar]
- Allada R, Chung BY (2010) Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72:605–624. 10.1146/annurev-physiol-021909-135815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani TS, Itoh TQ, Yildirim E, Hwangbo DS, Allada R (2015) Genetics of circadian rhythms. Sleep Med Clin 10:413–421. 10.1016/j.jsmc.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW (1999) Cycling vrille expression is required for a functional Drosophila clock. Cell 99:661–671. 10.1016/S0092-8674(00)81554-8 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a mean of altering cell fates and generating dominant phenotypes. Development 118:401–415. [DOI] [PubMed] [Google Scholar]
- Chang DC, Reppert SM (2003) A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr Biol 13:758–762. 10.1016/S0960-9822(03)00286-0 [DOI] [PubMed] [Google Scholar]
- Chen Y, Sasai N, Ma G, Yue T, Jia J, Briscoe J, Jiang J (2011) Sonic Hedgehog dependent phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of Smoothened. PLoS Biol 9:e1001083. 10.1371/journal.pbio.1001083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Vanselow JT, Kramer A, Edery I (2008) The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev 22:1758–1772. 10.1101/gad.1682708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Low KH, Pike DH, Yildirim E, Edery I (2010) Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. J Vis Exp 43:2157. 10.3791/2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Ko HW, Edery I (2011) NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145:357–370. 10.1016/j.cell.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin KD, Huang ZJ, Rosbash M (1995) Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14:365–372. 10.1016/0896-6273(95)90292-9 [DOI] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J (2005) The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci 25:5430–5437. 10.1523/JNEUROSCI.0263-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy C, Sehgal A (2017) Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205:1373–1397. 10.1534/genetics.115.185157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M (1994) Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A 91:2260–2264. 10.1073/pnas.91.6.2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Murphy KA, Zhou M, Li YH, Lam VH, Tabuloc CA, Chiu JC, Liu Y (2016) Codon usage affects the structure and function of the Drosophila circadian clock protein PERIOD. Genes Dev 30:1761–1775. 10.1101/gad.281030.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chélot E, Papin C, Limbourg-Bouchon B, Rouyer F (2002) The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420:178–182. 10.1038/nature01122 [DOI] [PubMed] [Google Scholar]
- Hara T, Koh K, Combs DJ, Sehgal A (2011) Post-translational regulation and nuclear entry of TIMELESS and PERIOD are affected in new timeless mutants. J Neurosci 31:9982–9990. 10.1523/JNEUROSCI.0993-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE, Panda S (2013) Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol 23:724–731. 10.1016/j.conb.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. (1998) Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol A 182:435–453. 10.1007/s003590050192 [DOI] [PubMed] [Google Scholar]
- Hirano A, Fu YH, Ptáček LJ (2016) The intricate dance of post-translational modifications in the rhythm of life. Nat Struct Mol Biol 23:1053–1060. 10.1038/nsmb.3326 [DOI] [PubMed] [Google Scholar]
- Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, Garcia M, Peters EC, Etchegaray JP, Traver D, Schultz PG, Kay SA (2010) High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol 8:e1000559. 10.1371/journal.pbio.1000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter-Ensor M, Ousley A, Sehgal A (1996) Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84:677–685. 10.1016/S0092-8674(00)81046-6 [DOI] [PubMed] [Google Scholar]
- Jang AR, Moravcevic K, Saez L, Young MW, Sehgal A (2015) Drosophila TIM binds importin α1, and acts as an adapter to transport PER to the nucleus. PLoS Genet 11:e1004974. 10.1371/journal.pgen.1004974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M (2007) WNT signaling pathway and stem cell signaling network. Clin Cancer Res 13:4042–4045. 10.1158/1078-0432.CCR-06-2316 [DOI] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I (2007) A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol 27:5014–5028. 10.1128/MCB.02339-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Takahashi M, Takeda H, Shiro M, Koike T (2004) Recognition of phosphate monoester dianion by an alkoxide-bridge dinuclear zinc (II) complex. Dalton Trans 8:1189–1193. 10.1039/b400269e [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T (2006) Phosphate-binding tag, a new tool to visualize phosphorylation proteins. Mol Cell Proteomics 5:749–757. 10.1074/mcp.T500024-MCP200 [DOI] [PubMed] [Google Scholar]
- Kivimäe S, Saez L, Young MW (2008) Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol 6:e183. 10.1371/journal.pbio.0060183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell 94:97–107. 10.1016/S0092-8674(00)81225-8 [DOI] [PubMed] [Google Scholar]
- Kloss B, Rothenfluh A, Young MW, Saez L (2001) Phosphorylation of PERIOD is influenced by cycling physical associations of DOUBLE-TIME, PERIOD, and TIMELESS in the Drosophila clock. Neuron 30:699–706. 10.1016/S0896-6273(01)00320-8 [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I (2002) Role for slimb in the degradation of Drosophila period protein phosphorylated by doubletime. Nature 420:673–678. 10.1038/nature01272 [DOI] [PubMed] [Google Scholar]
- Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I (2010) A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3β/SGG in circadian clocks. J Neurosci 30:12664–12675. 10.1523/JNEUROSCI.1586-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok RS, Li YH, Lei AJ, Edery I, Chiu JC (2015) The catalytic and non-catalytic functions of the Brahma chromatin-remodeling protein collaborates to fine-tune circadian transcription in Drosophila. PLoS Genet 11:e1005307. 10.1371/journal.pgen.1005307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R (2002) A role for casein kinase 2α in the Drosophila circadian clock. Nature 420:816–820. 10.1038/nature01235 [DOI] [PubMed] [Google Scholar]
- Lin JM, Schroeder A, Allada R (2005) In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J Neurosci 25:11175–11183. 10.1523/JNEUROSCI.2159-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Mahesh G, Yu W, Hardin PE (2017) CLOCK stabilizes CYCLE to initiate clock function in Drosophila. Proc Natl Acad Sci U S A 114:10972–10977. 10.1073/pnas.1707143114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105:769–779. 10.1016/S0092-8674(01)00383-X [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M (2001) Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107:567–578. 10.1016/S0092-8674(01)00545-1 [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302:1765–1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL (2007) Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol 27:8049–8064. 10.1128/MCB.00680-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M (2004) The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell 13:213–223. 10.1016/S1097-2765(03)00503-3 [DOI] [PubMed] [Google Scholar]
- Nawathean P, Stoleru D, Rosbash M (2007) A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol 27:5002–5013. 10.1128/MCB.02338-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A 97:3608–3613. 10.1073/pnas.97.7.3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N, Chen KF, Szabó G, Stanewsky R (2009) Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol 19:241–247. 10.1016/j.cub.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW (1998) Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94:83–95. 10.1016/S0092-8674(00)81224-6 [DOI] [PubMed] [Google Scholar]
- Rothenfluh A, Abodeely M, Young MW (2000) Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr Biol 10:1399–1402. 10.1016/S0960-9822(00)00786-7 [DOI] [PubMed] [Google Scholar]
- Saez L, Young MW (1996) Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron 17:911–920. [DOI] [PubMed] [Google Scholar]
- Smith EM, Lin JM, Meissner RA, Allada R (2008) Dominant-negative CK2alpha induces potent effects on circadian rhythmicity. PLoS Genet 4:e12. 10.1371/journal.pgen.0040012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri V, Hall JC, Rosbash M (2000) Two novel double-time mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila. J Neurosci 20:7547–7555. 10.1523/JNEUROSCI.20-20-07547.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó A, Papin C, Zorn D, Ponien P, Weber F, Raabe T, Rouyer F (2013) The CK2 kinase stabilizes CLOCK and represses its activity in the Drosophila circadian oscillator. PLoS Biol 11:e1001645. 10.1371/journal.pbio.1001645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top D, Harms E, Syed S, Adams EL, Saez L (2016) GSK-3 and CK2 kinases converge on timeless to regulate the master clock. Cell Rep 16:357–367. 10.1016/j.celrep.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top D, O'Neil JL, Merz GE, Dusad K, Crane BR, Young MW (2018) CK1/Doubletime activity delays transcription activation in the circadian clock. Elife 7:e32679. 10.7554/eLife.32679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE (2006) PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 20:723–733. 10.1101/gad.1404406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Houl JH, Hardin PE (2011) NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol 21:756–761. 10.1016/j.cub.2011.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Kay SA (2010) Clocks not winding down: unraveling circadian networks. Nat Rev Mol Cell Biol 11:764–776. 10.1038/nrm2995 [DOI] [PubMed] [Google Scholar]