A proper balance between the repair of DNA double-strand breaks (DSBs) by homologous recombination and nonhomologous end joining is critical for maintaining genome integrity and preventing tumorigenesis. This balance is regulated and fine-tuned by a variety of factors, including cell cycle and the chromatin environment.
KEYWORDS: 53BP1, BRCA1, DNA repair, homologous recombination, TIP60, acetylation
ABSTRACT
A proper balance between the repair of DNA double-strand breaks (DSBs) by homologous recombination and nonhomologous end joining is critical for maintaining genome integrity and preventing tumorigenesis. This balance is regulated and fine-tuned by a variety of factors, including cell cycle and the chromatin environment. The histone acetyltransferase TIP60 was previously shown to suppress pathological end joining and promote homologous recombination. However, it is unknown how regulatory posttranslational modifications impact TIP60 acetyltransferase activity to influence the outcome of DSB responses. In this study, we report that phosphorylation of TIP60 on serines 90 and 86 is important for limiting the accumulation of the pro-end joining factor 53BP1 at DSBs in S and G2 cell cycle phases. Mutation of these sites disrupts histone acetylation changes in response to DNA damage, BRCA1 localization to DSBs, and poly(ADP-ribose) polymerase (PARP) inhibitor resistance. These findings reveal that phosphorylation directs TIP60-dependent acetylation to promote homologous recombination and maintain genome stability.
INTRODUCTION
DNA double-strand break (DSB) repair is conducted by distinct protein complexes that either execute homology-directed repair or join free ends irrespective of homology. Myriad regulatory mechanisms dictate the balance between these mechanisms. Cell cycle-dependent phosphorylation of repair proteins is the most established determinant of DSB repair mechanism (1). The past decade has implicated histone modification and chromatin remodeling as another key component of repair balance (2–5). The pro-homologous recombination (pro-HR) breast cancer susceptibility type 1 protein (BRCA1) competes with p53-binding protein 1 (53BP1) for access to DSBs. Importantly, their relative loading on differentially modified chromatin regulates repair mechanism utilization, genome stability, cancer etiology, and response to poly(ADP-ribose) polymerase inhibition (PARPi) (6, 7).
The mammalian lysine acetyltransferase 60-kDa Tat-interactive protein (TIP60) may regulate the balance between the two canonical DSB repair pathways through histone acetylation. Loss of TIP60 or hypoacetylation on TIP60 histone targets results in reduced DSB association of BRCA1 and commensurate increases in 53BP1 at damage sites (8–12). The proximal effect of TIP60 is to limit the accumulation of 53BP1 at DSBs. 53BP1 deficiency was able to restore BRCA1 ionizing radiation-induced focus (IRIF) formation and PARP inhibitor resistance in TIP60-depleted cells, indicating that TIP60 alleviates a 53BP1 block of BRCA1 DSB recognition (8). While the underlying basis for these observations remains unknown, several putative TIP60 substrate acetylations have been proposed to antagonize 53BP1 binding to its cognate recognition marks on histone H4 dimethylated at lysine 20 (H4K20me2) and histone H2A ubiquitylated at lysine 15 (H2AK15ub) (8–10). Structural evidence provides additional insights into how TIP60-dependent histone acetylation may prevent 53BP1 recognition of damage-associated nucleosomes (8, 13). However, how regulation of TIP60 activity affects these damage-associated histone modifications is unknown. Additionally, whether the cell cycle influences TIP60-dependent repair is also not well understood.
Nonhomologous end joining (NHEJ) is active throughout interphase and is the dominant mode of DSB repair in mammalian cells. 53BP1 protects the broken ends from resection, allowing for downstream ligation of the minimally processed DSB termini. During the S and G2 phases of the cell cycle, a subset of DSBs are repaired by homologous recombination (HR) (14). During HR repair, the broken DNA ends are resected to generate a single-strand overhang, enabling BRCA1 and BRCA2 to nucleate Rad51 filaments for homology search and capture, followed by templated synthesis to complete repair. DSB localizations of the BRCA1 and 53BP1 proteins are inversely correlated, and BRCA1 levels at DSBs significantly increase in 53BP1-deficient cells (8, 15). 53BP1 displays a more-rapid association with DSB chromatin, followed by a transition to BRCA1 at damage sites over time. This has led to models postulating that pro-HR regulatory modifications counteract the hyperaccumulation of 53BP1 at DSBs to create a permissive environment for BRCA1 (16, 17).
While TIP60 HAT activity and substrates have been implicated in HR (8–12), it is unclear whether this activity regulates DNA repair mechanisms in a cell cycle-dependent manner. In this study, we report that phosphorylation of TIP60 at serines 90 and 86 (18, 19) is important for effective HR by suppressing 53BP1 accumulation at DSBs to promote BRCA1 loading in S and G2 cell cycle phases. Mutation of these phosphorylation sites caused chromosomal abnormalities, sensitivity to DNA damage, and altered histone acetylation dynamics after DNA damage. These findings support the importance of TIP60 for HR repair and provide insight into how its activity is fine-tuned during the cell cycle to regulate repair mechanism.
RESULTS
Conditional knockout of TIP60 increases 53BP1 at DSBs and NHEJ.
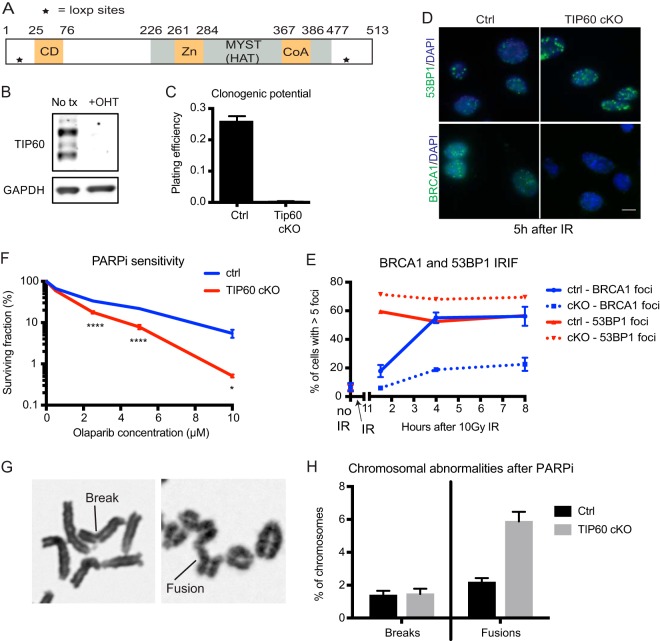
To study how TIP60 regulates 53BP1 accumulation at damage sites, we created a tamoxifen-inducible TIP60 knockout system. We crossed Tip60 f/f mice (20, 21) (schema in Fig. 1A) to ERT2-Cre mice (22) to generate Tip60 f/f; ERT2-Cre mice. Derivation of Tip60 f/f mouse embryonic fibroblasts (MEFs) expressing ERT2-Cre enabled efficient TIP60 deletion following 4-hydroxytamoxifen (4′OHT) treatment (Fig. 1B). TIP60 is essential in eukaryotes, and as expected, conditional knockout (cKO) of TIP60 resulted in long-term clonogenic failure of cells, even in the absence of damage (Fig. 1C). We thus used Tip60-cKO cells within 2 to 3 days after 4′OHT addition for our experiments. Consistent with prior reports (8–12), short-term TIP60 depletion led to increased 53BP1 and decreased BRCA1 focus formation after ionizing radiation (IR) damage (Fig. 1D and E). This perturbation in the BRCA1-53BP1 balance at DSBs correlated with sensitivity to PARPi, which induces DNA damage that requires repair by HR in S phase (Fig. 1F). TIP60 deficiency also increased the frequency of chromosomal fusion events (Fig. 1G and H), indicative of an HR defect coupled to an increase in toxic end joining. These data validate previous results that TIP60 suppresses 53BP1 at DSBs to promote BRCA1 loading and HR (8–12).
FIG 1.
Loss of TIP60 increases 53BP1 localization at DSBs and end joining. (A) Schematic of TIP60 domains and loxP sites on the Tip60 fl allele. For the targeting scheme, see reference 21. Functional domains are represented as color-filled rectangles. Numbers above the diagram correspond to amino acid positions. CD, chromodomain; Zn, zinc finger; CoA, acetyl coenzyme A-binding domain. (B) Western blot of TIP60 in Tip60 f/f, ERT2-Cre MEFs with no treatment (No tx) versus after 2 to 3 days of 4-hydroxytamoxifen (4′OHT) treatment. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is used as the loading control. (C) Plating efficiency of indicated MEFs in the absence of damage as measured by clonogenic assay. cKO, TIP60 conditional knockout by OHT treatment. n = 15. (D) Immunofluorescence images of 53BP1 and BRCA1 protein localization at 5 h following 10-Gy ionizing radiation (IR) in MEFs. Bar, 10 μm. (E) Quantification of percentage of cells with more than 5 foci at different time points post-IR. n = 3. (F) Survival of MEFs treated with different doses of the PARP inhibitor olaparib as measured by clonogenic assay. n = 4. ****, P < 0.0001. (G) Photomicrographs illustrating chromosomal breaks and fusions on MEF metaphase spreads. (H) Quantification of chromosomal abnormalities on metaphase spreads after olaparib treatment. n = 5. For panels C, E, F, and H, error bars show SEM.
Phosphorylation of TIP60 serines 90 and 86 is important for HR repair during S/G2.
HR is utilized for DSB repair during S and G2 phases of the cell cycle, whereas NHEJ is available throughout interphase (14). As a consequence, the increase in 53BP1 loading and the concomitant decrease in BRCA1 and HR observed upon TIP60 deletion may be due to the accumulation of cells in G1 phase of the cell cycle. To address this possibility, we assessed the cell cycle profile of Tip60-cKO cells at 2 to 3 days post-4′OHT addition, the time frame when immunofluorescence and cytogenetic analysis experiments were performed. At 2 to 3 days postdeletion, the Tip60-cKO cells did not demonstrate any significant differences in the percentage of cells in G1 but rather began to show accumulation in G2 (Fig. 2A and B). This revealed that TIP60 depletion-induced increase in 53BP1 foci and pathological end joining is not due to G1 arrest.
FIG 2.
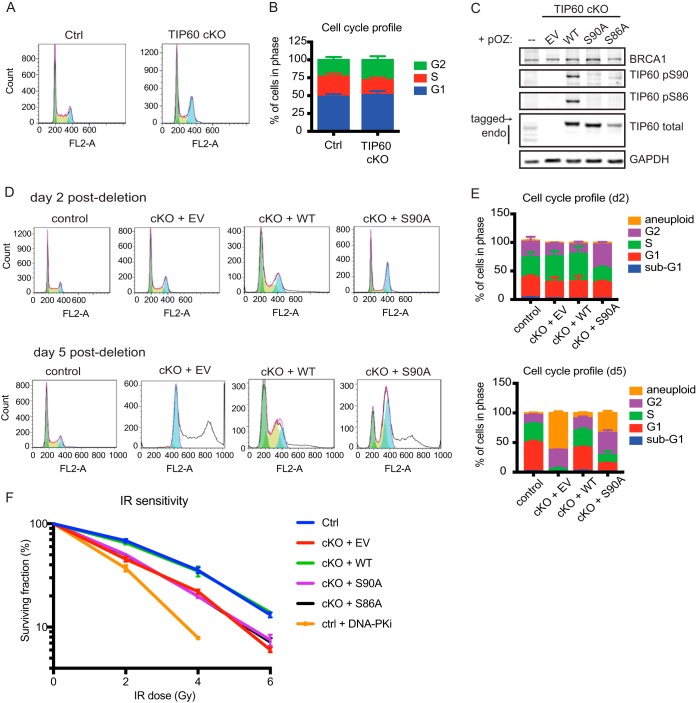
Serines 90 and 86 on TIP60 regulate DNA repair. (A) Representative cell cycle FLOW profiles of Tip60 f/f, ERT2-Cre MEFs at 2 to 3 days following no treatment versus OHT treatment. Green fill-in, G1; yellow-green, S; blue, G2. (B) Quantification of percentage of cells in each phase of cell cycle. n = 8. (C) Western blots of whole-cell lysate from Tip60 f/f, ERT2-Cre MEFs stably expressing FLAG-HA (FH)-tagged empty vector, wild-type TIP60, or mutant human TIP60. EV, empty vector; WT, wild type. Endogenous TIP60 (endo) and FLAG-HA-tagged exogenous TIP60 (tagged) are indicated. (D) Representative MEF cell cycle FLOW profiles at day 2 post-OHT addition (top panels) and day 5 post-OHT addition (bottom panels). Green fill-in, G1; yellow-green, S; blue, G2; uncolored, aneuploid. (E) Quantification of cell cycle distributions at day 2 post-OHT addition (top panel) and day 5 post-OHT addition (bottom panel). n = 3. (F) Survival of MEFs after different doses of IR as measured by clonogenic assay. DNA-PK inhibitor-treated cells are used as a negative control. n = 3. For panels B, E, and F, error bars show SEM.
These results suggested that TIP60 may be regulating 53BP1 focus formation during S and G2, a period of interphase that critically determines the competition between BRCA1- and 53BP1-dependent repair mechanisms. Interestingly, TIP60 activity has been suggested to be cell cycle regulated. Phosphorylation of TIP60 on serine 90 and/or serine 86 was reported to increase during late G2/M (18, 19). Serine 90 (S90) directs serine 86 (S86) phosphorylation, and both residues have been implicated in regulating TIP60 acetyltransferase activity on histone and nonhistone targets (18, 23–26). We hypothesized that these putative cell cycle-dependent phosphorylation sites on TIP60 may modulate its activity to limit the recruitment of 53BP1 to damage sites and regulate DNA repair.
To test this hypothesis, Tip60-cKO MEFs were reconstituted with FLAG-hemagglutinin (HA)-tagged empty vector (EV), wild-type (WT) TIP60, and phospho-dead serine to alanine mutants on S90 and S86 (S90A and S86A). Consistent with the literature (18, 23, 24, 26), mutation of S90 abolished S86 phosphorylation on epitope-tagged TIP60 (Fig. 2C). These results also validated the specificity of the phosphospecific TIP60 antibodies used. The S90A mutation has been reported to cause chromosomal segregation defects that lead to G2 arrest (19), which is corroborated by our results. S90A mutant-complemented Tip60-cKO cells accumulated in G2 over time, which correlated with aneuploidy (Fig. 2D and E).
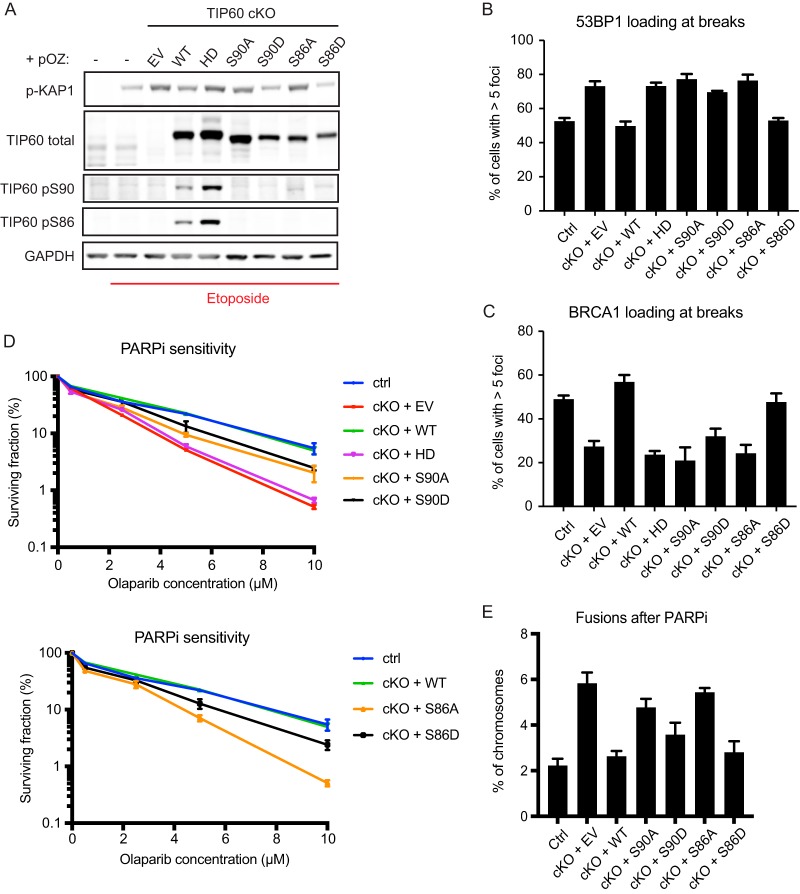
We then tested whether the phospho-dead mutants had impaired DNA repair capacity. The Tip60-cKO cells reconstituted with WT TIP60 showed resistance to IR damage, whereas those reconstituted with EV, S90A, or S86A remained sensitive to IR (Fig. 2F). To verify that the repair phenotype of S90A and S86A is due to defective phosphorylation, we created serine to aspartate phosphomimetic mutants (S90D and S86D). TIP60 depletion resulted in greater damage signaling after etoposide treatment, based on phosphorylated KAP1 (Fig. 3A). The increase in the amount of damage sensed by the Tip60-cKO cell is attenuated by reconstitution with WT, S90D, and S86D TIP60 but not by the HAT-deficient Q377E/G380E mutant “HD” (27) or S90A and S86A mutants. Taken together, these data indicated that phosphorylation at serines 90 and 86 contributes to TIP60-dependent DNA damage response functions.
FIG 3.
Phosphorylation of TIP60 S90 and S86 promotes HR repair. (A) Western blots of Tip60-cKO MEFs complemented with FLAG-HA-tagged TIP60 variants 5 h after treatment with damaging agent etoposide. HD, HAT-deficient Q377E/G380E mutant. Phosphorylated KAP1 is used as a marker of ATM activity. (B) Quantification of 53BP1 focus formation 5 h after IR. n = 3. (C) Quantification of BRCA1 focus formation 5 h after IR. n = 3. (D) Survival of MEFs treated with different doses of olaparib as measured by clonogenic assay. The same set of data is broken up into two different graphs for presentation clarity. n = 3. (E) Quantification of abnormal fusion events on MEF metaphase spreads. For panels B to E, error bars show SEM.
After establishing a role for TIP60 S90 and S86 phosphorylation in DNA damage responses, we turned to whether phosphorylation of these residues is responsible for TIP60 regulation of the HR-NHEJ balance. Similar to the HAT-deficient HD mutant, complementation with the S90A and S86A mutants failed to suppress 53BP1 focus formation after IR, whereas complementation with S90D and S86D partially rescued inhibition of 53BP1 foci (quantification data in Fig. 3B). Accordingly, despite having no effect on the BRCA1 protein level (Fig. 2C), S90A and S86A mutants failed to rescue BRCA1 foci, whereas S90D and S86D were able to partially restore BRCA1 foci (quantification data in Fig. 3C). Moreover, cells expressing either S90A or S86A TIP60 mutants demonstrated hypersensitivity to PARPi, while those expressing S90D or S86D mutants displayed relative resistance to PARPi (Fig. 3D). The PARPi sensitivity of S90A and S86A mutants correlated with increased frequency of chromosomal fusion events when these cells were treated with PARPi, indicative of pathological end joining. The S90D and S86D mutants, on the other hand, were able to partially suppress these toxic end joining events (Fig. 3E). Notably, in almost all cases, the S86D mutant showed a greater capacity for rescue than the S90D mutant. This discrepancy may be due to the inability of the S90D mutant to rescue S86 phosphorylation (Fig. 3A). S86 phosphorylation is dependent on S90 phosphorylation and is reportedly mediated by glycogen synthase kinase 3-beta recognition of an SXXS motif (23–25), which remains mutated in the S90D mutant. These results demonstrate that phosphorylation on S90 and S86 mediates TIP60-dependent suppression of 53BP1 localization at damage sites to inhibit toxic end joining and promote HR.
TIP60 phosphorylation suppresses 53BP1 focus formation in S/G2.
Given that S90 and S86 are important for HR repair, we were curious to know whether these phosphorylation events increase in response to damage. Exogenous FLAG-HA-tagged WT TIP60 did not show increased S90 or S86 phosphorylation in response to etoposide treatment (Fig. 4A), indicating that phosphorylation of these residues on the exogenous TIP60 is likely not damage induced.
FIG 4.
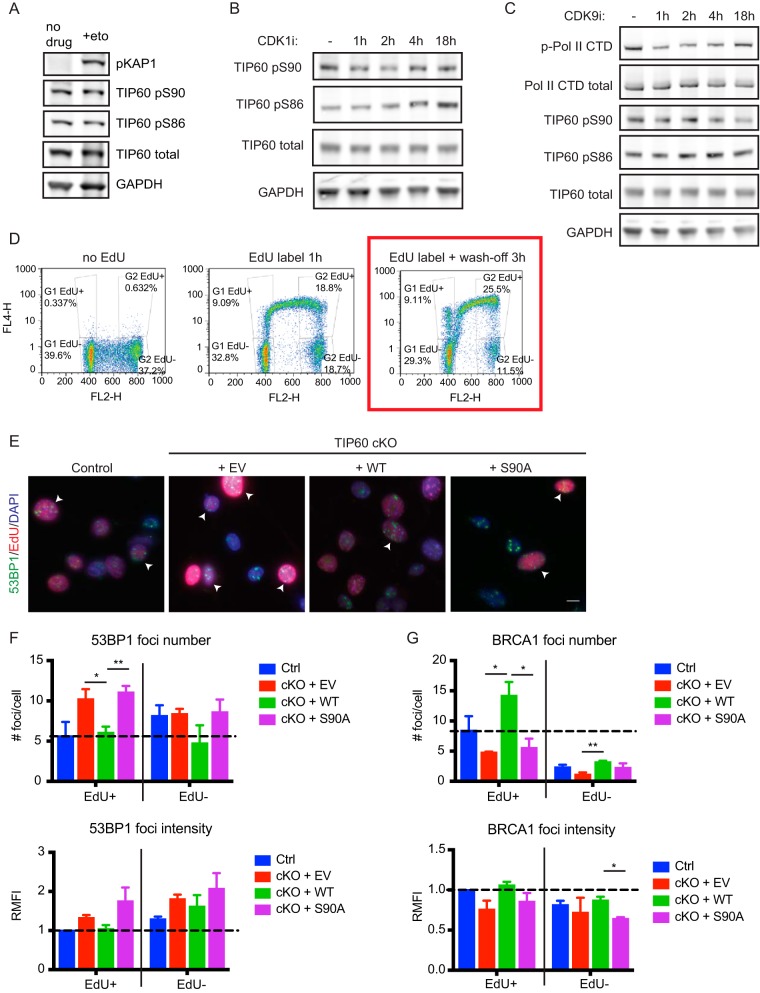
TIP60 S90 regulates 53BP1 localization during S-G2. (A) Western blots of pS90 and pS86 on the FLAG-HA-tagged WT TIP60 after 5 h of damage induction with etoposide. p-KAP1 is used as a marker of damage. (B) Western blots of pS90 and pS86 on FLAG-HA-tagged WT TIP60 in MEFs treated with 10 μM CDK1 inhibitor RO-3306 for various lengths of time. (C) Western blots of pS90 and pS86 on FLAG-HA-tagged WT TIP60 in MEFs treated with 2 μM CDK9 inhibitor SNS-032 for various lengths of time. p-Pol II CTD is used as a readout of CDK9 inhibition. (D) FLOW plots of cell cycle distributions of EdU− and EdU+ MEFs to illustrate enrichment in G2 of our scheme entailing EdU labeling and then wash-off. On the y axis, FL4-H indicates the intensity of EdU label (after conjugation with Alexa Fluor 647); on the x axis, FL2-H indicates propidium iodide (PI) stain intensity. Left panel, untreated cells; middle panel, cells have been labeled with EdU for 1 h with no wash-off; right panel, cells were EdU labeled for 1 h, washed, and then collected 3 h after wash-off. The labeling scheme corresponding to the right panel (highlighted by a red box) is the one that we employed for the experiments in panels E to G. Density is indicated by colors: red (highest density) > yellow > green > blue (lowest density). (E) Immunofluorescence images of 53BP1 ionizing radiation-induced focus (IRIF) formation in EdU+ (late S and G2) versus EdU− MEFs 5 h after IR and 3 h after EdU wash-off. Arrowheads point to EdU+ cells that have many bright 53BP1 foci. Bar, 10 μm. (F) ImageJ particle analysis quantification of 53BP1 foci per cell and relative mean focal intensity (RMFI) in EdU+ versus EdU− cells. The dotted line is set to the mean in EdU+ control cells. n = 3. (G) ImageJ particle analysis quantification of BRCA1 foci per cell and relative mean focal intensity (RMFI) in EdU+ cells versus EdU− cells. The dashed lines are set to the means in EdU+ control cells. n = 3. For panels F and G, error bars show SEM. *, P < 0.05; **, P < 0.01.
Both cyclin-dependent kinase 1 (CDK1) (18, 19) and CDK9 (26) have been reported to be the kinase responsible for phosphorylation of S90. CDK1 activity increases during S/G2, and CDK1 is proposed to promote end resection and HR via multiple mechanisms, including phosphorylation of the pro-HR endonuclease CtIP (15, 28). We postulated that CDK1 phosphorylation of TIP60 S90 during S/G2 may be another mechanism for inhibition of 53BP1 during these phases of the cell cycle. However, inhibition of CDK1 or CDK9 did not dramatically reduce pS90 on ectopically expressed WT TIP60 (Fig. 4B and C). This suggests that additional kinases may also be involved in maintaining phosphorylation of these sites.
Although CDK1 inhibition failed to abolish TIP60 pS90, we found that TIP60 pS86 increased in G2 (Fig. 4B). Prolonged CDK1 inhibition causes cells to accumulate in G2, and consistent with suggestions from a prior report (18), we observed that the exogenous epitope-tagged WT TIP60 becomes hyperphosphorylated at S86 after 18 h of CDK1 inhibition, when cells are enriched in phase G2 of the cell cycle (Fig. 4B). These data demonstrate that TIP60 phosphorylation is cell cycle regulated, albeit via a CDK1-independent mechanism.
Based on prior studies (18, 19), and given that pS86 increases in G2 (Fig. 4B), we postulated that TIP60 phosphorylation enhances its activity during S and G2. To determine whether TIP60 regulation of repair mechanism during S/G2 is mediated by phosphorylation, we devised an approach to identify late S and G2 cells in an asynchronous population for repair focus quantification (Fig. 4D; see also Materials and Methods). We were able to identify late S and G2 phases by transiently labeling cells with EdU for 1 h, followed by changing to medium lacking EdU and harvesting 3 h later. EdU positivity indicated cells that had transitioned from early S to late S/G2 (Fig. 4D). In Tip60-cKO cells, reconstitution with WT TIP60 reduced the number and intensity of 53BP1 foci, whereas reconstitution with EV or S90A resulted in elevated 53BP1 focus formation, particularly in EdU+ cells (Fig. 4F). In accordance, reconstituted WT TIP60 rescued the number and intensity of BRCA1 foci, whereas reconstituted EV or S90A failed to restore BRCA1 focus formation, especially in EdU+ cells (Fig. 4G). These results indicate that phosphorylation is important for TIP60 suppression of 53BP1 localization to damage sites during late S/G2.
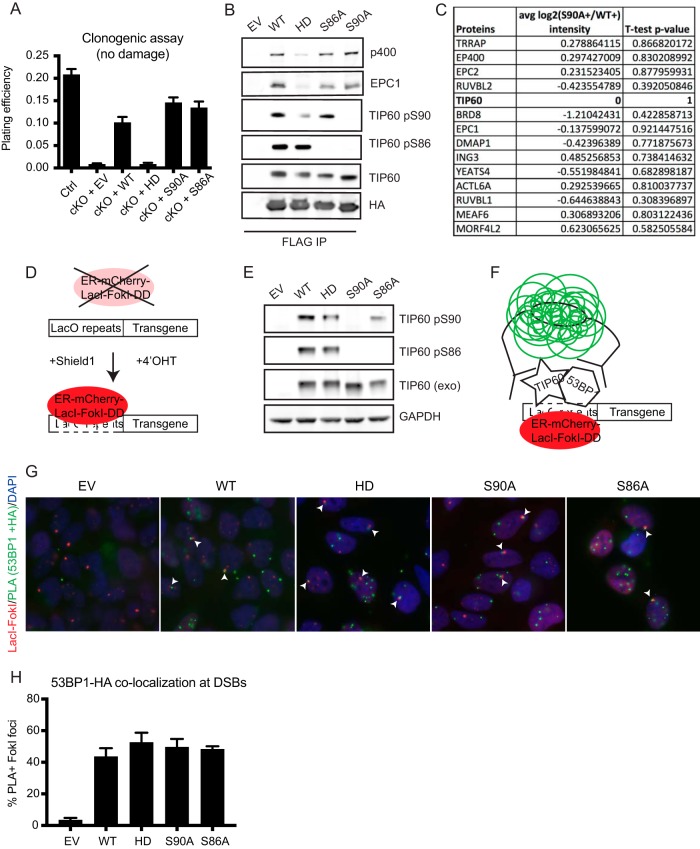
S90 and S86 are dispensable for viability, NuA4 complex formation, and recruitment to damage sites.
Although S90A and S86A have been reported to affect TIP60 HAT activity (18, 23–26), mutation of these phosphoserine residues does not affect cell viability over the long term (Fig. 5A) (26). Unlike the HD mutation, the S90A and S86A mutations did not significantly reduce TIP60 association with other members of the NuA4 remodeling complex by immunoprecipitation (Fig. 5B) or by complex purification-mass spectrometry (Fig. 5C). Thus, S90A and S86A are dispensable for cell viability and macromolecular complex formation despite being impaired in DSB repair activities.
FIG 5.
TIP60 S90 and S86 are dispensable for viability, complex formation, and recruitment to damage sites. (A) Plating efficiency of undamaged Tip60-cKO MEFs complemented with empty vector, wild-type TIP60, or mutant TIP60 as measured by clonogenic assay. n = 3. (B) Western blot of proteins immunoprecipitated with the indicated FLAG-HA-tagged constructs from HeLa S3 cells. (C) Table of analyses of mass spectrometry-identified canonical TIP60-p400 complex members copurified with FLAG-HA-tagged WT versus S90A TIP60 from HeLa S3 cells. Negative average log2 (S90A+/WT+) intensity means that the S90A mutant exhibits decreased association with the indicated complex partner compared to the WT. n = 3. (D) Schematic of mCherry-LacI-FokI damage induction by addition of Shield1 and 4′OHT (8). A LacO-transgene array is stably integrated into the U2OS genome. mCherry-LacI-FokI is constantly degraded at baseline, stabilized by the Shield1 ligand, and translocates to the nucleus after 4′OHT treatment. (E) Western blots of stably expressed FLAG-HA-tagged TIP60 variants in LacI-FokI reporter cells. TIP60 (exo), exogenous tagged TIP60. (F) Schematic of TIP60-53BP1 proximity ligation assay in the mCherry-LacI-FokI reporter cells. Colocalization between TIP60 and 53BP1 is visualized by green fluorescence. (G) Representative PLA images of mCherry-LacI-FokI reporter cells complemented with FLAG-HA-tagged TIP60 variants or empty vector (EV). mCherry marks where LacI-FokI is localized, and green fluorescence marks sites of PLA positivity, where there is colocalization between 53BP1 and the tagged TIP60 (HA tag). Arrowheads indicate damage sites (mCherry foci) where both TIP60 and 53BP1 are recruited (PLA+ green foci). (H) Quantification of the percentage of damage sites (mCherry foci) where both TIP60 and 53BP1 are recruited, as indicated by PLA+ green foci colocalizing with mCherry foci. n = 3. For panels A and H, error bars show SEM.
A recently published report suggests that the S90A mutant fails to localize to the chromatin (26), which accounts for its hypomorphic activity. To determine whether the S90A HR defect is due to failure to recruit to DSBs, we employed a proximity ligation assay (PLA) to assess colocalization between TIP60 variants and 53BP1 at defined DSBs. This approach allows us to detect weak interactions, which are magnified by the PCR step of the PLA, at a specific locus in the genome. We stably expressed epitope-tagged WT and mutant TIP60 in the previously published LacI-FokI cell line (8) (Fig. 5D and E), in which many DSBs can be inducibly created at a single locus in the cell and visualized by mCherry fluorescence (8, 29). Colocalization between TIP60 and 53BP1 is detected by the PLA reaction coupled to a green fluorophore (see cartoon in Fig. 5F). We chose 53BP1 as the PLA “bait,” as it is reliably recruited to damage sites, and the commercial 53BP1 antibody that we use has high sensitivity and specificity for imaging-based assays. Thus, an mCherry focus that lacks an overlapping green PLA focus indicates that TIP60 and 53BP1 do not colocalize to that damage site, whereas an mCherry focus that is also PLA positive (PLA+) represents a damage site where both TIP60 and 53BP1 are recruited. This approach ensures that we are detecting TIP60 localization specifically at damage sites rather than elsewhere in the genome by two means: (i) the site of the LacI-FokI DSBs is mCherry+, and (ii) a PLA+ signal indicates that 53BP1 is also recruited there. The PLA+ foci that do not overlap mCherry foci are either background signal or instances of TIP60-53BP1 interaction elsewhere in the genome and are excluded from the analysis.
While the reporter cells complemented with empty vector show minimal PLA signal, all the TIP60 variants show colocalization with 53BP1 at damage sites (Fig. 5G and H), indicating that the S90A and S86A mutants are competent to recruit to DSBs, making them useful separation-of-function mutants for studying TIP60 DNA repair activities.
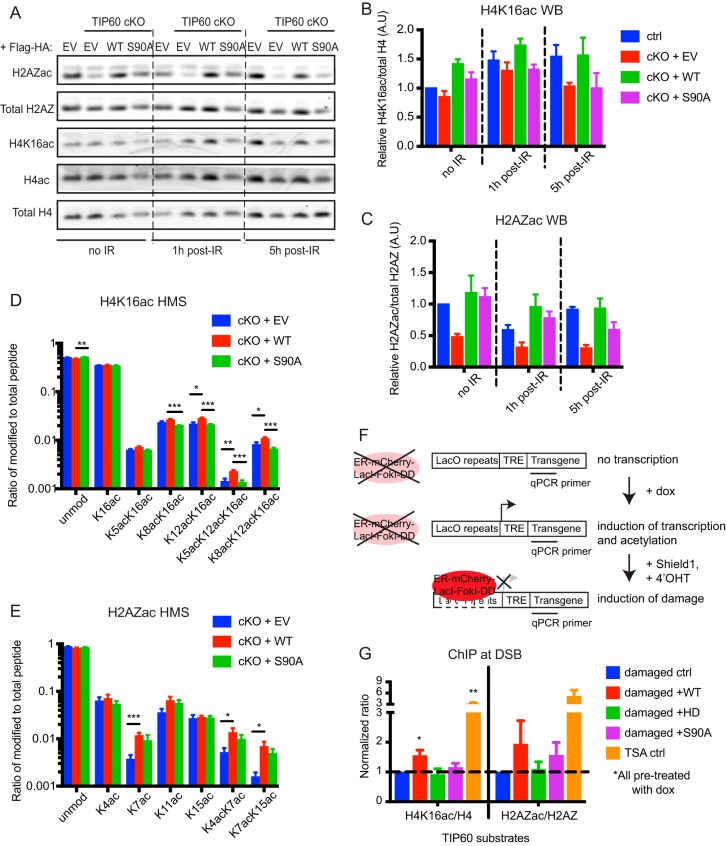
S90A phosphorylation regulates TIP60 acetylation of histones during the DNA damage response.
Given that S90 and S86 phosphorylation may be important for TIP60 HAT activity (18, 23–26), we sought to determine how mutations at these sites affect histone acetylation dynamics during the damage response. TIP60 reportedly acetylates nucleosomal histones H4, H2A, and the H2A variants H2A.X and H2A.Z (27, 30–32). In Tip60-cKO cells, we found that TIP60 loss affects both global H4K16ac and H2AZ K4/K7ac after IR damage and that the S90A mutant appears to partially rescue H2AZac but not H4K16ac (Fig. 6A to C). This result is corroborated by mass spectrometry analysis of bulk histone modifications after cells were damaged by etoposide treatment (Fig. 6D and E). Although reconstitution with WT TIP60 had a greater effect on H2AZK7ac than on H4K16ac, the S90A mutant had more-pronounced deficiency in H4K16ac than in H2AZac.
FIG 6.
S90 facilitates TIP60-dependent acetylation of histone H4 after damage. (A) Western blots of histones isolated from indicated MEFs at different time points after 10-Gy IR. (B and C) ImageJ quantification of Western blot band intensities of H4K16ac normalized to total H4 (B) and H2AZ K4/K7ac normalized to total H2AZ (C). n = 3. (D and E) Relative abundance of bulk unmodified and modified histone H4 peptide (amino acids [aa] 4 to 17) (D) and histone H2A.Z peptide (aa 1 to 19) (E) in etoposide-damaged MEFs as quantified by mass spectrometry. n = 10. (F) Schematic of ChIP-qPCR in the doxycycline (dox)-inducible LacI-FokI cell lines, in which pretreatment with doxycycline turns on transcription of the transgene and stimulates histone acetylation at the locus. (G) Normalized ratios of H4K16ac ChIP to total H4 ChIP and of H2AZ K4/K7ac ChIP to total H2AZ ChIP, with the control cell line expressing no exogenous TIP60 set to a normalized ratio of 1. n = 3. For panels B, C, D, E, and G, error bars show SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To test whether similar dynamics are occurring locally at defined DSBs, we stably expressed epitope-tagged versions of WT and mutant TIP60 in the doxycycline (dox)-inducible LacI-FokI nuclease reporter cell line (8) and performed chromatin immunoprecipitation (ChIP) followed by quantitative real-time PCR (qPCR) using primers near the nuclease cut site (Fig. 6F). In agreement with the bulk histone data, ChIP-qPCR showed that after normalizing to total H4 and total H2AZ, WT TIP60 overexpression increased the local levels of H4K16ac and H2AZac, respectively, at DSB chromatin. Overexpression of the HD mutant failed to increase either, and overexpression of the S90A mutant failed to increase H4K16ac but was able to partially increase H2AZac (Fig. 6G). This suggests that S90 phosphorylation is important for TIP60 HAT activity during the damage response.
DISCUSSION
Tip60-cKO cells recapitulate the phenotypes of earlier reports with TIP60 knockdown by small interfering RNA (siRNA) (Fig. 1) (8–12), corroborating TIP60-dependent histone acetylation and prevention of 53BP1 hyperaccumulation at DSBs. Our findings further reveal a previously unappreciated communication between cell cycle-dependent phosphorylation and acetylation during the damage response (Fig. 2, 3, 4, and 6) (18, 19). Similar to the findings of a recent report (26), S90A and S86A mutations did not affect TIP60 binding to other canonical complex members or long-term proliferation (Fig. 5). Thus, phosphorylation of TIP60 S90 and S86 controls a subset of TIP60-dependent functions, and the S90A and S86A mutants present useful tools for studying TIP60 with fewer confounding effects elicited by loss of viability in complete Tip60 null cells.
Multiple mechanisms ensure the timely activation of HR in S/G2, including CDK-dependent phosphorylation of end resection proteins, and ubiquitin-dependent association of BRCA1 and 53BP1 with key interacting partners (1). TIP60 pS86 increased when cells were enriched in G2 (Fig. 4B), consistent with a model of cell cycle regulation of TIP60 phosphorylation first proposed over 1 decade ago (18). Indeed, TIP60 loss had a greater effect on 53BP1 and BRCA1 focus formation in S/G2 cells than in G1 cells (Fig. 4D to G). Taken together, these data implicate phosphorylation-directed TIP60 HAT activity in the cell cycle control of DSB repair mechanism during S and G2.
The effect of S90 and S86 on histone acetylation is of particular interest, since these residues have been shown to regulate TIP60 activity on both histone and nonhistone targets (18, 19, 23, 24). However, it is unclear how these phosphorylations affect TIP60 function. Recent work proposes that S90A fails to bind chromatin, whereas S86A does bind chromatin yet intrinsically diminishes HAT activity (26). Although S90A slows down cell proliferation in the short term, cells recovered over the long term and had no viability or partner-binding issues (26), consistent with our observations (Fig. 5A and B). This suggests that it is unlikely that S90A cannot localize to chromatin, as TIP60 is essential. In agreement, S90A, similar to the other TIP60 variants, was capable of recruiting to damage sites (Fig. 5G and H) and partially rescued H2AZ acetylation (Fig. 6). These properties could explain why the Tip60-cKO plus S90A cells are viable whereas the HD mutant is not. Importantly, they support a model whereby S90A acts as a separation of function mutant, unlike the pan-HAT-deficient HD mutant, which fails to rescue survival, partner binding, or acetylation at damage sites (Fig. 5A and B and 6G).
Histone acetylation changes dynamically in response to DNA damage (2, 33, 34) (Fig. 6A to C). In comparison to 53BP1, BRCA1 exhibits delayed DSB recruitment, which maximizes 4 to 8 h after damage. Thus, changes in histone acetylation dynamics may offer insight into which acetylation marks are responsible for 53BP1 versus BRCA1 DSB association. Our finding that S90A mutation has a greater impact on acetylation of histone H4 and H2AZ at later times postdamage (Fig. 6A to C) is consistent with the involvement of these particular substrates in the temporal regulation of BRCA1 and 53BP1 localization. Further work on how acetylation of these histone tails prohibits 53BP1 binding could inform our understanding of how repair pathway choice is regulated in specific regions of the genome with different chromatin contexts.
MATERIALS AND METHODS
Tip60 f/f, ERT2-Cre MEF derivation.
Tip60 f/f mice (20, 21) (a gift from John Lough), with loxP sites flanking exons 3 and 11, were crossed to strain Cre-ERT2 B6.Cg-Tg(UBC-cre/ERT2)1EjB/J (22) (a gift from Eric J. Brown) until Tip60 f/+, ERT2-Cre mice were generated. Timed matings between Tip60 f/+, ERT2-Cre mice were set up, and pregnant females were sacrificed for day 13.5 embryos. Tip60 f/f, ERT2-Cre mouse embryonic fibroblasts were derived using standard procedures and immortalized by transfection with simian virus 40 (SV40) large T antigen followed by serial passage. All animal care and experimental procedures have been approved by the Hill Pavilion Institutional Animal Care and Use Committee (IACUC) at the University of Pennsylvania Veterinary School.
Genotyping.
Genotyping was performed by PCR on genomic DNA isolated from embryo heads. Primers used were TIP60 forward, 5′-AGGGAGTCAACGATCGCACGGGAGG-3′, and reverse, 5′-CACAGACAGGGAGTCTTAGCCAGGG-3′; the floxed allele yields a 359-bp band, and the endogenous allele yields a 258-bp band. Primers used were ERT2-Cre number 1, 5′-TACACCAAAATTTGCCTGCATTACCGG-3′, number 2, 5′-TTTCCATGAGTGAACGAACCTGGT-3′, number 3, 5′-GCATCAGCTAGCAGCAGGTCCAACT-3′, and number 4, 5′-GACGCCACACTGGGTCTTCATCAGT-3′; a 300-bp band serves as the internal control, and the ERT2-Cre positive genotype yields a 400-bp band.
Irradiation.
Irradiation of cells was performed using a Gammacell 40 irradiator (Nordion International) using a cesium-137 source.
MEF immunofluorescence.
MEFs plated on coverslips were rinsed with phosphate-buffered saline (PBS) and then preextracted with 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 0.5% Triton X-100 in PBS for 5 min at 4°C. They were then washed with PBS prior to fixation with 3% paraformaldehyde and 2% sucrose (pH 7.4) in PBS for 15 min at room temperature. After fixation, the coverslips were washed with PBS and then permeabilized with 150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl (pH 7.4), 0.05% NP-40, 0.25% gelatin, and 0.5% Triton X-100 in PBS for 10 min at 4°C. They were then washed with PBS with Tween 20 (PBS-T) and incubated with primary antibodies diluted in PBS-T at 37°C for 30 min. After primary incubation, the coverslips were rinsed with PBS-T and then incubated with Alexa Fluor-conjugated secondary antibodies diluted 1:500 in PBS-T at 37°C for 30 min. Finally, they were rinsed with PBS-T prior to mounting with Vectashield plus 4′,6-diamidino-2-phenylindole (DAPI). Foci per cell in EdU+ versus EdU− cells were quantified by particle analysis on thresholded images using ImageJ.
Whole-cell lysate extraction and Western blotting.
Whole-cell lysates were extracted from cell pellets using a high-salt lysis buffer (25 mM Tris-HCl [pH 7.4], 300 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF]).
The samples were run on gradient SDS-PAGE gels (Invitrogen) in morpholinepropanesulfonic acid (MOPS) buffer, wet transferred onto nitrocellulose membrane, and blocked with a 1:1 mix of Tris-buffered saline with Tween 20 (TBS-T) to LiCor Odyssey blocking buffer. They were then incubated with diluted primary antibodies in TBS-T overnight at 4°C, washed with TBS-T, and then incubated with secondary antibodies (LiCor anti-mouse antibody–IRDye 680 and anti-rabbit antibody–IRDye 800, 1:10,000) diluted in a 1:1 mix of TBS-T to LiCor Odyssey blocking buffer. The blots were then imaged using a LiCor Odyssey 675 machine, with scanning at 700-nm and 800-nm wavelengths. Brightness and contrast were adjusted in LiCor. Images were prepared for publication using ImageJ.
Clonogenic and PARPi sensitivity assays.
Forty-eight hours after 4′OHT addition or mock treatment, the cells were seeded at various densities in 6-well plates, depending on the cell line and intended drug treatment dose. Technical triplicates were plated for all conditions. Twenty-four hours later, the medium was removed, and fresh media containing various concentrations of olaparib were added to the cells. The plates were collected 10 to 12 days later and stained with crystal violet prior to quantification.
Metaphase spreads.
Forty-eight hours after 4′OHT addition or mock treatment, 5 μM olaparib was added to the cells. Twenty-one hours later, 0.5 μM nocodazole was added to arrest cells in metaphase. Three hours after nocodazole addition, the supernatant and adherent cells were collected. They were incubated with 75 mM KCl for 25 min at 37°C, spun down, and then fixed with a 3:1 mix of methanol-acetic acid at 4°C for more than 10 min prior to being spun down for spreading. Glass slides were soaked in methanol and then placed in 42°C humidity chambers. Small concentrated droplets of fixed cells were dropped from height onto the slides in humidity chambers and then air blown to spread. After multiple droplets were dropped and spread to cover the surface of the slide, the slides were removed from the humidity chambers to air dry at room temperature. They were then stained with Giemsa for 8 min at room temperature, washed with water, and air dried prior to mounting with Permount (Fisher).
Cell cycle analyses.
Cells were fixed in 70% ice-cold ethanol at −20°C for longer than an hour. They were then spun down and resuspended in 200 μl PBS containing RNase A and propidium iodide (PI) and stained at room temperature for longer than 30 min. The samples were then run through a flow cytometry machine.
Generation of FLAG-HA-tagged TIP60 constructs and cell lines.
Wild-type or “HAT-dead” Q377E/G380E (HD) human TIP60 (gifts from Brendan Price) was subcloned into the retroviral pOZ-N vector, with N-terminal FLAG-HA tag and interleukin-2 (IL-2) receptor selection. pOZ-TIP60 WT served as the template to generate S90A, S90D S86A, and S86D mutants using the Q5 site-directed mutagenesis kit (NEB). TIP60 f/f, ERT2-Cre MEFs, HeLa S3s, and mCherry-LacI-FokI U2OS reporter cells were infected with virus containing the different pOZ constructs per standard procedures and selected using IL-2-conjugated magnetic beads.
EdU labeling of MEFs for flow analysis.
Four hours prior to collection, 10 μM EdU was added to the cells. After 1 h of incubation, EdU was washed off, and fresh medium was added. Three hours later, the cells were collected for labeling and flow analysis. Cells were fixed in 70% ice-cold ethanol at −20°C for 1 h. They were then spun down and washed with 0.5% bovine serum albumin (BSA) and 0.5% Tween 20 in PBS. Then, they were blocked with 1% BSA–0.25% Triton X-100 in PBS for 15 min at 4°C. Subsequently, EdU was fluorescently labeled using the Click-iT EdU Alexa Fluor 647 flow cytometry assay kit (Thermo Fisher). After the Click-iT reaction, the cells were incubated with propidium iodide and RNase A in PBS for more than 30 min before flow analysis.
EdU labeling of irradiated MEFs for immunofluorescence.
MEFs seeded on coverslips were irradiated 5 h prior to collection. One hour later, 10 μM EdU was added to the cells. After 1 h of incubation, EdU was washed off and fresh medium was added. Three hours later, the cells were collected for Click-iT fluorescent EdU labeling and immunofluorescence. The MEFs were preextracted and permeabilized by the immunofluorescence procedure described above. They were then washed with 3% BSA–PBS-T and incubated with Click-iT cocktail per the Click-iT cell reaction buffer kit (Invitrogen) protocol. EdU was labeled with an azide-modified 488-nm fluorophore. The coverslips were then washed with 3% BSA–PBS-T and counterstained with BRCA1 or 53BP1 followed by an Alexa Fluor 568-conjugated secondary antibody. The colors of EdU versus protein were switched post hoc in ImageJ such that EdU was visualized in red and BRCA1 or 53BP1 was in green.
Immunoprecipitation.
HeLa S3 cells stably expressing FLAG-HA-tagged TIP60 constructs were lysed in a cell lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 1 mM EDTA, 0.05% NP-40, 1 mM PMSF) on ice for 10 min. Then, they were spun down, and the supernatant was removed, resuspended in a nuclear lysis buffer (25 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF), and lysed on ice for 10 min. The nuclear lysate was precleared with protein A-agarose beads (Fisher) for 2 h at 4°C. Precleared lysate was then incubated with FLAG-M2 beads (Sigma-Aldrich) overnight at 4°C. After multiple washes with nuclear lysis buffer, protein was eluted off the beads using 0.2 mg/ml FLAG peptide (Sigma) for 1 h at 4°C, and Western blotting was performed as described above.
Complex purification.
Two liters of HeLa S3 cells stably expressing FLAG-HA-tagged TIP60 constructs was pelleted and then resuspended in hypotonic buffer (10 mM Tris-HCl [pH 7.4], 7.5 mM KCl, 1.5 mM MgCl2) on ice for 15 min. The swollen cells were lysed by Dounce homogenization. Nuclei were spun down and then lysed in KETNG-400 buffer (400 mM KCl, 20 mM Tris-HCl [pH 7.4], 0.5 mM EDTA, 0.1% NP-40, 1.5 mM MgCl2, 10% glycerol) for 45 min at 4°C. The lysate was spun down at maximum speed, and the supernatant was dialyzed against two liters of KETNG-100 at 4°C for longer than 3 h. The dialyzed lysate was spun down at maximum speed again, and the supernatant was incubated with FLAG-M2 beads (Sigma-Aldrich) for longer than 3 h at 4°C. The beads were washed and then eluted with 0.2 mg/ml FLAG peptide (Sigma) for 1 h at 4°C. The eluate was incubated with EZView Red HA beads (Sigma-Aldrich) for longer than 2 h at 4°C. The HA beads were then washed and eluted with 100 mM glycine, pH 2.5. The eluate was neutralized with Tris (pH 8.8) and trichloroacetic acid (TCA) precipitated per standard procedures. The lyophilized eluate was submitted to the Taplin Mass Spectrometry Facility at Harvard University for mass spectrometry analysis.
Proximity ligation assay.
mCherry-LacI-FokI U2OS reporter cell lines (8) stably expressing FLAG-HA-tagged WT and mutant TIP60 (in pOZ-N vector) were seeded onto round coverslips in 24-well plates. On the day of the experiment, they were treated with Shield1 ligand and 4′OHT for 4 to 6 h to induce damage. The cells were then rinsed with PBS and fixed in 3% paraformaldehyde and 2% sucrose (pH 7.4) in PBS for 15 min at room temperature. The coverslips were then washed with PBS and permeabilized with 0.5% Triton X-100 in PBS for 5 min at 4°C. After permeabilization, they were washed twice with PBS-T (0.2% Tween), and PLA was performed using the Sigma-Aldrich Duolink in situ PLA kit in accordance with the manufacturer's instructions. Briefly, the cells on coverslips were blocked for 1 h at 37°C and then incubated with anti-HA and anti-53BP1 primary antibodies for 1 h at room temperature. The anti-rabbit MINUS probe and anti-mouse PLUS probe were then applied to the coverslips for 1 h at 37°C, followed by ligation of the probes for 30 min at 37°C. Rolling-circle amplification with fluorophore conjugation was then performed at 37°C for 100 min at 37°C. The coverslips were then mounted with Vectashield plus DAPI stain, and images were acquired on a fluorescence microscope.
Histone extraction and Western blotting.
MEFs were lysed with the NIB-250 buffer (15 mM Tris-HCl [pH 7.5], 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 250 mM sucrose) plus 0.3% NP-40 and inhibitors (PMSF, dithiothreitol [DTT], sodium butyrate, and sodium orthovanadate) at a ratio of 10:1. The nuclei were then rinsed with NIB-250 without NP-40, followed by acid extraction with 0.4 N sulfuric acid for longer than 3 h at 4°C. The histone-containing supernatant was then TCA precipitated on ice for 1 h, washed with acetone, and then air dried prior to solubilization with water and Western blot analysis. The histone samples were run on an SDS-PAGE gel (Invitrogen) in 2-(N-morpholino)ethanesulfonic acid (MES) buffer.
Histone preparation and mass spectrometry analysis.
Extracted histones were chemically derivatized and digested to tryptic peptides to make them amenable for bottom-up mass spectrometry as described earlier (35). The derivatized samples were desalted prior to liquid chromatography/mass spectrometry (LC-MS) analysis using C18 Stage-tips. For mass spectrometry, the peptides were separated using a 75-μm (inner diameter [ID]) by 17-cm Reprosil-Pur C18-AQ (3 μm; Maisch GmbH, Germany) nanocolumn fitted on an EASY-nLC nanoHPLC instrument (Thermo Scientific, San Jose, CA). The high-performance liquid chromatography (HPLC) gradient comprising 2% to 28% solvent B (A, 0.1% formic acid; B, 95% MeCN, 0.1% formic acid) over 45 min, from 28% to 80% solvent B in 5 min, 80% B for 10 min at a flow rate of 300 nl/min was used. This nLC was coupled online to an LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific), and data-independent acquisition (DIA) was used to acquire data (36). Briefly, full-scan MS (m/z 300 to 1,100) was acquired in the Orbitrap with a resolution of 120,000 (at 200 m/z) and an AGC target of 5 × 10e5. Tandem mass spectrometry (MS/MS) was done in in centroid mode in the ion trap with sequential isolation windows of 50 m/z with an automatic gain control (AGC) target of 3 × 104, a collision-induced dissociation (CID) collision energy of 35 and a maximum injection time of 50 ms. Data were analyzed using the in-house software, EpiProfile (37), wherein the peptide relative ratio was calculated using the total area under the extracted ion chromatograms of all peptides with the same amino acid sequence (including all of its modified forms) as 100%.
Chromatin immunoprecipitation-qPCR.
Dox-inducible LacI-FokI U2OS reporter cell lines (8) stably expressing FLAG-HA-tagged WT and mutant TIP60 were pretreated with doxycycline to induce transcription and histone acetylation at the LacO-transgene locus. After 3 h of doxycycline treatment, damage was induced with Shield1 ligand and 4′OHT addition. Alternatively, in the trichostatin A (TSA) condition, instead of inducing damage, the cells were treated with the histone deacetylase (HDAC) inhibitor trichostatin A instead. After 4 to 6 h of damage induction or TSA treatment, the cells were cross-linked with methanol-free formaldehyde in PBS (final concentration, 1%) for 5 min. Cross-linking was then stopped by incubation with glycine (final concentration, 0.125 M) for 5 min. Cells were then spun down, washed with PBS, and lysed in 10 mM Tris (pH 8.0), 10 mM NaCl, and 0.2% NP-40 plus PMSF and protease inhibitors on ice for 10 min. The isolated nuclei were then flash frozen and stored at −80°C. The next day, the nuclei were thawed on ice, resuspended in 10 mM Tris (pH 8.1), 1 mM EDTA, and 0.1% SDS and sonicated in a Covaris S-220 sonicator to obtain approximately 250- to 600-bp chromatin fragments. Chromatin fragments were diluted with 20 mM Tris (pH 8.1), 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, and 0.01% SDS to a final 1 ml buffer-IP and 10% leftover for input. The diluted chromatin was precleared with prebound rabbit IgG antibody-protein G magnetic beads (Pierce) for 2 h at 4°C and then incubated with prebound antibody-protein G magnetic beads overnight at 4°C. The next day, the beads were washed once in 20 mM Tris (pH 8.1), 2 mM EDTA, 50 mM NaCl, 1% Triton X-100, and 0.1% SDS, twice in 20 mM Tris (pH 8.1), 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, and 0.01% SDS, once in 10 mM Tris (pH 8.1), 1 mM EDTA, 0.25 M LiCl, 1% NP-40, and 1% deoxycholic acid, and twice in Tris-EDTA buffer (TE; 10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Washed beads were eluted twice with 100 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and reverse cross-linked with 0.1 mg/ml RNase, 0.3 M NaCl, and 0.3 mg/ml proteinase K at 65°C overnight. The DNA samples were purified by phenol-chloroform extraction, and qPCR with a standard curve protocol was carried out on an ABI 7900HT instrument using the SYBR green detection system. The primers used were as follows: forward, 5′-GGCATTTCAGTCAGTTGCTCAA-3′; reverse, 5′-TTGGCCGATTCATTAATGCA-3′.
Antibodies.
Antibodies for immunofluorescence assays were as follows: rabbit anti-53BP1 (Novus), 1:1,000; rabbit anti-BRCA1 (Millipore), 1:1,000; and mouse anti-BRCA1 C-9 (Santa Cruz), 1:100.
For PLA, the following antibodies were used: mouse anti-HA.11 (Covance), 1:1,000; rabbit anti-53BP1 (Novus), 1:1,000.
For Western blotting, the following antibodies were used: mouse anti-TIP60 (Santa Cruz), 1:500; rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; Cell Signaling), 1:5,000, or mouse anti-GAPDH (Cell Signaling), 1:2,000; custom-generated rabbit anti-mBRCA1 antibody; rabbit anti-TIP60 pS86 (Abcam), 1:500; rabbit anti-TIP60 pS90 (GeneTex), 1:500; rabbit anti-p400 (Abcam), 1:200; rabbit anti-EPC1 (Bethyl), 1:500; rabbit anti-HA (Cell Signaling), 1:1,000; rabbit anti-H2AZ K4/K7ac (Cell Signaling), 1:1,000; rabbit anti-H2AZ (Cell Signaling), 1:500; rabbit anti-H4K16ac (Active Motif), 1:2,000; H4 K5/K8/K12/K16ac (Millipore), 1:2,000; mouse anti-H4 (Cell Signaling), 1:200.
For ChIP assays, the following antibodies were used: rabbit anti-H4K16ac (Active Motif), rabbit anti-H4 (Millipore), rabbit anti-H2AZ K4/K7ac (Cell signaling), and rabbit anti-H2AZ (Abcam).
Statistics.
Mann-Whitney tests were performed using the GraphPad Prism 7 software. Error bars plotted are for standard errors of the means (SEM). Significance is marked by asterisks in the figures. Comparisons are made to control unless otherwise indicated.
ACKNOWLEDGMENTS
We thank members of the Greenberg lab for critical discussion, John Lough (Medical College of Wisconsin) for providing the Tip60 f/f mice, Eric Brown (University of Pennsylvania) for the Cre-ERT2 mice, and Brendan Price (Dana-Farber Cancer Institute) for TIP60 wild-type and HD mutant constructs.
This work was supported by National Institutes of Health grant R01-CA17494 and funds from the Basser Center for BRCA to R.A.G. M.L.L. was supported by NIH grants T32GM007170 and F30CA196115 and a Patel Scholars Award. B.A.G. acknowledges National Institutes of Health grants P01-GM110174 and P01-CA196539.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Hustedt N, Durocher D. 2016. The control of DNA repair by the cell cycle. Nat Cell Biol 19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 2.Price BD, D'Andrea AD. 2013. Chromatin remodeling at DNA double-strand breaks. Cell 152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeggo PA, Downs JA. 2014. Roles of chromatin remodellers in DNA double strand break repair. Exp Cell Res 329:69–77. doi: 10.1016/j.yexcr.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Hauer MH, Gasser SM. 2017. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev 31:2204–2221. doi: 10.1101/gad.307702.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhar S, Gursoy-Yuzugullu O, Parasuram R, Price BD. 2017. The tale of a tail: histone H4 acetylation and the repair of DNA breaks. Philos Trans R Soc Lond B Biol Sci 372:20160284. doi: 10.1098/rstb.2016.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunting SF, Callén E, Wong N, Chen H-T, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng C-X, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiao K-Y, Mizzen CA. 2013. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J Mol Cell Biol 5:157–165. doi: 10.1093/jmcb/mjs066. [DOI] [PubMed] [Google Scholar]
- 10.Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert J-P, Roques C, Pandita RK, Paquet E, Herst P, Gingras A-C, Pandita TK, Legube G, Doyon Y, Durocher D, Côté J. 2016. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol Cell 62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke TL, Sanchez-Bailon MP, Chiang K, Reynolds JJ, Herrero-Ruiz J, Bandeiras TM, Matias PM, Maslen SL, Skehel JM, Stewart GS, Davies CC. 2017. PRMT5-dependent methylation of the TIP60 coactivator RUVBL1 is a key regulator of homologous recombination. Mol Cell 65:900–916.e7. doi: 10.1016/j.molcel.2017.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sivanand S, Rhoades S, Jiang Q, Lee JV, Benci J, Zhang J, Yuan S, Viney I, Zhao S, Carrer A, Bennett MJ, Minn AJ, Weljie AM, Greenberg RA, Wellen KE. 2017. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol Cell 67:252–265.e6. doi: 10.1016/j.molcel.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien J-P, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, Durocher D. 2016. The structural basis of modified nucleosome recognition by 53BP1. Nature 536:100–103. doi: 10.1038/nature18951. [DOI] [PubMed] [Google Scholar]
- 14.Rothkamm K, Kruger I, Thompson LH, Lobrich M. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol 23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JTF, Tkáč J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell 49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Chapman JR, Sossick AJ, Boulton SJ, Jackson SP. 2012. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci 125:3529–3534. doi: 10.1242/jcs.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mok MTS, Henderson BR. 2012. Three-dimensional imaging reveals the spatial separation of γH2AX-MDC1-53BP1 and RNF8-RNF168-BRCA1-A complexes at ionizing radiation-induced foci. Radiother Oncol 103:415–420. doi: 10.1016/j.radonc.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Lemercier C, Legube G, Caron C, Louwagie M, Garin J, Trouche D, Khochbin S. 2003. Tip60 acetyltransferase activity is controlled by phosphorylation. J Biol Chem 278:4713–4718. doi: 10.1074/jbc.M211811200. [DOI] [PubMed] [Google Scholar]
- 19.Mo F, Zhuang X, Yao PY, Qin B, Su Z, Zang J, Wang Z, Zhang J, Dou Z, Tian C, Teng M, Niu L, Hill DL, Fang G, Ding X, Fu C, Liu X, Yao X. 2016. Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat Chem Biol 12:226–232. doi: 10.1038/nchembio.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao Y, Nagai Y, Deng G, Ohtani T, Zhu Z, Zhou Z, Zhang H, Ji MQ, Lough JW, Samanta A, Hancock WW, Greene MI. 2014. Dynamic interactions between TIP60 and p300 regulate FOXP3 function through a structural switch defined by a single lysine on TIP60. Cell Rep 7:1471–1480. doi: 10.1016/j.celrep.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher JB, Horst A, Wan T, Kim M-S, Auchampach J, Lough J. 2016. Depletion of Tip60 from in vivo cardiomyocytes increases myocyte density, followed by cardiac dysfunction, myocyte fallout and lethality. PLoS One 11:e0164855. doi: 10.1371/journal.pone.0164855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charvet C, Wissler M, Brauns-Schubert P, Wang S-J, Tang Y, Sigloch FC, Mellert H, Brandenburg M, Lindner SE, Breit B, Green DR, McMahon SB, Borner C, Gu W, Maurer U. 2011. Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol Cell 42:584–596. doi: 10.1016/j.molcel.2011.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin S-Y, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang S-M, Lian G, Liu Q, Ruan K, Wang Z, Zhang C-S, Chien K-Y, Wu J, Li Q, Han J, Lin S-C. 2012. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 25.Nie T, Yang S, Ma H, Zhang L, Lu F, Tao K, Wang R, Yang R, Huang L, Mao Z, Yang Q. 2016. Regulation of ER stress-induced autophagy by GSK3β-TIP60-ULK1 pathway. Cell Death Dis 7:e2563. doi: 10.1038/cddis.2016.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brauns-Schubert P, Schubert F, Wissler M, Weiss M, Schlicher L, Bessler S, Safavi M, Miething C, Borner C, Brummer T, Maurer U. 2018. CDK9-mediated phosphorylation controls the interaction of TIP60 with the transcriptional machinery. EMBO Rep 19:244–256. doi: 10.15252/embr.201744311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463–473. doi: 10.1016/S0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 28.Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 29.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keogh M-C, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev 20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol 27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, Bouchard N, Lacoste N, Utley RT, Gaudreau L, Côté J. 2010. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem 285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamburini BA, Tyler JK. 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sidoli S, Bhanu NV, Karch KR, Wang X, Garcia BA. 2016. Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J Vis Exp 2016:e54112. doi: 10.3791/54112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidoli S, Lin S, Xiong L, Bhanu NV, Karch KR, Johansen E, Hunter C, Mollah S, Garcia BA. 2015. Sequential window acquisition of all theoretical mass spectra (SWATH) analysis for characterization and quantification of histone post-translational modifications. Mol Cell Proteomics 14:2420–2428. doi: 10.1074/mcp.O114.046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan Z-F, Lin S, Molden RC, Cao X-J, Bhanu NV, Wang X, Sidoli S, Liu S, Garcia BA. 2015. EpiProfile quantifies histone peptides with modifications by extracting retention time and intensity in high-resolution mass spectra. Mol Cell Proteomics 14:1696–1707. doi: 10.1074/mcp.M114.046011. [DOI] [PMC free article] [PubMed] [Google Scholar]