Phorbol 12-myristate 13-acetate (PMA) promotes skin cancer in rodents. The mutations found in murine tumors are similar to those found in human skin cancers, and PMA promotes proliferation of human skin cells.
KEYWORDS: APOBEC3A, phorbol myristate acetate, psoriasis, replication fork and DNA damage, uracils in DNA
ABSTRACT
Phorbol 12-myristate 13-acetate (PMA) promotes skin cancer in rodents. The mutations found in murine tumors are similar to those found in human skin cancers, and PMA promotes proliferation of human skin cells. PMA treatment of human keratinocytes increases the synthesis of APOBEC3A, an enzyme that converts cytosines in single-stranded DNA to uracil, and mutations in a variety of human cancers are attributed to APOBEC3A or APOBEC3B expression. We tested here the possibility that induction of APOBEC3A by PMA causes genomic accumulation of uracils that may lead to such mutations. When a human keratinocyte cell line was treated with PMA, both APOBEC3A and APOBEC3B gene expression increased, anti-APOBEC3A/APOBEC3B antibody bound a protein(s) in the nucleus, and nuclear extracts displayed cytosine deamination activity. Surprisingly, there was little increase in genomic uracils in PMA-treated wild-type or uracil repair-defective cells. In contrast, cells transfected with a plasmid expressing APOBEC3A acquired more genomic uracils. Unexpectedly, PMA treatment, but not APOBEC3A plasmid transfection, caused a cessation in cell growth. Hence, a reduction in single-stranded DNA at replication forks may explain the inability of PMA-induced APOBEC3A/APOBEC3B to increase genomic uracils. These results suggest that the proinflammatory PMA is unlikely to promote extensive APOBEC3A/APOBEC3B-mediated cytosine deaminations in human keratinocytes.
INTRODUCTION
Phorbol 12-myristate 13-acetate (PMA; also called 12-O-tetradecanoylphorbol-13-acetate or TPA) causes inflammatory response in skin cells and promotes tumors. In the two-stage murine skin carcinogenesis protocol, a single application of subcarcinogenic dose of the mutagen 7,12-dimethylbenz[a]anthracene (DMBA), followed by repeated application of PMA, causes tumors (1). PMA is an analog of diacylglycerol and has the ability to bind and strongly activate protein kinase C family, which regulates signal transduction pathways through the activation of NF-κB (2, 3).
Skin carcinogenesis in mice and humans appears to follow common molecular mechanisms (4, 5). In both systems, mutations in the H-ras oncogene and tumor suppressor P53 are detected in early stages of the disease (5). Furthermore, a prolonged activation of protein kinase C leads to an inflammatory response through the synthesis of many growth factors and proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α) that cause a wide variety of cellular changes and promote oncogenic transformation (6). Inflammatory skin conditions in humans such as psoriasis are associated with increased risk for nonmelanoma skin cancers (7, 8). In keratinocytes this results in cell proliferation, increased production of reactive oxygen species, and inflammation (9–11). The ability of PMA to cause persistent inflammation and oxidative stress are thought to contribute to its function as a tumor promoter (4).
PMA is known to induce the expression of some members of the human APOBEC3 subfamily of DNA-cytosine deaminases (12–15). These seven enzymes (APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D/E, APOBEC3F, APOBEC3G, and APOBEC3H) belong to the AID/APOBEC family and convert cytosines in single-stranded DNA (ssDNA) to uracil (16). While error-free repair of these uracils restores the original C:G base pair, error-prone or incomplete repair of uracils, respectively, causes genome instability through the introduction of mutations or abasic sites and strand breaks (17). The main biological function of APOBEC3 subfamily of proteins is to provide innate immunity against viral infection and inhibit retrotransposition of endogenous retroelements partly through the creation of mutations and strand breaks (16, 18). In addition, it has been suggested that mutations caused by these enzymes in viral genomes may also benefit the viruses by allowing them to escape adaptive immunity (19). Recently, APOBEC3 mutational signature was also detected in multiple human cancers suggesting that these enzymes may play a role in tumor evolution (20, 21) and tumorigenesis (22–25). In particular, APOBEC3B (A3B) has been described as the major cancer genome mutator among the APOBEC3 proteins (26–30). APOBEC3A (A3A), which has a mutational signature similar to A3B, has also been implicated in cancer genome mutations in some studies (31, 32). Furthermore, when APOBEC3 mutational signature was split into A3A-like and A3B-like, mutations with an A3A-like signature were more common than those with A3B-like mutations (33).
A3A was initially discovered as a protein that is overexpressed when human skin keratinocytes were treated with PMA and was named phorbolin-1 (15). Elevated levels of A3A were also seen in skin inflammation conditions such as psoriasis and acne (15, 34). However, the impact of A3A protein on genome integrity may depend on the cell type and the subcellular localization of A3A. In monocytes, the endogenously expressed A3A is cytoplasmic and does not cause any significant genotoxicity when measured as cell viability (35). In contrast, several studies have shown that the protein made from stably or transiently transfected A3A gene is present in the nuclei, causes mutations and strand breaks, and disrupts the cell cycle (36–40). Therefore, the expression of A3A in keratinocyte cells in response to PMA treatment has the potential of causing mutations and genome instability, if it is transported to the nuclei.
To test this possibility, we treated a normal human oral keratinocyte cell line with PMA and confirmed the expression and nuclear localization of A3A. Surprisingly, the presence of A3A in the nucleus did not result in an increase in genomic uracil levels. We describe below the biochemical and biological response of this cell line to PMA treatment in the context of AID/APOBEC proteins and provide a possible explanation for why the nuclear A3A is not genotoxic to these cells.
RESULTS
Effects of PMA treatment on AID/APOBEC3 gene expression.
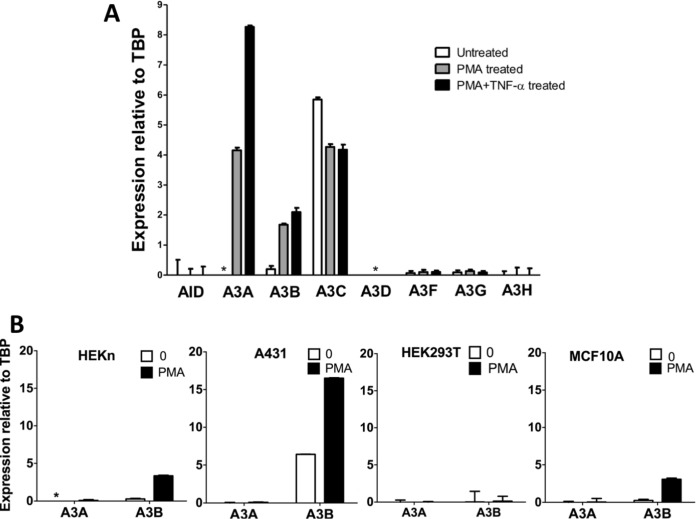
Normal oral keratinocyte (NOK) cells were treated with PMA and the changes in the expression of AID and APOBEC3 genes were determined using quantitative reverse transcription-PCR (qRT-PCR). The AID/APOBEC genes could be divided into three groups based on the effects of PMA treatment on their expression. A3C was the most expressed of all the eight genes in untreated NOK cells, and its expression decreased slightly following PMA treatment (Fig. 1A). In contrast, AID, A3D, A3F, A3G, and A3H genes were expressed at very low levels in the untreated cells, and these levels did not significantly increase in response to PMA treatment (Fig. 1A). The A3A and A3B genes were the only genes whose expression increased following PMA treatment, with the A3A expression increasing from undetectable levels to more than twice the level of A3B expression (Fig. 1A). These results suggest that the increased A3A protein expression reported in previous studies of PMA-treated normal keratinocytes (13, 15) is likely to be due to increased transcription, and find that PMA treatment increases A3B gene expression in addition to A3A. To determine whether the parallel increases in A3A and A3B gene expression following PMA treatment reflected some form of coordinated regulation of the two genes that would be common to other human cells, we tested the effects of PMA treatment in four other cell lines.
FIG 1.
Effects of PMA treatment on AID/APOBEC3 gene expression in different human cell lines. (A) qRT-PCR for mRNA expression of AID/APOBEC3s relative to TBP in NOK cells that were either untreated, treated with 25 ng/ml PMA, or treated with 25 ng/ml PMA and 10 ng/ml TNF-α for 24 h. (B) A3A and A3B mRNA expression relative to TBP without treatment (“0”) or after treatment with PMA for 24 h in HEKn, A431, and HEK293T cells and for 6 h in MCF10A cells. In all cases, the means and standard deviations (SD) are shown for triplicate qRT-PCRs. *, not detected.
None of the other cells tested expressed A3A at high levels with or without PMA treatment. This included two skin-derived cell lines, primary human neonatal keratinocyte cells (HEKn), and squamous cell carcinoma-derived cell line A431 (Fig. 1B). In contrast, A3B expression increased upon PMA treatment in three of the four cell types: HEKn, A431, and MCF10A (Fig. 1B). The increased expression of A3B gene following PMA treatment in MCF10A cells has been reported before (12). These results show that different types of primary and immortalized human cells have widely different responses to PMA treatment in terms of A3A and A3B gene expression and that the NOK cells are somewhat unusual in their ability to express A3A at high levels in response to PMA treatment.
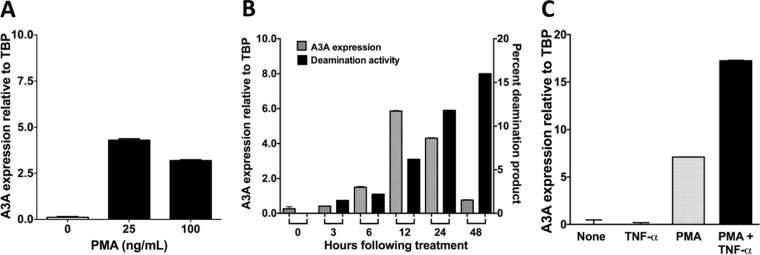
We optimized the expression of A3A in response to PMA treatment by varying the concentration of the phorbol ester, the length of treatment and combining it with cytokines. We found that the expression of A3A in NOK cells was higher at 25 ng/ml than at 100 ng/ml (Fig. 2A), and at the lower concentration it peaked around 12 h posttreatment (Fig. 2B). It is known that PMA causes changes in expression of a number of cytokines, including TNF-α, and we confirmed that PMA treatment of NOK cells increases the expression of TNF-α (S. U. Siriwardena and A. S. Bhagwat, unpublished data). TNF-α is known to activate NF-κB, which, in turn, is known to increase expression of A3B in some cell lines (12, 14), and hence we speculated that treatment of NOK cells with this cytokine may increase A3A/A3B expression. To test this, NOK cells were treated with a cocktail of PMA plus TNF-α (PMA+TNF-α), and the expression of AID/APOBEC genes was monitored. Consistent with the hypothesis, a combined treatment of NOK cells with PMA+TNF-α doubled the expression of A3A (Fig. 2C) and slightly increased A3B expression compared to treatment with PMA alone [Fig. 1A]. Combining PMA with TNF-α did not cause an increase in the expression of the other APOBEC3 genes or AID (Fig. 1A). We used the PMA+TNF-α combination in most subsequent experiments.
FIG 2.
Optimization of APOBEC3A upregulation by PMA. (A) qRT-PCR for A3A mRNA expression in NOK cells after 24 h of treatment with different doses of PMA. (B) Time-dependent change in A3A mRNA expression and cytosine deamination activity in whole-cell extracts following treatment with 25 ng/ml PMA. The deamination activity is shown as the percentage of substrate DNA made sensitive to strand cleavage by treatment with Ung, followed by NaOH. (C) A3A mRNA levels relative to TBP in NOK cells that were untreated or treated with 10 ng/ml TNF-α alone, 25 ng/ml PMA alone, or PMA+TNF-α for 24 h. For the quantification of gene expression using qRT-PCR, the means and SD are shown from triplicate samples.
Protein expression and DNA-cytosine deamination activity following PMA treatment.
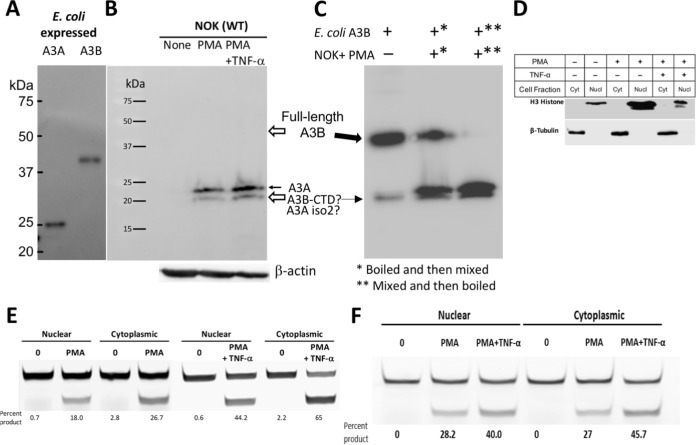
A monoclonal antibody against a polypeptide common to the carboxy-terminal domain of both A3A and A3B was used to detect these proteins (12). It readily detected the two proteins in a Western blot of cell extracts from E. coli cells expressing A3A or full-length A3B (Fig. 3A). The A3A protein was also detected in blots of whole-cell extracts of PMA treated NOK cells but not in untreated cells (Fig. 3B). Two bands were seen in this blot which may be the two known functional isoforms of A3A (23.0 and 21.7 kDa) (39, 76, 77). Furthermore, consistent with the qRT-PCR data, there was a further increase in the intensities of both the A3A bands when TNF-α was included with PMA during treatment (Fig. 3B). Surprisingly, there were no bands at the size of full-length A3B in the blot (Fig. 3B), suggesting that although the A3B gene is transcribed in NOK cells (Fig. 1A and 2C), posttranscriptional or posttranslational regulation prevents the accumulation of A3B protein in cells.
FIG 3.
Effect of PMA treatment on APOBEC3A and APOBEC3B protein expression and cytosine deamination activity. (A) Western blot of E. coli extract containing A3A or A3B using anti-A3A/A3B antibody. (B) Western blot analysis of whole-cell extracts of NOK cells either untreated (“None”) or treated with PMA or PMA+TNF-α for 24 h using anti-A3A/A3B antibody. The expression level of β-actin was used as a loading control. The position of full-length A3A is indicated by a closed arrow, and the expected positions of full-length A3B and the CTD of A3B are indicated by open arrows. (C) Mixing of E. coli extract containing full-length A3B with extracts of NOK cells treated with PMA. Overloading of E. coli extract (20 μg) in the first lane shows both the full-length A3B and a minor band consistent with the size of A3B-CTD. Both A3B forms are indicated by solid arrows. The order of mixing and boiling of the two extracts is indicated by asterisks in the panel footnotes. (D) Detection of organelle-specific protein markers using antibodies. Blots of cytoplasmic (Cyt) and nuclear (Nucl) extracts from NOK cells that were untreated or treated with PMA or PMA+TNF-α were probed using anti-histone H3 (nuclear marker) or anti-β-tubulin (cytoplasmic marker) antibodies. (E) In vitro cytosine deamination assay for nuclear and cytoplasmic fractions of NOK cells. A fluorescently labeled oligomer containing a single cytosine in 5′-TC context was incubated with the indicated cellular extract, and the uracils created by A3A/A3B were converted to strand breaks by successive treatment with E. coli Ung and NaOH (top band, substrate; bottom band, product). The percentages of cytosines converted to uracils were calculated based on band intensities and are shown below each lane. (F) In vitro cytosine deamination assay for nuclear and cytoplasmic fractions of UNGΔ/Δ NOK cells. The cell fractions were prepared, and the deamination assays were performed in the same manner as described for the UNG+/+ NOK cells.
To investigate this further, we mixed the cell extracts of E. coli expressing full-length A3B with PMA-treated NOK cell extracts and repeated the Western blot experiment. The blot was prepared under conditions that should increase the sensitivity of detection, using larger amounts of cell extracts and a longer exposure of the blot. Under these conditions, the full-length A3B expressed in E. coli was not only visible in the blot, but an additional band roughly the size of the A3B carboxy-terminal domain (CTD) was also visible (Fig. 3C). Another lane of the same gel contained proteins from preboiled E. coli and PMA-treated NOK cell extracts that were mixed together, and the blot showed all the bands seen with each extract separately, i.e., ∼46-kDa full-length A3B, ∼23-kDa A3A isoform 1, and the ∼22-kDa band which could be A3A isoform 2, A3B-CTD, or both (Fig. 3C). However, when the E. coli and NOK cell extracts were mixed together without boiling and then boiled, the full-length A3B band was no longer visible (Fig. 3C). This happened despite the presence of a protease cocktail in the lysis buffer (see Materials and Methods) and was reproducible (J. Stewart and A. S. Bhagwat, unpublished data). This suggests that the NOK cells contain a potent protease that eliminates the full-length A3B protein. However, because the putative A3B-CTD and the isoform 2 of A3A have roughly the same mobility on these gels (Fig. 3C), we cannot eliminate the possibility that the A3B-CTD persists in NOK cells after the proteolytic cleavage and contributes to the signal detected using anti-A3A/A3B antibody and the cytosine deamination activity in NOK cells.
The presence of DNA-cytosine deaminase activity in NOK cells was tested using a 40-mer containing a single cytosine, and the resulting uracils were excised using E. coli uracil DNA-glycosylase (Ung) creating a 25-nucleotide product. We monitored the activity in extracts from cells that were treated with PMA from 0 to 48 h and found that it increased continually during this time period (Fig. 2B, right y axis). Therefore, although A3A mRNA level peaks at 12 h post-PMA treatment and decreases thereafter (Fig. 2B, left y axis), A3A continues to accumulate in the cells. To determine whether this activity was confined to the cytoplasm, the NOK cells were treated with PMA or PMA+TNF-α for 24 h, their outer membranes were broken, and the cytoplasmic fraction was separated from the nuclear fraction. The selective presence of appropriate organelle-specific markers (H3 histone, nuclear; β-tubulin, cytoplasm) confirmed that both the fractions were fairly pure (Fig. 3D). We performed cytosine deamination assays using these fractions.
Little deamination activity was seen in untreated cells, and it increased >10-fold in both nuclear and cytoplasmic fractions of PMA-treated cells (Fig. 3E). Furthermore, the nuclear and cytoplasmic deamination activities were higher after PMA+TNF-α treatment compared to PMA alone in both of the the cellular compartments (Fig. 3E). Together, these results show that the treatment of NOK cells with PMA or PMA+TNF-α increases the transcription of both A3A and A3B genes, which results in a large increase in DNA-cytosine deamination activity in the nuclei of NOK cells.
Nuclear localization of anti-A3A/A3B antibody reactive protein.
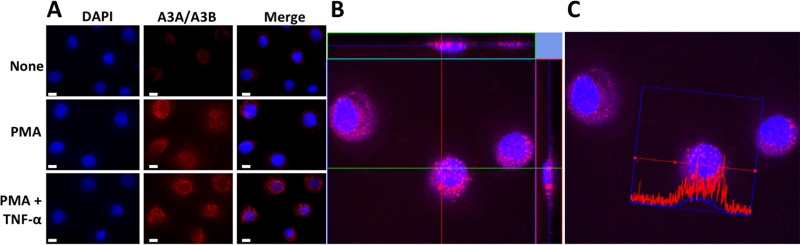
Although the appearance of nuclear deaminase activity following PMA and PMA+TNF-α treatment (Fig. 3E) was indicative of the presence of A3A/A3B-CTD in the nuclei, we sought to confirm this using an independent method. Furthermore, previous work with monocytes had shown that A3A is localized exclusively in the cytoplasm (35). To detect the presence of A3A in the nuclei, the stimulated NOK cells were fixed, treated with anti-A3A/A3B antibody, and visualized using fluorescence microscopy. The microscope images showed an increase in A3A/A3B signal following PMA or PMA+TNF-α treatments, and the protein(s) clearly appeared as foci within the nuclei and cytoplasm (Fig. 4A). Since full-length A3B is undetectable in the Western blots, it is reasonable to conclude that the new foci detected by the antibody are principally due to A3A with the possibility that A3B-CTD is also present in the nuclei.
FIG 4.
Subcellular localization of APOBEC3A using confocal microscopy. (A) Representative fluorescence microscopy images of untreated or treated NOK cells stained for A3A (red) and nuclei (blue). Size bars, 10 μm. (B) Confocal microscopy z-stacking images of NOK cells treated with PMA+TNF-α and immunostained for A3A (red) and nuclei (blue). An orthogonal analysis of foci in the nucleus in x-z and y-z planes shows overlap of red and blue. (C) Fluorescence intensity profile from a cell in the confocal image. The red (A3A) and blue (DAPI) intensity traces are shown across one cell.
To further establish the nuclear localization of A3A, z-sections were performed on the images of stained cells using a confocal microscope. A z-stack through the middle of a group of cells showed that A3A/A3B foci were found within both the nucleus and the cytoplasm of cells, and visualization of x-z and y-z planes showed that several of the A3A foci resided within the nucleus (Fig. 4B) (Siriwardena and Bhagwat, unpublished). Furthermore, quantification of DAPI (4′,6′-diamidino-2-phenylindole) and A3A/A3B-specific fluorescence signals across one cell showed that both the signals peaked over the nucleus (Fig. 4C). This confirms the results of cytosine deamination assays and clearly shows that a substantial amount of the A3A/A3B-CTD protein is present within the NOK nuclei.
Lack of increase in genomic uracils in PMA- and TNF-α-treated NOK cells.
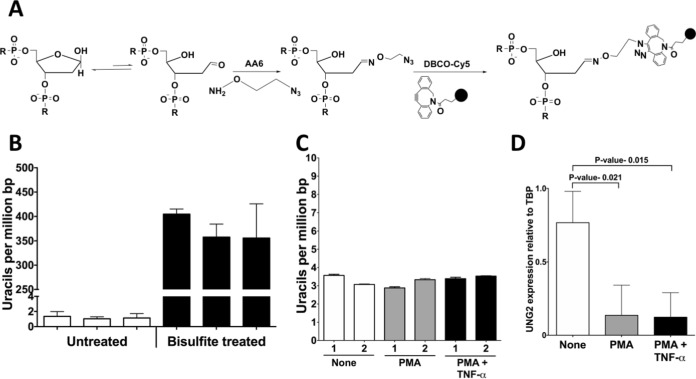
Since PMA or PMA+TNF-α treatment resulted in increased synthesis of A3A protein and its transport into NOK nuclei, we wanted to determine whether this caused cytosine deamination in DNA. We recently described assays to quantify genomic uracils using alkoxyamines ARP and AA3 (41, 42). These chemicals react with the abasic sites created by the excision of uracils in DNA by the Ung and are then tagged with a fluorescent label for quantification (41, 42). More recently, we also described another alkoxyamine, AA6, which also reacts with abasic sites and can be attached with a fluorescent tag using copper-free click chemistry (Fig. 5A) (43). We used AA6 for the quantification of uracils in NOK genome using a genomic DNA standard in which the uracils were quantified using liquid chromatography-tandem mass spectrometry (LC-MS/MS). This procedure provides reproducible quantification of genomic uracils over a wide range of uracil levels (Fig. 5B).
FIG 5.
Genomic uracil levels and UNG2 expression in NOK cells. (A) Principle of the method used for the quantification of genomic uracils. Treatment of DNA with E. coli Ung creates abasic sites which are reacted with an alkoxyamine, AA6, which contains a terminal azide group. This AA6-tagged DNA is reacted with the chemical DBCO-Cy5, and the fluorescence intensity of this DNA is interpolated in a standard plot created using DNA with known amounts of uracils to determine the uracil levels. (B) Reproducibility of uracil quantification. GM31, the DNA repair-proficient strain, contains few uracils and consistently shows about one uracil/106 bp. In contrast, bisulfite treatment of this DNA creates a large number of uracils, which are also quantified by this assay. The data represent three separate quantifications of each DNA sample using two or more replicates. (C) Quantification of genomic uracils in untreated, PMA-treated, or PMA+TNF-α-treated NOK cells after 24 h. The numbers 1 and 2 represent results from two independent experiments. (D) qRT-PCR for mRNA expression of UNG2 (nuclear isoform) relative to TBP in PMA- or PMA+TNF-α-treated NOK cells. In panels B and C, the means and SD are shown.
When this method was used to quantify genomic uracils in cells treated with PMA or PMA+TNF-α, little increase in the uracil levels was detected and repetition of the experiment gave essentially the same result (Fig. 5C). Although there was a slight increase in uracils following PMA treatment in one experiment, there was a decrease in genomic uracils following the phorbol ester treatment in the other experiment (Fig. 5C). We previously showed that when murine splenocytes are stimulated ex vivo, the transcription of both AID and UNG1 genes are upregulated, resulting in no net increase in genomic uracil levels (42). This observation suggested the possibility that PMA causes an increase in uracil excision activity by upregulating the UNG gene and the resulting higher activity levels are able to eliminate any excess uracils created by A3A. This was tested by quantifying of UNG2 transcripts using qRT-PCR. However, the UNG2 gene expression decreased instead of increasing 24 h after PMA+TNF-α treatments (Fig. 5D). Hence, upregulation of the UNG2 gene is not the cause of the lack of uracil level increase following PMA treatment.
Transfection of NOK cells with A3A-expressing plasmid increases genomic uracils.
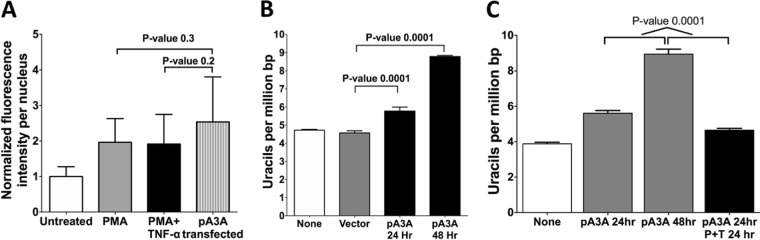
Despite this decrease in Ung2 transcription levels, when the uracil excision activity was tested in whole-cell extracts of untreated and treated cells, substantial levels of uracil-DNA glycosylase inhibitor (UGI)-sensitive uracil excision activity was detected in cells after PMA and PMA+TNF-α treatment (Siriwardena and Bhagwat, unpublished). This raised the possibility that the UNG2 activity in NOK cells was so high that, even after the reduction of UNG2 expression following PMA treatment, the residual uracil excision activity was able to eliminate the newly created uracils by A3A and A3B. To show that high levels of A3A expression can lead to increased genomic uracil levels, NOK cells were transfected with a plasmid expressing a EGFP-A3A fusion gene from a cytomegalovirus (CMV) promoter. The transfected cells were reacted with the anti-A3A/A3B antibody, and the resulting nuclear fluorescence was quantified for representative cells. The result showed that while the treatment of NOK cells with PMA or PMA+TNF-α doubled the A3A/A3B-specific signal, transfection of the cells with A3A-expressing plasmid caused only an ∼50% further increase in fluorescence, and this change was not statistically significant (Fig. 6A). This shows that treatment of cells with PMA creates roughly comparable levels of nuclear A3A/A3B-CTD as plasmid transfection.
FIG 6.
Effects of transfection of NOK cells with a plasmid expressing A3A. (A) Quantification of the fluorescence intensity in the nuclei of NOK cells that were untreated, treated with PMA, treated with PMA+TNF-α, or transfected with A3A plasmid. The cells were fixed and stained with anti-A3A/A3B antibody. (B) Quantification of genomic uracils in NOK cells transfected with a plasmid expressing A3A or a control plasmid lacking the A3A gene (vector). Uracil levels in A3A-transfected cells harvested at 24 or 48 h are shown. (C) Quantification of genomic uracils in NOK cells that were untreated, transfected with the A3A plasmid, or treated with PMA+TNF-α for 24 h after transfection. The means and SD are shown. The P value was calculated using a two-tailed t test.
When the genomic uracils were quantified in cells transfected with A3A-expressing plasmid, there was a statistically significant increase in uracil levels after 24 h, and the level increased further in the next 24 h (Fig. 6B). This shows that the expression of A3A protein as a result of transfection of NOK cells with a plasmid was capable of deaminating cytosines in the genome. Furthermore, the uracils generated by the enzyme could not be completely excised by UNG2, causing an increase in genomic uracil levels.
Lack of increase in genomic uracils in PMA- and TNF-α-treated UNGΔ/Δ NOK cells.
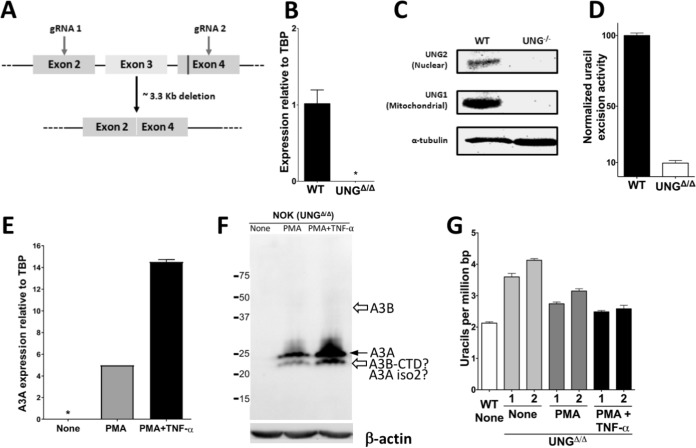
To determine further whether UNG2 in NOK cells was eliminating uracils created by A3A after PMA or PMA+TNF-α treatment, we constructed an UNGΔ/Δ derivative of NOK cells using CRISPR/Cas9 technology. The strategy involved deleting all of exon 3 and parts of exons 2 and 4, eliminating several residues critical for catalysis, and is outlined in Fig. 7A. The resulting cells showed elimination of UNG-specific transcripts (Fig. 7B), complete loss of both UNG1 and UNG2 isoforms of the protein (Fig. 7C), and a 90% loss in uracil excision activity (Fig. 7D). The remaining uracil excision activity is likely to be due to backup DNA glycosylases such as SMUG1 (44–46). UNGΔ/Δ NOK cells are still responsive to PMA treatment, overexpressing A3A (Fig. 7E and F) and increasing cytosine deamination activity in nuclear and cytoplasmic fractions (Fig. 3F).
FIG 7.
Generation, validation, and use of UNGΔ/Δ NOK cell lines. (A) Schematic representation of a part of the human UNG gene and the gRNAs designed to cause a large deletion. The region of exon 4 coding for catalytically important residues of UNG is shown as a vertical line. (B) Quantification of mRNA expression of UNG2 in WT and UNGΔ/Δ NOK cells relative to TBP. *, not detected. (C) Western blot analysis of whole-cell extracts for the expression of two isoforms of the UNG protein in WT and UNG-knockout (UNG–/–) NOK cells. The expression level of α-tubulin was used as a loading control. (D) Normalized uracil excision activity of UNGΔ/Δ NOK cells relative to WT (set to 100). The means and SD are shown. The activity was determined by incubating an oligomer containing a single uracil with whole-cell extracts, followed by cleavage at the resulting abasic sites and gel electrophoresis. (E) Expression of A3A in UNGΔ/Δ NOK cells that were either untreated, treated with PMA, or treated with PMA+TNF-α as determined by qRT-PCR. (F) Western blot analysis of whole-cell extracts of UNGΔ/Δ NOK cells that were either untreated (“None”) or treated with PMA or PMA+TNF-α for 24 h using anti-A3A/A3B antibody. The expression level of β-actin was used as a loading control. The position of full-length A3A is shown by a closed arrow, and the expected positions of full-length A3B and the CTD of A3B are indicated by open arrows. (G) Comparison of genomic uracils in untreated WT cells to those in untreated, PMA-treated, or PMA+TNF-α-treated UNGΔ/Δ NOK cells for 24 h. The numbers 1 and 2 indicate two independent UNGΔ/Δ clones. The means and SD are shown.
Two independent clones of UNGΔ/Δ cells were characterized further and had, respectively, 69 and 94% higher levels of genomic uracils than wild-type (WT) NOK cells in the absence of PMA treatment (Fig. 7G). This suggests that the backup DNA repair pathways are unable to remove all the uracils introduced in cellular DNA through spontaneous or A3B/A3C-mediated (Fig. 1A) deamination of cytosines or the incorporation of dUTP during replication (44, 47). However, treatment of these cells with PMA or PMA+TNF-α did not further increase the uracil levels and instead slightly decreased them (Fig. 7G). Since the expression of A3A peaks around 12 h after PMA treatment (Fig. 2B), we determined the uracil levels at multiple time points for 24 h after PMA treatment and found no increase in uracils at any of these time points (Siriwardena and Bhagwat, unpublished). This finding is in contrast to the expression of A3A from a transfected plasmid, which increases the uracil levels in both WT NOK cells (Fig. 6B) and UNGΔ/Δ cells (Siriwardena and Bhagwat, unpublished). These data, together with the uracil quantification data with WT NOK cells (Fig. 5C), suggest that the phorbol ester treatment does not generate measurable levels of excess uracils and may instead cause fewer uracils to accumulate in the genome.
PMA treatment causes reversible arrest in cell growth.
We and others have shown that a major target for the APOBEC3 subfamily of enzymes in the genome is the lagging strand template within replication forks (48–51). Based on this observation, we reasoned that the lack of increase in genomic uracils following phorbol ester treatment could be explained if this treatment were to reduce replication. To test this, we monitored the growth of NOK cells following PMA and PMA+TNF-α treatments.
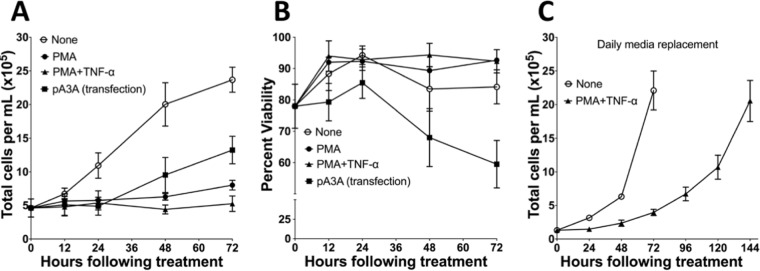
The treatment of cells with PMA alone stopped cell growth for at least 24 h, and this was followed by a slow growth resulting in an increase in cell number of only ∼70% over 3 days. The treatment with PMA+TNF-α had an even stronger effect on cell growth, causing its complete cessation for at least 72 h (Fig. 8A). This was in contrast to untreated cells, which increased 5-fold in number, and cells transfected with a plasmid expressing A3A tripled in number during the same time period (Fig. 8A). These data show that although both PMA treatment and plasmid transfection result in increased A3A expression, the former treatment has a much stronger negative effect on cell growth.
FIG 8.
Effects of PMA treatment or A3A transfection on NOK cell growth and viability. (A and B) Comparison of the change in total cell number (A) and the viability (B) of NOK cells that were untreated, PMA treated, PMA+TNF-α treated, or transfected with a plasmid expressing A3A. (C) Comparison of the change in total cell number of untreated or PMA+TNF-α-treated NOK cells with daily medium replacement. The means and SD are shown for each data point.
The two experimental conditions also had different effects on cell viability. While the PMA or PMA+TNF-α treatments had little effect on the viability of cells even after 72 h of treatment, the cells transfected with plasmid expressing A3A lost viability after 24 h, decreasing viability by about 20% over 72 h (Fig. 8B). Similarly, transfection of UNGΔ/Δ NOK cells with A3A-expressing plasmid also yielded a loss in viability, but little loss of viability occurred when the same cells were treated with PMA or PMA+TNF-α (Siriwardena and Bhagwat, unpublished). To confirm that the PMA+TNF-α-treated cells had not lost their ability to grow, we replaced the growth medium in treated and untreated cells every 24 h and found that the density of treated cells increased gradually, reaching the same density as untreated cells after 6 days (Fig. 8C).
We wondered whether PMA treatment could overcome the ability of plasmid-coded A3A to deaminate cytosines in the genome. To determine this, NOK cells were transfected with A3A-expressing plasmid and, after 24 h, half of the cells were treated with PMA+TNF-α. When genomic uracils were quantified from both untreated and treated cells, PMA+TNF-α was found to inhibit uracil creation by A3A. While the plasmid-transfected cells that were not treated with PMA+TNF-α continued to accumulate uracils 24 to 48 h posttransfection, the transfected cells treated with PMA+TNF-α at 24 h did not accumulate more uracils but may have slightly decreased the uracil levels (Fig. 6C). Together, these results show that PMA treatment of NOK cells stops cell growth (Fig. 8A) without a loss of cell viability (Fig. 8B) and does not result in the accumulation of uracils in the genome (Fig. 5C). In contrast, NOK cells transfected with a plasmid expressing A3A continue to grow after an initial pause (Fig. 8A), accumulate uracils in their genome (Fig. 6B), and lose viability (Fig. 8B).
DISCUSSION
The discovery of mutations caused by AID in immunoglobulin genes (52) and APOBEC3G in the HIV genome (53) has raised the possibility that the AID/APOBEC family of proteins could cause mutations in many cellular genes. Sequencing of individual genes (54) and whole exomes and genomes (24, 26) in cells expressing these enzymes offers strong support for this notion. In particular, the discovery that a substantial fraction of breast cancer cell lines (27) and patient tumors harbor elevated levels of A3B protein, genomic uracils, and mutations with a signature characteristic of this deaminase suggested that these enzymes may make a major contribution to cancer genome mutations (24, 26, 27). Later, when the mutational signatures of A3A and A3B were distinguished from each other, mutations with an A3A-like signature were found to be common in patient tumors (33). Subsequent demonstration that B cell lymphoma cell lines and tumors also accumulate uracils and mutations characteristic of AID (42, 55) and that other types of tumors contain certain mutations in a sequence context preferred by A3A (33) or A3B (22, 23, 26, 31, 33) suggests that this is a general phenomenon in human cancer. Since the causal relationship between the expression of A3A/A3B genes with such mutations in human tumors is sometimes difficult to establish, it is important to do so in human tissue culture models.
We have shown here that, in contrast to the correlation between high A3A or A3B expression and APOBEC signature mutations in many cancer genomes, the greatly elevated expression of A3A and the moderately high expression of A3B genes as a result of the treatment of a human keratinocyte cell line with PMA does not result in increased damage to DNA in the form of uracils. This is true despite the fact that the PMA treatment creates a large increase in DNA-cytosine deamination activity in the nuclear extracts and an increase in foci inside the nuclei that bind anti-A3A/A3B antibody. While our data cannot rule out cytosine-to-uracil conversions in a small number of genomic regions, creating localized mutations, these findings suggest that the expression of APOBEC genes and their presence in nuclei may not always lead to genome-wide mutations and instability.
Our results were unexpected because previous experiments in which A3A or A3B genes were expressed in human cells via plasmid transfection resulted in mutations (27, 56) and strand breaks (38). The expression of A3A from a plasmid-based Tet-On system also resulted in increased uracil levels, the induction of DNA damage response, and cell cycle arrest in the S phase (37). We also found that expression of a plasmid-based A3A gene from a CMV promoter in NOK cells results in increased levels of genomic uracils (Fig. 6B), confirming that the expression of exogenously introduced A3A gene causes damage to the cellular genome.
A possible trivial explanation for the lack of increase in genomic uracils in UNG+/+ (Fig. 6B) and UNGΔ/Δ (Fig. 7G) NOK cells following PMA or PMA+TNF-α treatment is that the expressed A3A is not transported into the nucleus or is efficiently exported out of it. In fact, A3A expressed in monocytes and macrophages following type I interferon treatment is predominantly cytoplasmic in two-dimensional (2D) images of cells stained with anti-A3A antibodies (57) and does not cause mutations in the cellular thymidine kinase gene (58). We eliminated this possibility by demonstrating that some A3A foci are seen in the nuclei of PMA-treated NOK cells in both 2D images and z-sections across nuclei (Fig. 4), and the nuclear cytosine deamination activity increases following PMA treatment (Fig. 3E and F).
Consequently, the likely explanation for the lack of increase in genomic uracils following the expression of PMA-induced A3A is that the substrate for the enzyme, ssDNA, is largely absent in the treated cells. This happens because PMA-treated NOK cells are severely inhibited in cell growth (Fig. 8A). Depending on the cell type, PMA can cause either cell cycle progression or cell cycle arrest (59–65). When PMA causes cell cycle arrest, some cells arrest in G2 (59, 62), while others arrest at the G1 boundary (66) by a process referred to as hypermitogenic arrest (67). In either case, the cells do not enter S phase.
In contrast, cells that have been transfected with A3A-expressing plasmid continue to divide after a 24-h pause. We and others have shown that the APOBEC3 subfamily proteins, when expressed in E. coli (48) or yeast (50) target the lagging strand template within replication forks. In other experiments, the induction of chromosomal doxycycline-regulated A3A gene in a hepatic cell line resulted in a significant increase in genomic uracils, which was dependent on the ability of cells to enter S phase (37). Furthermore, analysis of human tumors with strong A3A/A3B mutational signatures have shown that mutations are strand biased around replication origins in a manner that suggests that the source of the mutations was cytosine deamination in the lagging strand template (49, 51). Therefore, while A3A expression through nonphysiological means (plasmid transfection or induction with doxycycline) introduces DNA damage in a replication-dependent manner, PMA treatment of NOK cells inhibits cell growth and prevents the enzyme from generating uracils at detectable levels.
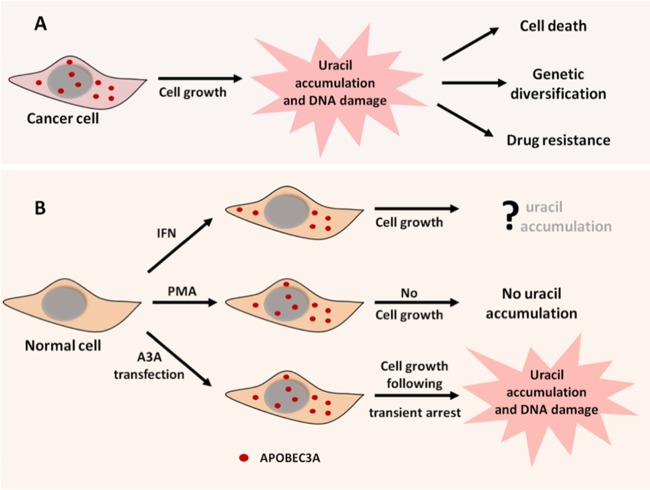
The AID/APOBEC family enzymes are potentially hazardous to cells. They can cause mutations, strand breaks, and translocations (27, 31, 39, 68) and have been implicated in causing cancer (24, 26, 27, 69), as well as in the diversification of tumors through mutations promoting drug resistance (20, 21) (Fig. 9A). Such genomic instability is also be demonstrated by expressing A3A in human cells under nonphysiological conditions (37, 38) (Fig. 9, lower branch). Consequently, normal cells that express these enzymes must have elaborate mechanisms to protect their genomes against them. One of these mechanisms operates on A3F and A3G expressed in T cells and A3A expressed in monocytes. In these cases, the proteins remain overwhelmingly in the cytoplasm (35, 70, 71) and thus do not cause significant damage to the cellular genome (35) (Fig. 9B, upper branch). This does not apply to PMA-treated NOK cells because A3A localizes to both the cytoplasm and the nucleus (Fig. 4) but still does not increase the levels of genomic uracils (Fig. 5C). Another proposed mechanism for protecting human genome against APOBECs is through proteins that bind the deaminase. TRIB3 protein binds A3A and A3C, causing the degradation of latter protein and protecting the genome against cytosine deamination and strand breaks (36). This mechanism is unlikely to be operative in NOK cells because we found robust levels of A3A protein and cytosine deamination activity following stimulation with PMA.
FIG 9.
Consequences of APOBEC3A expression in cancer cells and normal cells. (A) In rapidly dividing cancer cells, A3A expression causes an increase in genomic uracils, resulting in genome instability. (B) The outcomes of A3A expression in normal cells depend on the mode of A3A induction and the state of cell growth. The upper branch of the diagram shows A3A localized exclusively in the cytoplasm and preventing DNA damage. The middle branch shows A3A in both the cytoplasm and the nucleus and shows how growth arrest protects the genome from A3A. The lower branch shows the results of the nonphysiological expression of A3A, resulting in extensive DNA damage due to continued DNA replication or growth arrest in the S phase.
The reversible cessation of DNA replication seen in NOK cells is an attractive alternative mechanism for the protection of cellular genome from APOBEC enzymes. Since these enzymes function to protect against viruses and foreign DNA, their synthesis is likely to be upregulated in a cell in response to cytokines released by neighboring infected cells. In this situation, the chances of cell survival would be enhanced by the upregulation of APOBEC enzymes as part of a broad innate immune response and the damage to the cellular genome would be minimized by a block in DNA replication (Fig. 9B, middle branch). In contrast, tumor cells often divide rapidly, and hence the overexpression of AID/APOBEC proteins and their transport to the nucleus will result in mutations and strand breaks in the cellular genome. This leads to the DNA damage response (37, 38, 72) and cell death through apoptosis (39). This could also lead to genetic diversification of the tumor, allowing it to adapt to its environment and escape antitumor chemotherapy (21). Therefore, the consequences of the expression of APOBEC enzymes in different cells are likely to be determined by a complex set of physiological responses involving intracellular protein transport, protein degradation, and cell growth.
MATERIALS AND METHODS
Growth and treatment of cell lines.
The human oral keratinocyte cell line, NOK, was prepared from gingival tissue by transfecting it with the hTERT gene (73) and was kindly provided by Karl Munger (Harvard Medical School). Cells were cultured in serum-free medium (Keratinocyte-SFM; Life Technologies) supplemented with epidermal growth factor and bovine pituitary extract. The human cell lines HEK293T (immortalized embryonic kidney) and MCF-10A (breast epithelial) were obtained from the American Type Culture Collection, and the primary human epidermal keratinocytes (HEKn) were obtained from Thermo-Fisher. The epidermoid carcinoma cell line A431 was kindly provided by Young-Hoon Ahn (Wayne State University). HEK293T and A431 cells were cultured in Dulbecco’s modifed Eagle medium (HyClone) with 10% fetal bovine serum and 1% penicillin-streptomycin (Pen-Strep) (HyClone). MCF-10A cells were grown in HuMEC basal serum-free medium (Gibco) with a HuMEC supplement kit and 100 ng/ml cholera toxin (Sigma). HEKn cells were grown in EpiLife medium (Gibco) supplemented with EpiLife defined growth supplement (EDGS) and 1% Pen-Strep. All cultures were incubated at 37°C in a humidified incubator with 5% CO2 in T25 or T75 flasks or on 24-well plates (CytoOne; USA Scientific). All of the cell lines were used within 12 months of their short tandem repeat (STR)-based validation (Wayne State University BioBank). One million cells were grown in T25 cell culture flasks for 24 h. Different concentrations of PMA (Sigma) alone or in combination with 10 ng/ml TNF-α (Stem Cell Technologies) were added to the media, and the cells grown further for the indicated lengths of time.
Generation of UNGΔ/Δ NOK cell line using CRISPR/Cas9 technology.
Two guide RNAs were designed using CRISPR DESIGN (http://crispr.mit.edu/) tool to delete a part of exon 2, all of exon 3, and part of exon 4 of UNG2 gene (GenBank accession number NM_080911) in NOK cells. Exon 2 was targeted by using the oligomers 5′-CACCGCGGCCCGCAACGTGCCCGT and 5′-AAACACGGGCACGTTGCGGGCCGC (PAM sequence GGG), while exon 4 was targeted using the oligomers 5′-CACCGCTTGATTAGGTCCATGATA and 5′-AAACTATCATGGACCTAATCAAGC (PAM sequence TGG). Annealed oligonucleotides were cloned into pSpCas9 (BB)-2A-GFP vector (kindly provided by Kefei Yu, Michigan State University) and verified by DNA sequencing. The two CRISPR/Cas9 vectors with guide DNAs were transfected into NOK cells using Xfect transfection reagent (Clontech). After 48 h, green fluorescent protein (GFP)-positive cells were sorted by fluorescence-activated cell sorting (Microscopy, Imaging, and Cytometry Resources Core at Wayne State University), and single cells were seeded into a 96-well plate. DNA was obtained from single cell subclones using QuickExtract DNA extraction solution (Epicentre) for PCR-based screening for deletions with primers specific for the UNG2 gene (forward primer 5′-TGTTCCACAAATGGGCGTCT and reverse primer 5′-CTGCAGTCACCTGTAAAGCAAC). The clones with apparent gene deletions were further analyzed by Sanger sequencing, qRT-PCR for mRNA expression, Western blotting for UNG protein expression, and uracil excision activity assay (see below) to confirm the deletions.
RNA extraction and qRT-PCR.
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription of RNA was carried out with oligo-d(T)23 primers and a ProtoScript II first-strand cDNA synthesis kit (New England Biolabs). To determine the expression levels of genes for AID, A3A, A3B, A3C, A3D, A3F, and A3H, cDNAs were amplified using previously described primers (27) and the PowerUP SYBR Green Mix (Applied Biosystems), along with primers for TATA box-binding protein (TBP). The expression levels of TNF-α and UNG were determined using the following primers: TNF-α (forward, 5′-GGAGAAGGGTGACCGACTCA; reverse, 5′-CTGCCCAGACTCGGCAA), UNG2 (forward, 5′-CCTCCTCAGCTCCAGGATGA; reverse, 5′-TCGCTTCCTGGCGGG), and Ung1 and UNG2 (forward, 5′-CCCCACACCAAGTCTTCACC; reverse, 5′-TTGAACACTAAAGCAGAGCCC). The amplification was performed using an Applied Biosystems 7500 Fast Real-Time PCR system. The data were analyzed using the ΔΔCT method.
Quantification of genomic uracils.
The cells were harvested by centrifugation and lysed by incubation for 1 h at 37°C in Tris-EDTA buffer (TE) containing 2 μg/ml of RNase A and 0.5% sodium dodecyl sulfate (SDS), followed by incubation with proteinase K (100 μg/ml; Qiagen) at 56°C for 3 h. The DNA was isolated by phenol-chloroform extraction and ethanol precipitation and then dissolved in TE.
The uracils in the genomic DNA were quantified using an assay based on AA6, an alkoxyamine that can be coupled with a fluorescent dye using copper-free click chemistry (43). This assay is a modification of previously described assay using an alkoxyamine called AA3 (41). Briefly, HaeIII-digested genomic DNA was incubated with methoxyamine (Fisher Scientific) at a 10 mM final concentration to block the endogenous abasic sites. The DNA was treated with E. coli uracil DNA-glycosylase (Ung; New England Biolabs) and 2 mM AA6. AA6-tagged DNA was reacted with DBCO-Cy5 (1.7 μM; Click Chemistry Tools) for 2 h at 37°C with shaking. Labeled DNA was purified using a DNA Clean and Concentrator kit (Zymo Research). DBCO-Cy5-labeled DNA was transferred onto a positively charged Zeta probe membrane (Bio-Rad), and the membrane was scanned using a Typhoon 9210 phosphorimager (GE Healthcare). The images were analyzed using the ImageQuant software to quantify the fluorescence intensities.
Quantification of uracils in CJ236 DNA using LC-MS/MS.
Uracils in CJ236 DNA were quantified using Shimadzu LC/MS 8040, which has a Nexera X2 ultraperformance liquid chromatography (UPLC) system coupled to a triple-quadrupole mass spectrometer operated under multiple-reaction-monitoring mode. For the first stage of the separation, Acquity UPLC HSS T3 reversed-phase column (1.8 μm, 2.1 by 100 mm) was used at 30°C. Uracils were extracted from CJ236 genomic DNA according to a previously described procedure (27) and separated on the UPLC column using a gradient containing solvent A (100 mM acetic acid) and solvent B (methanol). The solvent B was ramped up from 0 to 10% in 0.25 min, brought back to 0% in 3.5 min, and then held at 0% for up to 8.75 min at a flow rate of 0.3 ml/min. Stock standard solutions of both uracil and the uracil isotope were made at a 1-mg/ml concentration from uracil (Sigma) and uracil (13C4, 15N2; Cambridge Isotope Laboratories, Inc.) in water containing 0.1% formic acid and stored at –20°C. The standard curve for uracil was prepared using concentrations of the base ranging from 100 to 500 ng/ml, each spiked with 1,000 ng/ml of the uracil isotope. Mass spectrometry detection was performed using positive electrospray ionization, monitoring the mass transitions 113.08 [MH+] > 70.08 [M-CONH]+ and 113.08 [MH+] > 96.08 [M-NH2]+ for uracil and 119.08 [MH+] > 74.08 [M-CONH]+ for isotopically labeled uracil, respectively. The area under the curve for the fragmentation peaks of uracil and isotopically labeled uracil were obtained from the LC-MS/MS analysis. The ratios of areas under the curve were plotted against the respective concentration ratios of uracil and isotopically labeled uracil to generate a standard plot.
To obtain the amount of uracils in CJ236 DNA, an amount of isotopically labeled uracil was added to DNA during the uracil extraction procedure. After LC-MS/MS analysis, the ratio of areas under the curve for the fragmentation peaks was obtained as described above and interpolated onto the standard plot to obtain the concentration ratio of uracil and isotopically labeled uracil. This number was used to calculate the number of uracils in CJ236 genomic DNA (3,754 uracils per 106 bp).
Western blot analysis.
To test the ability of the rabbit anti-human A3A/A3B monoclonal antibody (12) (kindly provided by R. Harris, University of Minnesota) to detect both A3A and full-length A3B, E. coli cells containing the plasmid pSU-A3A (74) or pSU-A3B-full (75) were grown to mid-log phase, and A3B expression was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM and growing the cells further for 4 h. The cells were collected by centrifugation and broken by sonication in a buffer (20 mM Tris [pH 8.0], 50 mM NaCl), and the cell debris was removed by centrifugation. One microgram of cell extract from A3A- and A3B-expressing cells was loaded onto a 15% SDS-PAGE gel and transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore).
In another experiment, whole-cell extracts were prepared by incubating treated and untreated NOK cells in lysis buffer (50 mM Tris-HCl, 150 mM NaCl [pH 7.4], 1% Triton X-100, 1× Halt protease inhibitor cocktail) for 1 h at 4°C, followed by centrifugation at 12,000 × g for 10 min to clarify the lysate. The total protein concentration was measured using Bio-Rad protein assay dye reagent concentrate (Bio-Rad). Protein samples (30 to 50 µg) were separated on an SDS-PAGE gel and transferred to a PVDF membrane. In another experiment, 20 µg of E. coli extract containing full-length A3B protein and 50 µg of extract from PMA-treated NOK cells were either boiled separately and then mixed together or mixed first and then boiled before loading them onto an SDS-PAGE gel and transferring them to a PVDF membrane.
After the transfer of proteins, the membranes were blocked with 5% (wt/vol) nonfat milk and probed with the anti-human A3A/A3B antibody (12) at a 1:1,000 dilution or rabbit anti-β-actin monoclonal antibody (Cell Signaling Technology; 1:1,000 dilution). The β-actin served as the loading control. In other experiments the membrane was probed with rabbit anti-UNG polyclonal antibody (Abcam; 1:1,000 dilution) or mouse anti-α-tubulin monoclonal antibody. The protein bands were visualized by the addition of Super Signal West Pico Plus chemiluminescence substrate (Thermo Fisher) and detected using FluorChemQ scanner (Cell Biosciences, Inc.).
In vitro cytosine deamination or uracil excision assays.
For the cytosine deamination assay, nuclear and cytoplasmic proteins were extracted from cells using a NucBuster protein extraction kit (Millipore), and the protein concentrations were measured using Bio-Rad protein assay dye reagent. To confirm that there was little cross-contamination of the cellular fractions, nuclear and cytoplasmic protein extracts (30 µg) were analyzed by separating them on an SDS-PAGE gel, transferring the proteins onto Immobilon-P PVDF membrane, and probing the membrane with rabbit anti-histone H3 antibody (nuclear marker; Cell Signaling Technology) at a 1:500 dilution or with mouse anti-β-tubulin antibody (cytoplasmic marker; Santa Cruz) at a 1:500 dilution, followed by the appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology; 1:1,000). The resulting signal was detected as described above.
The deamination reactions were performed with 3 µg of protein and 2 pmol of ssDNA containing a single cytosine (5′-ATTATTATTATTATCGATTTATTTATTTATTTATTTATTT-3′-6-carboxyfluorescein (FAM) [indicated in boldface]) was performed in the reaction buffer (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1 mM dithiothreitol) for 1 h at 37°C. The reactions were terminated by heating them to 95°C for 10 min. Five units of E. coli Ung (New England Biolabs) were added to the reaction, and incubation was continued at 37°C for 30 min. The reactions were stopped by adding NaOH to 0.1 M and heating to 95°C for 10 min. The reaction products were separated on a 15% denaturing polyacrylamide gel and visualized using Typhoon 9210 phosphorimager (GE Healthcare). ImageJ software was used to quantify the intensities of the substrate and the deamination product.
For the uracil excision assay, whole-cell extracts were prepared as described above, and 3 µg of protein was incubated with 2 pmol of ssDNA containing a single uracil (5′-ATTATTATTATTATUGATTTATTTATTTATTTATTTATTT-FAM) for 1 h at 37°C. In some reactions, UGI (New England Biolabs) was added to the cell lysate to inhibit Ung prior to incubation with the substrate. The reactions were stopped by adding NaOH to 0.1 M and heating to 95°C for 10 min. The products were processed and analyzed in a similar way as the C deamination reactions.
Transfection of cells with A3A-expressing plasmid.
One million WT or UNGΔ/Δ NOK cells were transiently transfected with pEGFP-N3-A3A, an A3A expression vector (58) (kindly provided by R. Harris) or empty vector (pEGFP-N3) using Xfect transfection reagent according to manufacturer’s instructions (Clontech). After 24 h, GFP fluorescence was used to confirm the efficiency of transfection, and typically >70% cells were transfected with the plasmid. The cells were harvested for DNA extraction and uracil quantification.
Cell viability assay.
A total of 1 × 105 cells (or 0.5 × 105 cells for experiments lasting several days) of WT and UNGΔ/Δ NOK were seeded in 24-well tissue culture plates. The following day, the cells were treated with PMA or PMA+TNF-α or transfected with A3A-expressing plasmid. The cells were harvested by trypsinization at different time points. The viability and the total numbers of cells were determined by a trypan blue exclusion assay using a TC20 automated cell counter (Bio-Rad).
Visualization of A3A by immunofluorescence.
To compare the levels of A3A expression following plasmid transfection with PMA or PMA+TNF-α treatment, the cells were washed 24 or 48 h posttreatment and fixed with 4% paraformaldehyde for 10 min at room temperature. The cells were then permeabilized with 0.3% Triton X-100 in phosphate-buffered saline and blocked with 10% goat serum for 1 h. The cells were then stained with the same primary antibody against A3A/A3B mentioned above, followed by goat anti-rabbit secondary antibody conjugated with Cy3 (Thermo Fisher, 1:2,000). Coverslips were mounted onto the slide using ProLong Gold antifade mountant with DAPI (Invitrogen) and visualized using a fluorescence microscope (Nikon E800). The visual fields with multiple nuclei were photographed, and the Cy3 fluorescence per nucleus was quantified using the ImageJ software. The fluorescence intensities of differentially treated NOK cells were compared using an unpaired two-tailed t test, and the statistical significance of any observed difference was evaluated by calculating the P value. Confocal microscopy was performed using a Zeiss LSM 780 microscope (Microscopy, Imaging, and Cytometry Resources Core, Wayne State University).
ACKNOWLEDGMENTS
We thank members of our research group, Sophia Shalhout and Shanqiao Wei for useful discussions and suggestions.
This study was supported by a grant from the National Institutes of Health (grant GM57200) to A.S.B. and funds from Wayne State University.
REFERENCES
- 1.Furstenberger G, Berry DL, Sorg B, Marks F. 1981. Skin tumor promotion by phorbol esters is a two-stage process. Proc Natl Acad Sci U S A 78:7722–7726. doi: 10.1073/pnas.78.12.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griner EM, Kazanietz MG. 2007. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer 7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 3.Spitaler M, Cantrell DA. 2004. Protein kinase C and beyond. Nat Immunol 5:785–790. doi: 10.1038/ni1097. [DOI] [PubMed] [Google Scholar]
- 4.Rundhaug JE, Fischer SM. 2010. Molecular mechanisms of mouse skin tumor promotion. Cancers (Basel) 2:436–482. doi: 10.3390/cancers2020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarz M, Munzel PA, Braeuning A. 2013. Non-melanoma skin cancer in mouse and man. Arch Toxicol 87:783–798. doi: 10.1007/s00204-012-0998-9. [DOI] [PubMed] [Google Scholar]
- 6.Giorgi C, Agnoletto C, Baldini C, Bononi A, Bonora M, Marchi S, Missiroli S, Patergnani S, Poletti F, Rimessi A, Zavan B, Pinton P. 2010. Redox control of protein kinase C: cell- and disease-specific aspects. Antioxid Redox Signal 13:1051–1085. doi: 10.1089/ars.2009.2825. [DOI] [PubMed] [Google Scholar]
- 7.Peleva E, Exton LS, Kelley K, Kleyn CE, Mason KJ, Smith CH. 2018. Risk of cancer in patients with psoriasis on biological therapies: a systematic review. Br J Dermatol 178:103–113. doi: 10.1111/bjd.15830. [DOI] [PubMed] [Google Scholar]
- 8.Pouplard C, Brenaut E, Horreau C, Barnetche T, Misery L, Richard MA, Aractingi S, Aubin F, Cribier B, Joly P, Jullien D, Le Maitre M, Ortonne JP, Paul C. 2013. Risk of cancer in psoriasis: a systematic review and meta-analysis of epidemiological studies. J Eur Acad Dermatol Venereol 27(Suppl 3):36–46. doi: 10.1111/jdv.12165. [DOI] [PubMed] [Google Scholar]
- 9.Murakawa M, Yamaoka K, Tanaka Y, Fukuda Y. 2006. Involvement of tumor necrosis factor (TNF)-alpha in phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced skin edema in mice. Biochem Pharmacol 71:1331–1336. doi: 10.1016/j.bcp.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Fischer SM, Baldwin JK, Adams LM. 1986. Effects of anti-promoters and strain of mouse on tumor promoter-induced oxidants in murine epidermal cells. Carcinogenesis 7:915–918. doi: 10.1093/carcin/7.6.915. [DOI] [PubMed] [Google Scholar]
- 11.Perchellet EM, Perchellet JP. 1989. Characterization of the hydroperoxide response observed in mouse skin treated with tumor promoters in vivo. Cancer Res 49:6193–6201. [PubMed] [Google Scholar]
- 12.Leonard B, McCann JL, Starrett GJ, Kosyakovsky L, Luengas EM, Molan AM, Burns MB, McDougle RM, Parker PJ, Brown WL, Harris RS. 2015. The PKC/NF-κB signaling pathway induces APOBEC3B expression in multiple human cancers. Cancer Res 75:4538–4547. doi: 10.1158/0008-5472.CAN-15-2171-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madsen P, Anant S, Rasmussen HH, Gromov P, Vorum H, Dumanski JP, Tommerup N, Collins JE, Wright CL, Dunham I, MacGinnitie AJ, Davidson NO, Celis JE. 1999. Psoriasis upregulated phorbolin-1 shares structural but not functional similarity to the mRNA-editing protein apobec-1. J Invest Dermatol 113:162–169. doi: 10.1046/j.1523-1747.1999.00682.x. [DOI] [PubMed] [Google Scholar]
- 14.Maruyama W, Shirakawa K, Matsui H, Matsumoto T, Yamazaki H, Sarca AD, Kazuma Y, Kobayashi M, Shindo K, Takaori-Kondo A. 2016. Classical NF-κB pathway is responsible for APOBEC3B expression in cancer cells. Biochem Biophys Res Commun 478:1466–1471. doi: 10.1016/j.bbrc.2016.08.148. [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen HH, Celis JE. 1993. Evidence for an altered protein kinase C (PKC) signaling pathway in psoriasis. J Invest Dermatol 101:560–566. doi: 10.1111/1523-1747.ep12365986. [DOI] [PubMed] [Google Scholar]
- 16.Siriwardena SU, Chen K, Bhagwat AS. 2016. Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem Rev 116:12688–12710. doi: 10.1021/acs.chemrev.6b00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krokan HE, Drablos F, Slupphaug G. 2002. Uracil in DNA–occurrence, consequences and repair. Oncogene 21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 18.Stavrou S, Ross SR. 2015. APOBEC3 proteins in viral immunity. J Immunol 195:4565–4570. doi: 10.4049/jimmunol.1501504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borzooee F, Asgharpour M, Quinlan E, Grant MD, Larijani M. 2018. Viral subversion of APOBEC3s: lessons for anti-tumor immunity and tumor immunotherapy. Int Rev Immunol 37:151–164. doi: 10.1080/08830185.2017.1403596. [DOI] [PubMed] [Google Scholar]
- 20.Swanton C, McGranahan N, Starrett GJ, Harris RS. 2015. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov 5:704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatesan S, Rosenthal R, Kanu N, McGranahan N, Bartek J, Quezada SA, Hare J, Harris RS, Swanton C. 2018. APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann Oncol doi: 10.1093/annonc/mdy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jönsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerød A, Tutt A, Martens JWM, Aparicio SAJR, Borg Å, Salomon AV, Thomas G, Børresen-Dale A-L, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR. 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Imielinsk M, Jäger N, Jones DTW, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt ANJ, Valdés-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. 2013. Signatures of mutational processes in human cancer. Nature 500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA. 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell 46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burns MB, Temiz NA, Harris RS. 2013. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. 2013. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burns MB, Leonard B, Harris RS. 2015. APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed J 38:102–110. doi: 10.4103/2319-4170.148904. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki H, Suzuki A, Tatematsu T, Shitara M, Hikosaka Y, Okuda K, Moriyama S, Yano M, Fujii Y. 2014. APOBEC3B gene overexpression in non-small-cell lung cancer. Biomed Rep 2:392–395. doi: 10.3892/br.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonard B, Hart SN, Burns MB, Carpenter MA, Temiz NA, Rathore A, Vogel RI, Nikas JB, Law EK, Brown WL, Li Y, Zhang Y, Maurer MJ, Oberg AL, Cunningham JM, Shridhar V, Bell DA, April C, Bentley D, Bibikova M, Cheetham RK, Fan JB, Grocock R, Humphray S, Kingsbury Z, Peden J, Chien J, Swisher EM, Hartmann LC, Kalli KR, Goode EL, Sicotte H, Kaufmann SH, Harris RS. 2013. APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res 73:7222–7231. doi: 10.1158/0008-5472.CAN-13-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. 2013. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife 2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogerd HP, Kornepati AV, Marshall JB, Kennedy EM, Cullen BR. 2015. Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proc Natl Acad Sci U S A 112:E7249–E7256. doi: 10.1073/pnas.1516305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, Kim J, Kwiatkowski DJ, Fargo DC, Mieczkowski PA, Getz G, Gordenin DA. 2015. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet 47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trivedi NR, Gilliland KL, Zhao W, Liu W, Thiboutot DM. 2006. Gene array expression profiling in acne lesions reveals marked upregulation of genes involved in inflammation and matrix remodeling. J InvestDermatol 126:1071–1079. doi: 10.1038/sj.jid.5700213. [DOI] [PubMed] [Google Scholar]
- 35.Land AM, Law EK, Carpenter MA, Lackey L, Brown WL, Harris RS. 2013. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J Biol Chem 288:17253–17260. doi: 10.1074/jbc.M113.458661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aynaud MM, Suspene R, Vidalain PO, Mussil B, Guetard D, Tangy F, Wain-Hobson S, Vartanian JP. 2012. Human Tribbles 3 protects nuclear DNA from cytidine deamination by APOBEC3A. J Biol Chem 287:39182–39192. doi: 10.1074/jbc.M112.372722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green AM, Landry S, Budagyan K, Avgousti DC, Shalhout S, Bhagwat AS, Weitzman MD. 2016. APOBEC3A damages the cellular genome during DNA replication. Cell Cycle 15:998–1008. doi: 10.1080/15384101.2016.1152426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. 2011. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep 12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mussil B, Suspene R, Aynaud MM, Gauvrit A, Vartanian JP, Wain-Hobson S. 2013. Human APOBEC3A isoforms translocate to the nucleus and induce DNA double strand breaks leading to cell stress and death. PLoS One One 8:e73641. doi: 10.1371/journal.pone.0073641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suspène R, Mussil B, Laude H, Caval V, Berry N, Bouzidi MS, Thiers V, Wain-Hobson S, Vartanian J-P. 2017. Self-cytoplasmic DNA upregulates the mutator enzyme APOBEC3A leading to chromosomal DNA damage. Nucleic Acids Res 45:3231–3241. doi: 10.1093/nar/gkx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei S, Shalhout S, Ahn YH, Bhagwat AS. 2015. A versatile new tool to quantify abasic sites in DNA and inhibit base excision repair. DNA Repair (Amst) 27:9–18. doi: 10.1016/j.dnarep.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shalhout S, Haddad D, Sosin A, Holland TC, Al-Katib A, Martin A, Bhagwat AS. 2014. Genomic uracil homeostasis during normal B cell maturation and loss of this balance during B cell cancer development. Mol Cell Biol 34:4019–4032. doi: 10.1128/MCB.00589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei S, Perera MLW, Sakhtemani R, Bhagwat AS. 2017. A novel class of chemicals that react with abasic sites in DNA and specifically kill B cell cancers. PLoS One One 12:e0185010. doi: 10.1371/journal.pone.0185010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. 2002. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem 277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 45.Kemmerich K, Dingler FA, Rada C, Neuberger MS. 2012. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung-/-Msh2-/- mice. Nucleic Acids Res 40:6016–6025. doi: 10.1093/nar/gks259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T. 2001. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J 20:4278–4286. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alsoe L, Sarno A, Carracedo S, Domanska D, Dingler F, Lirussi L, SenGupta T, Tekin NB, Jobert L, Alexandrov LB, Galashevskaya A, Rada C, Sandve GK, Rognes T, Krokan HE, Nilsen H. 2017. Uracil accumulation and mutagenesis dominated by cytosine deamination in CpG dinucleotides in mice lacking UNG and SMUG1. Sci Rep 7:7199. doi: 10.1038/s41598-017-07314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhagwat AS, Hao W, Townes JP, Lee H, Tang H, Foster PL. 2016. Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli. Proc Natl Acad Sci U S A 113:2176–2181. doi: 10.1073/pnas.1522325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haradhvala NJ, Polak P, Stojanov P, Covington KR, Shinbrot E, Hess JM, Rheinbay E, Kim J, Maruvka YE, Braunstein LZ, Kamburov A, Hanawalt PC, Wheeler DA, Koren A, Lawrence MS, Getz G. 2016. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 164:538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoopes JI, Cortez LM, Mertz TM, Malc EP, Mieczkowski PA, Roberts SA. 2016. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep 14:1273–1282. doi: 10.1016/j.celrep.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seplyarskiy VB, Soldatov RA, Popadin KY, Antonarakis SE, Bazykin GA, Nikolaev SI. 2016. APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome Res 26:174–182. doi: 10.1101/gr.197046.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pham P, Bransteitter R, Petruska J, Goodman MF. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 53.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 54.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. 2014. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep 7:1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Pettersen HS, Galashevskaya A, Doseth B, Sousa MML, Sarno A, Visnes T, Aas PA, Liabakk N-B, Slupphaug G, Sætrom P, Kavli B, Krokan HE. 2015. AID expression in B-cell lymphomas causes accumulation of genomic uracil and a distinct AID mutational signature. DNA Repair (Amst) 25:60–71. doi: 10.1016/j.dnarep.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 56.Suspene R, Aynaud MM, Guetard D, Henry M, Eckhoff G, Marchio A, Pineau P, Dejean A, Vartanian JP, Wain-Hobson S. 2011. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc Natl Acad Sci U S A 108:4858–4863. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Land AM, Wang J, Law EK, Aberle R, Kirmaier A, Krupp A, Johnson WE, Harris RS. 2015. Degradation of the cancer genomic DNA deaminase APOBEC3B by SIV Vif. Oncotarget 6:39969–39979. doi: 10.18632/oncotarget.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. 2010. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol 17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han J, Kim S, Yang JH, Nam SJ, Lee JE. 2012. TPA-induced p21 expression augments G2/M arrest through a p53-independent mechanism in human breast cancer cells. Oncol Rep 27:517–522. doi: 10.3892/or.2011.1511. [DOI] [PubMed] [Google Scholar]
- 60.Kikkawa U, Takai Y, Tanaka Y, Miyake R, Nishizuka Y. 1983. Protein kinase C as a possible receptor protein of tumor-promoting phorbol esters. J Biol Chem 258:11442–11445. [PubMed] [Google Scholar]
- 61.Koeffler HP. 1981. Human myelogenous leukemia: enhanced clonal proliferation in the presence of phorbol diesters. Blood 57:256–260. [PubMed] [Google Scholar]
- 62.Kosaka C, Sasaguri T, Ishida A, Ogata J. 1996. Cell cycle arrest in the G2 phase induced by phorbol ester and diacylglycerol in vascular endothelial cells. Am J Physiol 270:C170–C178. doi: 10.1152/ajpcell.1996.270.1.C170. [DOI] [PubMed] [Google Scholar]
- 63.Parkinson EK, Emmerson A. 1982. The effects of tumour promoters on the multiplication and morphology of cultured human epidermal keratinocytes. Carcinogenesis 3:525–531. doi: 10.1093/carcin/3.5.525. [DOI] [PubMed] [Google Scholar]
- 64.Tahara E, Kadara H, Lacroix L, Lotan D, Lotan R. 2009. Activation of protein kinase C by phorbol 12-myristate 13-acetate suppresses the growth of lung cancer cells through KLF6 induction. Cancer Biol Ther 8:801–807. doi: 10.4161/cbt.8.9.8186. [DOI] [PubMed] [Google Scholar]
- 65.Wille JJ Jr, Pittelkow MR, Scott RE. 1985. Normal and transformed human prokeratinocytes express divergent effects of a tumor promoter on cell cycle-mediated control of proliferation and differentiation. Carcinogenesis 6:1181–1187. doi: 10.1093/carcin/6.8.1181. [DOI] [PubMed] [Google Scholar]
- 66.Nakagawa M, Oliva JL, Kothapalli D, Fournier A, Assoian RK, Kazanietz MG. 2005. Phorbol ester-induced G1 phase arrest selectively mediated by protein kinase Cδ-dependent induction of p21. J Biol Chem 280:33926–33934. doi: 10.1074/jbc.M505748200. [DOI] [PubMed] [Google Scholar]
- 67.Blagosklonny MV. 2003. Cell senescence and hypermitogenic arrest. EMBO Rep 4:358–362. doi: 10.1038/sj.embor.embor806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, Nussenzweig MC. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rebhandl S, Huemer M, Greil R, Geisberger R. 2015. AID/APOBEC deaminases and cancer. Oncoscience 2:320–333. doi: 10.18632/oncoscience.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han Y, Wang X, Dang Y, Zheng YH. 2008. APOBEC3G and APOBEC3F require an endogenous cofactor to block HIV-1 replication. PLoS Pathog 4:e1000095. doi: 10.1371/journal.ppat.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wichroski MJ, Robb GB, Rana TM. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog 2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nikkila J, Kumar R, Campbell J, Brandsma I, Pemberton HN, Wallberg F, Nagy K, Scheer I, Vertessy BG, Serebrenik AA, Monni V, Harris RS, Pettitt SJ, Ashworth A, Lord CJ. 2017. Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. Br J Cancer 117:113–123. doi: 10.1038/bjc.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Piboonniyom SO, Duensing S, Swilling NW, Hasskarl J, Hinds PW, Munger K. 2003. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res 63:476–483. [PubMed] [Google Scholar]
- 74.Wijesinghe P, Bhagwat AS. 2012. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res 40:9206–9217. doi: 10.1093/nar/gks685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siriwardena SU, Guruge TA, Bhagwat AS. 2015. Characterization of the catalytic domain of human APOBEC3B and the critical structural role for a conserved methionine. J Mol Biol 427:3042–3055. doi: 10.1016/j.jmb.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thielen BK, McNevin JP, McElrath MJ, Hunt BV, Klein KC, Lingappa JR. 2010. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J Biol Chem 285:27753–27766. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, Brown WL, Harris RS. 2012. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem 287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]