Abstract
BACKGROUND:
MicroRNAs (miRNAs) are small, non-coding RNAs that are important for post-transcriptional gene regulation in both healthy and morbid conditions. Numerous miRNAs promote tumorigenesis, while others have a tumour suppressive effects. Acute myeloid leukaemia (AML) is a heterogeneous group of genetically diverse hematopoietic malignancies with variable response to treatment.
AIM:
Our study aimed to investigate the possible role of miR-150 in de novo adult AML and the impact of its level on survival, and we used in the silicon analysis to predict the main target genes involved in miR-150 mediated cancer pathway.
MATERIAL AND METHODS:
We evaluated miR-150 expression profiling assay using TaqMan primer probes RT-PCR in the plasma of 50 adult AML patients, before the start of treatment and at day 28 of treatment, along with 20 normal adult control samples. miR-16 was used as an endogenous reference for standardisation. Follow-up of patients during treatment at day 28 of induction chemotherapy and after one year was done.
RESULTS:
In this study, we found a significantly lower level of miR-150 in AML patients when compared to controls (p = 0.005) with 0.62 fold change than in healthy controls. Patients were divided into two groups: the low miR-150 group (miR-150 < 1) and the high miR-150 group (miR-150 > 1). A statistically significant difference was found between the two groups regarding initial total leukocytic count and initial PB blast count while for the TLC, HB and PLT count at follow up. No difference in the overall survival between the low and the high miR-150 groups could be demonstrated.
CONCLUSION:
Our results suggest that miR-150 functions as a tumour suppressor and gatekeeper in inhibiting cell transformation and that its downregulation is required for leukemogenesis.
Keywords: Acute myeloid leukaemia, microRNA, biomarkers, miR-150, leukemogenesis
Introduction
Acute myeloid leukaemia (AML) is a clonal disorder of hematopoietic progenitor cells which is characterised by diverse heterogeneity regarding genotypic, phenotypic and clinical features [1], [2]. AML is the most common acute leukaemia in adult patients and can arise “de novo” or as a secondary event [3]. Among the genetic aberrations that control disease development, there are microRNAs.
MicroRNAs (miRNAs) are short non-coding RNAs (~20-24 nucleotides) that are involved in post-transcriptional regulation of gene expression in multicellular organisms. This is achieved by affecting both the stability and translation of mRNAs [4]. miRNAs play an important role in many biological processes in the body such as cell growth, proliferation, differentiation, and apoptosis. They also act as both oncogenes and tumour suppressors contributing to a malignant transformation in solid and haematological tumours, including AML [5].
MiR-150 is a family of microRNA precursors found in mammals, including humans. The mature miRNA sequence is a 22 nucleotide, which is excised from the precursor hairpin by the enzyme Dicer [6]. This sequence then associates with RISC (RNA-induced silencing complex), that directly bind to the potential target site in the 3’ untranslated region (3’UTRs) of specific target mRNA, leading to the repression of mRNA translation or the degradation of target mRNAs (7). In normal hematopoiesis, miR-150 regulates genes whose downstream products encourage the differentiation of stem cells towards becoming megakaryocytes rather than erythrocytes [8]. It is also thought that together with miR-155, control B and T cell differentiation [9].
MiR-150 has been linked to the development of some cancers [10]. Mraz et al. reported that the expression of miR-150 was shown to regulate levels of GAB1 and FOXP1 proteins in malignant and normal B cells, which influences their BCR signalling [11].
Aberrant miRNA expression is a feature of different cancers including haematological malignancies. In a study by Morris et al., they identified that the expression of miR-150 is low or absent in blastic crisis (BC) of chronic myeloid leukaemia (CML) and acute myeloid leukaemia (AML) patients’ samples and also in cell lines [12]. They found that the expression of miR-150 in AML cell lines, CD34+ progenitor cells from healthy individuals and primary BC CML and AML patients’ samples at levels similar to miR-150 expression in normal bone marrow promotes myeloid differentiation of these cells.
They also reported that in AML cell lines, differentiation of miR-150 expressing cells occurs independently of retinoic acid receptor α (RARA) signalling and that the high throughput gene expression profiling (GEP) studies of the AML cell lines HL60, PL21 and THP-1 suggest that activation of CEPBA, CEBPE and cytokines associated with myeloid differentiation in miR-150 expressing cells, as compared to control cells, contributes to myeloid differentiation. They concluded from these data that miR-150 promotes myeloid differentiation, a previously uncharacterized role for this miRNA and that absent or low miR-150 expression contributes to down-regulated myeloid differentiation in acute leukaemia cells [12].
From two genomes wide large-scale miRNA expression profiling assays of various subtypes of primary AML samples and normal controls, Jiang et al., (13) identified miR-150 as one of the most significantly and consistently down-regulated miRNAs in most of the AML cases they have studied. They furthermore confirmed this significant down-regulation by a subsequent study [14].
Material and Methods
This is a matched case-control study included 50 adults de novo AML cases that were recruited from the Medical Oncology Department of the National Cancer Institute, Cairo University, Egypt, from January 2015 to March 2016. As well as 20 healthy volunteers were included as a control group. Approval from the Ethical Committee was obtained (Medical Ethical Research Committee—National Research Centre, Number P100510) to carry over this study and informed consents were signed by the patients by the Helsinki declaration. The demographic characteristics were documented at a presentation in addition to morphological, cytochemical, immunophenotypic and genetic analyses which were done at initial diagnosis. The follow-up data including the response to treatment and survival were documented for all patients.
Sampling and Extraction of micro-RNA
Two ml of peripheral blood were collected on EDTA from the newly diagnosed adult AML cases (n = 50) before starting therapy, follow up samples were collected at day 28 of treatment for 31 of them. Samples were also collected from the twenty healthy adults.
miRNA was isolated from plasma samples using miRNeasy Mini Kit (Cat number# 217004, Qiagen, USA) as recommended by the manufacturer’s instructions, the purity and the concentration of the purified miRNA was detected using spectrophotometer Nano-drop (Maestrogen, Taiwan, MN-913) and stored at -80 °C till further assessments.
Detection of miRNA expression using Real-Time PCR
Gene-specific complementary DNA (cDNA) was prepared from miRNA by using reversing TaqMan microRNA RT-Kit, Cat number # 4366596, (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions. miRNA expression for enrolled samples was quantified using TaqMan 2x universal master mix II Cat number # 4440043, (Applied Biosystems, Foster City, CA, USA) and TaqMan microRNA Assay Mix containing PCR primers and TaqMan probes for miR-150. MiR-16 was used as endogenous control for normalisation. Fluorescence was acquired and detected by ABI step one- Applied Biosystems. To determine miRNA relative expression, it was reported as fold change (ΔCt and ΔΔCt calculations).
The overall survival (OS) was calculated from the date of diagnosis to the date of death or last follow up visit. Disease-free survival was calculated from the date of complete remission to the date of relapse, death or last follow up visit.
Statistical analysis
The data were presented as mean ± SD or median. The results were calculated using the Student’s t-test, χ2, paired sample T-test, Mann-Whitney U test or Kruskal-Wallis test where appropriate. Categorical variables were described with count and percentage. Kaplan-Meier method was used to calculate the survival rates and the log-rank test was used to test the significance in the difference in the patients’ survival. Statistical analyses were performed using SPSS software (version 16.0 for Windows; SPSS INC., Chicago, IL, USA) where P values were two-tailed and considered statistically significant when less than 0.05.
Bioinformatics analysis
In a silico analysis for the miR-150 was performed to find target genes regulated by miR-150 and identifying the possible cellular pathways in which these target genes are involved. The miRNA target genes were predicted with the miRBase (www.mirbase.org). MicroRNA target gene analysis was performed using miRWalk 2.0 server [http://mirwalk.uni-hd.de/] which is a database that gives both predicted and experimentally validated miRNA-targets [15], [16]. The predicted target genes were obtained with cut off p-value 0.05, in addition to the validated ones. Both predicted and validated target genes were combined to furtherly undergo functional enrichment analysis.
Functional enrichment analysis for the miRNA target genes was done using the DAVID server [Database for Annotation, Visualization and Integrated Discovery], (https://david.ncifcrf.gov) [17], [18]. Pathway enrichment analyses of the predicted miRNA target genes were performed with KEGG pathway (www.genome.jp/kegg) [19].
Results
We included in our study 50 de novo adult AML patients (before starting treatment and at D28 of treatment). Thirty two of them were males (64.7%) and 18 were females (35.3%); mean age was (37.48 ± 12.38), as well as 20 age and sex-matched controls, as well as 20 age and sex-matched controls. We estimated the expression level of miR-150 in both the patient group initially and at D28 and the control group using TaqMan primer-probe assay Real Time PCR. We also evaluated parameters of clinical importance in the AML group such as; total leukocytic count (TLC), haemoglobin concentration (HB), platelet count (PLTs), bone marrow (BM) cellularity and blast % in both peripheral blood (PB) and bone marrow, initially and at D28. Immunophenotypic markers, cytogenetics and FLT-3 mutational status, were investigated too.
The expression level of miR-150 in both the patients group initially and the control group was estimated using Real-Time PCR. A significant difference between the initial level of miR-150 in patients and controls (p = 0.005) was found.
Plasma initial miR-150 was down-regulated in adult AML with 0.62 fold change than in healthy controls as demonstrated in Figure 1. Table 1 and Fig. 2 showed statistically significant lower values of relative expression of miR-150 in patients initially and at D28 compared to controls (p = 0.004).
Figure 1.
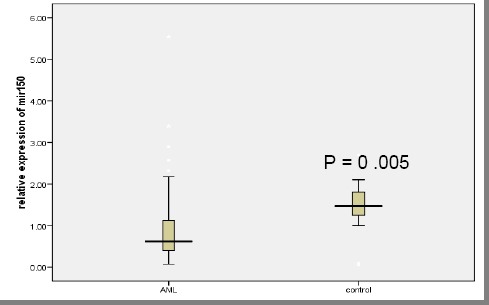
Relative initial plasma miR-150 levels in adult AML patient and control population. Expression levels of miR-150 were normalised to miR-16. Data were represented as the median value, and the Mann-Whitney U test was used to define statistical significance
Table 1.
Demographic and clinical characteristics of patients before treatment and at D28 of treatment
| Parameters | Initial value | D28 value | P value |
|---|---|---|---|
| miR-150 n=31 | 1.1 ± 1.18 | 0.85 ± 0.92 | 0.3 |
| TLC x109/L | 34.5 ± 49.05 | 3.8 ± 4.1 | 0.001** |
| HB gm/dL | 7.9 ± 1.5 | 7.9 ± 1.2 | 0.9 |
| PLTs x109/L | 57.96 ± 68.3 | 1.85x102± 172. | <0.001** |
TLC: total leucocytic count, HB: haemoglobin, PLTs: platelet count,
highly significant statistical difference.
Table 2.
Various clinical parameters in low and high groups when miR-150 was estimated initially
| Parameters | Mir-150 initial ≤ 1 N = 35 | Mir-150 initial > 1 N = 15 | P value | |
|---|---|---|---|---|
| Age (years) | Mean ± S.D | 33.9 ± 10.5 | 38.85 ± 10.27 | 0.262 |
| Sex n (%) | Male | 10 (45.5) | 5 (62.5) | 0.409 |
| Female | 13 (54.5) | 3 (37.5) | ||
| TLC initial x109/L | Median | 9.5 | 36 | 0.023* |
| Range | 0.7- 42.3 | 1.68-112.58 | ||
| Plts initial x109/L | Median | 40 | 33 | 0.649 |
| Range | (2.0-211) | (2.0-290) | ||
| HB initial gm/dl | Median | 7.9 | 7.3 | 0.094 |
| Range | 5.6-12.1 | 4.8-11.5 | ||
| PB initial Blasts% | (mean ± S.D) | 40.06 ± 27.85 | 58.2 ± 25.87 | 0.036* |
| BM initial cellularity n(%) | Normocellular | 9 (25.7) | 3 (20.0) | 0.362 |
| Hypocellular | 3 (8.6) | 2 (13.3) | ||
| Hypercellular | 14 (40) | 9 (60.0) | ||
| Extrahypercellular | 9 (25.7) | 1 (6.7) | ||
| BM initial blast% | (mean ± S.D.) | 62.6 ± 20.07 | 67.93 ± 20.0 | 0.393 |
| FAB classification n(%) | M0 | 1 (2.9) | 0 | 0.185 |
| M1 | 7 (20) | 1 (6.7) | ||
| M2 | 13 (37.1) | 5 (33.3) | ||
| M3 | 1 (2.9) | 4 (26.7) | ||
| M4 | 8 (22.9) | 2 (13.3) | ||
| M5 | 2 (5.7) | 0 | ||
| M7 | 2 (5.7) | 2 (13.3) | ||
| Others (phenotypic) | 1 (2.9) | 1 (6.7) | ||
| IPT n(%) | Myeloid | 20(57.1) | 9 (60) | 0.353 |
| Mono | 1(2.9) | 0 | ||
| Myeloma | 6(17.1) | 0 | ||
| Myeloid with aberrant | 5(14.3) | 2 (13.3) | ||
| Myeloid-B | 1(2.9) | 1 (6.7) | ||
| Others | 2(5.7) | 3 (20) | ||
| FLT3 n(%) | Wild | 26(74.3) | 8 (53.3) | 0.346 |
| Mutant | 5(14.3) | 4 (26.7) | ||
| NA | 4(11.8) | 3 (20) | ||
| Cytogenetics n(%) | T (9, 22) | 1(4.8) | 0 | 0.138 |
| T (8, 21) | 2(9.5) | 0 | ||
| T (15, 17) | 1(4.8) | 3 (27.3) | ||
| Normal karyotype | 7(33.3) | 6 (54.2) | ||
| Others | 10(47.6) | 2 (18.2) | ||
| BM Cellularity N(%) D28 | Normocellular | 13(52) | 6 (50) | 0.067 |
| Hypocellular | 9(36) | 2 (16.7) | ||
| Hypercellular | 1(4) | 4 (33.3) | ||
| NA | 2(8) | 0 | ||
| Blast n(%) D28 | Less than or equal 5 | 18(75.0) | 9 (75) | 0.05 |
| More than 5 | 6(25) | 3 (25) | ||
| TLC x109/L D28 | Median | 2.0 | 4.27 | 0.023* |
| Range | (0.1-16.3) | (0.56-16) | ||
| HB gm/dl D28 | Median | 7.7 | 8.7 | 0.013* |
| Range | (3.3-10.3) | (7-10) | ||
| Plts x109/L D28 | Median | 84.5 | 333 | 0.014* |
| Range | (2.00-579) | (5-558) | ||
| PB blasts n(%) D28 | Equal zero | 16(76.2) | 7 (87.5) | 0.50 |
| More than 1 | 5(23.8) | 1 (12.5) | ||
| Response to treatment n(%) | CR | 10(62.5) | 22 (64.7) | 0.98 |
| Refractory | 3(18.8) | 6 (17.6) | ||
| NA | 3(18.8) | 6 (17.6) | ||
| Relapse | Relapse | 2(16.7) | 5 (19.2) | 0.79 |
| No relapse | 9(75) | 17 (65.4) | ||
| NA | 1(8.3) | 4 (15.4) | ||
| Early death | Before D28 | 3(50) | 7 (53.8) | 0.87 |
| After D28 | 3(50) | 6 (46.2) | ||
TLC: total leucocytic count, HB: haemoglobin, PLT: platelet count, PB: peripheral blood, BM: bone marrow, FAB: French American British classification, IPT: immunophenotyping, D28: day 28 of treatment, NA: non-available, CR: complete remission, OS: overall survival.
Figure 2.
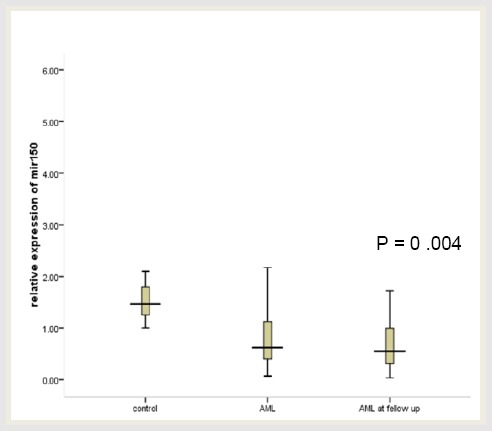
Relative plasma miR-150 levels in AML patients initially, at D28 of treatment and control population. Expression levels of miR-150 were normalised to miR-16. Data were represented as the median value, and Kruskal Wallis Test was used to define statistical significance
We followed up our patients for their miR-150 at D28 of treatment. We found a significant decrease in the TLC and increase PLT count when measured initially in patients compared to its measurement at D28. No significant difference was found regarding HB concentration and miR-150 level (Table 3).
Table 3.
Various clinical parameters in low and high groups when miR-150 was estimated at D28
| Parameter | miR-150 at D28< 1 n=23 | miR-150 at D28> 1 n=8 | P value | |
|---|---|---|---|---|
| TLC x109/L initial | Median | 10 | 15.95 | 0.963 |
| Range | (1.2-242.3) | (1.68-65.35) | ||
| PLT x109/L | Median | 33 | 18 | 0.37 |
| Range | (2.0-208) | (9-290) | ||
| HB gm/dL | Median | 8.1 | 7.9 | 0.1 |
| Range | (5.9-12.1) | (4.8-9.1) | ||
| PB Blasts% | Median | 40 | 18.5 | 0.139 |
| Range | (4-94) | (0-65) | ||
| BM cellularity n(%) | Normocellular | 5 (21.7) | 1 (12.5) | 0.089 |
| Hypocellular | 2 (8.7) | 3 (37.5) | ||
| Hypercellular | 10 (43.5) | 4 (50) | ||
| Extrahypercellular | 6 (26.1) | 0 (0) | ||
| FAB n (%) | M0 | 1 (4.3) | 0 (0) | 0.084 |
| M1 | 4 (17.4) | 1 (12.5) | ||
| M2 | 10 (43.5) | 1 (12.5) | ||
| M3 | 2 (8.7) | 1 (12.5) | ||
| M4 | 5 (21.7) | 2 (25.0)0 (0) | ||
| M7 | 0 (0) | 0 (0) | ||
| Other | 1 (4.3) | 0 (0) | ||
| IPT n (%) | Myeloid | 15 (65.2) | 4 (50) | 0.030* |
| Myelomonocytic | 2 (8.7) | 0 (0) | ||
| Myeloid with aberrant | 4 (17.4) | 0 (0) | ||
| Myeloid B | 1 (4.3) | 0 (0) | ||
| Other | 1 (4.4) | 4 (50) | ||
| FLT-3 n (%) | Wild | 19 (82.6) | 7 (87.5) | 0.117 |
| Mutant | 4 (17.4) | 0 (0) | ||
| NA | 0 (0) | 1 (12.5) | ||
| Cytogenetics n (%) | t(8,21) | 2 (13.3) | 0 (0) | 0.524 |
| t(15,17) | 2 (13.3) | 0 (0) | ||
| Normal karyotype | 6 (40) | 4 (66.7) | ||
| Others | 5 (33.3) | 2 (33.3) | ||
| BM D28 cellularity n (%) | Normocellular | 9 (45) | 3 (37.5) | 0.817 |
| Hypocellular | 8 (40) | 3 (37.5) | ||
| Hypercellular | 3 (15) | 2 (25) | ||
| BM Blasts% D28 | Median | 25 | 2.5 | 0.69 |
| Range | (0-67) | (0-30) | ||
| Less than or equal 5 | 15 (75) | 6 (75) | ||
| More than or equal 6 | 5 (25) | 2 (25) | ||
| TLC x109/L at D28 | Median | 2.6 | 2.94 | 0.663 |
| Range | (0.1-16.3) | (0.38-16) | ||
| HB gm/dL at D28 | Median | 7.9 | 8 | 0.788 |
| Range | (3.3-10) | (6.9-10.3) | ||
| PLT x109/L at D28 | Median | 111.5 | 110 | 0.826 |
| Range | (2-558) | (5-579) | ||
| PB blasts % at D28 | Equal zero | 15 (71.4) | 7 (100) | 0.1 |
| More than or equal 1 | 6 (28.6) | 0 | ||
| Response to treatment | Refractory | 4 (18.2) | 2 (25) | 0.98 |
| CR | 18 (81.8) | 6 (71.475) | ||
| Relapse status | No relapse | 16 (84.2) | 3 (42.9) | 0.035* |
| Relapse | 3 (15.8) | 4 (57.1) | ||
| Early death | Before D28 | 2 (22.2) | 1 (100) | 0.13 |
| After D28 | 6 (75) | 0 (100) | ||
TLC: total leucocytic count, HB: hemoglobin, PLT: platelet count, PB: peripheral blood, BM: bone marrow, FAB: French American British classification, IPT: immunophenotyping, D28: day 28 of treatment, NA: non-available, CR: complete remission, OS: overall survival, RFS: Relapse-free survival.
AML patients who achieved complete remission (CR) after induction chemotherapy at D28 were 32 cases (64%), while 7 cases were relapsed (21%) later on. No statistically significant difference was found in the relative expression of miR-150 before and after treatment in patients undergo CR (p = 0.59).
Table 2 shows a comparison between different clinical parameters when miR-150 was measured before the start of treatment. Patients were divided into two groups; the low expressers for the miR-150 group (miRNA-150 < 1) and the high expressers for the miR-150 group (miR-150 > 1). A statistically significant correlation was found between high miR-150 and higher initial TLC and PB blast %, while this significant correlation was observed with higher TLC, HB concentration and PLT count at D28 (p = 0.023, 0.036, 0.023, 0.013, 0.014 respectively). A relation also was found between initial low miR-150 and normocellular marrow at D28 samples but not reach a statistical significance (p = 0.067).
Table 3 shows a comparison between different clinical and laboratory parameters in the low miR-150 group and the high miR-150 group when miR-150 was measured at D28. A statistically significant relation was found between low miR-150 and myeloid phenotype (p = 0.030) and also with no relapse status for the patients (p = 0.035).
Survival analysis
Kaplan-Meier survival curves were used to estimate overall survival (OS) and relapse-free survival (RFS) in the AML patients which are shown in figures (3A and 3B) and are summarised in Table 4.
Figure 3.
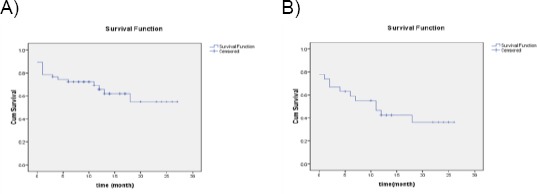
A) Overall survival of AML patients; B) relapse-free survival of AML patients
Table 4.
Overall survival and disease/relapse-free survival
| Parameter | AML patients | Mean | No of dead/relapse n (%) | No of alive/ relapse-free n (%) |
|---|---|---|---|---|
| Overall survival (time /month) (95%CI) | N = 47 | 17.73 (14.2-21.21) | 17 (36.17) | 30 (63.8) |
| Disease/relapse free survival (95%CI) | N = 27 | 12.82 (8.56-17.07) | 16 (59.3) | 11 (40.7) |
Moreover, there was no difference in the overall survival between the low and the high miR-150 groups when measured initially before the start of treatment (Figure 4A). Also, there was no difference in the overall survival between the low and the high miR-150 groups when measured at D28 as illustrated in (Figure 4B).
Figure 4.
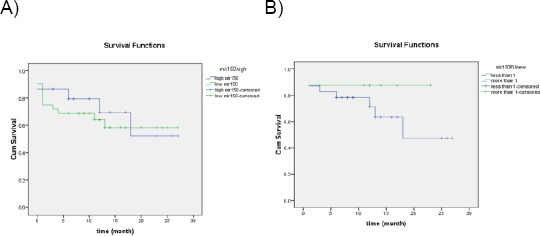
A) Overall survival compared between high and low miR-150 when measured initially; B) Overall survival compared between high and low miR-150 when measured at follow up
We used in the silico analysis to predict the target genes, as shown in Table 5. We identified the most promising potential target genes and highly significant enrichment pathways for miR-150. The main target genes are ACOX1, BDNF, RAPGEF3, FGF7, FGF12, PIK3R3, TNF, E2F3, SMAD4, STAT5B which are involved in cAMP, MAPK, ErbB, mTOR, chronic myeloid leukaemia signalling pathway and pathway in cancer.
Table 5.
Prediction of the target genes miR-150 and its pathways
| Target genes | Pathways | |
|---|---|---|
| miR-150 | ACOX1, PPARA, ADCY1, ADCY2, ADCY5, ADCY6, GABBR1, CNGB1, ADORA1, ATP2B1, BDNF, ATP2B3, ATP2B4, GRIN2B, TIAM1, PDE4A, PAK1, RAPGEF3, | cAMP signaling pathway |
| FGF7, ZAK, PDGFB, FGF9, GNA12, FGF12, MAX, BDNF, MAPT, PAK1, FGF1, IL1A, AKT3, AKT2, PRKCA, BRAF, CACNG8, TP53, CACNG4, CACNG2, PRKCB, | MAPK signaling pathway | |
| GRB2, CAMK2G, STAT5B, ELK1, PAK3, CAMK2B, SHC1, PAK1, SHC3, PIK3R3, AKT3, PIK3R1, PIK3R2, SHC4, AKT2 | ErbB signaling pathway | |
| PRKCA, TNF, BRAF, STK11, PIK3CB, IGF1, RICTOR, PRKCB, EIF4B, RPS6KA6, RPS6KA3, AKT1S1, TSC1, ULK1, ULK2, ULK3, PRKAA2, PIK3R3, AKT3, PIK3R1, AKT2, PIK3R2 | mTOR signaling pathway | |
| E2F3, BRAF, GRB2, PIK3CB, TGFBR1, CBL, STAT5B, TP53, SMAD4, CDK4, CBLB, CCND1, CDKN1B, ARAF, MDM2, SHC1, PIK3R3, SHC3, CRK, PIK3R1, AKT3, AKT2, PIK3R2, SHC4 | Chronic myeloid leukemia signaling pathway | |
| ADCY1, PPARD, E2F3, ADCY2, FGF7, PDGFB, FGF9, ADCY5, ADCY6, STAT5B, GNA12, SPI1, FGF12, CTNNB1, MAX, PAX8, SLC2A1, FGF1 | Pathway in cancer | |
Most of the gene ontology (GO) annotations were associated with cell process, regulation of biological process, regulation of cell communication, intracellular signalling cascade, localisation and others Figure 5.
Figure 5.
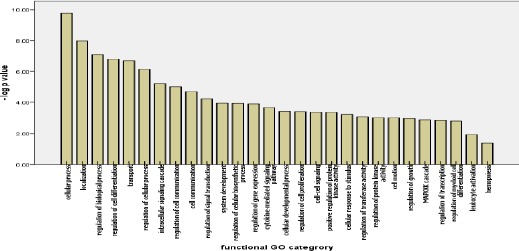
Functional gene ontology terms for miR-150 target genes
Discussion
The miRNA network is highly redundant, as a single miRNA could have multiple target mRNAs, and on the other hand, a single mRNA may be targeted by many miRNAs [20]. Many miRNAs have been shown to be involved in a myriad of cellular processes which include differentiation, apoptosis, metabolism and development [21]. Physiologically, as well as pathologically, miRNAs have been reported to play roles in cancers, inflammatory responses, diabetes and autoimmune diseases [22].
In this work, we investigated the role of miR-150 level in de novo adult AML; where we found a significantly lower level in patients when compared to controls (P = 0.012). These results are supported by the clarification of a previously uncharacterized role for miR-150 in promoting myeloid differentiation, where Morris et al. [25], demonstrated that low miR-150 expression contributes to the leukemic phenotype in various AML subtypes and BC of CML. Also, Fayyad-Kazan et al. [14] and Wang et al. [23], reported similar results to ours, where they found lower miR-150 expression in AML patients as compared to controls (P = 0.0026, P = 0.006 respectively). Also, Xu DD et al. reported that miR-150 was downregulated in leukaemia stem cells (LSCs; CD34+CD38- cells) and clinical samples. Ectopic expression of miR-150 suppressed the LSCs biological behaviours, both in vitro and in vivo by its effect on Nanog signalling pathway [24].
Similarly, the expression levels of miR-150 were decreased in AML cases as in a study by Morris et al. [12], and overexpression of miR-150 promoted myeloid differentiation of cancer cells and suppressed their proliferation [25]. Also, Fayyad-Kazan et al. [14] reported that in a recent study, plasma levels of tumour necrosis factor alpha, interleukin-10, and interleukin-18, which all have a sequence, complementary to miR-150, were negatively correlated with the plasma levels of miR-150. This could impact on the immune system response to leukaemias. These data revealed that miR-150 could be a promising biomarker for AML at diagnosis and suggest that microRNA expression signature in plasma can serve as a valuable diagnostic and potential prognostic marker for human AML.
MiR-150 was measured twice, first; before the start of treatment, then; at D28 after induction chemotherapy for the 31 cases that they’re follow up samples could be reached, the comparison in between did not reveal any statistical significance. Also, the comparison between levels of MiR-150 before treatment and at D28 for cases that undergo complete remission did not reveal any statistical significance. So, our results showed no difference between miRNA levels initially and at D28 with and without discrimination of the response to treatment. This was in contrast to Wang et al. [23] and Fayyad-Kazan et al. [14], who reported that there was up-regulation of miR-150 level in AML patients who achieved CR reaching the level of controls (P < 0.01) compared to its down-regulation before treatment. In our study, this could be explained by the fact that there was no difference in miRNA level between the cytogenetically good and bad prognostic groups of patients in our study, in contrast to Wang et al. [23] and Morris et al. [25], who reported a significant decrease in miRNA level in the poor and intermediate risk cytogenetics groups as compared to the favorable group.
Our patients were divided into two groups; the low expressers for the miR-150 group (miR-150 < 1) and the high expressers for the miR-150 group (miR-150 > 1). The correlation between plasma miR-150 levels and clinicopathological data were verified. A statistically significant correlation was found between high miR-150 and both a higher initial TLC and PB blast % and with a higher TLC, HB and PLT count at D28 (p = 0.023, 0.036, 0.023, 0.013, 0.014 respectively). A relation was also found between low miR-150 and normocellular marrow at D28 samples but could not reach a statistical significance (p = 0.067). However, no significance could be recorded with others. Wang et al., [23] reported a significant correlation with the percentage of initial BM blast% (P = 0.020). However no significant difference was found with initial TLC, PLT number (P > 0.05). He also found a significant relation with FAB classification (P = 0.013) and cytogenetics (P = 0.012). However, no significant difference was found between the level of miR-150 and gender, age and extramedullary disease (P > 0.05)
Wang et al. [23], reported that AML patients with M5 subtype had a lower serum miR-150 level than those with other subtypes including M0, M1, M2 and M4 (P < 0.01). Our study included only 2 patients with M5, so, monocytic leukaemias (M4 and M5) in our study were summed up together and showed lower miR-150 level than other FAB subtypes, although with no statistically significant difference. We accordingly expect that with increasing the number of patients, comparable results could be obtained.
We found no statistically significant relation regarding Flt-3 mutational status between the high and low miR-150 groups; either initially or at D28. Our results are supported by Jiang et al. [13], who reported that forced expression of miR-150, reduced the levels of Flt-3 to 40-65%. Their results indicated that Flt3 functions as a direct target of miR-150 in regulating leukemic cell self-renewal and at least in part, responsible for the inhibitory effects of forced expression of miR-150 on leukemogenesis [30].
As regarding the prognostic value of initial plasma miR-150 in AML adult patients, our follow up data did not reveal any relation between different levels of our target miR-150 and either; the spectrum of response to treatment involving CR and refractory cases; those undergo relapse or not, or those undergo early death before D28 or not. But revealed a statistically significant relationship between lower levels of miR-150 at D28 and cases that did not show relapse (P = 0.035). Also, our survival analysis data revealed no significant relation between levels of miR-150 and overall or relapse-free survival, which was inconsistent to Wang et al. [23], who showed that AML patients in his low miR-150 group had significantly shorter five-year overall survival (P = 0.009) and event-free survival (P = 0.004) than patients in his high miR-150 group.
As regard target genes pathway analysis, it showed that the predicted target genes might play their roles through cellular pathways. Those target genes are implicated in proliferation, adhesion, and apoptosis. The main target gene of miR-150 included AKT2, CBL, and PRKCA [29].
Mir-150 targets and downregulates AKT2 gene that is involved in Erb, MAPK, CML and mTOR signalling pathway, resulting in reduced the phosphorylated levels of AKtser473/4. Subsequently, this increases the levels of tumour suppressor genes such as Bim and P53, which leads to telomerase activation and immortalisation of cancer cells [26]. Also, CBL gene is implicated in AML signalling pathway and downregulated by miR-150 [27], [28].
The α isoform of Protein kinase C (PKCα), has been recognised as a tumour growth regulator in different types of cancers. Targeting PKCα-mediated signal transduction induces cell death in AML cells inhibiting BCL-2 phosphorylation and blocking ERK activation [29]. Overexpression of miR-150 significantly suppressed the endogenous expression of PKCα, and luciferase reporter/mutagenesis assays confirmed that PRKCA is a transcriptional target of miR-150 [31].
In conclusion, miRNAs have emerged as a class of gene expression important regulators contributing to AML pathogenesis and as potential biomarkers [32].
We conclude that miR-150 functions as a pivotal gatekeeper in inhibiting cell transformation and functions as a tumour suppressor and its repression are required for leukemogenesis. Moreover, miRNAs regulate different mRNA targets, and their modulation can represent a potential therapeutic target in combination with current chemotherapy in the eradication of leukemic progenitors.
Footnotes
Funding: This research did not receive any financial support
Competing Interests: The authors have declared that no competing interests exist
References
- 1.Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. https://doi.org/10.1056/NEJMra1406184 PMid:26376137. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Dohner H, Campbell PJ. Genomic classification in acute myeloid leukemia. N Engl J Med. 2016;375:900–901. doi: 10.1056/NEJMc1608739. https://doi.org/10.1056/NEJMc1608739. [DOI] [PubMed] [Google Scholar]
- 3.De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J. 2016;6:e441. doi: 10.1038/bcj.2016.50. https://doi.org/10.1038/bcj.2016.50 PMid:27367478 PMCid: PMC5030376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vitsios DM, Davis MP, van Dongen S, Enright AJ. Large-scale analysis of microRNA expression, epi-transcriptomic features and biogenesis. Nucleic Acids Res. 2017;45:1079–1090. doi: 10.1093/nar/gkw1031. https://doi.org/10.1093/nar/gkw1031 PMid:28180281 PMCid: PMC5388392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: Therapeutic implications and emerging concepts. Blood. 2017;130:1290–1301. doi: 10.1182/blood-2016-10-697698. https://doi.org/10.1182/blood-2016-10-697698 PMid:28751524 PMCid: PMC5600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–6. doi: 10.1016/s0092-8674(01)00616-x. https://doi.org/10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 7.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–40. doi: 10.1016/j.cell.2005.10.022. https://doi.org/10.1016/j.cell.2005.10.022 PMid:16271387. [DOI] [PubMed] [Google Scholar]
- 8.Edelstein LC, Bray PF. MicroRNAs in platelet production and activation. Blood. 2011;117(20):5289–96. doi: 10.1182/blood-2011-01-292011. https://doi.org/10.1182/blood-2011-01-292011 PMid:21364189 PMCid: PMC3109704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasilatou D, Papageorgiou S, Pappa V, Papageorgiou E, Dervenoulas J. The role of microRNAs in normal and malignant hematopoiesis. European Journal of Haematology. 2010;84(1):1–16. doi: 10.1111/j.1600-0609.2009.01348.x. https://doi.org/10.1111/j.1600-0609.2009.01348.x PMid:19744129. [DOI] [PubMed] [Google Scholar]
- 10.Lulla RR, Costa FF, Bischof JM, Chou PM, Bonaldo MF, Vanin EF, Soares MB. Identification of Differentially Expressed MicroRNAs in Osteosarcoma. Sarcoma. 2011;732690 doi: 10.1155/2011/732690. https://doi.org/10.1155/2011/732690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mraz M, Chen L, Rassenti LZ, Ghia EM, Li H, Jepsen K, Smith EN, Messer K, Frazer KA, Kipps TJ. miR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood. 2014;124(1):84–95. doi: 10.1182/blood-2013-09-527234. https://doi.org/10.1182/blood-2013-09-527234 PMid:24787006 PMCid: PMC4125356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris V, Zhang A, Yang T, Derek L, Stirewalt Ramamurthy R, Meshinchi S, Vivian G. Oehler MicroRNA-150 expression induces myeloid differentiation of human acute leukemia cells and normal hematopoietic precursors. PLoS One. 2013;8(9):e75815. doi: 10.1371/journal.pone.0075815. https://doi.org/10.1371/journal.pone.0075815 PMid:2408663;9 PMCid: PMC3782459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22(4):524–535. doi: 10.1016/j.ccr.2012.08.028. https://doi.org/10.1016/j.ccr.2012.08.028 PMid:23079661 PMCid: PMC3480215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fayyad-Kazan H, Bitar N, Najar M, Lewalle P, Fayyad-Kazan M, Badran R, et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J Transl Med. 2013;11:31. doi: 10.1186/1479-5876-11-31. https://doi.org/10.1186/1479-5876-11-31 PMid:23391324 PMCid: PMC3579719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. JBI. 2011;44(5):839–47. doi: 10.1016/j.jbi.2011.05.002. https://doi.org/10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Dweep H, Gretz N. miRWalk2.0. A comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. https://doi.org/10.1038/nmeth.3485 PMid:26226356. [DOI] [PubMed] [Google Scholar]
- 17.Dennis Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. “DAVID: Database for Annotation, Visualization, and Integrated Discovery”. Genome Biology. 2003;4(5):P3. https://doi.org/10.1186/gb-2003-4-5-p3. [PubMed] [Google Scholar]
- 18.Huang DW, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. https://doi.org/10.1038/nprot.2008.211 PMid:19131956. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. “KEGG: Kyoto Encyclopedia of Genes and Genomes”. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. https://doi.org/10.1093/nar/28.1.27 PMid:10592173 PMCid: PMC102409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang XS, Gong JN, Yu J, Wang F, Zhang XH, et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood. 2012;119:4992–5004. doi: 10.1182/blood-2011-10-385716. https://doi.org/10.1182/blood-2011-10-385716 PMid:22493297. [DOI] [PubMed] [Google Scholar]
- 21.Vitsios DM, Davis MP, van Dongen S, Enright AJ. Large-scale analysis of microRNA expression, epi-transcriptomic features and biogenesis. Nucleic (2013) Acids Res. 2017;45:1079–1090. doi: 10.1093/nar/gkw1031. https://doi.org/10.1093/nar/gkw1031 PMid:28180281 PMCid: PMC5388392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Gao L, Luo X, Wang L, Gao X, et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t (8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 121:499–509. doi: 10.1182/blood-2012-07-444729. https://doi.org/10.1182/blood-2012-07-444729 PMid:23223432. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Wang J, Yin Z, Zhang W, Hu X, Wang Y. Serum miR-150 as a novel prognostic biomarker for acute myeloid leukemia. Int J Clin Exp Pathol. 2017;10(6):6906–6911. [Google Scholar]
- 24.Xu DD, Zhou PJ, Wang Y, Zhang Y, Zhang R, Zhang L, Chen SH, Fu WY, Ruan BB, Xu HP, et al. miR-150 suppresses the proliferation and tumorigenicity of leukemia stem cells by targeting the Nanog signaling pathway. Front Pharmacol. 2016;7:439. doi: 10.3389/fphar.2016.00439. https://doi.org/10.3389/fphar.2016.00439 PMid:27917123 PMCid: PMC5114241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris VA, Cummings CL, Korb B, Boaglio S, Oehler VG. Deregulated KLF4 expression in myeloid leukemias alters cell proliferation and differentiation through microRNA and gene targets. Mol Cell Biol. 2015;36:559–573. doi: 10.1128/MCB.00712-15. https://doi.org/10.1128/MCB.00712-15 PMid:26644403 PMCid: PMC4751692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. https://doi.org/10.1038/leu.2011.81 PMid:21502955. [DOI] [PubMed] [Google Scholar]
- 27.Bousquet Marina, et al. miR-150 blocks MLL-AF9–associated leukemia through oncogene repression. Molecular Cancer Research. 2013;11(8):912–922. doi: 10.1158/1541-7786.MCR-13-0002-T. https://doi.org/10.1158/1541-7786.MCR-13-0002-T PMid:23604034. [DOI] [PubMed] [Google Scholar]
- 28.Fleischmann K, Pagel P, von Frowein J, Magg T, Roscher AA, Schmid I. The leukemogenic fusion gene MLL-AF9 alters microRNA expression pattern and inhibits monoblastic differentiation via miR511 repression. Journal of Experimental & Clinical Cancer Research. 2016;35:9. doi: 10.1186/s13046-016-0283-5. https://doi.org/10.1186/s13046-016-0283-5 PMid:26762252 PMCid: PMC4712549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang ZH, Wang SL, Zhao JT, Lin ZJ, et al. miR-150 exerts antileukemia activity in vitro and in vivo through regulating genes in multiple pathways. Cell death & disease. 2016;7(9):e2371. doi: 10.1038/cddis.2016.256. https://doi.org/10.1038/cddis.2016.256 PMid:27899822 PMCid: PMC5059860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez N, Cordiner RA, Young RS, Hug N, Macias S, Caceres JF. Genetic variation and RNA structure regulate microRNA biogenesis. Nat Commun. 2017;8:15114. doi: 10.1038/ncomms15114. https://doi.org/10.1038/ncomms15114 PMid:28466845 PMCid: PMC5418625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: Therapeutic implications and emerging concepts. Blood. 2017;130:1290–1301. doi: 10.1182/blood-2016-10-697698. https://doi.org/10.1182/blood-2016-10-697698 PMid:28751524 PMCid: PMC5600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trino S, Lamorte D, Caivano A, Laurenzana I, Tagliaferri D, Falco G, et al. MicroRNAs as New Biomarkers for Diagnosis and Prognosis, and as Potential Therapeutic Targets in Acute Myeloid Leukemia. Int J Mol Sci. 2018;19:460. doi: 10.3390/ijms19020460. https://doi.org/10.3390/ijms19020460 PMid:29401684 PMCid: PMC5855682. [DOI] [PMC free article] [PubMed] [Google Scholar]


