Abstract
BACKGROUND:
HCV infection and its complications are among the leading public health challenges; the emergence of drug-resistant variants are expected to be a major problem. A novel combinatorial small interfering RNA (siRNA) could be a novel triple therapy that could be suitable for genotype 4. Although HCV is a hepatotropic virus, there is reliable evidence about its replication in peripheral blood mononuclear cells (PBMC) of chronically infected patients; these cells act as an extra-hepatic reservoir for viral recurrence and persistence. The patients with HCV-RNA in PBMC showed a significantly lower response to therapy that supports to be one of the factors influencing therapeutic response. Almost all regions of HCV show potential for siRNA target with relative efficiencies of individual siRNA sequences.
AIM:
This study aims to test the efficacy of siRNA against HCV-4 replication in PBMC in vitro, to introduce an alternative therapeutic option for HCV-4 suitable to eradicate it from both hepatic and extra-hepatic reservoirs.
METHODS:
Efficacy of synthesised siRNA molecule that targets 5/UTR of domain IIIC within IRES of HCV RNA to eradicate HCV intra-PBMC in vitro was tested and compared with IFN/RBV in vitro, by using both qRT-PCR and western blot. Sixty genotype-4 chronic HCV patients who are naïve for any HCV treatment were enrolled and tested for the presence of HCV intra-PBMC using qRT-PCR before and after siRNA treatment in vitro.
RESULTS:
Real-time PCR analysis showed a significant reduction of HCV RNA levels after 24hr post-HCV-positive-PBMCs treatment by siRNA with cell vitality reached up to 98%. Besides a complete inhibition of NS5A viral protein expression, that is functionally essential for viral assembly, replication and egress.
CONCLUSION:
So, Targeting HCV infection using RNA interference technology might be a reliable therapeutic option for chronic HCV patients with HCV minus strand within PBMCs.
Keywords: IRES, PBMC, siRNA, HCV, NR, UTR
Introduction
Hepatitis C virus (HCV) infection is a global blood-borne disease that affects almost 3% of the world population with morbidity and mortality that is second to HIV among the emerging infections [1]. Egypt has been reported as the highest in HCV prevalence worldwide, that ranged from 6% to exceed 40% among region and demographic groups with 11-14% of the population chronically infected with the virus [2]. The infection may get worse to cirrhosis with the subsequent development of complications including hepatocellular carcinoma [3], [4].
HCV is a positive-sense single-stranded RNA virus, classified as the sole member of the genus Hepacivirus in the family Flaviviridae. The HCV RNA genome is roughly 9.6 kb long with an open Reading Frame (ORF) that encode a vast viral polyprotein of around 3,01 thousand amino acids [4]. Viral translation is mediated through the internal ribosome entry site (IRES) found within the 5’ untranslated region (5’UTR) which comprise of ~341 bp long that is profoundly conserved even between various HCV isolates. The 5’UTR does not encode for functional protein and contains the IRES that initiate viral polyprotein translation in a cap-independent manner [5]. The IRES is basic for translational as it independently binds to the 40S ribosomal subunit and guides the ribosome to the initiation codon of the HCV mRNA with a specific end goal to facilitate its translation in a cap-independent manner. It contains four profoundly organised stem-looped domains that named I-IV which facilitate HCV RNA translation. Domain I is not essential for IRES activity but rather essential for HCV replication, IRES in domains II-IV play a role in viral genetic replication in a cap-independent manner while Domain III contains subdomains that are critical for the binding of 40S ribosomal subunit [6]. Translation of HCV mediated by a profoundly conserved internal ribosomal Entry site (IRES) within the 5’UTR making it a significant focus for new medication development [5].
Despite hepatotropic nature of HCV, many studies proved that PBMC represents another extrahepatic reservoir where HCV can replicate, not only that, but it may also be the real culprit in viral recurrence after liver transplantation [7]. The substantial goal of viral treatment is to attain a sustained virological response (SVR) that is defined as undetectable HCV-RNA in peripheral blood determined 24 weeks after the end of treatment (ETR). Nonresponders (NR) to therapy are sometimes obstinate to retreatment and do not necessarily benefit from escalating treatment dose [8].
Despite the great success of the current therapies, there are many reported concerns that may increase the demand for new therapy and diagnostic approach to cover it [4], for example; Human Biliverdin reductase (HBVr) reported to be upregulated in many studies upon Direct-acting antivirals (DAAs) therapy, HBVr usually elevated at mean of 52 days after first DAA dose and reported to cause ten DAA-therapy discontinuations [9]. These patients cannot make a benefit from DAAs, so alternative options are urgently necessary, out of this alternative is RNAi that represent an effective silencing approach for molecular therapeutics through a particular succession RNA degradation procedure to diminish infection or replication of its invaders [5].
siRNA is promising silencing mechanism, vector-based or synthetic siRNAs directed against 5’ UTR, NS3, NS5B, NS4B and core represented significant specific ability in reducing both viral infection and replication. The 5/UTR of HCV-IRES has also been targeted by siRNA resulted in inhibition of HCV by more than 80% at a concentration of 2.5 nM. Moreover, consensus siRNAs against 5/UTR diminished viral replication level in vitro on infected Huh-7 cell-line [10]. While the dominant part of HCV-siRNAs is integral to the positive strand, a lessening in both strands replication has been reported [11]. It is conceivable that targeting the positive strand leads to a decline in negative strands synthesis [12].
Interferon (IFN) pathway can be initiated by dsRNAs, so it was important to address whether HCV-particular siRNAs could trigger IFN pathway. It was exhibited that hindrance of viral replication by HCV RNAi in vitro was not related to an up-regulation of IFN-stimulated genes. Targeting HCV by siRNAs were better at diminishing HCV-RNA levels than an oscillating dose of IFN-α [11].
All these possibilities motivate us to test the efficacy of specific synthetic-siRNA on clearance of HCV from PBMCs of chronic HCV-infected patients aiming to get a new therapeutic recommendation for inconvenient DAAs therapy patients, non-responders or relapse’s patients. In addition to scavenging viral infection from PBMCs that act as a real culprit in HCV relapses after liver transplantation.
Patients and Methods
Patients and Samples collection
Total of sixty patients was enrolled in this study; all were naïve and newly diagnosed HCV patients, from National Hepatology and Tropical Medicine Research Institute (NHTMRI) Cairo, Egypt. All patients were pre-diagnosed HCV antibody positive by ELISA technique. HCV detected in serum and PBMC for all samples. Ethical committee approval was taken from National Hepatology and Tropical Medicine Research Institute in addition to a signed consent form from each patient. Patient’s inclusion criteria were; first, they should be treatment naïve patients, then they should not have any other neurological disorders or undergo conservative treatment, also not to be co-infected by any hepatic viral infection than HCV that was detected by the presence of its antibodies (Ab) in serum. About 12 mL blood was withdrawn from each patient on heparin.
siRNA Choice
siRNA targeting domain IIIC within IRES was chosen that showed 100% alignment with HCV sequences in the gene bank (database), in addition to its ability to align specifically against domain IIIC within IRES. This siRNA was specially synthesised and ordered from thermo-scientific U.S.A (cat# K-005000-G1-01, E-007500 -01), to be HPLC purified and sterilised using ultra-filtration methods to assure removal of any materials that might interfere with transfection or might be lethal to cells.
Optimising siRNA Transfection Procedure
Since there is a great variation in the capacity of transfection according to each cell type, so the transfection procedure must be determined and adjusted for each cell line before transfection. Also, it is well known that PBMCs is hard to transfect cells. Therefore, the GAPDH siRNA (Thermo, USA) was used to optimise: a) optimal cell plating density; b) optimal amount of siRNA to be transfected; and c) either to transfect it in serum-free or serum-containing medium (FCS) and determine optimal serum concentration.
Self-delivery Accell™-siRNAs were synthesised by Dharmacon® that allows effective delivery in wide-ranging cell lines including primary cells. Ficoll-hypaque density gradient technique was used to isolate PBMC from the blood. PBMC was washed at least four times with PBS, re-suspended in Accell delivery media®, and counted; then 1 × 106 cells were used for detection of presence of HCV and 0.6 ×106 cells/well were plated in 48 well-plate for testing efficacy of different siRNA concentrations (50-100-200 µM) on silencing HCV replication and compare its effect with IFN/RBV in separate wells were concomitantly treated with 110 IU/ml IFN plus100 µM Ribavirin in vitro [13]. Total RNA was harvested at different times post-siRNA transfection using Viral RNA extraction kit (QIAamp-viral RNA mini kit) from QIAGEN according to the manufacturer’s manual. Also, control for HCV-siRNA was used that was Human GAPDH siRNA in addition to positive and negative (non-targeting NTC) control for each.
Determination of HCV in Patient’s Sera
Two hundred (µL) serum were used for RNA extraction by viral RNA extraction kit (QIAamp-viral RNA mini kit- QIAGEN-USA) consistent with the manufacturer’s manual. 200 ng of RNA utilised to detect virus C using one-step RT-PCR kit (QIAGEN, USA) and used as stated by the manufacturer’s protocol, and then the PCR amplicon was photographed on 2% agarose gel.
HCV Genotyping
Genotyping was done using the VERSANT HCV genotype 2.0 Assay (LiPA - Bayer).
Uncovering HCV Strands in Patient’s PBMC
Qualitative analysis
Cellular RNA was extracted from counted uncultured PBMC, collected from HCV serum positive patients, using Viral RNA extraction kit (QIAamp-viral RNA mini kit- QIAGEN, USA) consistent with the manufacturer’s manual. HCV strands were detected by RT-PCR; briefly, 200 ng of extracted RNA was reversely transcribed to cDNA in 50 µL mixture PCR beads (GE healthcare life science, USA) and 25 pmoles antisense primer 5’GGTGCACGGTCTACGAGACCTC3’ for a positive strand or 5’AACTACTGTCTTCACGCAGAA3’ for a negative one. 50 µL was the first round PCR volume, using 50 pmoles from each primer and 5’TGCTCATGGTGCACGGTCTA3’. Then the second round uses 20% of this volume for its amplification with the internal pair of primers 5CGCAGAAAGCGTCTAGCCAT3 in addition to 5ACTCGGCTAGCAGTCTCGCG3. The thermal cycler conditions were the same as the first round, then PCR amplicon was analysed on 2% agarose gel electrophoresis and documented.
Quantitative analysis
HCV quantitation was performed on a Piko-Real-Time PCR System (Thermo-Scientific) using sensifast SYBR kit (Bioline) according to the manufacturer procedure.
In vitro treatment of infected PBMC by siT and IFN/RBV
Patients proved to be infected with HCV in both serum and PBMCs were enrolled in this step. PBMCs were separated using ficoll, washed 4 times with PBS to avoid any attached viral particles on cell membrane, then resuspended in Accell delivery media® For each patient, cells were plated on a 48-well plate at a concentration of 0.6 × 106/mL, and cultured at 37°C, 5% CO2 for 72 hr in absence or presence different siRNA concentrations, siNTC, GAPDH siRNA and IFN/RBV separately (triplicate for each treatment). Cultured cells without any treatment were used as a control. Cells were harvested, washed 4 times with PBS, and subjected to total cellular RNA extraction. Total RNA extracted from cultured PBMCs was reverse transcribed and amplified using the same assay described previously for both qualitative and quantitative measurement [14].
Western blot analysis of HCV NS5A antigens in PBMC with and without siRNA
Cell lysates of PBMC either with or without siRNA treatment were subjected to SDS-PAGE. Then washed three times, after that membrane was incubated with diluted peroxidase-labelled anti-human IgG/IgM antibody mixture at 1:5000 in phosphate buffer saline (PBS) 3 g/L for previously treated strips with the anti-NS5A (Novocastra Laboratories, UK) for at least two hr at room temperature. Then Visualize of the immune complexes on the nitrocellulose membranes by promoting the strips with 0.01 mol/L PBS (pH 7.4) that contain 40 mg 3,3’,5,5 tetramethylbenzidine and 100 μl of 30 ml/L hydrogen peroxide (Immunopure TMB substrate Kit, Rockford, USA).
Statistical Analysis
All experiments were performed in triplicates on three independent cultures. Data were shown in means ± SD. P < 0.05 was considered statistically significant.
Results
Clinical Aspects of HCV patients
Patients showed a significant difference (P<0.001) in liver function test ALT, AST, liver synthetic function assays; prothrombin and albumin (P<0.0001) and total leukocyte count (P=0.0002), on the other hand Age, BMI, gender, and total bilirubin showed no significant difference between normal and chronic HCV infected patients Table (1).
Table 1.
Clinical data of chronic-HCV patients’ vs healthy control
| Variable | Chronic HCV patients (mean ± SD) | Healthy persons (mean ± SD) | P value |
|---|---|---|---|
| Age | 49 ± 10 | 47±8 | 0.674 (NS) |
| BMI (Kg/m2) | 26.8 ± 5 | 26.2±5 | 0.7464 (NS) |
| Gender | |||
| Male | 22 (55%) | 5 (50%) | 0.5719 (NS) |
| Female | 18 (45%) | 5 (50%) | 0.5687 (NS) |
| Platelet count (103/µL) | 216 ± 68 | 265±120 | 0.1006 (NS) |
| ALT (IU/L) | 68.1 ± 41.7 | 13±6 | 0.0003 (S) |
| AST (IU/L) | 52.3 ± 32.6 | 12±8 | 0.0006 (S) |
| Bilirubin Total (mg/dl) | 0.43 ± 0.16 | 0.39±0.19 | 0.1087 (NS) |
| Bilirubin Direct (mg/dl) | 0.85 ± 0.37 | 0.29±0.16 | 0.086 (NS) |
| Prothrombin concentration % | 69.9 ± 19.3 | 103±4.7 | <0.0001 (S) |
| Albumin (g/dl) | 3.28 ± 0.6 | 4.7±0.8 | <0.0001 (S) |
| Total leukocyte count (103/µL) | 6.08 ± 2.18 | 3.0±1.5 | =0.0002 (S) |
| Serum HCV viral load | |||
| <500,000 IU/ml | 25 (62.5%) | 0 | |
| ≥500,000 IU/ml | 15 (37.5%) | 0 | |
| PBMC viral load | |||
| PBMC + HCV | 19 (47.5%) | 0 | |
| PBMC -HCV | 21 (52.5%) | 0 | |
| Fibrosis level | |||
| F (n: 0/1/2/3/4) | 16/14/9/1 | 0/0/0/0 | |
NS= no significant difference; S = significant difference.
HCV RNA detectability in Patient’s Serum and PBMC
All the 60 patients were HCV RNA positive in serum but only 19 out of them (31.7%) showed detectable HCV -RNA in PBMC using RT-PCR for HCV strands, from clinical data observations, 17(89.5%) out of these 19 PBMC positive HCV patient show non-responder state to PEG-IFN and 11(64.7%) showed non-response to sovaldi therapy at week 12.
Results of optimisation of siRNA concentration and delivery method in vitro
To assess the potentiality and efficacy of siRNA in Accell delivery media, optimisation of the various parameters carried out using GAPDH siRNA as a positive internal control. The results represented a significant silencing of GAPDH mRNA expression in transfected PBMCs when Accell delivery reagent was used (p < 0.05) (Figure 1). But there was no observed inhibition when lipofectamine nor electroporation was used compared to using Accell delivery with no siRNA as a negative control.
Figure 1.
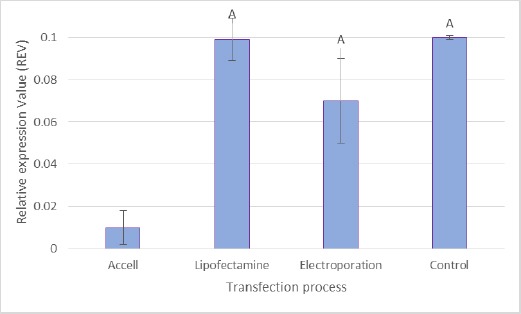
Effect of different delivery methods on GAPDH expression
Two other transfection reagents were tested for transfection optimisation. Accell delivery media showed a significant knockdown of GAPDH compared to Lipofectamine and Electroporation; (A); p < 0.05. Values expressed as relative expression value (REV) with mean ± SD, n = 19.
Results of in vitro inhibition of HCV replication by siRNA in PBMC
Current study demonstrates that the introduction of siRNA targeting domain IIIC within 5’UTR IRES site into PBMC, that previously confirmed to be infected by HCV using quantitative PCR assay, caused a dramatic sharp decrease of HCV RNA by more than 94% compared to non-treated cells using qRT-PCR (Table 2), also NS5A protein expression levels detected by western blot assay was completely inhibited (Figure 3). was also noticed that the effect of siRNA on HCV replication and life-cycle occurs very early after 24 hours post siRNA transfection with variable treatment concentrations (Figure 2).
Table 2.
The HCV RNA levels (%*) in HCV-infected PBMCs inhibited by siRNA for 72 h
| Doses of siRNA (nM) | ||||
|---|---|---|---|---|
| 10 | 50 | 100 | 200 | |
| Untreated | 100 ± 11.74 | 100 ± 8.45 | 100 ± 12.47 | 100 ± 6.23 |
| siGAPDH | 96 ± 19.47 | 86 ± 9.24 | 80 ± 8.96 | 76 ± 10.04 |
| siNTC | 92 ± 8.25 | 92 ± 14.63 | 111 ± 16.28 | 108 ± 8.06 |
| siT | 29 ± 12.50a | 18 ± 8.50a | 6 ± 1.11a,b | 8 ± 2.83a,b |
| IFN/RBV (110 IU/ml IFN plus100 µM Ribavirin) | 57±11.8a | |||
| RBV (100 µM Ribavirin) | 8 ± 2.7a | |||
*data Presented as viral load (mean ± SD); a P<0.01, as compared with untreated control; bP<0.01, as compared with the 10nM group; cP<0.01, as compared with the 50nM group.
Figure 3.
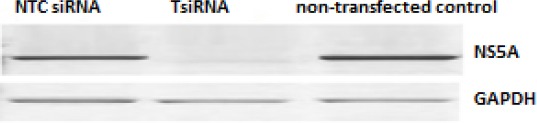
Composite picture for Western blot for HCV-NS5A compared to intracellular GAPDH
Figure 2.
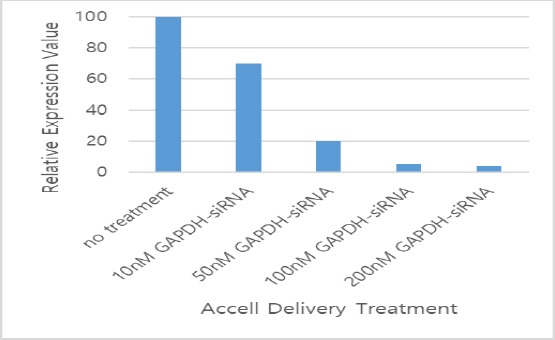
Concentration optimisation using GAPDH siRNA. All values were expressed as relative expression value (REV) with mean ± SD, n = 9
siRNA were applied to HCV-infected PBMCs cell-culture to assess the effects of siRNA on the HCV replication. Infected PBMCs that were untreated and transfected non-targeting (scrambled) siRNA (siNTC) served as negative controls while PBMCs that were untreated and transfected with GAPDH siRNA served as positive controls, Figure 3 and Table 2 illustrate viral RNA levels and protein expression for both transfected and control PBMCs, as quantitated by qPCR at 72 h after transfection. siRNA was significantly able to suppress HCV replication at 100 nM compared with untreated PBMCs (P < 0.01).
The result of the quantitative PCR of samples after these in vitro treatments shows significant viral reduction with siT compared to IFN/RBV (P = 0.001). As shown in Table 2, SiT decreased the viral load by mean ± SD 94.0 ± 1.11% while the combination therapy decreased it by mean 57 ± 11.8% and finally ribavirin alone decreases it by mean 8 ± 2.7% only that later complied with in-vivo response results for same patients of IFN/RBV and DAAs therapies.
NS5A levels were significantly decreased after 24 hr. of the transfection of 100 nM of SiT (Fig. 3). And maximum inhibition levels were observed on 72 hr. post transfection (mean silencing 94.0 ± 1.11%). Also, there are no significant inhibition levels- for HCV in cells transfected with either the NTC siRNA nor those with GAPDH siRNA- was detected.
Upper row showed HCV-NS5A expression in different transfection for PBMC positive HCV cells after 24hr post-treatment. Lower row showed the GAPDH expression in same transfected cells.
Results of SiRNA cytotoxicity detection
To detect possible cytotoxic effects that may be induced by the siRNA, both trypan-blue exclusion method and MTT assay were performed. The cell viabilities reached up to 98% (Table 3) for both cells transfected by siRNA or for cells grown in Accell delivery media (transfection media). Thus, neither the transfection media (on another way transfection process) nor selected siRNA in the current study induced toxicity against PBMCs cells.
Table 3.
Cell viability (%*) of PBMCs exposed to either siRNA for 72 h or IFN/RBV
| Doses of siRNA (nM) | ||||
|---|---|---|---|---|
| 10 | 50 | 100 | 200 | |
| Untreated | 100 ± 4.47 | 100 ± 3.72 | 100 ± 2.46 | 100 ± 1.16 |
| siGAPDH | 96 ± 19.47 | 86 ± 9.24 | 80 ± 8.96 | 76 ± 10.04 |
| siNTC | 100 ± 9.86 | 100 ± 5.72 | 108 ± 4.28 | 105 ± 5.2 |
| siT | 102 ± 3.50 | 108 ± 6.50 | 116 ± 11.11 | 118 ±1 2.53 |
| IFN/RBV (110 IU/ml IFN plus100 µM Ribavirin) | 57±11.8a | |||
| RBV (100 µM Ribavirin) | 68±18.2a | |||
data Presented as viral load (mean ± SD) siNTC= siRNA non targeting pool control.
Discussion
In the current study, 60 genotypes 4 chronic HCV infected patients were enrolled. They are naïve for HCV treatment, HCV RNA was detected in PBMCs of 19 patients out of 60 (about 31.7%), and the remainder show no HCV in its PBMCs, similarly this finding complies with our result that was published 2014 [14] that represent presence of HCV intra-PBMC in 10 out of 25 HCV infected patients that represent 40% of total study population.
Viral translation is mediated through IRES site found within the 5/UTR that consists of ~341 bp in length with highly conserved sequence even between different HCV isolates. IRES can initiate viral polyprotein translation in a cap-independent manner via independently binds to the 40S ribosomal subunit and directs the ribosome to the initiation codon of mRNA of HCV to expedite translation in a cap-independent manner and it is a vital target for the development of new antiviral compounds [15].
It is well known that viruses, particularly retroviruses and HCV, are notoriously prone to the high rate of errors during their replication process, and continuously undergoes mutation and mostly produce mutated viral proteins to escape immune-system defence mechanisms. These mutations may also escape attacking by various siRNAs [16]. The protein-coding sequence of the HCV genome that was targeted in the study by McCaffrey et al., 2002 varies considerably among different HCV genotypes, and even among strains of the same genotype [17]. Also, the high error rate of the non-proofreading HCV RNA-dependent RNA polymerase greatly showed that was called “siRNA escape mutants” which have silent mutations in the protein-coding sequence [18]. In contrast, the 5’UTR especially IRES IIIC, that was selected as a target in the current study, is almost identical among the known strains of HCV[7]. Moreover, structural fetters on the 5’UTR, regarding its ability to direct internal ribosome entry and translation of viral proteins, would not riddance escape mutations [19].
For this reason, the IRES domains within 5’UTR of the HCV genome appears to be an ideal target for siRNA in clinical applications. Not all 5’UTR-directed siRNAs have the same efficacy; there are many siRNA were detected based on the consensus sequences of Egyptian HCV 5’UTR. In this study siT was chosen that showed 100% alignment with HCV sequences in the gene bank (database) and also can align in 5’UTR specifically against domain IIIC within IRES in silico then we validate this hypothesis in the wet lab.
In the current study, we were able to demonstrate that the introduction of SiT targeting domain IIIC within IRES in 5’UTR into PBMC that previously confirmed its infection by HCV using quantitative PCR assay, caused a dramatic sharp decrease of HCV RNA by more than 94% compared to non-treated cells, HCV RNA levels decreased 25-fold (P = 0.0005) in Huh-7 cells too after transfection with siRNA (data not shown), in addition to modulation of NS5A viral protein expression. It was noticed that the effect of siRNA on HCV replication and life-cycle occurs very early after 24 hours post siRNA transfection. Our data is in agreement with both McCaffrey et al., 2002 [17] who showed in his study that, HCV NS5B RNA polymerase gene fragment, which was transiently co-transfected with siRNA into mouse liver by hydrodynamic injection, was cleaved after treatment with specific siRNA [17]. Our data are also consistent with those of Randall et al., 2003 who demonstrated that siRNA could target and cleaves the HCV 5’UTR efficiently and specifically. More importantly, Randall and his associates showed that the cleavage of HCV-RNA not only suppressed viral protein synthesis but also blocked the replication of sub-genomic viral RNA [20] that was supported by our data from PBMCs too.
Up to our Knowledge, it’s the first time for a comparative study between the effect of either ribavirin alone or with combination with IFN at recommended dose against 100 µM siRNA as optimal dose and we clearly observe the superior effect of siRNA over other treatment on PBMC in vitro, as the same pattern, Table 2 showed the superiority of siT that reached 94% viral clearance while the combination therapy decreased it by 57% and finally ribavirin decreased it by 8% only, the finding support siRNA efficiency on HCV silencing either in hepatic cell or extra-hepatic reservoir like PBMCs, Also it was the first time to test efficacy of siRNA to eradicate HCV replication on PBMCs that are hard to transfect cells acts as HCV reservoir.
As a safe and efficient way of delivery of siRNAs to cells that can suppress HCV replication in all infected cells in vivo have not been discovered yet, chemically specially modified synthetic siRNA might easily be made and delivered into cells on their own. In our study we compared three different ways of transfection and we conclude that Accell siRNA delivery method® is specially modified for use without a transfection reagent and works at a higher concentration than classical siRNA with minimal disruption of the expression profile and approved to transfect hard to transfect cells like PBMCs with no off-target effect (Figure 1).
Results showed that the siT that we selected, siT- (nt 59–79 from the 5’UTR beside nt 109–129 from the HCV core area), successfully inhibit HCV replication in PBMC culture without affecting housekeeping genes (Figure 3), it was clearly observed safety profile of siT on PBMCs in addition to cell vitality that exceeded 98% using trypan blue exclusion method and cytotoxicity MTT assay, thus confirming earlier reports on siRNA and suggesting the potential therapeutic value of siRNA in HCV type-4.
To confirm our result and data both Q-PCR of viral RNA, western blot for NS5A protein were performed to assess the effect of siT at 24 hr on such protein expression. NS5A may be an RNA binding protein because of its ability to bind to 3’UTR of the plus and minus HCV RNA strands with a key modulator role of host cell function and activity, especially IFN response of the host cell, despite siRNA showed superior efficacy in modulating NS5A gene expression in this study, results need more validation by comparing its efficacy to DAAs targeting the same NS5A like Declatasvir® and Elbasvir®.
Our preliminary data showed that siT efficiently suppresses HCV replication in PBMC in vitro. The result of this study overcomes our result of another study previously published in 2014; antisense SODN1 for inhibition of HCV in both PBMC and hepatoma cells because of failure of antisense to eradicate HCV within PBMCs compared to hepatoma cells [14]. Besides over-coming IFN+RBV too making it promising molecule for either non-responders or relapses’ patients beside those non-suitable for current therapies.
Our data suggest that siRNAs targeting 5’UTR can induce an anti-HCV response in cell culture of PBMC harbouring the virus. Therefore, it represents a future therapy that could eradicate viral RNA from either hepatic cells or PBMCs, and consequently, it can potentially cure patients with HCV by eradicating it from its reservoir. The efficiency of siT in inhibiting HCV replication in PBMCs potentially suggests that this RNA-targeting approach might support and provide an effective therapeutic option for HCV infection especially for those hard to treated patients.
Acknowledgements
This work was supported in part by a grant from the Academy of Scientific research and technology (ASRT/TRG/H/2010-4) and National Research Centre (NRC), Egypt.
Footnotes
Funding: This work was supported in part by a grant from the Academy of Scientific research and technology (ASRT/TRG/H/2010-4) and National Research Centre (NRC), Egypt
Competing Interests: The authors have declared that no competing interests exist
References
- 1.Organization WHO. WHO fact sheet 164-hepatitis C. 2014 [Google Scholar]
- 2.Lehman EM, Wilson ML. Epidemic hepatitis C virus infection in Egypt: estimates of past incidence and future morbidity and mortality. J Viral Hepat. 2009;16(9):650–8. doi: 10.1111/j.1365-2893.2009.01115.x. https://doi.org/10.1111/j.1365-2893.2009.01115.x PMid:19413698. [DOI] [PubMed] [Google Scholar]
- 3.Itskowitz MS. Hepatitis C: epidemiology, diagnosis, and management. Compr Ther. 2007;33(2):87–93. doi: 10.1007/s12019-007-8005-8. https://doi.org/10.1007/s12019-007-8005-8 PMid:18004020. [DOI] [PubMed] [Google Scholar]
- 4.Elemeery MN, et al. Validation of a serum microRNA panel as biomarkers for early diagnosis of hepatocellular carcinoma post-hepatitis C infection in Egyptian patients. World J Gastroenterol. 2017;23(21):3864–3875. doi: 10.3748/wjg.v23.i21.3864. https://doi.org/10.3748/wjg.v23.i21.3864 PMid:28638226 PMCid: PMC5467072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penin F, et al. Structural biology of hepatitis C virus. Hepatology. 2004;39(1):5–19. doi: 10.1002/hep.20032. https://doi.org/10.1002/hep.20032 PMid:14752815. [DOI] [PubMed] [Google Scholar]
- 6.Khaliq S, et al. Down-regulation of IRES containing 5'UTR of HCV genotype 3a using siRNAs. Virology Journal. 2011;8:221–221. doi: 10.1186/1743-422X-8-221. https://doi.org/10.1186/1743-422X-8-221 PMid:21569449 PMCid: PMC3116492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi ST, Lai MM. HCV 5′and 3′UTR: when translation meets replication. Hepatitis C Viruses: Genomes and Molecular Biology. 2006:49–87. [Google Scholar]
- 8.Bare P. Hepatitis C virus and peripheral blood mononuclear cell reservoirs Patricia Bare. World J Hepatol. 2009;1(1):67–71. doi: 10.4254/wjh.v1.i1.67. https://doi.org/10.4254/wjh.v1.i1.67 PMid:21160967 PMCid: PMC2999261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearlman BL, Traub N. Sustained virologic response to antiviral therapy for chronic hepatitis C virus infection: a cure and so much more. Clin Infect Dis. 2011;52(7):889–900. doi: 10.1093/cid/cir076. https://doi.org/10.1093/cid/cir076 PMid:21427396. [DOI] [PubMed] [Google Scholar]
- 10.Ansar M, et al. Inhibition of full length hepatitis C virus particles of 1a genotype through small interference RNA. Virol J. 2011;8:203. doi: 10.1186/1743-422X-8-203. https://doi.org/10.1186/1743-422X-8-203 PMid:21535893 PMCid: PMC3094304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashfaq UA, et al. siRNAs: potential therapeutic agents against hepatitis C virus. Virol J. 2011;8:276. doi: 10.1186/1743-422X-8-276. https://doi.org/10.1186/1743-422X-8-276 PMid:21645341 PMCid: PMC3118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger KL, Randall G. Possibilities for RNA interference in developing hepatitis C virus therapeutics. Viruses. 2010;2(8):1647–65. doi: 10.3390/v2081647. https://doi.org/10.3390/v2081647 PMid:21994699 PMCid: PMC3185727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buckwold VE, et al. Synergistic In Vitro Interactions between Alpha Interferon and Ribavirin against Bovine Viral Diarrhea Virus and Yellow Fever Virus as Surrogate Models of Hepatitis C Virus Replication. Antimicrobial Agents and Chemotherapy. 2003;47(7):2293–2298. doi: 10.1128/AAC.47.7.2293-2298.2003. https://doi.org/10.1128/AAC.47.7.2293-2298.2003 PMid:12821481 PMCid: PMC161860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youssef SS, et al. In vitro inhibition of hepatitis C virus by antisense oligonucleotides in PBMC compared to hepatoma cells. Biomed Res Int. 2014;2014:196712. doi: 10.1155/2014/196712. https://doi.org/10.1155/2014/196712 PMid:24991538 PMCid: PMC4058683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khaliq S, et al. Down-regulation of IRES containing 5'UTR of HCV genotype 3a using siRNAs. Virol J. 2011;8:221. doi: 10.1186/1743-422X-8-221. https://doi.org/10.1186/1743-422X-8-221 PMid:21569449 PMCid: PMC3116492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carmichael GG. Medicine: silencing viruses with RNA. Nature. 2002;418(6896):379–80. doi: 10.1038/418379a. https://doi.org/10.1038/418379a PMid:12140542. [DOI] [PubMed] [Google Scholar]
- 17.McCaffrey AP, et al. RNA interference in adult mice. Nature. 2002;418(6893):38–9. doi: 10.1038/418038a. https://doi.org/10.1038/418038a PMid:12097900. [DOI] [PubMed] [Google Scholar]
- 18.Zekri ARN, et al. Consensus siRNA for inhibition of HCV genotype-4 replication. Virology Journal. 2009;6:13–13. doi: 10.1186/1743-422X-6-13. https://doi.org/10.1186/1743-422X-6-13 PMid:19173711 PMCid: PMC2661880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokota T, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003;4(6):602–8. doi: 10.1038/sj.embor.embor840. https://doi.org/10.1038/sj.embor.embor840 PMid:12740604 PMCid: PMC1319196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci U S A. 2003;100(1):235–40. doi: 10.1073/pnas.0235524100. https://doi.org/10.1073/pnas.0235524100 PMid:12518066 PMCid: PMC140937. [DOI] [PMC free article] [PubMed] [Google Scholar]


