Abstract
Tumour heterogeneity poses a substantial problem for the clinical management of cancer. Somatic evolution of the cancer genome results in genetically distinct subclones in the primary tumour with different biological properties and therapeutic sensitivities. The problem of heterogeneity is compounded in metastatic disease owing to the complexity of the metastatic process and the multiple biological hurdles that the tumour cell must overcome to establish a clinically overt metastatic lesion. New advances in sequencing technology and clinical sample acquisition are providing insights into the phylogenetic relationship of metastases and primary tumours at the level of somatic tumour genetics while also illuminating fundamental mechanisms of the metastatic process. In addition to somatically acquired genetic heterogeneity in the tumour cells, inherited population-based genetic heterogeneity can profoundly modify metastatic biology and further complicate the development of effective, broadly applicable antimetastatic therapies. Here, we examine how genetic heterogeneity impacts metastatic disease and the implications of current knowledge for future research endeavours and therapeutic interventions.
Metastasis continues to be an enormous problem for the clinical management and treatment of cancer. More than 600,000 cancer-associated deaths are estimated for 2017 in the United States alone1, and up to 90% of cancer-related mortality for solid tumours is due to sequelae of metastasis2. In addition, with improved therapeutic and management strategies, the number of patients living with metastatic disease has been rising3. Considering only breast cancer, in the US, an estimated 155,000 women are currently living with metastatic disease, and this number is anticipated to continue to increase4. Although improvements in cancer screening and adjuvant therapy have increased patient survival by preventing metastases in at least some cancers (for example, breast cancer5), little improvement in survival has been observed once patients advance to the metastatic state6. These statistics highlight the heavy public health burden of metastatic disease and emphasize the critical need to understand and more effectively intervene clinically in the late stages of cancer progression.
Metastasis is an extremely complex process in which tumour cells escape from the primary site, disseminate to a secondary location, survive and adapt to the ectopic site and finally colonize and proliferate to form clinically relevant lesions while evading immune surveillance. Each of these steps is a point of selection for different biological properties in the tumour cell, a process that has the potential to introduce considerable heterogeneity between the final successful metastatic cell and the primary tumour as well as between successful metastatic cells at different sites within a patient. The effect of these selection pressures is to diversify the original complex but localized disease at the primary site into multiple separate diseases that are spread throughout the body. The resulting genetic and epigenetic heterogeneity substantially contributes to the current inability to successfully eradicate established meta static disease. Advances in next-generation sequencing that enable cost-effective genome sequencing from moderately small amounts of material and their application to difficult-to-acquire clinical metastatic samples, coupled with genomic complex trait approaches in mouse models, are yielding new insights into the genetic underpinnings of metastasis. In this Review, we specifically address the potential roles and evidence for somatic tumour genetics and the germline genetics of an individual as sources of heterogeneity in the metastatic setting and their implications for effective therapeutic targeting. However, it should be recognized that superimposed on this genetic heterogeneity will be additional phenotypic heterogeneity owing to tumour cell interactions with other host cells and their microenvironment as well as the hierarchical subclonal organization of many tumours into subpopulations of tumorigenic cancer stem cells and their more differentiated, nontumorigenic progeny7–9.
Early versus late dissemination
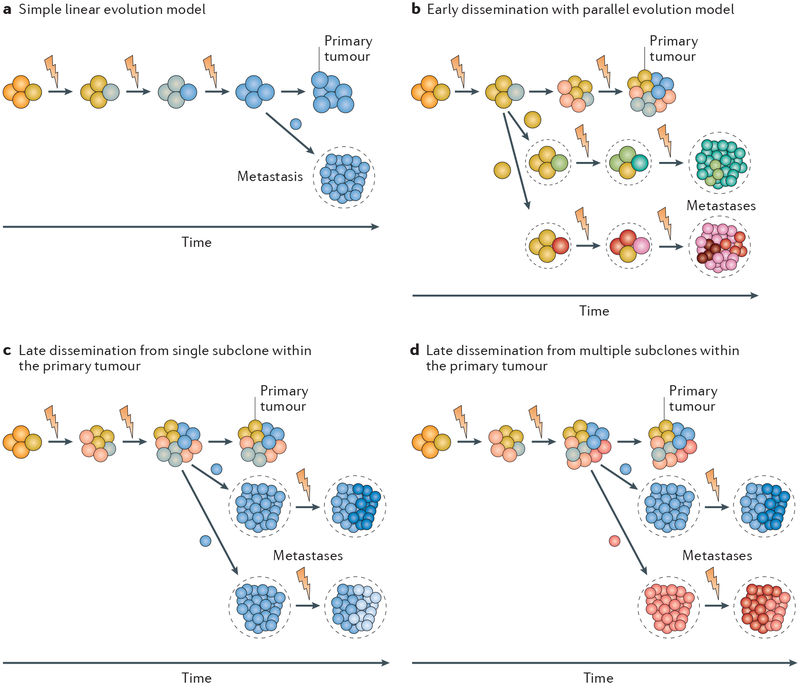
Somatic genomic heterogeneity arises in the primary tumour through the continual accumulation of genomic alterations during tumour growth10. This cumulative DNA damage results in the formation of clonal populations with different biological properties depending on the specific collection of genetic alterations that they harbour. The relative fraction of any clone within the primary tumour is dictated by the growth and survival advantage endowed by the unique genomic constitution of that clone. In the most extreme case, primary tumours would consist of single dominant clones that arise sequentially and, by successive selective sweeps, replace less-fit ancestral tumour clones. In this scenario, metastasis occurs when a late-arising clone finally acquires all the necessary properties to disseminate and successfully colonize a secondary site (FIG. 1a). Historically, this simple linear model was thought to explain the metastatic process11.
Fig. 1 |. Models of metastasis evolution and implications of genetic heterogeneity.
a | The classical simple linear model, where clones sequentially arise that dominate the primary tumour owing to survival and proliferative advantages. In this model, metastases arise late in evolution from the most advanced primary tumour clone. b | The early dissemination and parallel evolution model, where tumour cells begin to disseminate early in the primary tumour lifespan and continue to somatically evolve in parallel with the primary tumour during clinical dormancy until they acquire metastatic capacity and proliferate into a clinically relevant lesion. Owing to the independent evolution of the disseminated tumour cells, this model suggests that metastases and primary tumours share only the early tumorigenic driver events. c | The late dissemination model, where tumours evolve over time until a late-arising subclone is able to successfully seed multiple metastases. This model predicts that independent metastases would share the somatic events that occurred during the evolution of the metastatic primary subclone. Subsequently, owing to continuing evolution, individual metastases may diverge somewhat by acquisition of additional subclonal somatic events at the distant site. d | Late dissemination from multiple metastatically competent subclones within the primary tumour. Metastases seeded by this mechanism would share all the somatic events acquired by the tumour preceding the divergence of the different metastatically competent primary tumour subclones. The resulting metastases from the different subclones would be distinguished from each other by the presence of unique somatic events. Blizzard symbol indicates somatic genetic alterations.
However, accumulating evidence has demonstrated that tumour evolution and metastatic progression are much more complicated processes. Primary tumours are composed of multiple genetically distinct subclones, some of which can be mutually supportive12, and these subclones arise at different times and either persist, expand or become extinct as tumours grow and evolve12–14. Furthermore, instead of dissemination being exclusively a late event, some recent studies demonstrate that tumour cell dissemination can begin early during tumorigenesis. Tumour cell dissemination in the preneoplastic phase has been seen in preclinical models of breast15 and pancreatic cancer16, and in a clinical setting, circulating pancreatic epithelial cells were found in patients with pancreatic cystic lesions but without overt cancer16.
These observations have led to two updated paradigms for metastatic spread: the early and late dissemination models, each with different implications for the extent of heterogeneity between metastases and the primary tumours as well as between metastases within a patient. In the early dissemination model, multiple subclones in the primary tumour may distribute tumour cells throughout the body during the period before primary tumour resection or successful therapy in situ. After a variable period of dormancy at the distant site, some of these disseminated cells may begin to evolve independently and in parallel with the primary tumour, eventually acquiring the ability to form clinically relevant metastatic lesions (FIG. 1b). Such metastases would share few mutations with the bulk of the primary tumour tissue, potentially only the original tumour-initiating driver mutations. In addition, as the individual disseminated tumour cells acquire full metastatic capacity independently of each other in the early dissemination model, there will likely be considerable genetic divergence among individual metastatic lesions within a patient. In the late dissemination model, the degree of relatedness depends on the time at which the metastatically competent subclones diverged within the primary tumour. Metastases arising from metastatically competent subclones that diverged late in the development of the primary tumour would retain substantial genomic similarity to each other and to the primary tumour (FIG. 1c). By contrast, metastases arising from subclones that diverged early but disseminated late would show greater genomic differences between metastases but individually would retain similarities to the primary tumour (FIG. 1d).
Recently, increasingly sophisticated sequencing and computational analysis tools are enabling construction of phylogenetic relationships between primary and secondary tumours (from locoregional recurrences, lymph node metastases and distant metastases) that illuminate the tumour evolutionary process and provide varying degrees of support for each of these models17. On balance, the small numbers of studies performed to date tend to favour the late dissemination model. For example, one phylogenetic analysis of matched primary colorectal tumours and metastases, which was based on exome sequencing and copy number profiling, revealed that the metastatic samples were most highly related to each other while retaining substantial similarities to the primary tumour18, as predicted by the late dissemination model. Analysis of somatic mutations in pancreatic cancers and their patient-matched metastases has revealed the presence of presumed metastasis-seeding subclones within primary tumours on the basis of their high degree of genetic relatedness to the metastases19, which also supports a late dissemination model. Similarly, a recent study using 17 sets of matched primary and metastatic breast cancer lesions showed that there were very few mutations private to the primary tumours when compared with matched distant metastases, indicating that the tumour cells disseminated late during primary tumour evolution, with dissemination occurring on average at a timepoint representing 87% of the molecular age of the primary tumour at diagnosis20. By contrast, a study of colorectal cancer found evidence of early and late dissemination in different patients21, raising the possibility that the timing of dissemination may vary between patients within a given disease subtype. A similar conclusion was found in a study of carcinogen-induced skin cancer in a mouse model22. Early dissemination was seen in HER2-driven breast cancer mouse models23,24, where, intriguingly, the early disseminating cells were found to be more meta-statically competent than cells that disseminated later23. However, the disseminated cancer cells in patients with breast cancer that had overt metastases (M1 stage) were genomically similar to the primary tumours23, consistent with a late dissemination model. Overall, it is clear that tumour cells are capable of disseminating early, but it remains to be established whether the formation of clinically relevant metastases from such cells is a frequent occurrence.
The more complex pattern of late dissemination from multiple independent subclones within the primary tumour (FIG. 1d) has also been observed in a few cases. In a study of melanoma, a combination of exome and targeted sequencing of eight patients revealed that although metastasis was a late-evolving trait, on the basis of the segregation of independent mutations of CTNNB1 (encoding β-catenin) in the primary tumour, a loco regional metastasis was seeded by a single primary tumour subclone, while a distant metastatic lesion appeared to be seeded by multiple subclones from the primary tumour25. Thus, at least in melanoma, some fraction of the heterogeneity observed between lesions may be the result of independent clonal evolution within the primary tumour rather than subsequent somatic evolution of metastasis ‘seeds’ after dissemination from a common primary tumour parent clone. Similarly, exome-sequencing data of tumours from patients with pancreatic and lung cancer provided support for early genetic divergence of multiple meta-static lineages within the primary tumour in a subset of patients26. Interestingly, in a recent study of colorectal cancer that used polyguanine tract insertion and deletion analysis to examine primary tumour and metastasis phylogenies, lymph node metastases and distant (mostly liver) metastases were shown to originate from distinct clones within the primary tumour in nearly two-thirds of the patients examined27. A similar result was observed in a skin tumour mouse model22, suggesting that lymph nodes do not simply represent ‘way stations’ for disseminating cells en route to more distant sites.
The genetic evidence for late dissemination is also consistent with clinical data in some tumour types. For example, tumour size has been found to be an independent predictor of cancer-specific survival in breast cancer and non-small-cell lung cancer (NSCLC)28. One would not necessarily expect a correlation between tumour size and survival if the early dissemination and parallel evolution model was the dominant metastatic pathway. Interestingly, a similar phenomenon was also observed in a transgenic mouse model of HER2-driven breast cancer, in which disseminated tumour cells from older BALB-neuT mice with larger tumours were found to be more tumorigenic than tumour cells from younger animals with smaller tumours29. Taken together, these observations suggest that despite the potential for early and continuous shedding of tumour cells from the primary site, the environment of the primary tumour continues to function as a metastatic ‘incubator’ that somehow generates or selects for subclones of cells with increased ability to complete the metastatic cascade30.
Overall, the studies suggest that no single model of metastatic spread applies universally. Indeed, these two models for metastatic dissemination likely represent extremes on a biological continuum. The validity and relative frequency of the different metastatic mechanisms will need to be established by additional large-scale studies of patient-matched primary tumours and metastases across multiple tumour types.
Other sources of heterogeneity
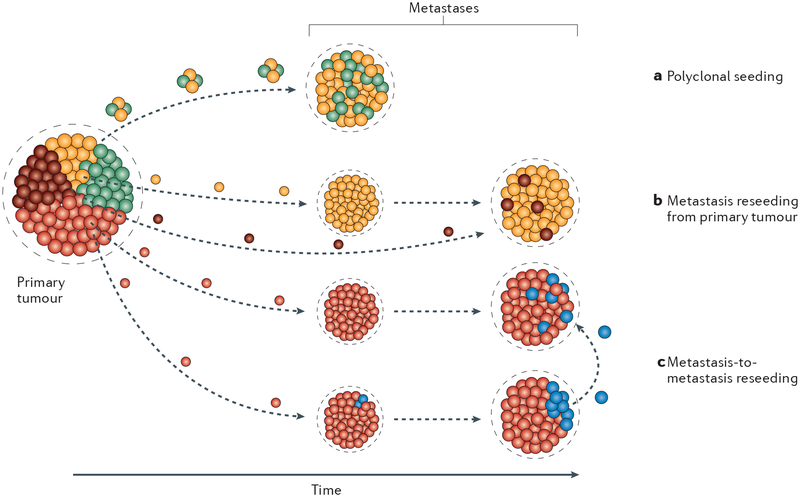
The extent of metastatic genetic heterogeneity can also be modified through the phenomena of polyclonal seeding, metastasis reseeding and metastasis-to-metastasis reseeding (FIG. 2). The possible occurrence of these processes needs to be taken into account in the interpretation of phylogenetic analyses of metastasis.
Fig. 2 |. Metastatic heterogeneity owing to alternative seeding mechanisms.
a | Polyclonal seeding, where multiple tumour cells from different subclones of the primary tumour disseminate to the secondary site and proliferate in parallel during metastasis evolution. b | Primary tumour reseeding, where metastases are initially generated by a single subclone within the primary tumour. Subsequently, disseminated tumour cells from a different subclone of the primary tumour colonize and proliferate within the already established metastatic lesion. Somatic heterogeneity introduced into the metastasis by this reseeding mechanism would also be present within the primary tumour. c | Metastasis-to-metastasis reseeding, where independent metastases are founded and continue to evolve. A subsequently arising subclone within a metastasis then seeds tumour cells to other metastases. The somatic events that define the metastasis reseeding subclone would not be present in the primary tumour in this mechanism.
Polyclonal seeding.
The classic work by Fidler, Wolman and Talmadge31 in which they intravenously co-injected karyotypically distinct metastatic cells into mice showed that all cells within a given metastasis had the same chromo somal aberrations, indicating that individual metastases are seeded by single cells. Subsequent cytogenetic studies31 and analysis of experimental32 and clinical19,33 samples were mostly consistent with this interpretation. However, recent experimental work has challenged this concept of a monoclonal origin for all metastases by demonstrating that tumour cell clusters are much more efficient than single cells at forming meta-static lesions25,34–38 (FIG. 2a). Metastasis seeding by mutually supportive cells from different subclones of the primary tumour could add to the somatic genetic heterogeneity within metastases and increase metastatic fitness. The interaction of genetically distinct clones has been shown to enhance tumour progression at the primary tumour site through effects on tumour cell proliferation and the microenvironment12 and could presumably have similar effects at the metastatic site. In several patients with metastatic prostate cancer, multiple metastases showed the same subclonal clusters of mutations, suggesting that polyclonal seeding had occurred repeatedly38. Similarly, sequencing studies of a patient with melanoma also revealed the same deletion of CTNNB1 in subclonal populations of two independent metastases25, suggesting that these lesions were seeded by more than one tumour cell. How common this phenomenon is will have to be determined by further more extensive studies.
Tumour reseeding.
As noted earlier, dissemination of tumour cells can begin early in tumour evolution39–41, and large numbers of tumour cells are thought to be continuously shed into the circulation from advanced tumours42, and potentially from metastatic lesions, on a daily basis. This continual circulation of tumour cells raises another potential mechanism for both intermetastasis and intrametastasis heterogeneity. As metastases are already permissive environments for tumour cell survival and growth, they might represent preferential sites for circulating tumour cells to recolonize. It has been suggested that exchange of tumour cells between anatomically distant tumour masses contributes to both metastasis heterogeneity and progression by distributing cells with different biological properties, such as greater proliferative capa city or drug resistance43, into spatially distinct metastases (FIG. 2b,c). Experimental evidence in support of the possibility of cell transfer between distant tumour masses was shown by engrafting either fluorescently labelled or unlabel led tumour cells into the contralateral flanks of mice. The tumours resulting from the unlabelled inoculum contained fluorescent cells, indicating colonization by circulating tumour cells from the contralateral fluorescently labelled tumour43. Furthermore, a second study using fluorescently labelled osteosarcoma cells to establish lung metastases demonstrated that metastasis-derived cells can repopulate experimentally implanted tumours in bone44, indicating that at least in experimental settings, reseeding may occur bidirectionally between primary tumours and metastases. Widespread and efficient reseeding would result in increased intralesion heterogeneity by exchange of novel subclones arising in individual lesions. However, it would have the potential for decreasing interlesion heterogeneity by reducing the number of subclones private to that lesion of origin. Nevertheless, clinical evidence for reseeding is currently limited and is difficult to distinguish from polyclonal seeding. Whole-genome sequencing of multiple metastases from patients in one study of prostate cancer revealed the presence of several tumour clones that were shared among different metastatic sites, consistent with either polyclonal seeding or reseeding between metastatic lesions38. By contrast, investigations of acquired BRAF inhibitor resistance in patients with melanoma have indicated that the majority of individual lesions within a patient have unique resistance mechanisms45, which would argue against reseeding as a major mechanism of spreading resistant subclones, at least for this tumour type.
Metastasis-specific driver events
Genetic heterogeneity between primary tumours and metastases provides a potential window into the biology of the metastatic process by highlighting genetic events that are enriched in metastases and may play a causal role in their successful establishment. Such tumour-to-metastasis heterogeneity is also an important consideration for the design of improved antimetastatic therapies because to date, clinical trials testing novel cancer therapeutics or strategies are based on information obtained mostly from primary tumour tissue. The ability to perform deep sequencing on hundreds of primary tumour samples has identified genes that are recurrently mutated in different tissue types that likely represent ‘tumour drivers’, the mutations that cause neoplastic transformation46.
One major unanswered question at this point is whether there are analogous metastasis-specific genetic driver events that contribute to the later stages of tumour progression and could be targeted in the development of metastasis-specific therapies. In the past, metastasis-specific loss of heterozygosity or loss of gene expression47 had been used to identify a class of genes known as ‘metastasis suppressors’ in experimental studies that were largely based on the differential metastatic ability of tumour cell lines following chromosome transfer or functional genomic screens (reviewed in REF. 48). For example, the breast cancer metastasis-suppressor 1 (BRMS1) gene was identified as a gene on chromo-some 11 that suppressed metastasis of highly meta-static MDA-MB-435 cells following microcell-mediated chromosome transfer49. The distinguishing characteristic of these genes is that re-expression results in suppression of metastatic potential without substantial effects on primary tumour growth. More than 20 such metastasis-suppressor genes have been identified50 and shown to function in a wide variety of cellular processes and pathways, including cell adhesion, chromatin remodelling, transcriptional regulation and MAPK signalling, indicating that there may be a number of opportunities to disrupt metastatic biology to improve patient outcomes.
While these experimental studies suggest that metastasis-specific genetic driver events exist, the data thus far in clinical samples are equivocal. Like primary tumour drivers, metastasis driver mutations would be expected to be present in all the tumour cells within a given meta static lesion and to be recurrently mutated in more than one patient. In addition, as the existing data support the late dissemination model as the most common mechanism of metastatic spread, metastasis driver mutations would be expected to be present in a subclonal fraction of the primary tumour but to be clearly enriched in the metastasis. Ideally, metastasis-specific driver events would be identified by large-scale comparative sequencing of patient-matched primary tumour and metastatic lesions, but that goal has yet to be achieved.
Recently, a large exome-sequencing study of 500 biopsied metastatic lesions across multiple tumour types, which used The Cancer Genome Atlas (TCGA) primary tumour database as a historical control, indicated that for most cancer types, the metastatic lesions had a higher number of potential driver mutations than did the corresponding but unmatched primary tumours51. Thus, either metastatic competency is a feature of subclones with a higher driver mutational load in the primary tumour or metastatic lesions continue to evolve genetically at the distant site as they adapt to their new environment. A similar study in breast cancer applying targeted sequencing of 365 known cancer genes to 227 samples of distant metastases or loco regional relapses, which also used the TCGA data set as a comparator, came to a similar conclusion20. By further comparing the driver mutation profile of primary tumours with matched metastases or loco regional recurrences in a smaller subset of 51 patients with breast cancer, it was shown that 50% of patients had additional driver mutations in the recurrent lesions, many of which were rarely mutated in primary breast cancer20. Intriguingly, inactivation of the SWI/SNF chromatin remodelling complex, particularly through mutations in AT-rich interactive domain (ARID) family members, was a frequent event20. Mutations in this complex have also been seen in meta-static endometrial cancer33. This observation raises the possibility that genetic events driving widespread modulation of the epigenetic landscape may play a particularly important role in metastatic evolution. Neither of these two large studies identified mutations in the classical metastasis-suppressor genes previously identified by experimental approaches, suggesting that if these are important clinically, they will be epigenetically rather than genetically modified in metastases.
In addition to these relatively large studies, a number of smaller-scale sequencing studies of matched primary and metastatic tumours have also been performed. For skin, pancreatic and breast cancers, such studies have so far not identified any genes that recurrently acquire point mutations in metastatic lesions19,22,52,53. However, in colorectal cancer, evidence is emerging that suggests that KRAS mutations are associated with metastasis to the lung54 and liver55. Furthermore, TP53 mutations appeared to be enriched in prostate cancer metastases compared with the matched primary tumours56. Thus, the existing data suggest that in some tumour types, metastatic progression is driven by oncogenic alterations in genes that serve as primary tumour drivers in other tumour types. Additional studies with larger numbers of samples will be required to fully understand whether metastasis-specific driver mutations contribute in a major way to the differences seen between primary tumour and metastasis biology or whether epigenetic differences are quantitatively more important57. Conceivably, as the metastatic seed cell must successfully overcome many different biological hurdles to establish a clinically overt metastasis, phenotypic plasticity conferred by a flexible epigenome may be more important than hardwired genetic events in driving metastatic progression.
Population-level germline heterogeneity
Generation and/or selection of different constellations of somatic genetic events in metastatic lesions clearly contribute appreciably to metastatic heterogeneity between and within patients. However, there is another understudied level of genetic heterogeneity that will profoundly impact precision medicine strategies, namely, inherited genetic diversity at the population level. The millions of polymorphisms that segregate among the human population are responsible for the variations not only in visible traits such as height but also in disease susceptibility, and they make every human unique58. Furthermore, it is now appreciated that the same mutation on different genetic backgrounds can have markedly different effects on individual pathophysiology, examples of which can be found in both humans and model systems59,60.
This phenomenon is most clearly demonstrable in mouse models. For example, knockout of the epidermal growth factor receptor (Egfr) on a CF-1 mouse genetic background results in peri-implantation neonatal death owing to degeneration of the inner cell mass of the blasto cyst. By contrast, on a 129/Sv background, animals died at mid-gestation owing to defects in the placenta, while on a CD-1 background, animals survived up to three weeks postnatally before succumbing to multiple system abnormalities59. As all the animals inherited the identical mutation by breeding, these results indicate that polymorphisms within the different genetic backgrounds were capable of modulating the deleterious effects of the Egfr knockout to different degrees. In humans, a good example of the effect of inherited polymorphism on cancer phenotypes is seen with the BRCA1 gene. Patients who carry a single mutated copy of BRCA1 are at substantial risk of developing breast cancer in their lifetime. However, not every patient carrying the same mutation will develop disease61,62, and cancer risk has been found to vary between populations63, suggesting that there are contributions of inherited genetic variants that modify the penetrance of the disease, some of which are being uncovered by genome-wide association studies (GWAS)64.
Insights from mouse models.
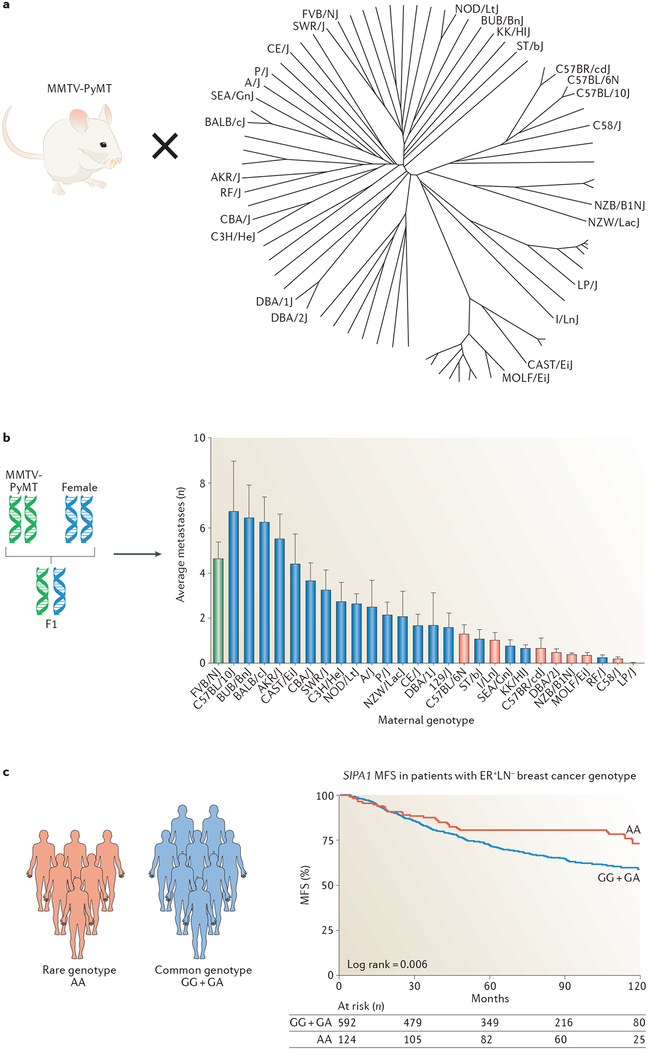
With respect to metastasis, profound effects of genetic background in modulating phenotypic expression of metastatic disease have been clearly demonstrated in animal model systems65–71. Owing to the inherent variability of the metastatic phenotype, animal models provide a powerful method to identify modifier genes because of the higher signal:noise ratio afforded by the increased control of potential confounding variables such as environment. In the most intensely studied case, the mouse mammary tumour virus promoter-driven polyoma middle T oncogene (MMTV-PyMT) transgenic mouse model of breast cancer72 was used, in which expression of the powerful PyMT is sufficient to induce rapidly developing metastatic disease without the need for further extensive genetic evolution in the tumour cells. When this model was bred to different inbred strains of mice to vary the genetic background72 on which the disease developed (FIG. 3a), examination of the progeny of these crosses revealed a significant variation in the efficiency of forming pulmonary metastases (FIG. 3b), with metastatic burden varying over a 40-fold range65. As the primary oncogenic driver event — expression of the PyMT transgene — was identical in all the animals, the phenotypic variation in metastasis was primarily due to inherited polymorphisms modifying the various steps of the metastatic process.
Fig. 3 |. The effect of polymorphism on metastatic progression.
a | Mouse mammary tumour virus promoter-driven polyoma middle T oncogene (MMTV-PyMT) male mice were bred to female mice from many different branches of the mouse phylogenetic tree. b | The subsequent F1 progeny have different genetic backgrounds owing to the introduction of a different haploid genome from the maternal strain. Subsequent analysis of the average number of metastases (y axis in the bar chart) in the transgene-positive F1 female progeny revealed that the metastatic capacity of mammary tumours across the different genetic backgrounds varied substantially, with eight strains showing statistically significant differences (red bars) when compared with the original FVB/NJ homozygous genetic background (green bar). c | A minor allele in the promoter of signal-induced proliferation-associated 1 (SIPA1) (homozygous for the allele denoted by the genotype AA (in red)) predicts distant metastasis-free survival (MFS) in the oestrogen receptor-positive (ER+) lymph node-negative (LN−) subtype of human breast cancer. Parts a and b are adapted from REF. 70, CC0 1.0. Part c is adapted from REF. 97, Macmillan Publishers Limited.
Further evidence for the existence of metastasis susceptibility loci and the identification of candidate modifier genes was obtained by quantitative trait locus mapping in mouse genetic experiments. Genetic backcross mapping panels73 and a specialized mapping strategy known as a recombinant inbred backcross74 demonstrated a reproducible association of the proximal end of mouse chromosome 19 with the metastatic efficiency of the MMTV-PyMT mouse breast cancer model75. Comparisons of the haplotypes across this region of mouse chromosome 19 to find haplotypes that correlated with metastatic capacity across the inbred strains of interest reduced the number of potential candidate genes76. Expression and direct sequencing analysis suggested that signal-induced proliferation-associated 1 (Sipa1), which encodes a RAP1 GTPase-activating protein (GAP) was a promising metastasis susceptibility gene candidate, which was subsequently validated in knockdown and overexpression studies in metastatic mouse mammary tumour cell lines67. Similar studies revealed that polymorphisms that result in amino acid substitutions in candidate metastasis susceptibility genes (for example, Sipa1)67 or generate differences in transcriptional programmes in either tumour cells (for example, ribosomal RNA processing 1 homologue B (Rrp1b))68 and/or stroma (for example, zinc-finger and BTB domain containing 16 (Zbtb16))70 can alter the metastatic capacity of both cell-line-based67–69,71,77–81 and genetically engineered mouse models (GEMMs) of metastatic mammary disease70,71,81. Similar studies in a mouse model of prostate cancer have also identified candidate metastasis susceptibility genes, indicating that inherited susceptibility to metastasis may be a general phenomenon82–85, although the specific genes identified to date are not shared between the two tissue types.
A strength of the animal modelling approaches is that the cellular targets of these metastatic susceptibility variants can be identified through transplantation studies in which the candidate susceptibility gene is modified independently in the host or tumour cells. The results of such studies show that either tissue compartment may be the prime target, depending on the variant in question70. For example, variants in the circadian rhythm gene aryl hydrocarbon receptor nuclear trans-locator-like 2 (Arntl2) were found to affect metastasis in a tumour-autonomous fashion in mouse models of human breast cancer71. By contrast, variation in Zbtb16 levels in tumour cells had no effect on metastatic capacity, but haploinsufficiency of the gene in host tissue increased the metastatic capacity of injected mammary tumour cells70. A recent study using a large panel of mutant knockout mice has also demonstrated an important role of stromal genes in metastatic susceptibility86. Intravenous injection of the B16 mouse melanoma cell line into more than 810 knockout models identified 19 novel genes that played a role in stromal control of metastasis. Deletion of spinster homologue 2 (Spns2), which encodes a sphingosine-1-phosphate (S1P) transporter, had a particularly profound effect, resulting in more effective immune cell trafficking to the lung and reduced metastatic burden86.
As more metastasis susceptibility alleles are identified through such approaches, they are starting to converge around the involvement of a core set of biological processes that are implicated in affecting metastatic efficiency. These include broad processes such as transcriptional regulation71,77,79,87, chromatin biology80 and RNA metabolism81,88, which are presumably involved in metastasis-specific rewiring of the transcriptome, as well as metastasis-specific processes such as adhesion and migration67,69 and antitumour defences such as immune surveillance69,86. Moreover, inherited genetic susceptibility is not limited to the nuclear genome. Recent studies have demonstrated that polymorphisms in the mitochondrial genome also affect metastatic efficiency and implicate an important role for metabolism in tumour progression89.
The identification of metastasis susceptibility genes not only sheds new light on the biology of metastatic progression but may also have important clinical implications for prognosis and treatment. Various gene-expression-based prognostic tests are currently used in the clinic (for example, MammaPrint and Oncotype DX) that are based on the pioneering work demonstrating that transcriptional profiles from bulk primary tumour tissue could discriminate patient outcome90,91. With the exception of a strong proliferation component, the biology underlying these prognostic signatures has not been clearly defined. Intriguingly, analysis of the transcriptomic effects of experimental manipulation in the mouse has generated gene signatures that are capable of discriminating outcome in human patient data sets77,92. Even more intriguingly, gene expression profiles from both tumours and normal tissues from mouse strains with high and low metastatic propensity can also discriminate patient outcome88,93, suggesting that the existing prognostic gene signatures include biomarkers of inherited susceptibility in addition to somatic drivers that may contribute to the prognostic signal.
Beyond contributing to the development of more powerful prognostic gene signatures, the identification of metastasis susceptibility genes may also provide novel strategies to prevent or treat metastasis through modulation of the susceptibility phenotype. The gene signatures derived from the mouse strains with high and low metastatic propensity implicate some pathways that are already targeted by US Food and Drug Administration (FDA)-approved drugs. For example, the diabetes mellitus pathway shows differential activation between mice with high and low metastatic capa city, and experimental targeting of this pathway with the peroxisome proliferator-activated receptor-γ (PPARγ) agonist rosiglitazone reduced meta static capacity in a metastatic breast cancer model70. Similarly, other commonly used agents in the treatment of diseases other than cancer have been shown to suppress metastatic disease, including both caffeine94 and cannabinoids70,95. Further investigation of such agents and their incorporation into existing therapeutic strategies might therefore provide substantial benefit to certain subsets of patients.
Evidence for metastasis susceptibility alleles in the human population.
Crucially, polymorphisms in the human orthologues of some of these mouse metastasis susceptibility genes have been associated with meta-static progression or disease outcome in patients68,71,96–102. A polymorphism in the proximal promoter region of SIPA1 that reduces SIPA1 transcriptional activity in humans101 was found to be associated with improved breast cancer survival97, which is consistent with the experimental mouse studies. Similarly, a polymorphism that encodes an amino acid substitution in RRP1B was found to be associated with survival, with patients with breast cancer carrying this rarer allelic variant being less likely to develop distant metastatic disease92,100. The results of these studies suggest that polymorphisms in the human population contribute to the metastatic susceptibility and heterogeneity observed across the human population and validate the experimental approach in mice.
Epidemiology studies to interrogate metastatic susceptibility in patients have found that cancer survival, like cancer incidence, is familial in multiple tumour types, including breast, pancreatic, prostate, bladder and renal cancers103–106, further supporting a role for inherited polymorphism in metastatic disease. However, despite strong evidence in the mouse systems, GWAS in humans so far have provided only suggestive but not conclusive evidence that inherited factors are associated with progression to metastatic disease or survival96,98,99,101,102,107,108. There are several reasons why definitive proof of metastasis susceptibility genes has not yet been found in humans, all converging around signal-to-noise issues. One possible explanation is that metastasis susceptibility varies by tumour subtype. For example, association studies of metastasis-free survival for the human orthologues of the mouse susceptibility genes SIPA167 (for example, see FIG. 3c) and RRP1B67,68 demonstrated that polymorphisms could discriminate outcome only in patients with oestrogen receptor-positive, lymph node-negative breast cancer97. Therefore, investigations in unstratified cohorts would dilute the power to detect subtype specific associations. Similarly, it is likely that some fraction of the metastasis susceptibility component may vary depending on the oncogenic driver. A third possibility is the potential influence of the environment. Genetic studies of meta-static susceptibility are done in therapy-naive animals housed in carefully controlled environments that permit mostly robust and reproducible metastatic phenotypes. Human patients are exposed to a vast array of environmental variables that likely alter the natural course of the disease and add noise to association studies. For example, diet has been shown to influence pulmonary metastasis in the MMTV-PyMT model109,110.
Another potential confound for patients is exposure to therapy. As adjuvant therapy is given to patients specifically to try to eliminate occult metastatic disease and because 20–40% of treated patients benefit111, inclusion of those patients whose metastatic disease was prevented by adjuvant therapy would further reduce the power of association studies to detect metastasis susceptibility genes. All these confounds can be at least partially compensated for by increasing the size and annotation of the human cohorts used in GWAS but with increasing costs and effort. For example, a recent study required genotyping of more than 250,000 individuals to identify 4% of the population-wide twofold familial risk of breast cancer112. Therefore, animal models, for the time being at least, continue to provide a cost-effective, efficient platform to identify putative metastasis susceptibility candidates that can be validated in focused epidemiology studies in human cohorts.
Nongenetic, nonenvironmental heterogeneity
While genes (somatic and germline), tumour micro-environment and more systemic environmental effects undoubtedly contribute to interindividual heterogeneity in metastasis, there is likely a third factor that contributes a substantial fraction of the human population variation in metastatic propensity. In preclinical settings, it is consistently observed that implantation of genetically identical tumour cells into genetically identical inbred mouse hosts, housed under identical conditions, gives a wide range of metastatic burden between individual animals113. Extensive work by Gartner114 and others115,116 has shown that for other quantitative traits such as body weight, resistance to infection, stress response and various behavioural phenomena, inbred animals display an irreducible phenotypic variation that is unaffected by experimental standardization. This third component of interindividual variation is referred to as ‘intangible variation’ or ‘phenotypic noise’ and is the largest nongenetic contributor to phenotypic variance, dominating over environmental effects117,118. It appears to be established stochastically early in development, within the first three cell divisions of the zygote, and is likely epigenetically driven117,118. Gartner117 has speculated that one of the evolutionary objectives of this component of variation is to resist the inbreeding effects caused by selection of the genetically fittest. The net result is a somatic epigenome that varies between individuals and impacts phenotypic expression of many traits. This phenomenon likely also contributes to interindividual heterogeneity in phenotypic expression of the metastatic trait in human populations. Systematic analysis of the transcriptomic and biological variation between individual inbred animals in experimental metastasis cohorts and their correlation with metastatic burden is an underexplored avenue that could lead to novel interventions to reduce the probability and extent of metastatic development in patient populations.
Development of metastatic therapeutics
Emergent principles of genetic heterogeneity of metastases.
It is still early in the application of new genetic analysis tools to metastatic disease, and there are still few studies that have compared primary tumours with patient-matched metastatic lesions in clinical samples. While the picture may change with additional studies and may vary in detail between tumour types, a few consistent themes seem to be emerging from the studies described above.
First, multiple metastases in a given patient are usually genetically more similar to each other than to the matched primary tumour33,38,53. This feature probably reflects purifying selection from genetically heterogeneous primary tumours for biological properties specifically necessary for successful metastasis and is also consistent with the prevalence of a late dissemination mechanism. The issue of whether there are recurrent metastasis-specific genetic driver mutations that are selected for in this process needs to be resolved by larger-scale studies.
Second, metastases within a given organ are genetically more similar to each other than they are to metastases in different organs in the same patient33,57,119. This suggests that different target organ microenvironments select for or require different adaptive mechanisms for the metastatic tumour cell to successfully colonize. Furthermore, metastases in close proximity to each other in a given organ are genetically more similar than metastases from more distant sites in the same organ38.
Third, inherited genetic variation between individual patients is likely to profoundly affect the metastatic phenotype through effects on both the tumour cell and the microenvironment.
Implications for therapy from what we have learned so far.
The inherited genomic variation and acquired somatic mutation described above, when combined with variation induced by cellular plasticity and the influence of the distant organ microenvironment (not discussed in this Review), generate an enormous degree of heterogeneity in metastatic disease and contribute to making the biological properties of metastases distinct from those of the originating primary tumour. Mortality in the cancer setting is generally due to failure to effectively treat metastatic disease, and the new molecular insights that are emerging need to be translated into improved therapeutics. Metastases generated via the early dissemination with parallel evolution mechanism may be so different from each other and from the primary tumour as to need treatment as independent tumours. Conversely, metastases arising from a late dissemination mechanism are more likely to share common genetic vulnerabilities that might be treatable by a common therapeutic strategy. However, even though the genetics of the primary tumour may be a reasonable surrogate for the genetics of synchronously diagnosed metastases arising from late dissemination, metachronous metastases are more genetically divergent, presumably owing to continuing genomic evolution15. Furthermore, ongoing epigenomic evolution in the metastases20,120, in combination with microenvironmental effects introduced by the different secondary sites121, likely also contributes to our inability to completely eradicate metastatic disease.
Importantly, the genetic studies suggest that therapeutic strategies need to be organ-specific. For example, while characterization of intracranial metastasis has shown a high degree of genetic homogeneity between brain metastases from a given patient, substantial differences were observed when these were compared with the genomes of extracranial lesions from the same patient53. Similar results have been observed in prostate cancer38, pancreatic cancer119 and colon cancer27. As the authors of the brain metastasis study noted, such observations suggest that biopsy of a single intracranial lesion will be much more informative for guiding therapy selection for successful treatment of the brain metastases than any information generated from the primary tumour or extracranial lesions. The data also strongly suggest that different microenvironmental influences in the secondary sites select for survival and outgrowth of genetically different metastatic variants. Again, it should be noted that this selection process will not only be limited to somatic genetic variation but may also occur at the epigenetic level. Experimental metastasis systems have demonstrated a reversible suppression of PTEN in breast cancer brain metastatic lesions, which is facilitated by exosome-mediated microRNA transfer from surrounding tissues122. Similarly, comparison of hormone receptor expression in breast cancer has demonstrated both loss and gain of expression not only between the primary tumour and metastases123 but also between metastases124. Thus, information on the somatic genetics of the metastasis will ideally need to be complemented by additional ‘omics’ analyses to generate a more complete picture of targetable vulnerabilities. Clinical strategies for treating metastatic disease may have to include combinations of therapies to target metastases in multiple organs or potentially be performed in series to clear metastatic lesions in different target organs sequentially.
What still needs to be done.
Despite recent advances in our knowledge of the genetics of metastasis, progress in developing antimetastatic therapies is currently hampered on two main fronts. The first is that we still have an inadequate understanding of the natural history of the progression to metastasis. The second is that the preclinical drug development process has historically been geared towards effectiveness against the primary tumour rather than against metastatic disease, and it has also failed to capture even a fraction of the hetero geneity of the disease phenotype. These deficiencies need to be addressed.
In terms of enhancing our understanding of the processes and pathways that contribute to the generation of metastatic heterogeneity, a number of tools will be helpful. Historically, it has been difficult to obtain meta static material in clinical settings, and nearly all the phylogenetic studies done to date have involved small sample numbers. However, warm autopsy programmes and increased biopsy sampling of metastatic lesions51 coupled with sophisticated advances in genomic125 and single-cell analysis tools126 are already illuminating how primary tumour subclones seed and evolve during the establishment of multiple metastases within a given patient. More recently, direct-to-patient outreach programmes using social media are enabling the collection and analysis of samples from a large number of patients across the US outside of the conventional clinical trial structure, which will greatly increase the numerical power of these studies (www.mbcproject.org)127. This pioneering approach will have a major impact on our understanding of the genomics of metastatic disease in the years to come.
Complementing approaches that use clinical mat erial, mouse models of metastatic disease provide a unique window onto all stages of the metastatic process41,128–131. Importantly, GEMMs of metastatic disease enable access to the very early stages of the process that cannot be readily queried in humans and also permit study of the natural history of disease progression in the untreated state. However, intelligent use of mouse models requires the recognition of features that differ from human disease, with no models accurately reflecting all aspects of human disease under study. Instead, each model tends to highlight and amplify an important component of the relevant biology under consideration. For example, GEMMs with strong drivers such as the MMTV-PyMT model do not accumulate as many somatic variants as observed in chemically induced tumour model systems or naturally arising adult human tumours, which can be considered a limitation132. However, this simplified somatic tumour genome provides a greater signal:noise ratio than is possible in the more genomically complex human tumours, enabling detection of important causal events in smaller numbers of samples. Data generated from animal models therefore provide an important vehicle for hypothesis generation and testing that can subsequently be examined and verified across the broader human population.
Continued investigations into these model systems will help determine whether metastasis driver mutations can be identified, functionally validated and targeted using precision medicine strategies. Identifying metastasis modifier genes and their associated biology can also lead to novel therapeutic strategies based on mitigating the modifier effects. These genetic approaches can be further complemented by approaches targeting the metastatic epigenome or the metastasis-supportive microenvironments, which can also be well studied in mouse models. Organoid models of metastatic progression, derived from GEMMs or patient samples, are showing promise for rapid functional screening of genes and pathways that can influence metastatic progression and for analysis of aspects of epigenomics and chromatin landscape evolution that is currently difficult to do using clinical material120,133,134.
A better understanding of the biology of the meta-static process and the steps that influence metastatic heterogeneity will help inform the drug development process. However, many improvements also need to be incorporated into the preclinical drug development pipeline. First and foremost, antimetastatic drugs should be tested for efficacy against metastatic disease rather than against primary tumours. Mouse models should be used in formats that mimic the clinical situation as closely as possible. Thus, GEMMs or orthotopic transplant models, which incorporate the systemic conditioning effects of the primary tumour in shaping immune responses and metastatic niche development, are preferable to intravenous tumour cell administration, and surgical resection of the primary tumour is desirable. Where possible, models should include an intact immune system, and attempts should be made to incorporate genetic heterogeneity in study designs by using multiple different model systems on diverse genetic backgrounds. While these approaches are costly and labour-intensive, they have a greater probability of generating effective antimetastatic therapeutics.
Conclusion
The complexity of metastatic disease and the difficulty in treating it are due in no small part to the hetero geneity of metastatic lesions. In this Review, we have described the various origins and influences of genetic sources of heterogeneity both between primary tumours and metastases and between and within the metastases themselves. While many of the data indicate quite close genetic similarity between metastases in a given patient, thus supporting the hypothesis of metastasis seeding by late dissemination of a single metastatic primary tumour subclone, a few more recent studies provide some evidence of the possible contribution of more complex polyclonal seeding to metastatic disease. In addition to these phenomena, an expanding understanding of the contribution of organ micro-environments to successful metastatic colonization is revealing the extensive impact of the secondary site in shaping the genetics of new tumours as they spread throughout the body and continue to evolve. However, studies of inherited susceptibility to metastasis are indicating that all this information needs to be applied in the context of the personal genetic landscape of the patient, which can profoundly impact how the somatic genetic changes in the tumour cells and influences of the local microenvironment affect the biology and therapeutic responsiveness of disseminated tumour cells. New mechanisms uncovered by such studies could be exploited in the clinic to disrupt the ‘soil’, an approach that may complement the current focus on targeting the metastatic ‘seed’. To achieve these goals, additional deep characterization of the genomes of matched primary and metastatic tumours of larger numbers of patient and experimental tissue samples will be neces sary to resolve many of the unanswered questions regarding the aetiology of metastatic disease and the impact of heterogeneity on the biology of the lethal terminal stages of cancer progression.
Acknowledgements
The authors wish to apologize to the many colleagues whose work may have been inadvertently omitted or not included owing to space constraints. This research was supported by the Intramural Research Program of the US National Institutes of Health (NIH), National Cancer Institute (K.W.H., L.W.).
Reviewer information
Nature Reviews Cancer thanks Y. Kang and the other anonymous reviewer, for their contribution to the peer review of this work.
Glossary
- Microcell-mediated chromosome transfer
A method of chromosomal transfer by fusion of membrane-encapsulated donor chromosomes with recipient cells.
- Polymorphisms
Naturally occurring DNA variants that are passed down through different generations in populations.
- Modifier genes
Genes that contribute to or affect the distribution of continuous traits, such as human height.
- Quantitative trait locus mapping
Genetic mapping to identify genomic intervals that contain genes that contribute to continuously distributed traits, such as human height.
- Genetic backcross mapping panels
A population of animals used for genetic mapping that are generated by breeding two strains to generate F1 progeny, which are then bred back to one of the parental strains.
- Recombinant inbred backcross
A genetic mapping study that results from breeding a panel of recombinant inbred strains to a mouse strain of interest.
- Haplotypes
Collections of specific DNA sequences of single nucleotide polymorphisms that are clustered and frequently inherited together.
- Warm autopsy programmes
Autopsies and tissue collection that occur as soon as possible after patient demise (also known as rapid autopsy programmes).
Footnotes
Competing interests
The authors declare no competing financial interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD & Jemal A Cancer statistics, 2017. CA Cancer J. Clin 67, 7–30 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Spano D, Heck C, De Antonellis P, Christofori G & Zollo M Molecular networks that regulate cancer metastasis. Semin. Cancer Biol 22, 234–249 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Sundquist M, Brudin L & Tejler G Improved survival in metastatic breast cancer 1985–2016. Breast 31, 46–50 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Mariotto AB, Etzioni R, Hurlbert M, Penberthy L & Mayer M Estimation of the number of women living with metastatic breast cancer in the United States. Cancer Epidemiol. Biomarkers Prev 26, 809–815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry DA et al. Effect of screening and adjuvant therapy on mortality from breast cancer. N. Engl. J. Med 353, 1784–1792 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Tevaarwerk AJ et al. Survival in patients with metastatic recurrent breast cancer after adjuvant chemotherapy: little evidence of improvement over the past 30 years. Cancer 119, 1140–1148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGranahan N & Swanton C Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Magee JA, Piskounova E & Morrison SJ Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 21, 283–296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Natrajan R et al. Microenvironmental heterogeneity parallels breast cancer progression: a histologygenomic integration analysis. PLoS Med. 13, e1001961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greaves M & Maley CC Clonal evolution in cancer. Nature 481, 306–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowell PC The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).This is the classic paper that describes the linear model of cancer progression.
- 12.Marusyk A et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514, 54–58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yates LR & Campbell PJ Evolution of the cancer genome. Nat. Rev. Genet 13, 795–806 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calbo J et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19, 244–256 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Weng D et al. Metastasis is an early event in mouse mammary carcinomas and is associated with cells bearing stem cell markers. Breast Cancer Res. 14, R18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhim AD et al. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology 146, 647–651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faison WJ et al. Whole genome single-nucleotide variation profile-based phylogenetic tree building methods for analysis of viral, bacterial and human genomes. Genomics 104, 1–7 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Kim TM et al. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin. Cancer Res 21, 4461–4472 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Yachida S et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates LR et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 32, 169–184. e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie T et al. Patterns of somatic alterations between matched primary and metastatic colorectal tumors characterized by whole-genome sequencing. Genomics 104, 234–241 (2014). [DOI] [PubMed] [Google Scholar]
- 22.McCreery MQ et al. Evolution of metastasis revealed by mutational landscapes of chemically induced skin cancers. Nat. Med 21, 1514–1520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosseini H et al. Early dissemination seeds metastasis in breast cancer. Nature 540, 552–558 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harper KL et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 540, 588–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanborn JZ et al. Phylogenetic analyses of melanoma reveal complex patterns of metastatic dissemination. Proc. Natl Acad. Sci. USA 112, 10995–11000 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao ZM et al. Early and multiple origins of metastatic lineages within primary tumors. Proc. Natl Acad. Sci. USA 113, 2140–2145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naxerova K et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Sun Y & Chen H Effect of tumor size on prognosis of node-negative lung cancer with sufficient lymph node examination and no disease extension. Onco Targets Ther. 9, 649–653 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Msaki A et al. A hypoxic signature marks tumors formed by disseminated tumor cells in the BALB-neuT mammary cancer model. Oncotarget 7, 33081–33095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joosse SA & Pantel K Genetic traits for hematogeneous tumor cell dissemination in cancer patients. Cancer Metastasis Rev. 35, 41–48 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Talmadge JE, Wolman SR & Fidler IJ Evidence for the clonal origin of spontaneous metastases. Science 217, 361–363 (1982).This paper provides experimental evidence indicating that metastatic lesions arise from a single ‘seed’ cell.
- 32.Wu X et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature 482, 529–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson WJ et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat. Genet 48, 848–855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aceto N et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).This study suggests that clusters of tumour cells are more efficient at establishing metastatic lesions than single cells.
- 35.Cheung KJ et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl Acad. Sci. USA 113, E854–863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maddipati R & Stanger BZ Pancreatic cancer metastases harbor evidence of polyclonality. Cancer Discov. 5, 1086–1097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McFadden DG et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 156, 1298–1311 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gundem G et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deryugina EI & Kiosses WB Intratumoral cancer cell intravasation can occur independent of invasion into the adjacent stroma. Cell Rep. 19, 601–616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Husemann Y et al. Systemic spread is an early step in breast cancer. Cancer Cell 13, 58–68 (2008).This paper demonstrates that tumour cells can begin to disseminate very early in primary tumour evolution.
- 41.Riethmuller G & Klein CA Early cancer cell dissemination and late metastatic relapse: clinical reflections and biological approaches to the dormancy problem in patients. Semin. Cancer Biol 11, 307–311 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Butler TP & Gullino PM Quantitation of cell shedding into efferent blood of mammary adenocarcinoma. Cancer Res. 35, 512–516 (1975). [PubMed] [Google Scholar]
- 43.Kim MY et al. Tumor self-seeding by circulating cancer cells. Cell 139, 1315–1326 (2009).This is the first experimental demonstration of the possibility for transfer of cells between independent tumours.
- 44.Zhang Y et al. Tumor self-seeding by circulating tumor cells in nude mouse models of human osteosarcoma and a preliminary study of its mechanisms. J. Cancer Res. Clin. Oncol 140, 329–340 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Johnson DB et al. Acquired BRAF inhibitor resistance: a multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur. J. Cancer 51, 2792–2799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomczak K, Czerwinska P & Wiznerowicz M The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Pozn.) 19, A68–A77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steeg PS, Bevilacqua G, Pozzatti R, Liotta LA & Sobel ME Altered expression of NM23, a gene associated with low tumor metastatic potential, during adenovirus 2 Ela inhibition of experimental metastasis. Cancer Res. 48, 6550–6554 (1988). [PubMed] [Google Scholar]
- 48.Yan J, Yang Q & Huang Q Metastasis suppressor genes. Histol. Histopathol 28, 285–292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seraj MJ, Samant RS, Verderame MF & Welch DR Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 60, 2764–2769 (2000). [PubMed] [Google Scholar]
- 50.Stafford LJ, Vaidya KS & Welch DR Metastasis suppressors genes in cancer. Int. J. Biochem. Cell Biol 40, 874–891 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Robinson DR et al. Integrative clinical genomics of metastatic cancer. Nature 548, 297–303 (2017).This paper describes the largest genomic analysis to date of metastatic lesions from a variety of tumour sites.
- 52.Makohon-Moore AP et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet 49, 358–366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brastianos PK et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 5, 1164–1177 (2015).The genomic studies performed in this manuscript highlight the similarities of metastases within an organ and the dissimilarities between organs.
- 54.Pereira AA et al. Association between KRAS mutation and lung metastasis in advanced colorectal cancer. Br. J. Cancer 112, 424–428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margonis GA et al. Association between specific mutations in KRAS codon 12 and colorectal liver metastasis. JAMA Surg. 150, 722–729 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong MK et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat. Commun 6, 6605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDonald OG et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet 49, 367–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Genomes Project C et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Threadgill DW et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269, 230–234 (1995).This paper highlights the substantial effect that genetic background can have on the expression of germline mutations.
- 60.Antoniou AC et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet 42, 885–892 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Struewing JP et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N. Engl. J. Med 336, 1401–1408 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Ford D et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am. J. Hum. Genet 62, 676–689 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Milne RL & Antoniou AC Modifiers of breast and ovarian cancer risks for BRCA1 and BRCA2 mutation carriers. Endocr. Relat. Cancer 23, T69–T84 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Hamdi Y et al. Association of breast cancer risk in BRCA1 and BRCA2 mutation carriers with genetic variants showing differential allelic expression: identification of a modifier of breast cancer risk at locus 11q22.3. Breast Cancer Res. Treat 161, 117–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lifsted T et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int. J. Cancer 77, 640–644 (1998).This study is the first demonstration that inherited polymorphism is an important factor for metastatic progression.
- 66.Lancaster M, Rouse J & Hunter K Modifiers for mammary tumor latency, progression and metastasis are present on mouse chromosomes 7, 9 and 17. Mamm. Genome 16, 120–126 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Park YG et al. Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Nat. Genet 37, 1055–1062 (2005).This study describes the identification of the first inherited metastasis susceptibility gene.
- 68.Crawford NP et al. Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis. PLOS Genet. 3, e214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Faraji F et al. Cadm1 is a metastasis susceptibility gene that suppresses metastasis by modifying tumor interaction with the cell-mediated immunity. PLoS Genet. 8, e1002926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bai L et al. An integrated genome-wide systems genetics screen for breast cancer metastasis susceptibility genes. PLoS Genet. 12, e1005989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ha NH, Long J, Cai Q, Shu XO & Hunter KW The circadian rhythm gene Arntl2 is a metastasis susceptibility gene for estrogen receptor-negative breast cancer. PLoS Genet. 12, e1006267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guy CT, Cardiff RD & Muller WJ Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol 12, 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Darvasi A Experimental strategies for the genetic dissection of complex traits in animal models. Nat. Genet 18, 19–24 (1998). [DOI] [PubMed] [Google Scholar]
- 74.Hunter KW & Williams RW Complexities of cancer research: mouse genetic models. ILAR J. 43, 80–88 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Hunter KW et al. Predisposition to efficient mammary tumor metastatic progression is linked to the breast cancer metastasis suppressor gene Brms1. Cancer Res. 61, 8866–8872 (2001). [PubMed] [Google Scholar]
- 76.Park YG, Clifford R, Buetow KH & Hunter KW Multiple cross and inbred strain haplotype mapping of complex-trait candidate genes. Genome Res. 13, 118–121 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crawford NP et al. Bromodomain 4 activation predicts breast cancer survival. Proc. Natl Acad. Sci. USA 105, 6380–6385 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crawford NP et al. The diasporin pathway: a tumor progression-related transcriptional network that predicts breast cancer survival. Clin. Exp. Metastasis 25, 357–369 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goldberger N, Walker RC, Kim CH, Winter S & Hunter KW Inherited variation in miR-290 expression suppresses breast cancer progression by targeting the metastasis susceptibility gene Arid4b. Cancer Res. 73, 2671–2681 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Winter SF, Lukes L, Walker RC, Welch DR & Hunter KW Allelic variation and differential expression of the mSIN3A histone deacetylase complex gene Arid4b promote mammary tumor growth and metastasis. PLoS Genet. 8, e1002735 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Faraji F et al. Post-transcriptional control of tumor cell autonomous metastatic potential by CCR4-NOT deadenylase CNOT7. PLoS Genet. 12, e1005820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee M et al. GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin. Exp. Metastasis 32, 769–782 (2015). [DOI] [PubMed] [Google Scholar]
- 83.Ono M et al. WISP1/CCN4: a potential target for inhibiting prostate cancer growth and spread to bone. PLoS ONE 8, e71709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patel SJ, Molinolo AA, Gutkind S & Crawford NP Germline genetic variation modulates tumor progression and metastasis in a mouse model of neuroendocrine prostate carcinoma. PLoS ONE 8, e61848 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Winter JM et al. Mapping complex traits in a diversity outbred F1 mouse population identifies germline modifiers of metastasis in human prostate cancer. Cell Syst. 4, 31–45. e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van der Weyden L et al. Genome-wide in vivo screen identifies novel host regulators of metastatic colonization. Nature 541, 233–236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alsarraj J et al. BRD4 short isoform interacts with RRP1B, SIPA1 and components of the LINC complex at the inner face of the nuclear membrane. PLoS ONE 8, e80746 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Faraji F et al. An integrated systems genetics screen reveals the transcriptional structure of inherited predisposition to metastatic disease. Genome Res. 24, 227–240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vivian CJ et al. Mitochondrial genomic backgrounds affect nuclear DNA methylation and gene expression. Cancer Res. 77, 6202–6214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van ‘t Veer LJ et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002). [DOI] [PubMed] [Google Scholar]
- 91.van de Vijver MJ et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med 347, 1999–2009 (2002). [DOI] [PubMed] [Google Scholar]
- 92.Crawford NP, Yang H, Mattaini KR & Hunter KW The metastasis efficiency modifier Ribosomal RNA Processing 1 Homolog B (RRP1B) is a chromatin-associated factor. J. Biol. Chem 284, 28660–28673 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lukes L, Crawford NP, Walker R & Hunter KW The origins of breast cancer prognostic gene expression profiles. Cancer Res. 69, 310–318 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang H et al. Caffeine suppresses metastasis in a transgenic mouse model: a prototype molecule for prophylaxis of metastasis. Clin. Exp. Metastasis 21, 719–735 (2005). [DOI] [PubMed] [Google Scholar]
- 95.Qamri Z et al. Synthetic cannabinoid receptor agonists inhibit tumor growth and metastasis of breast cancer. Mol. Cancer Ther 8, 3117–3129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crawford NP et al. Germline polymorphisms in SIPA1 are associated with metastasis and other indicators of poor prognosis in breast cancer. Breast Cancer Res. 8, R16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hsieh SM, Look MP, Sieuwerts AM, Foekens JA & Hunter KW Distinct inherited metastasis susceptibility exists for different breast cancer subtypes: a prognosis study. Breast Cancer Res. 11, R75 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pei R et al. Association of SIPA1 545 C > T polymorphism with survival in Chinese women with metastatic breast cancer. Front. Med 7, 138–142 (2013). [DOI] [PubMed] [Google Scholar]
- 99.Gdowicz-Klosok A, Giglok M, Drosik A, Suwinski R & Butkiewicz D The SIPA1 −313A>G polymorphism is associated with prognosis in inoperable non-small cell lung cancer. Tumour Biol. 36, 1273–1278 (2015). [DOI] [PubMed] [Google Scholar]
- 100.Nanchari SR et al. Rrp1B gene polymorphism (1307T>C) in metastatic progression of breast cancer. Tumour Biol. 36, 615–621 (2015). [DOI] [PubMed] [Google Scholar]
- 101.Xie C et al. Sipa1 promoter polymorphism predicts risk and metastasis of lung cancer in Chinese. Mol. Carcinog 52 (Suppl. 1), E110–E117 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Brooks R et al. Polymorphisms in MMP9 and SIPA1 are associated with increased risk of nodal metastases in early-stage cervical cancer. Gynecol. Oncol 116, 539–543 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ji J, Forsti A, Sundquist J, Lenner P & Hemminki K Survival in familial pancreatic cancer. Pancreatology 8, 252–256 (2008). [DOI] [PubMed] [Google Scholar]
- 104.Ji J, Forsti A, Sundquist J, Lenner P & Hemminki K Survival in bladder and renal cell cancers is familial. J. Am. Soc. Nephrol 19, 985–991 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hemminki K, Ji J, Forsti A, Sundquist J & Lenner P Survival in breast cancer is familial. Breast Cancer Res. Treat 110, 177–182 (2008). [DOI] [PubMed] [Google Scholar]
- 106.Hemminki K, Ji J, Forsti A, Sundquist J & Lenner P Concordance of survival in family members with prostate cancer. J. Clin. Oncol 26, 1705–1709 (2008). [DOI] [PubMed] [Google Scholar]
- 107.Pirie A et al. Common germline polymorphisms associated with breast cancer-specific survival. Breast Cancer Res. 17, 58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gaudet MM et al. Genetic variation in SIPA1 in relation to breast cancer risk and survival after breast cancer diagnosis. Int. J. Cancer 124, 1716–1720 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.La Merrill M, Gordon RR, Hunter KW, Threadgill DW & Pomp D Dietary fat alters pulmonary metastasis of mammary cancers through cancer autonomous and non-autonomous changes in gene expression. Clin. Exp. Metastasis 27, 107–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gordon RR et al. Genotype X diet interactions in mice predisposed to mammary cancer: II. Tumors and metastasis. Mamm. Genome 19, 179–189 (2008). [DOI] [PubMed] [Google Scholar]
- 111.Singh AK, Loscalzo J (eds) The Brigham Intensive Review of Internal Medicine (Oxford Univ. Press, 2012). [Google Scholar]
- 112.Michailidou K et al. Association analysis identifies 65 new breast cancer risk loci. Nature 551, 92–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nam JS et al. An anti-transforming growth factor beta antibody suppresses metastasis via cooperative effects on multiple cell compartments. Cancer Res. 68, 3835–3843 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gartner K A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab. Anim 24, 71–77 (1990). [DOI] [PubMed] [Google Scholar]
- 115.Wright S The relative importance of heredity and environment in determining the piebald pattern of guinea-pigs. Proc. Natl Acad. Sci. USA 6, 320–332 (1920). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kirkwood TB et al. What accounts for the wide variation in life span of genetically identical organisms reared in a constant environment? Mech. Ageing Dev 126, 439–443 (2005). [DOI] [PubMed] [Google Scholar]
- 117.Gartner K Commentary: random variability of quantitative characteristics, an intangible epigenomic product, supporting adaptation. Int. J. Epidemiol 41, 342–346 (2012). [DOI] [PubMed] [Google Scholar]
- 118.Blewitt ME, Chong S & Whitelaw E How the mouse got its spots. Trends Genet. 20, 550–554 (2004). [DOI] [PubMed] [Google Scholar]
- 119.Campbell PJ et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roe JS et al. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell 170, 875–888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alsaggar M, Yao Q, Cai H & Liu D Differential growth and responsiveness to cancer therapy of tumor cells in different environments. Clin. Exp. Metastasis 33, 115–124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang L et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aurilio G et al. Discordant hormone receptor and human epidermal growth factor receptor 2 status in bone metastases compared to primary breast cancer. Acta Oncol. 52, 1649–1656 (2013). [DOI] [PubMed] [Google Scholar]
- 124.Hoefnagel LD et al. Discordance in ERalpha, PR and HER2 receptor status across different distant breast cancer metastases within the same patient. Ann. Oncol 24, 3017–3023 (2013). [DOI] [PubMed] [Google Scholar]
- 125.Almendro V et al. Genetic and phenotypic diversity in breast tumor metastases. Cancer Res. 74, 1338–1348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Navin N et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Painter C et al. The metastatic breast cancer project: translational genomics through direct patient engagement [abstract P1–05–13]. San Antonio Breast Cancer Symposium https://www.sabcs.org/Portals/SABCS2016/Documents/SABCS-2016-Abstracts.pdf?v=1 (2016). [Google Scholar]
- 128.Condeelis JS, Wyckoff J & Segall JE Imaging of cancer invasion and metastasis using green fluorescent protein. Eur. J. Cancer 36, 1671–1680 (2000). [DOI] [PubMed] [Google Scholar]
- 129.Bravo-Cordero JJ, Hodgson L & Condeelis J Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol 24, 277–283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dovas A, Patsialou A, Harney AS, Condeelis J & Cox D Imaging interactions between macrophages and tumour cells that are involved in metastasis in vivo and in vitro. J. Microsc 251, 261–269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sosa MS, Bragado P & Aguirre-Ghiso JA Mechanisms of disseminated cancer cell dormancy: anawakening field. Nat. Rev. Cancer 14, 611–622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Westcott PM et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature 517, 489–492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.O’Rourke KP et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol 35, 577–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roper J et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol 35, 569–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]