Haibe-Kains et al.1 reported inconsistency between two large-scale pharmacogenomic studies (the Cancer Cell Line Encyclopedia2 (CCLE) and the Cancer Genome Project3 (CGP)). Upon careful analysis of the same data we arrived at much more positive conclusions. Here, we highlight the most important reasons for this.
To assess the concordance of two large studies of the efficacy of cancer drugs, Haibe-Kains et al. compared the correlation in drug sensitivities with correlation in gene expression values measured on the same human cancer cell lines. The authors reported correlation “between” cell lines for gene expression but, inconsistently, “across” cell lines for drug sensitivity (see Methods). On reanalysis, we found much higher correlations between cell lines than across cell lines for both gene expression and drug sensitivity measures (median rs = 0.88 between cell lines, rs = 0.56 across cell lines for expression; median rs = 0.62 between cell lines and rs = 0.35 across cell lines for AUC, a drug sensitivity measure). Thus, by correcting this inconsistency, the correlations for expression and drug sensitivity data were far more similar than was originally reported, severely undermining the authors’ interpretation of the relative quality of expression and drug sensitivity datasets.
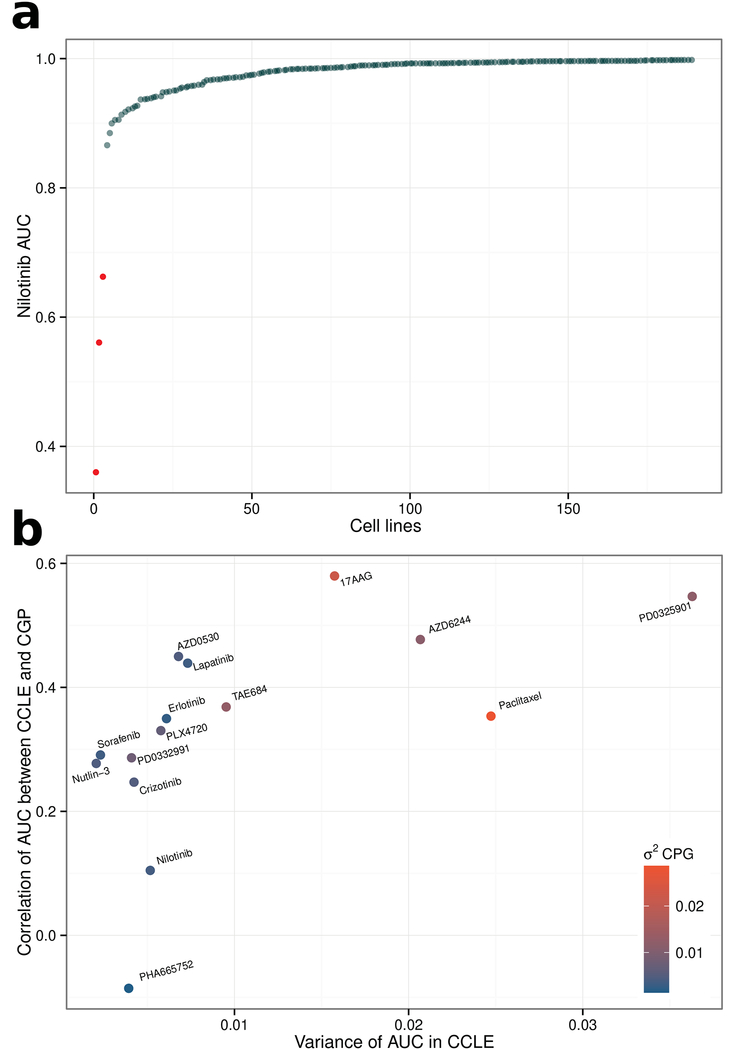
However, the fundamental issue is that the authors’ reported Spearman correlation coefficients do not fairly reflect the concordance of drug sensitivity between the studies, because of the lack of variability in drug response, which arises due to the highly-targeted nature of many of the drugs assessed. To see why correlation is not an appropriate measure of biological concordance for these data, consider the hypothetical example of a drug that is not effective against any cell lines (a possibility for an experimental drug): In such a case randomly-fluctuating measurement error, inherent in biological assays, will dominate over the (non-existent) biological variability, meaning that there could be no expectation of correlation between repeated measures of drug sensitivity (assuming other experimental variables are held constant). In this study, many of the drugs were highly-targeted agents, which by design require specific (and often rare) molecular targets for response (see Supplementary Table). Consider nilotinib, which the authors reported as exhibiting “poor consistency” between CGP and CCLE (rs = 0.1 for AUC); nilotinib targets BCR-ABL1, a fusion-gene. In CGP, BCR-ABL1 status was reported strongly associated with drug sensitivity (P = 2.54 × 10−65), accurately reflecting the known biology. BCR-ABL1 status was not reported by CCLE, however, upon re-analysis we identified three BCR-ABL1-positive cell lines among the 189 nilotinib-treated cell lines that overlapped CGP and these were also the three most sensitive samples (P = 9 × 10−7). Hence, despite the fact that these drug sensitivity data were accurately recapitulating biological expectations in both studies, the authors’ criteria incorrectly classify nilotinib sensitivity as discordant. Of the 577 cell lines screened in CGP 573 do not harbour the nilotinib target, i.e. the BCR-ABL1 fusion gene. Thus, given (as expected) no drug response in almost all cell lines screened (median AUC across all cell lines = 0.99; AUC of 1 represents no drug response; Fig. 1(a); Supplementary Table), there was little biological variability across the vast majority of cell lines, resulting in low correlation between the repeated measurements made by CCLE and CGP, despite clearly concordant results. Similarly, most other drugs that the authors compared were also targeted agents, meaning this lack of drug response was common; indeed, for 10 of the 15 drugs, median AUC was greater than 0.90 in CGP and 8 of these 10 also have median AUC greater than 0.9 in CCLE, resulting in little variability across most cell lines when treated with these drugs. Indeed, we identified a systematic relationship between variability in drug response in either study and correlation between the two studies (Fig. 1(b)). A valid comparison of CGP and CCLE must consider the pharmacology of the drugs screened and in particular the differences in the variability induced by different drugs. Nilotinib was not an isolated case; in fact, despite the highly experimental nature of many of the compounds screened by CCLE/CGP, we still identified multiple expected associations that were consistently reported by both studies including ERBB2 for lapatinib4, NQ01 expression for 17-AAG5, BRAF mutation for PD-03259016, AZD62447, and PLX47208, MDM2 for Nutlin-3a9, and MET for Crizotinib10 (Supplementary Table). Finally, the utility of these pharmacogenomic datasets is now further supported by findings that models fit using data from CGP could reliably predict drug response in multiple clinical trials11,12.
Fig 1.
Highly targeted agents (nilotinib) highlight a major limitation of the authors’ test for concordance. Scatterplot showing the nilotinib AUC values (in CGP) for the 189 cell lines that were screened by both CGP and CCLE. Only a very small proportion of cell lines achieve a response, e.g. the three BCR-ABL1 positive cell lines highlighted in red. This almost complete lack of biological variability renders a Spearman correlation ineffective as a means to assess concordance.
(b) The authors’ test for concordance is confounded by variability in drug response. Scatterplot showing the strong association between “Spearman’s correlation of AUC between CCLE and CGP” and “variance of AUC in CCLE”. Drugs whose AUC is more variable are more likely to be highly correlated between CCLE and CGP (rs = 0.83, P = 1.9 × 10−4). The points have been color coded by their “variance of AUC in CGP”, which is also significantly associated with both “variance of AUC in CCLE” and “Spearman’s correlation of AUC between CCLE and CGP”.
In summary, our analysis shows that Haibe-Kains et al.’s conclusions are unsubstantiated and we propose that a fair assessment of concordance between large pharmacogenomic datasets will require the development or adaptation of methods that account for the issues raised here, although great care will be required to ensure that such methods do not introduce their own unforeseen biases.
Methods
In CGP and CCLE, using ordered data common to both studies, gene expression and drug sensitivity (AUC) values can be arranged in n1 × m and n2 × m matrices respectively where m is the number of cell lines, n1 is the number of genes and n2 is the number of drugs common to both studies. Correlations “between” cell lines are calculated by the correlation of matching columns of CGP and CCLE matrices (vectors of length n1 for expression or n2 for AUC). Correlations “across” cell lines are the correlations of matching rows (vectors of length m for both data).
To achieve easy reproduction of our results, we have made the source code for our analysis available in a GitHub repository (https://github.com/paulgeeleher/nature_bca).
Supplementary Material
References:
- 1.Haibe-Kains B et al. Inconsistency in large pharmacogenomic studies. Nature 504, 389–93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretina J et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garnett MJ et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konecny GE et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 66, 1630–9 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Kelland LR, Sharp SY, Rogers PM, Myers TG & Workman P DT-Diaphorase expression and tumor cell sensitivity to 17-allylamino, 17-demethoxygeldanamycin, an inhibitor of heat shock protein 90. J. Natl. Cancer Inst. 91, 1940–9 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Solit DB et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature 439, 358–62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dry JR et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244). Cancer Res. 70, 2264–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai J et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. U. S. A 105, 3041–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller CR et al. Potential for treatment of liposarcomas with the MDM2 antagonist Nutlin-3A. Int. J. Cancer 121, 199–205 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Timm A & Kolesar JM Crizotinib for the treatment of non-small-cell lung cancer. Am. J. Health. Syst. Pharm 70, 943–7 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Geeleher P, Cox NJ & Huang RS Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 15, R47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falgreen S et al. Predicting response to multidrug regimens in cancer patients using cell line experiments and regularised regression models. BMC Cancer 15, 235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.