Abstract
A simultaneous comparison of the RNA molecules expressed by Salmonella bacteria and human cells during infection reveals how a bacterial small RNA alters the transcript profiles of both the bacteria and the host cells.
What happens when bacteria encounter or enter host cells? How does each of the species respond, the bacteria to survive in their new environment and the host cells to either tolerate non-harmful bacteria or defend against pathogenic ones? To answer these questions, it is imperative to understand how gene transcription in both cells changes during the encounter. Over the years, approaches applied to this problem have ranged from in vivo gene-expression technology1 to sequencing the full complement of bacterial or host-cell transcripts2,3 (the transcriptome). However, such analyses have largely focused on messenger RNAs and have profiled either the bacteria or the host, not both at once. In this issue, Westermann et al.4 (page 496) go beyond the individual organisms by using dual RNA-seq, an approach that simultaneously profiles bacterial and host transcriptomes throughout the course of an infection.
The RNA-seq method takes advantage of the ever-increasing depth of sequencing (the number of reads for a particular sample) now possible. Westermann et al. first assessed whether the dual RNA-seq approach accurately reflected known gene regulation in human HeLa cells and in the bacterium Salmonella enterica serovar Typhimurium (hereafter Salmonella), a common cause of food poisoning, during infection. The authors’ data confirmed that, as previously reported5, transcription of invasion-related genes in the genomic region known as Salmonella pathogenicity island 1 (SPI-1) was reduced after bacterial internalization, whereas transcription of SPI-2 genes, which promote intracellular survival, increased.
Having validated the sensitivity of the approach, Westermann et al. focused on mRNAs and regulatory RNAs whose expression changed during the course of the 24-hour infection. In bacteria, small regulatory RNAs (sRNAs) that base-pair with target mRNAs to modulate the mRNA’s stability or translation are integral to a wide range of stress responses, including the response to host cells6. Thus, the authors were intrigued by an 80-nucleotide sRNA, which they denoted PinT, whose expression was highly induced during infection, and which was activated by the bacterial PhoP/Q system, known to be crucial for Salmonella survival in the intracellular environment.
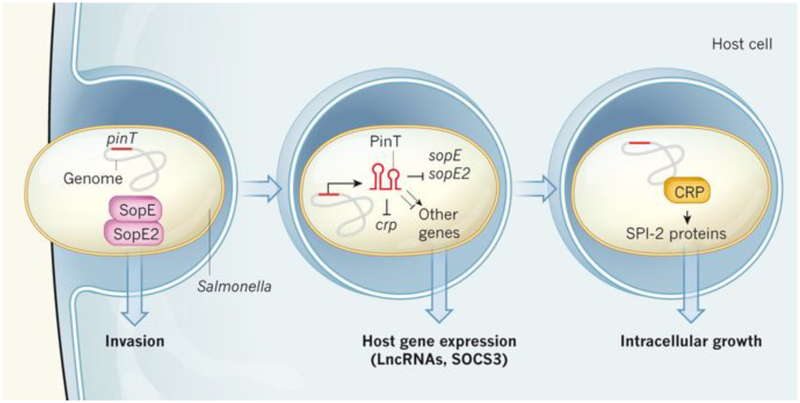
A striking finding of the dual RNA-seq analysis was that tens of bacterial and hundreds of host-cell transcripts were affected merely by the presence or absence of PinT (Fig. 1). On the bacterial side, overproduction of PinT led to reduced levels of the mRNAs encoding SopE and SopE2, two SPI-1 effector proteins that mediate host-cell invasion by Salmonella. These mRNAs were elevated in strains lacking the pinT gene. By mutating the pinT, sopE and sopE2 sequences, the authors revealed that the inhibitory effect of PinT occurred through direct base-pairing with the mRNAs. Dual RNA-seq also revealed a role for PinT in repressing SPI-2 genes later in infection. However, control of these genes was indirect and occurred through PinT base-pairing with the mRNA that encodes the cyclic AMP receptor protein (CRP), an activator of transcription of SPI-2 genes. These data indicate that PinT, on bacterial internalization, controls the temporal expression of both SPI-1 effectors and SPI-2 virulence genes, thus facilitating the bacterium’s transition from an invasive state to a state of intracellular replication.
Figure 1: PinT orchestrates gene expression in Salmonella and its host cells.
When Salmonella bacteria invade cells, expression of the genes sopE and sopE2 facilitates bacterial invasion. After the bacteria are internalized, the levels of the transcripts from these genes fall. By simultaneously monitoring the RNA molecules present in both Salmonella and host cells over the course of an infection, Westermann et al.4 found that a small regulatory RNA expressed in Salmonella, which they name PinT, induces this repression by base-pairing with the sopE and sopE2 messenger RNAs. PinT also base-pairs with the mRNA that encodes CRP, a protein that activates transcription of genes encoding SPI-2 proteins. This repression is reduced later in infection, allowing the SPI-2 proteins to regulate the bacterium’s intracellular growth. The authors also observed differences in host-cell transcripts when the cells were infected with Salmonella mutants lacking PinT, including altered levels of long non-coding RNAs (lncRNAs) and the mRNA for SOCS3. This suggests that PinT targets other bacterial genes that influence host-cell gene expression.
Westermann et al. then compared the transcriptomes of the host HeLa cells challenged with either wild-type Salmonella or a strain lacking PinT. They discovered numerous changes in cells infected with the PinT-lacking mutant, including altered levels of many long non-coding RNAs (lncRNAs), hyperactivation of mitochondrial genes, increased abundance of mRNAs for proteins involved in innate immune pathways (such as the interleukin-8 mRNA) and accelerated activation of SOCS3, a protein that regulates the inflammatory JAK–STAT signalling pathway. The last finding is of particular interest, because properly balanced JAK–STAT signalling is essential for optimal Salmonella infection. Too little inflammation reduces the ability of Salmonella to compete with the intestinal microbiota, whereas too much inflammation might result in the bacteria being killed by the host7,8,9.
The dual RNA-seq approach of simultaneously interrogating the bacteria and host transcriptomes gives unprecedented insight into the dynamic RNA-expression landscape during the bacterium–host interaction. This method is particularly useful for analysing genes such as pinT that, when deleted, cause little or no detectable change in standard bacterial virulence assays, but nevertheless have a strong impact on mRNA levels. The authors’ study of PinT, however, also demonstrates some of the ongoing challenges in studying bacterium–host interactions. For example, although the effect of PinT on host gene expression such as JAK–STAT signalling is interesting, it remains unclear which PinT-controlled bacterial gene(s) are responsible. In addition, it has become evident that there is substantial variation in gene expression even between individual bacteria in single-species cultures; these differences are not captured by approaches in which cell samples are sequenced in bulk.
Finally, although dual RNA-seq eliminates the need to separate bacteria from the host cell for investigation, a comprehensive transcriptome analysis over the course of an infection still demands the analysis of a series of time points in short succession. Westermann et al. conducted a time series for their analysis of HeLa cells infected with wild-type and PinT-lacking Salmonella, but this approach is impractical for the analysis of large collections of bacterial mutants or isolates. Continued improvement in deep-sequencing technologies, including single-cell sequencing, will undoubtedly circumvent some of these limitations. In the meantime, the application of Westermann and colleagues’ dual RNA-seq approach to a wide range of bacterium–host interactions will open a treasure trove of insight into transcriptional actions and reactions as bacteria enter and proliferate in host cells.
References
- 1.Slauch JM, Mahan MJ & Mekalanos JJ Meth. Enzymol 235, 481–492 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Mandlik A et al. Cell Host Microbe 10, 165–174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiu L et al. Genet. Mol. Res 14, 16948–16965 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Westermann AJ et al. Nature 529, 496–501 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Hautefort I et al. Cell. Microbiol 10, 958–984 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner EGH & Romby P Adv. Genet 90, 133–208 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Uchiya K & Nikai T Infect Immun 73, 5587–5594 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannemann S, Gao B & Galán JE PLoS Pathog 9, e1003668 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stecher B et al. PLoS Biol 5, 2177–2189 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]