Abstract
Aims/hypothesis:
Inflammation is associated with increased body mass, and purportedly, increased size of adipose cells. Our goal was to determine if increased size of adipose cells, is associated with localized inflammation in weight-stable, moderately-obese humans.
Subjects/methods:
49 healthy, moderately-obese individuals were recruited for quantification of insulin resistance (modified insulin suppression test) and subcutaneous abdominal adipose tissue biopsy. Cell size distribution was analyzed with Beckman Coulter Multisizer III, and inflammatory gene expression with rtPCR. Correlations between inflammatory gene expression and cell size parameters, with adjustment for gender and insulin resistance, were calculated.
Results:
Adipose cells were bimodally distributed, with 47% in a “large” cell population, and the remainder in a “small” cell population. The median diameter of the large adipose cells was not associated with expression of inflammatory genes. Rather, the fraction of small adipose cells was associated with inflammatory gene expression, independent of gender, insulin resistance, and BMI. This association was more pronounced in insulin-resistant than insulin-sensitive individuals. Insulin resistance was also independently associated with expression of inflammatory genes.
Conclusions:
Ths study demonstrates that among moderately-obese, weight-stable individuals, an increased proportion of small adipose cells is associated with inflammation in subcutaneous adipose tissue, whereas size of mature adipose cells is not. The observed association between small adipose cells and inflammation may reflect impaired adipogenesis and/or terminal differentiation, but whether this is a cause or consequence of inflammation is unclear. This question and whether the small, large, or total adipose cell population contribute to the inflammation are topics for future research.
Keywords: adipocyte, inflammation, insulin resistance, obesity, adipose tissue
Introduction
Obesity is associated with insulin resistance, which is the precursor to type 2 diabetes and cardiovascular disease. The mechanistic link between excess body weight and insulin resistance is unclear. Furthermore, not all moderately-obese individuals are insulin resistant (IR) – indeed, insulin sensitivity can vary 6-fold in this population (1). We have taken advantage of this observation to compare molecular and cellular characteristics of subcutaneous abdominal adipose tissue from equally-obese individuals who were classified as either IR or insulin sensitive (IS), by the modified insulin suppression test.
It has been suggested that the biological characteristics of adipose tissue, rather than mass per se, might lead to insulin resistance. Specifically, large adipose cells are thought to contribute to insulin resistance via increased lipolysis (2, 3), and more recently, by inciting an inflammatory cascade (4,5). Published literature shows that increasing BMI and larger mean adipose cell size in humans and rodents is associated with greater density of macrophages in adipose tissue (6), and that large adipose cells appear necrotic and are surrounded by macrophages (4), but these studies are non-quantitative, not done in a weight-stable state, and the single study with humans included very obese subjects without clear evidence of weight stability, and biopsies from different body regions (4).
We have previously shown that simply measuring the mean diameter of adipose cells, or estimating size by dividing total lipid by total number of cells is inadequate to describe the size of adipose cells because the size distribution is essentially bimodal, with cells primarily residing in either a large or a small cell fraction, and few in-between (7). After taking this into consideration, we have been able to perform more complex analyses of the relationship between cell size characteristics and both molecular and clinical characteristics of human subjects. Using this technique, we addressed the hypothesis that larger adipose cell size is associated with inflammation in subcutaneous adipose tissue.
Methods
Subjects
Subjects included 49 moderately-obese, otherwise healthy adults selected from a larger subject pool recruited via newspaper advertisement in the cities surrounding Stanford University. Subjects were required to be 35–65 years of age, free of major organ disease, nondiabetic as defined by fasting plasma glucose concentration < 126 mg/dL, with BMI 26–36 kg/m2 and stable body weight for 3 months. Subjects could not be engaged in a weight loss program, or taking lipid-lowering medications, including fish oil/alpha omega fatty acids, steroid preparations, or medications for weight loss. Subjects with a history of eating disorder, bariatric surgery, or liposuction were excluded, as were pregnant or lactating subjects. All subjects underwent quantification of insulin-mediated glucose disposal via a modified insulin suppression test (IST, see below) and a subcutaneous periumbilical adipose tissue biopsy. The study was approved by the Stanford University Human Subjects Committee and all subjects gave written, informed consent.
Quantification of Insulin-Mediated Glucose Disposal and Other Clinical Measurements
Insulin-mediated glucose disposal was quantified by a modification (8) of the IST as originally described and validated (9,10). Briefly, subjects were infused for 240 min with octreotide (0.27 μg/m2•min) (to suppress endogenous insulin secretion), insulin (25 mU/m2 •min), and glucose (240 mg/m2 •min). Blood was drawn at 10-min intervals from 210 to 240 min of the infusion to measure plasma glucose and insulin concentrations, and the mean of these four values was used as the steady-state plasma insulin (SSPI) and glucose (SSPG) concentrations for each individual. As SSPI concentrations are similar in all subjects during these tests, the SSPG concentration provides a direct measure of the ability of insulin to mediate disposal of an infused glucose load; the higher the SSPG concentration, the more insulin-resistant the individual. Subjects whose SSPG results were in the top or bottom 40th percentile were included, and defined as IR or IS, respectively. Individuals in the middle 20th percentile were excluded, as they were not clearly IR or IS. This procedure allows for identification of subjects who are relatively homogeneous with respect to degree of obesity and general health, but who differ substantially with respect to insulin sensitivity.
Other clinical and laboratory measurements
After a 12-h overnight fast, plasma glucose, insulin, and lipid/lipoprotein concentrations were measured as previously described (9,11). Other experimental measurements included weight, height, calculated BMI, and waist circumference, measured at end-expiration as the point midway between the iliac crest and lower costal margin, race/ethnicity, and blood pressure (average of 6 readings taken after sitting for 5 minutes).
Subcutaneous Abdominal Fat Biopsy and Determination of Cell Size Distribution
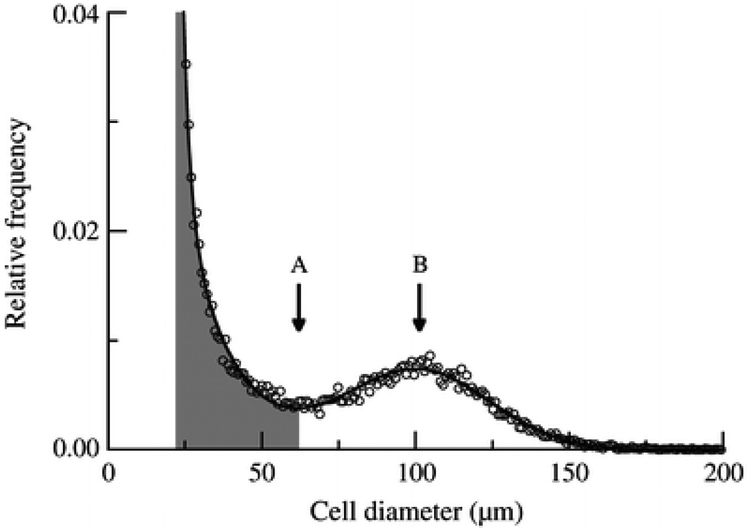
Subcutaneous abdominal fat biopsies were performed under sterile conditions by scalpel incision inferior to the umbilicus, as previously described (6). Two samples of 20–30 mg of tissue were immediately fixed in osmium tetroxide and incubated in a water bath at 37°C for 48-h as previously described (12), for cell size distribution analysis using a Beckman Coulter (Miami, FL, USA) Multisizer III with a 400-um aperture. The effective cell-size range using this aperture is 20 to 240-um. The instrument was set to count 6,000 particles and the fixed-cell suspension was diluted so that coincident counting was less than 10%. After collection of pulse sizes, the data were expressed as particle diameters and displayed as histograms of counts against diameter using linear bins and a linear scale for the x-axis. All subjects displayed a roughly bimodal distribution of cells with a large cell “hump” and a “tail” of small cells, separated by a cell count “nadir” as described previously (6). Quantitative analyses to describe the cell size distribution were done via mathematical modeling (non-linear least squares function nls, R 1.9 (http://r-project.org)), in which a single formula using 7 cell-size parameters could describe the individual data points of each subject:
where x = cell diameter and x0 = the smallest diameter; h1 and w1 = height and width of the first exponential; h2 and w2 = height and width of the second exponential; and hp, cp, and wp = height, center, and width of the Gaussian curve. In the curve formulated by this equation, the small cell “tail” is represented by the sum of 2 exponentials, and the large cell “hump” is represented by a Gaussian curve (Figure 1). Endpoints of interest included cp (peak center), which represents the median diameter of the large cells, and the fraction of small cells, defined as the percent of cells below the nadir.
Figure 1.
Multisizer analysis of cell size distribution in a single patient with large cells represented by a Gaussian formula and small cells represented by a double-exponential formula. Fraction of small cells (shaded area) includes all cells below the nadir (black arrow). Peak center is the cell diameter where the Gaussian hump peaks (white arrow).
Quantitative RT-PCR for Markers of Inflammation
Total RNA was extracted from flash-frozen biopsy samples using TRIzol (Life Technologies, Gaithersburg, MD, USA) and Adipose Tissue RNAeasy kits (Qiagen, Valencia, CA, USA) according to the manufacturers’ instructions. After DNase treatment, cDNA was synthesized from 5 mg of total RNA using MMLV reverse transcriptase (SuperScript II kit, Invitrogen, Carlsbad, CA, USA). Taqman® primer/probe sets for markers of inflammatory cell infiltrate (CD14, CD45, CD68, and egf-like module containing, mucin-like, hormone receptor-like 1 (EMR1)) and inflammation (interleukin (IL)-6, IL-8, inducible nitric oxide synthase (iNOS), monocyte chemotactic protein-1 (MCP-1), and tumor necrosis factor (TNF)-α), as well as for 18S ribosomal RNA were obtained from Applied Biosystems (Foster City, CA).
Amplification was carried out in triplicate on an ABI Prism 7700 at 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. A threshold cycle (CT value) was obtained from each amplification curve and a ΔCT value was first calculated by subtracting the CT value for 18S ribosomal RNA from the CT value for each sample. A ΔΔCT value was then calculated by subtracting the ΔCT value of a single insulin-sensitive subject (control). Fold-changes compared with the control were then determined by raising 2 to the ΔΔCT power.
Statistical Analyses
Data are presented as mean + standard deviation from the mean. Non-normally distributed variables (all inflammation genes except MCP-1) were log transformed for statistical tests. Insulin sensitivity was treated as a categorical variable, with subjects classified as being from either the IR or IS group. Multiple linear regression analysis with each inflammatory marker as the dependent variable was performed with the fraction of small cells, gender, and insulin resistance group as predictors. Formal testing for interactions between fraction of small cells and both gender and insulin resistance group was done. Similar multiple regression models were constructed for prediction of each inflammatory gene with the peak diameter of the large cells as a predictor, along with gender and insulin resistance group. Again, formal testing for interactions between peak diameter and both gender and insulin resistance was done. P< 0.05 was considered statistically significant.
Results
Anthropometric and laboratory characteristics of 49 moderately-obese individuals are shown in Table 1. The range of BMI in this study was 25.3 to 34.8 kg/m2. Approximately 60% of subjects were female, and 78% were Caucasian. Of the 49, 28 were IR and 21 were IS, quantitated by the modified IST as described. Mean peak adipose cell diameter was 111 ± 15 um, and mean fraction of small adipose cells was 53 ± 10%. Table 2 demonstrates the independent associations between fraction of small adipose cells and relative expression of eight inflammation genes in adipose tissue. Standardized correlation coefficients (β) are shown for each predictor in the model (predictors were fraction of small cells, insulin resistance group, and gender). Statistical adjustment for BMI did not alter results and it was removed from the final models.
Table 1.
Demographic and Clinical Characteristics of Study Subjects
| Characteristic | Mean ± SD (N=49) |
|---|---|
| Age (yrs) | 52 ± 9 |
| Race (Cauc/Hisp/Black/Asian) | 38/4/2/5 |
| Gender (F/M) | 30/19 |
| BMI (kg/m2) | 30.0 ± 2.7 |
| Weight (kg) | 88.6 ± 15.0 |
| Waist Circumference (cm) | 102 ± 11 |
| Fasting Blood Glucose (mg/dL) | 97 ± 8 |
| Systolic Blood Pressure (mmHg) | 125 ± 14 |
| Diastolic Blood Pressure (mmHg) | 74 ± 8 |
| SSPG (mg/dL) | 157 ± 78 |
| Cholesterol (mg/dL) | 192 ± 34 |
| HDL-C (mg/dL) | 49 ± 18 |
| LDL-C (mg/dL) | 122 ± 28 |
| Triglycerides (mg/dL) | 121 ± 69 |
Table 2.
Independent Associations Between Fraction of Small Adipose Cells and Inflammatory Genes, Adjusted for Insulin-Resistance Group and Gender*
| Gene | Predictors | β ± SE | Standardized β | P value |
|---|---|---|---|---|
| TNF-α | Fraction small cells | 0.002 ± 0.10 | −0.003 | 0.98 |
| IR group | 0.03 ± 0.02 | 0.19 | 0.22 | |
| Gender | 0.03 ± 0.02 | 0.20 | 0.20 | |
| iNOS | Fraction small cells | −0.44 ± 1.36 | −0.05 | 0.74 |
| IR group | 0.06 ± 0.28 | 0.03 | 0.84 | |
| Gender | 0.01 ± 0.29 | 0.008 | 0.96 | |
| CD14 | Fraction small cells | −1.62 ± 0.63 | 0.33 | 0.01 |
| IR group | 0.35 ± 0.13 | 0.37 | 0.009 | |
| Gender | 0.07 ± 0.13 | 0.08 | .58 | |
| CD45 | Fraction small cells | 2.29 ± 0.87 | 0.34 | 0.01 |
| IR group | 0.46 ± 0.18 | 0.36 | 0.01 | |
| Gender | −0.18 ± 0.18 | −0.14 | 0.31 | |
| CD68 | Fraction small cells | 0.21 ± 0.64 | 0.04 | 0.74 |
| IR group | 0.53 ± 0.73 | 0.55 | 0.0002 | |
| Gender | −0.08 ± 0.14 | −0.08 | 0.58 | |
| EMR-1 | Fraction small cells | 1.89 ± 0.74 | 0.33 | 0.01 |
| IR group | 0.43 ± 0.15 | 0.39 | 0.007 | |
| Gender | −0.14 ± 0.16 | −0.12 | 0.38 | |
| IL-6 | Fraction small cells | 3.29 ± 1.51 | 0.28 | 0.04 |
| IR group | 1.03 ± 0.31 | 0.45 | 0.002 | |
| Gender | −0.05 ± 0.32 | −0.02 | 0.88 | |
| IL-8 | Fraction small cells | 1.24 ± 1.14 | 0.13 | 0.28 |
| IR group | 1.19 ± 0.23 | 0.63 | <0.0001 | |
| Gender | −0.41 ± 0.24 | −0.22 | 0.09 | |
| MCP-1 | Fraction small cells | 0.20 ± 0.06 | 0.38 | 0.003 |
| IR group | 0.04 ± 0.01 | 0.40 | 0.003 | |
| Gender | 0.003 ± 0.01 | 0.03 | 0.78 |
P-values are adjusted for gender and insulin resistance group via multiple linear regression analysis. Adjustment for BMI did not alter results.
It is clear from this table that CD14, CD45, EMR-1, IL-6, and MCP-1 were associated with fraction of small cells, independent of insulin resistance group and gender. Insulin resistance was also independently associated with these same genes, as well as CD68 and IL-8. Gender was NS associated with expression of any of the inflammation genes. Formal testing for interaction between insulin resistance group and fraction of small cells in predicting inflammation genes revealed statistically significant or borderline significant interactions for EMR-1 (p<0.02) and CD14 (p=0.09), but not for the rest of the genes.
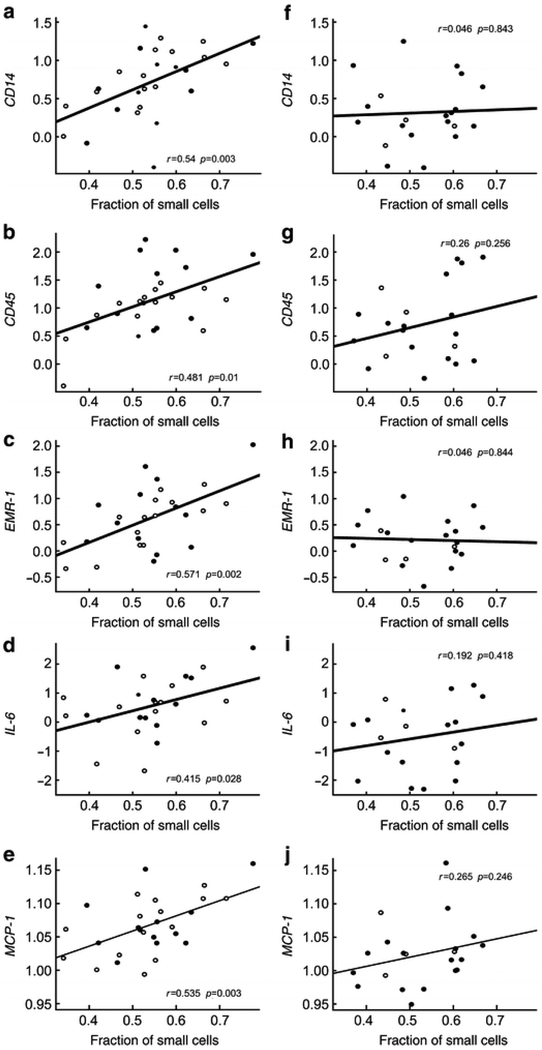
Because insulin resistance group modified the relationship between fraction of small cells and inflammatory gene expression in some cases, correlation coefficients between inflammation genes and fraction of small adipose cells are shown separately for IR and IS subjects in Figure 2. It is apparent that correlations were stronger for the IR versus the IS subgroup for all genes, most dramatically so for CD14 and EMR-1, the two genes for which the interaction between IR group and fraction of small cells in predicting inflammation was statistically significant.
Figure 2.
Relationship between fraction of small cells and inflammatory genes in insulin resistant (left panels) and insulin sensitive (right panels) males (open circles) and females (closed circles).
Table 3 shows the independent relationships between peak center (diameter of large adipose cells) and inflammatory gene expression. Peak center was not significantly associated with any inflammatory genes. As before, insulin resistance independently predicted expression of 7 genes (CD14, CD45, CD68, EMR-1, IL-6, IL-8, MCP-1). Again, gender was not a significant predictor of inflammatory genes, although male gender bore an inverse association with IL-8 that was of borderline significance (p=0.05).
Table 3.
Independent Associations Between Peak Center of Large Adipose Cells and Inflammatory Genes, Adjusted for Insulin-Resistance Group and Gender*
| Gene | Predictors | β ± SE | Standardized β | P value |
|---|---|---|---|---|
| TNF-α | Peak center | −0.006 ± 0.0008 | −0.11 | 0.49 |
| IR group | 0.03 ± 0.02 | 0.22 | 0.16 | |
| Gender | 0.021 ± 0.026 | 0.14 | 0.44 | |
| iNOS | Peak center | −0.01 ± 0.01 | −0.20 | 0.26 |
| IR group | 0.22 ± 0.29 | 0.08 | 0.61 | |
| Gender | −0.16 ± 0.33 | −0.09 | 0.62 | |
| CD14 | Peak center | 0.007 ± 0.007 | 0.22 | 0.16 |
| IR group | 0.256 ± 0.14 | 0.34 | 0.03 | |
| Gender | 0.24 ± 0.16 | 0.15 | 0.36 | |
| CD45 | Peak center | 0.004 ± 0.007 | 0.11 | 0.50 |
| IR group | 0.47 ± 0.19 | 0.36 | 0.02 | |
| Gender | −0.17 ± 0.22 | −0.13 | 0.45 | |
| CD68 | Peak center | 0.0002 ± 0.004 | −0.08 | 0.60 |
| IR group | 0.50 ± 0.13 | 0.58 | 0.0002 | |
| Gender | −0.07 ± 0.15 | −0.15 | 0.34 | |
| EMR-1 | Peak center | 0.001 ± 0.006 | 0.04 | 0.98 |
| IR group | 0.46 ± 0.17 | 0.42 | 0.009 | |
| Gender | −0.19 ± 0.19 | −0.16 | 0.34 | |
| IL-6 | Peak center | −0.004 ± 0.01 | −0.06 | 0.70 |
| IR group | 1.15 ± 0.34 | 0.50 | 0.002 | |
| Gender | −0.21 ± 0.38 | −0.09 | 0.59 | |
| IL-8 | Peak center | 0.0007 ± 0.008 | −0.11 | 0.42 |
| IR group | 1.27 ± 0.25 | 0.68 | <0.0001 | |
| Gender | −0.55 ± 0.28 | −0.29 | 0.05 | |
| MCP-1 | Peak center | 0.00004 ± 0.0005 | −0.04 | 0.80 |
| IR group | 0.04 ± 0.01 | 0.45 | 0.004 | |
| Gender | −0.004 ± 0.017 | −0.03 | 0.83 |
P-values are adjusted for gender and insulin resistance group via multiple linear regression analysis. Adjustment for BMI did not alter results.
Discussion
The results of this study demonstrate that among a group of moderately-obese healthy individuals, increased size of mature adipose cells was not associated with inflammation in subcutaneous adipose tissue. Rather, an increased proportion of small (relative-to-large) adipose cells independently predicted expression of five inflammatory genes. This relationship was independent of insulin resistance, BMI, and gender. These findings yield a new perspective on the purported relationship between adipose cell size characteristics and inflammation.
The finding that large adipose cell diameter is not associated with increased expression of inflammatory markers is at odds with prior cross-sectional data from 2000–2001, showing that both BMI and mean adipocyte size correlated with CD68 expression in adipose tissue in humans and mice (6). Evaluation of genetically obese mice, including those deficient in hormone-sensitive lipase which develop large adipose cells in the absence of obesity, demonstrated increased density of macrophages localized to sites of single apparently necrotic adipose cells, termed “crown-like structures”, which were increased 30-fold in the former, and 15-fold in the latter (4). In 12 obese (BMI 30–45 kg/m2) and nine normal or overweight (BMI 20–29.9 kg/m2) humans with biopsies from gluteal and abdominal sites mean adipocyte size correlated with electron microscopically-defined features of adipocyte death and “crown-like structures”, primarily in the obese subjects (13/19 biopsy samples vs 2/9 biopsy samples in lean subjects) (4). Larger adipose cell size has also been reported to predict insulin resistance and increased risk for type 2 diabetes, independent of body fat (13). Thus, while it has been generally accepted that larger adipose cells may contribute to insulin resistance via secretion of inflammatory cytokines and attraction of inflammatory macrophages, (5), our results do not confirm this hypothesis.
Differences between our findings and those others are most likely due to differences in subject populations. Our relatively homogeneous group of subjects had a relatively narrow BMI range, excluding both normal weight and morbidly obese subjects. Importantly, they were all required to have stable body weight. Prior studies on overfed mice may not be representative of weight-stable, moderately-obese or overweight humans, as these mice were extremely-obese and in a state of weight gain, with mean adipocyte diameter increased many-fold in comparison to the controls, whereas our moderately-obese individuals had large cell diameters that were only about 40% greater than those described in lean individuals (14). The prior human study showed inflammation surrounding large adipocytes primarily in individuals with BMI 30–45 kg/m2. Indeed, in a previous study evaluating inflammation in subcutaneous adipose tissue of moderately-obese humans, we found only one subject out of 16 to have crown like structures, and this individual was IS rather than IR (15). Another potential reason for discordant findings is the use of the Beckman Multisizer methodology, which allowed us to look beyond cell size as estimated from a small portion of tissue visualized microscopically, or mean cell size estimated from total lipid extracted and total cell count, which could be expected to yield erroneous conclusions if cell size is distributed in a bimodal fashion, as we have previously shown (7). Our results were adjusted statistically for insulin sensitivity, gender and BMI, which changed the results to only a small degree, but importantly, confirmed that the observed lack of association between peak cell diameter and inflammation, was free of confounding.
Turning now to the second major finding of this study, there was a consistent positive association between increased fraction of small adipose cells and inflammation in adipose tissue. This association was independent of insulin resistance, although it appears that insulin resistance status modified the relationship for at least two of the five associated genes. Specifically, formal testing for interaction was significant or borderline for EMR-1 and CD14, respectively, and when IR and IS subgroups were analyzed separately, the relationship between fraction of small cells and inflammation was significant only among the IR subgroup for all five associated genes. If the study had been larger, it is possible that the interaction terms would have reached statistical significance for CD14 and possibly other genes. This relationship should be interpreted in the context of prior data showing that among moderately-obese individuals, inflammatory markers and macrophage density are higher in the IR than the equally-obese IS subgroup (15). Thus, it is important that we not only confirmed our prior findings in a smaller subject sample, but that we were able to ascertain that the relationship between inflammation and small adipose cell fraction was independent of insulin resistance. Thus, it appears that an increased proportion of small adipose cells is associated not only with insulin resistance (7), but also with inflammation, which in turn, is associated with insulin resistance (15).
The significance of the increased proportion of small cells is not clear. In our prior analysis showing that an IR subgroup of moderately-obese subjects was characterized by an enhanced proportion of small adipose cells as compared with IS BMI-matched controls (7), gene expression of differentiation markers was also decreased in the IR group, leading us to conclude that the small cells represented immature cells with impaired ability to store triglyceride and function properly in energy storage/mobilization. Other human studies also provide support for the notion that impaired adipogenesis characterizes the insulin resistant state. First, two studies have demonstrated decreased expression of differentiation and/or fat storage genes in individuals with type 2 diabetes (16) or insulin resistance (17) as compared with healthy controls. Second, two studies conducted in patients with HIV lipodystrophy showed that compared to non-lipodystrophic controls, subcutaneous adipose tissue was characterized by decreased expression of differentiation genes and increased expression of inflammatory genes, which in turn, were significantly inversely associated (18,19). Thus, one interpretation of the current results is that impaired differentiation of adipocytes is associated with inflammation in subcutaneous adipose tissue, which in turn is related to inflammation, possibly reflected in the relatively stronger associations observed in our IR versus IS subjects.
In-vitro studies also support a relationship between inflammation and impaired adipocyte differentiation and fat storage. For example, incubation of 3T3-L1 preadipocytes with IL-6 or TNF-α impairs terminal differentiation and results in decreased triglyceride storage. Preadipocytes accumulate little lipid in the setting of IL-6 and almost no lipid in the setting of TNF- α, maintaining a fibroblast-like appearance and exhibiting a higher migration rate leading to cluster formations, consistent with a proliferative state (20). In a study by Gustafson et al, CEBP-α was significantly decreased, consistent with cells remaining in a proliferative state, while markers of terminal differentiation were decreased and inflammatory cytokines (MCP-1, IL-6) were increased. These results suggest that inflammation in adipose tissue may incite a cascade leading not only to decreased terminal differentiation, but also to increased inflammatory activity of adipocytes. This notion is supported by the demonstration that when 3T3-L1 adipocytes are co-cultured with monocytes, secretion of TNF- α from the monocytes stimulates elaboration of IL-6, IL-8, MCP-1, and TNF- α from the adipocytes, a process that is prevented in the presence of TNF-α blocking antibody (21).
An alternative explanation for the association between small adipose cells and inflammation in subcutaneous adipose tissue is that either the small or large adipose cells are acting in a proinflammatory manner. While one might conclude that the small cells may be dysfunctional and inflammatory in nature, given the observed association between proportion of small cells and inflammatory gene expression, it is also possible that increased proportion of small cells reflects impaired adipogenesis, placing storage stress on the existing large cells, which in turn incites inflammation. In this context it is important to note that the small cells are adipose cells and not macrophages, as evidenced from a prior investigation in which we isolated adipose cells by flotation and continued to observe the bimodal distribution by Multisizer analysis (7). Furthermore, the aperture range for Multisizer analysis excludes cells with diameter less than 20 um, essentially excluding cells of monocyte lineage, and electron microscopy demonstrates even the small cells to be smooth and round, with contrasting ruggated-appearing [Is ruggated a word? Do you mean corrugated? - AS] macrophages adherent to the surfaces of the larger cells. While our results do not make clear whether inflammation in adipose tissue is a product of monocyte/macrophages or adipocytokines, the data show an association between increased fraction of small adipose cells and inflammatory markers both nonspecific (IL-6, IL-8, MCP-1, EMR-1) and specific (CD14, CD45) for monocyte lineage cells. These findings complement a prior study in which we found upregulation of inflammatory genes, both specific and nonspecific for monocyte-lineage cells in association with insulin resistance (15). In histologic sections in the prior study, even the IR subjects exhibited relatively sparse macrophages, and only rare partial crown-like structures. On the other hand, Weisberg et al showed that inflammation in adipose tissue could be traced to cells of bone marrow origin, and thus were of monocyte lineage. In all, it is most likely that at least part of the inflammation in subcutaneous adipose tissue is attributable to adipose cells themselves. At this point, however, it is unclear whether large or small adipose cells are functioning differently in this respect, and to what extent monocyte-lineage cells are contributing. Further research is needed to clarify this possibility.
Finally, it is important to consider how the observed relationships in the current study relate to insulin resistance. In this regard, the current findings corroborate and extend our prior findings in 29 women, that inflammation in subcutaneous adipose tissue is associated with insulin resistance independent of obesity (15). The current study includes men, again demonstrating a consistent relationship between insulin resistance and inflammation that is independent of BMI, and also of gender. Importantly, our prior study did not include cell size data, whereas the current study, by adjusting for potential confounding by adipose cell size or fraction of small cells, demonstrates that the relationship between insulin resistance and inflammation is not merely a function of differences in these cell size parameters. Conversely, we have found a relationship between cell-size parameters and inflammation, independent of insulin resistance. We have thus now demonstrated relationships between two characteristics of adipose tissue – increased proportion of small cells, and inflammation, that are each significantly associated with insulin resistance (7,15). The current results show that these two variables are also independently associated with each other. A triad of insulin resistance, inflammation, and increased proportion of small cells is thus suggested, although the directionality of the relationships is not clear.
This study has several limitations. First, it would have been instructive to compare inflammatory markers in small versus large adipose cells, but separation of cells by size requires larger amounts of tissue than we were able to obtain in this study. There is one published paper, however, in which the authors were able to separate large and small adipose cells, showing differential gene expression, supporting the notion that gene expression, and presumably function, vary with size of adipose cells (22). To our knowledge, no studies of this nature have addressed inflammatory or differentiation genes specifically. Secondly, we did not measure differentiation markers in this analysis, because we did not set out to evaluate differentiation. In a prior study, however, we showed that compared to IS subjects, IR subjects demonstrated decreased expression of differentiation markers as well as an increased proportion of small adipose cells, indicating that impairment of adipose cell differentiation characterizes the IR state. Finally, our study is largely limited to Caucasian subjects, and it would be interesting to determine whether the observed relationships persist in larger cohorts of other races. Importantly, we were able to assess both females and males, and adjust for rigorously quantified level of insulin resistance, demonstrating that gender does not appear to play a role in the relationships between adipose cell size and inflammatory gene expression, and that both small cell fraction and insulin resistance are independently associated with inflammation.
In conclusion, inflammation in subcutaneous adipose tissue appears not to be associated with increased size of mature adipose cells, but rather, with a relative predominance of cells in the “small-cell” phase of development. When considered in the context of prior findings that inflammation is associated with insulin resistance, and that a predominance of small cells is also associated with insulin resistance, we can begin to formulate hypotheses about the biological properties of adipose tissue that contribute to the development of insulin resistance. Specifically, our results support the possibilities that 1) inflammation impairs differentiation and/or maturation of preadipocytes into mature adipose cells; 2) that individuals with impaired adipose cell differentiation develop inflammation; 3) that the small adipose cells demonstrate biological activity similar to inflammatory cells. Future work addressing these possibilities, including studies separating large and small adipose cells, will shed further light on these emerging concepts regarding the metabolic and morphologic mechanisms by which subcutaneous adipose tissue can promote insulin resistance independent of total body mass.
Acknowledgements
Funding for this study was provided by study grants: NIH/NIDDK 1 R01 DK071309–01, 5RO1DK071333, 5K23 RR16071, and NIH RR 000070, and by the NIDDK Intramural Research Program.
ClinicalTrials.gov Identifier: NCT00285844
Abbreviations:
- BMI
body mass index
- CD
cluster of differentiation
- CT
cycle threshold
- EMR1
egf-like module containing mulin-like, hormone receptor-like 1
- IL-6
interleukin-6
- IL-8
interleukin-8
- iNOS
inducible nitric oxide synthase
- IR
insulin resistant
- IS
insulin sensitive
- IST
insulin suppression test
- MMLV
Maloney Murine Leukemia Virus
- MCP-1
monocyte chemotactic protein-1
- rtPCR
real-time polymerase chain reaction
- SSPG
steady-state plasma glucose
- SSPI
steady-state plasma insulin
- TNF-α
tumor necrosis factor-alpha
Footnotes
Duality of Interest Statement
There are no dualities of interest on the part of the authors.
References
- 1.McLaughlin T, Abbasi F, Lamendola C, Reaven G. (2006) Heterogeneity in prevalence of risk factors for cardiovascular disease and type 2 diabetes in obese individuals: impact of differences in insulin sensitivity. Arch Intern Med 167:642–8. [DOI] [PubMed] [Google Scholar]
- 2.Jacobsson B, Smith U (1972) Effect of cell size on lipolysis and antilipolytic action of insulin in human fat cells. J Lipid Res 13:651–656. [PubMed] [Google Scholar]
- 3.Ostman J, Backman L, Hallberg D (1975) Cell size and the antilipolytic effect of insulin in human subcutaneous adipose tissue. Diabetologia 11:159–164. [DOI] [PubMed] [Google Scholar]
- 4.Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, Wang S, Fortier M, Greenberg AS, Obin MS (2005) Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res. 46:2347–55. [DOI] [PubMed] [Google Scholar]
- 5.Weller KE, Hotamisligil GS (2003) Obesity-induced inflammatory changes in adipose tissue. J Clin Invest 112:1785–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisberg S, McCann D, Desai M, Rosenbaum M, Leibel R, and Ferrante A. (2003). Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112: 1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLaughlin T, Sherman A, Tsao P, Gonzales O, Yee G, Lamendola C, Reaven G, Cushman S. (2007). Diabetologia 50:1707–15. [DOI] [PubMed] [Google Scholar]
- 8.Pei D, Jones CNO, Bhargava R, Chen Y-DI, Reaven GM (1994) Evaluation of octreotide to assess insulin-mediated glucose disposal by the insulin suppression test. Diabetologia 37:843–5. [DOI] [PubMed] [Google Scholar]
- 9.Shen S-W, Reaven GM, and Farquhar JW (1970) Comparison of impedance to insulin mediated glucose uptake in normal and diabetic subjects. J Clin Invest; 49:2151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenfield MS, Doberne L, Kraemer FB, Tobey TA, and Reaven GM (1981) Assessment of insulin resistance with the insulin suppression test and the euglycemic clamp. Diabetes; 30:387–392.24. [DOI] [PubMed] [Google Scholar]
- 11.Carantoni M, Abbasi F, Chu L, Chen YD, Reaven GM, Tsao PS, Varasteh B, Cooke JP. (1997) Adherence of mononuclear cells to endothelium in vitro is increased in patients with NIDDM. Diabetes Care; 20:1462–1465. [DOI] [PubMed] [Google Scholar]
- 12.Stern JS, Batchelor BR, Hollander N, Cohn CK, Hirsch J (1972) Adipose cell size and immunoreactive insulin levels in obese and normal weight adults. Lancet; ii:948–51. [DOI] [PubMed] [Google Scholar]
- 13.Weyer C, Foley JE, Bogardus C, Tataranni PA, Pratley RE. (2000) Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia; 43: 1498–506. [DOI] [PubMed] [Google Scholar]
- 14.Krotkiewski M, Bjorntorp P, Sjostrom L, Smith U. (1983) Impact of obesity on metabolism in men and women: importance of regional adipose tissue distribution. J Clin Invest 1983; 72:1150–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin T, Deng A, Gonzales O, Aillaud M, Yee G, Lamendola C, Abbasi F, Connolly AJ, Sherman A, Cushman SW, Reaven G, Tsao PS. (2008) Insulin resistance is associated with a modest increase in inflammation in subcutaneous adipose tissue of moderately obese women. Diabetologia; 51:2303–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Jansson PA, Nagaev I, et al. (2004) Evidence of impaired adipogenesis in insulin resistance. BBRC 317: 1045–1051. [DOI] [PubMed] [Google Scholar]
- 17.Dubois S, Heilbronn LK, Smith SR, Albu JB, Kelley ER. (2006) Decreased expression of adipogenic genes in obese subjects with type 2 diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jan V, Cervera P, Maachi M, et al. (2004) Altered fat differentiation and adipocytokine expression are inter-related and linked to morphological changes and insulin resistance in HIV-1 infected lipodystrophic patients. Antivir Ther. 9:555–564. [PubMed] [Google Scholar]
- 19.Bastard JP, Caron M, Vidal H, et al. (2002) Association between altered expression of adipogenic factor SREBP1 in lipoatrophic adipose tissue from HIV-1-infected patients and abnormal adipocyte differentiation and insulin resistance. Lancet 359:1026–1031. [DOI] [PubMed] [Google Scholar]
- 20.Gustafson B, Smith U (2006) Cytokines promote WNT signaling and inflammation and impair the normal differentiation and lipid accumulation in 3T3-L1 preadipocytes. J Biol Chem. 281: 9507–16. [DOI] [PubMed] [Google Scholar]
- 21.Suganami T, Nishida J, Ogawa Y. (2005) A paracrine loop between adipocytes and macrophages aggravatesinflammatory changes. Arterioscler Thromb Vasc Biol. 25:2062–2068. [DOI] [PubMed] [Google Scholar]
- 22.Jernas M, Palming J, Sjoholm K, Jennische E, et al. (2006) Separation of human adipocytes by size: hypertrophic fat cells display distinct gene expression. FASEB J. 20:E832–E839. [DOI] [PubMed] [Google Scholar]