Abstract
Diminished microRNA-29b levels have recently been revealed to provoke increased expression and accumulation of extracellular matrix molecules, such as collagens in fibrotic remodeling. Subsequently, the aim of this study was to find out whether microRNA-29b might also regulate human xylosyltransferase (XT)-I expression. XT-I has been characterized previously as a fibrosis biomarker catalyzing the key step of proteoglycan biosynthesis. While we demonstrate that XYLT1 is neither a target of microRNA-29b identified in silico nor a direct 3′ untranslated region binding partner of microRNA-29b, transfection of normal human dermal fibroblasts with microRNA-29b inhibitor strongly increased XYLT1 mRNA expression and XT activity. Combined results of the target prediction analysis and additional transfection experiments pointed out that microRNA-29b exerts indirect influence on XT-I by targeting the transcription factor specificity protein 1 (Sp1). We could confirm our hypothesis due to the decrease in XYLT1 promoter activity after Sp1 binding site mutation and the approval of occupancy of these binding sites by Sp1 in vitro. Taken together, a hitherto unidentified pathway of XT-I regulation via microRNA-29b/Sp1 was determined in this study. Our observations will facilitate the understanding of complex molecular fibrotic pathways and provide new opportunities to investigate microRNA-based antifibrotic tools.
Introduction
MicroRNAs (miRNAs) are small endogenous noncoding RNA molecules, which consist of 18 to 25 nucleotides. Together with an RNA-induced silencing protein complex, a miRNA interacts with the 3′ untranslated region (3′UTR) of its target mRNAs. In the case of perfect base pairing between the miRNA and target sequence, the mRNA is degraded. Contrarily, a repression of translation takes place in the case of imperfect base pairing. Therefore, miRNAs represent a cellular tool for posttranscriptional regulation1,2.
As published earlier, 1 to 5% of the human genome code for miRNAs, whereby each miRNA can bind several target mRNAs3. Many diseases have been associated with abnormal miRNA expression profiles, so that miRNAs are not only used as biomarkers, but also handled as potential therapeutic targets to exert influence on dysregulated molecular pathways4. Several miRNA mimics or antimiRs have currently reached clinical trials, whereby recent advances in technologies to deliver miRNAs in vivo will assure a safe and targeted application of miRNA therapeutics in the future5.
One type of disease associated with abnormal miRNA expression is fibrosis6,7. Fibrotic remodeling occurs in different tissues and is characterized by an excessive cellular synthesis of extracellular matrix (ECM) molecules promoting scar tissue formation. Although many underlying molecular processes are well described to date, a therapeutic approach has not yet been established. Among others, current studies deal with the differentiation of fibroblasts to matrix synthesizing contractile myofibroblasts, which were identified as the key mediator cells of fibrotization. In comparison to physiological wound healing processes, myofibroblasts in fibrotic tissues do not become apoptotic after scar tissue formation but persist producing ECM due to the uninterrupted stimulation with fibrotic cytokines, such as transforming growth factor-β1 (TGF-β1). Consequently, an imbalance of ECM synthesis promotes tissue stiffening, damage and functional organ deficit8,9.
It has been revealed recently that several miRNAs, such as miRNA-21, miRNA-145 or miRNA-29b, are expressed aberrantly in fibrotic diseases. Zhou et al.10 found that the miRNA-21 level in the serum of systemic sclerosis (SSc) is highly induced, while they identified 21 miRNAs which are differentially expressed in SSc skin tissues. In line with these observations, an attenuation of unilateral ureteral obstruction-induced renal fibrosis after blocking miRNA-21 in vivo was reported11. miRNA-145 expression was demonstrated to be upregulated in the tissue of idiopathic pulmonary fibrosis patients and in lung fibroblasts treated with TGF-β1. A miRNA-145 deficiency was further evaluated to be protective against bleomycin-induced lung fibrosis in a mouse model12.
In this study, we focus on miRNA-29b, which is strongly downregulated in various fibrotic tissues13–15. The miRNA-29 family decreases the expression of collagens and other ECM components directly; consequently, these are increased in fibrotic tissues, such as skin sections of SSc patients16. miRNA-29b was also shown to be diminished in SSc serum exosomes, which is why miRNA-29b was recently defined as a key regulator of SSc17.
The challenge in developing new anti-fibrotic tools is to understand the underlying molecular pathways of fibrotization and translate these molecular discoveries into targeted therapeutic drugs18. One putative downstream target of miRNA-29b we consider here is human xylosyltransferase (XT)-I (EC 2.4.2.26). XT-I is one of two human isoenzymes which catalyzes the rate-limiting step of proteoglycan (PG) glycosylation by transferring UDP-xylose to defined serine residues of a PG core protein19,20. Subsequent sugars are added by other glycosyltransferases, resulting finally in the formation of glycosaminoglycans21. PG and their linked glycosaminoglycan residues define, together with collagens and other structural components, the architecture of the ECM and are, therefore, involved in a wide range of cellular functions, including proliferation, signaling, development and adhesion22. Since XT are shed from the golgi membrane in the extracellular space, XT activity is not only the pacemaker of PG synthesis, but also a suitable fibrosis biomarker. Due to the shedding, 90% of XT activity is found in cell culture supernatants and only a minority of the enzyme remains in the cellular membrane. Thus XT activity quantified in cell culture supernatants is a useful link to XYLT mRNA expression quantified in cell lysates. However, at present it remains unclear whether an increase in XT activity is solely caused by increased translation and protein expression or whether other mechanisms as for instance mRNA stability or posttranslational modifications are involved, too.
Quantification of serum XT activity of liver fibrosis or SSc patients revealed a significantly increased XT activity, which goes along with the upregulated PG synthesis in fibrotic tissues23,24. On top of that, TGF-β1-induced upregulated XYLT1 mRNA expression and XT activity are associated with dermal myofibroblast differentiation and, therefore, qualify XT as an excellent cellular fibrosis biomarker similar to alpha smooth muscle actin expression25. Surprisingly, the coexistence of two XT isoenzymes has not yet been fully elucidated. In addition to several biochemical property differences described earlier, we only identified XT-I, but not XT-II, as being involved in fibrotic remodeling. However, we assume that XT-I expression is not only inducible by TGF-β1, but also might be regulated by hitherto unidentified molecular pathways providing the opportunity for therapeutic intervention.
Regarding this, no description of a XT-I regulation by miRNAs has been described or postulated to date. Therefore, the aim of this study was to find out whether one of the most important fibrosis-associated miRNAs, miRNA-29b, exerts influence on XYLT1 mRNA expression. We used normal human dermal fibroblasts (NHDF) of healthy controls, which have been shown previously to be an adequate cellular model system for the investigation of molecular processes in dermal fibrosis, to evaluate this issue. The results of our study reveal for the first time that XT-I is indirectly upregulated via the miRNA-29b/Sp1 pathway in fibrosis.
Results
TGF-β1 supplementation downregulates miRNA-29b expression in NHDF
According to the literature, miRNA-29b expression is downregulated in various cell culture models after fibrotic supplementation of TGF-β126. Gene expression analysis using a miRNA PCR array was performed to verify this in our myofibroblast cultivation system. We found that miRNA-29b expression was 0.4-fold diminished in NHDF treated with TGF-β1 compared to controls (data not shown). This result was confirmed by a Taqman-based real-time PCR assay, which revealed a 0.5-fold reduction of miRNA-29b expression in NHDF treated with TGF-β1 compared to untreated controls (Fig. 1). Thus, our cell culture system represents a reliable experimental setting to study a putative linkage between TGF-β1-induced miRNA-29b downregulation and TGF-β1-induced XT-I upregulation. The latter has been described recently25.
Figure 1.
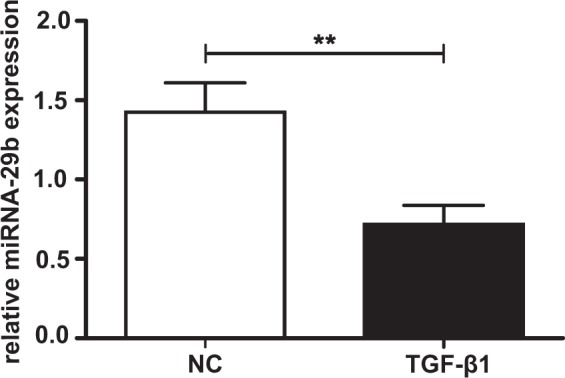
Effect of TGF-β1 supplementation on miRNA-29b expression in NHDF. NHDF cell lines (n = 2) were seeded (50 cells/mm²) and incubated for 24 h. Thereafter, a serum withdrawal (0.1% FCS) lasting 24 h was performed. Finally, the cell culture medium was supplemented with TGF-β1 (5 ng/mL) or PBS (negative control, NC), respectively. After 48 h, miRNA-29b expression was quantified by using Taqman probes in a quantitative real-time PCR. miRNA-29b expression was normalized to the appropriate expression level of the housekeeping gene miRNA-191 and is expressed as a ratio to one cell line. Data are presented as means +/− SEM with corresponding standard error for three biological replicates per cell line. **p < 0.01 (Mann-Whitney U test).
In silico analysis depicts a miRNA-29b-mediated mRNA expression regulation of various fibrosis-associated genes, except XYLT1
Since fibrotic tissue remodeling is accompanied by TGF-β1-induced changes in miRNA-29b and XYLT1 expression, our aim was to investigate whether XYLT1 might be regulated directly by miRNA-29b. This was evaluated by analyzing the predicted targets of miRNA-29b in silico. The comparison of results of three independent databases indicated several congruent target mRNAs (Table 1). Due to the high number of putative targets predicted by the software tools, a manual selection of ECM- and fibrosis-related targets was accomplished. All of the targets listed in Table 1 have been associated with fibrotic remodeling previously, such as collagens, fibrillin 1 and transcription factors (Sp1 and Krueppel-like factors)27,28. Remarkably, none of the in silico analyses predicted XYLT1 mRNA to be a target of miRNA-29b. However, it was shown earlier that the XYLT1 promoter region harbors several Sp1 binding sites29,30. Thus, a putative XYLT1 regulation via the miRNA-29b/Sp1 pathway had to be elucidated.
Table 1.
List of targets of miRNA-29b identified in silico.
| Target | PicTar | TargetScan | DIANA mT |
|---|---|---|---|
| COL11A1 | 17.93 | −1.24 | 0.98924 |
| COL1A1 | 10.92 | 0.96684 | |
| COL3A1 | 8.24 | −1.71 | 0.99989 |
| COL4A1 | 10.03 | −0.70 | 0.99999 |
| COL5A2 | 7.07 | 0.98480 | |
| COL7A1 | 7.47 | −0.59 | 0.99990 |
| FBN1 | 8.23 | ||
| KLF13 | 17.00 | ||
| SP1 | 15.00 | −0.18 |
Three independent software tools (Target Scan, PicTar and DIANA mT) were used to identify the targets of miRNA-29b. A manual selection of ECM- or fibrosis-related targets is listed (left column: COL11A1 collagen type 11 α 1, COL1A1 collagen type 1 α 1, COL3A1 collagen type 3 α 1, COL4A1 collagen type 4 α 1, COL5A1 collagen type 5 α 1, COL7A1 collagen type 7 α 1, FBN1 fibrillin 1, KLF13 Krueppel-like factor 13, SP1 spec ificity protein 1. Each target prediction tool uses its own mathematical algorithm which describes the binding intensity of miRNA to mRNA in terms of an indicated score63–65. Concerning Target Scan, a more negative score represents the best binding conditions, while a high score of PicTar and DIANA mT suggests strong miRNA binding to the appropriate mRNA.
miRNA-29b does not regulate XYLT1 mRNA expression via binding to the XYLT1 3′UTR
SW1353 chondrosarcoma cells were transfected with miRNA-29b and luciferase plasmids coding for the 3′UTR of XYLT1 to further examine whether miRNA-29b binds directly to the 3′UTR of XYLT1 in vitro. Quantified firefly luciferase activity, which is influenced by miRNA-29b binding to the XYLT1 3′UTR, was normalized to constantly expressed renilla luciferase activity. By comparing the relative luciferase activity of cell lysates transfected with miRNA-29b or a negative control miRNA, it could be demonstrated that miRNA-29b transfection did not exert any influence on relative luciferase activity. Thus, miRNA-29b does not bind directly to the XYLT1 3′UTR (Fig. 2).
Figure 2.
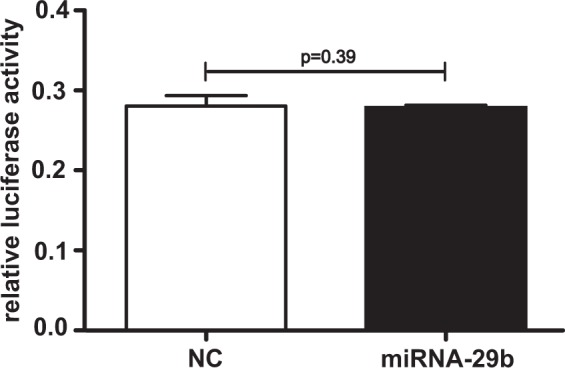
Evaluation of direct miRNA-29b binding to the XYLT1 3′UTR. SW1353 cells were seeded and incubated for 24 h. Cells were transfected with XYLT1 3′UTR coding luciferase vectors and miRNA-29b or a negative control miRNA (NC). Cell lysates were harvested after 48 h, and firefly luciferase activity and renilla luciferase activity were determined. Changes in relative luciferase activity resemble the putative interaction of miRNA-29b with the 3′UTR of XYLT1 mRNA. Results are expressed as means +/− SEM for six biological replicates (Mann-Whitney U test).
XYLT1 mRNA expression and XT activity are regulated indirectly by miRNA-29b
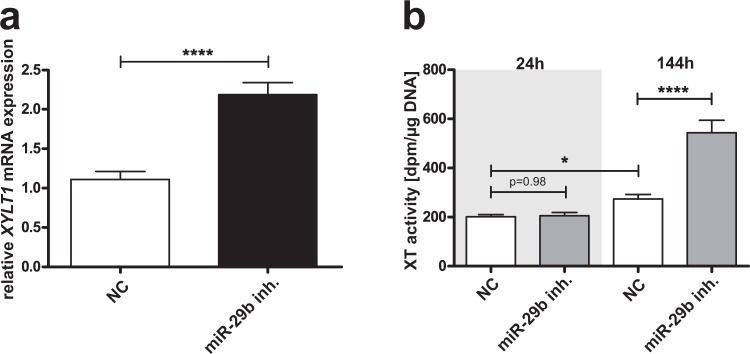
Although no direct binding of miRNA-29b to the XYLT1 mRNA 3′UTR could be observed, we analyzed whether an indirect regulation might participate in abnormal XT regulation in fibrosis. We conducted several cell culture experiments to determine this. At first, NHDF were transfected with a miRNA-29b inhibitor to resemble the miRNA suppression in fibrosis. XYLT1 mRNA expression was 2.0-fold increased after treatment in comparison to controls (NC, Fig. 3a). In accordance with the changes in XYLT1 mRNA expression detected, XT activity was also 2.0-fold increased in cell culture supernatants after 144 h of miRNA-29b inhibitor treatment compared to controls (Fig. 3b). No difference in XT activity between treated and untreated cells could be detected after 48 h. A slight 1.4-fold time-dependent increase in the XT activity of the controls originated from enzyme accumulation in the cell culture supernatant.
Figure 3.
Influence of miRNA-29b inhibitor on the XYLT1 mRNA expression level and XT activity in NHDF. NHDF cell lines (n = 2) were seeded and transfected with miRNA-29b inhibitor (miR-29b inh.) or a control miRNA (NC). The relative XYLT1 mRNA expression level was determined by quantitative real-time PCR after 48 h. Data were normalized to a normalization factor calculated from the geometric mean of HPRT, GAPDH and B2M mRNA expression levels. The values indicated are expressed as ratio to one cell line (a). The XT activity (b) was quantified after 48 and 144 h in cell culture supernatants by using a radiochemical enzymatic assay. The XT activity in dpm was referred to the DNA content of the appropriate cell lysate. Values are means +/− SEM for three biological replicates per cell line; *p < 0.05; ****p < 0.0001 (Mann-Whitney U test).
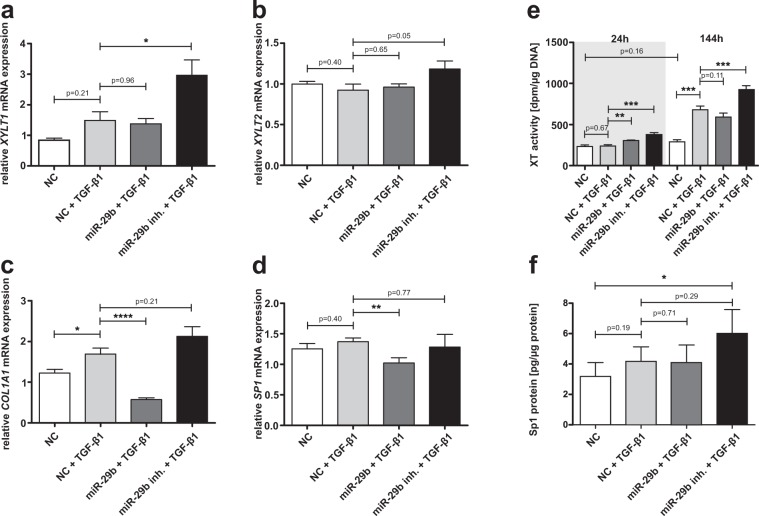
In a second experimental setting, we analyzed XYLT1 (Fig. 4a), XYLT2 (Fig. 4b), COL1A1 (Fig. 4c) and SP1 (Fig. 4d) mRNA expression levels as well as XT activity (Fig. 4e) and Sp1 protein content (Fig. 4f) after treating NHDF with miRNA-29b or its inhibitor under supplementation of TGF-β1 or a vehicle, respectively. It is possible using this approach to figure out any regulatory effects of miRNA-29b or its inhibitor which might strengthen or weaken the regulatory effect of TGF-β1 compared with a fibrotic state.
Figure 4.
Influence of the combinatory effect of miRNA-29b or miRNA-29b inhibitor with TGF-β1 on NHDF mRNA expression levels, XT activity and Sp1 protein expression. NHDF cell lines (n = 2) were seeded and after 24 h, transfection with a negative control miRNA (NC), miRNA-29b (miR-29b) or its inhibitor (miR-29b inh.) was performed. The cell culture medium was changed 6 h later and supplemented with TGF-β1 or vehicle (NC), as indicated. The cells were harvested 24 h after TGF-β1 supplementation for relative quantification of the mRNA expression levels of XYLT1 (a), XYLT2 (b), COL1A1 (c) and SP1 (d). Data were normalized to a normalization factor, determined by calculating the geometric mean of HPRT, GAPDH and B2M mRNA expression levels, and expressed as a ratio to one cell line. The XT activity (e) was quantified after 24 and 144 h in cell culture supernatants by using a radiochemical enzymatic assay. The XT activity in dpm was referred to the DNA content of the appropriate cell lysate. The Sp1 protein concentration (f) was measured in nuclear extracts via ELISA and referred to total nuclear protein content. Values are means ± SEM for three biological replicates per cell line *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 (Mann-Whitney U-test).
XYLT1 mRNA expression showed a slight 1.8-fold but not significant increase after supplementation of TGF-β1 compared to untreated cells. An additional supply of miRNA-29b did not exert any influence, but transfection with miRNA-29b inhibitor resulted in a significant 2.0-fold upregulation of XYLT1 mRNA expression. XYLT2 mRNA expression was not influenced by any treatment. Incubation of NHDF with TGF-β1 resulted in a significantly 1.4-fold upregulated COL1A1 mRNA expression level. TGF-β1 supplementation in parallel with miRNA-29b or miRNA-29b inhibitor transfection entailed a strong 0.3-fold reduction or a slight but not significant increase of the COL1A1 mRNA expression, respectively. Determination of SP1 mRNA expression only depicted a 0.8-fold downregulation after miRNA-29b transfection under supplementation of TGF-β1. Treatment solely with TGF-β1 or combined incubation with TGF-β1 and miRNA-29b inhibitor did not affect SP1 mRNA expression levels significantly. Changes in the XT activity resembled the effects observed in the XYLT1 mRNA expression level. Differences between the treatment groups were more pronounced after 144 h, because XT accumulate in the cell culture supernatant. We detected a strong 2.3-fold increase in XT activity after cultivating NHDF in the presence of TGF-β1 compared to controls. Additional transfection with miRNA-29b or its inhibitor was found to result in a slight but not significant decrease or a 1.4-fold significant induction of XT activity, respectively.
An ELISA was performed to determine differences in the Sp1 protein level. TGF-β1 treatment induced Sp1 protein content tendentially, but not significantly. Combined supplementation of TGF-β1 and miRNA-29b inhibitor resulted in a significant 1.9-fold increase of the Sp1 protein concentration in comparison to untreated cells. According to our hypothesis, this observation suggests an involvement of Sp1 in the miRNA-29b-mediated XYLT1 upregulation. It has been shown previously that Sp1 plays a crucial role in XYLT1 regulation and that miRNA-29b targets Sp1 in a fibrotic context30,31. However, we addressed the relevance of Sp1 transcription factor binding sites for XYLT1 expression and the occupancy of these transcription factor binding sites in vitro to further support our hypothesis of a fibrotic interplay of miRNA-29b, Sp1 and XT-I.
XYLT1 promoter activity is strongly influenced by the deletion of Sp1 transcription factor binding sites
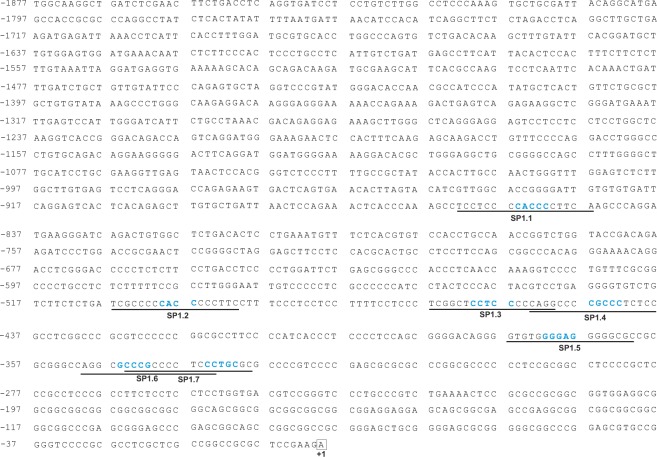
Since an indirect XYLT1 regulation through miRNA-29b was described here for the first time, we further evaluated whether Sp1 might be involved in this regulatory mechanism. Seven Sp1 binding sites (named SP1.1 to SP1.7), harboring a GC-rich binding motive, were identified in the XYLT1 promoter region, as shown in Fig. 5. Individual deletion of each Sp1 transcription factor binding site via site-directed mutagenesis was assessed by mutating the GC-rich binding motive to an AT-rich sequence in existing XYLT1 promoter coding luciferase vectors, as has been described previously32. SW1353 cells were transfected with the appropriate XYLT1 promoter construct and a dual luciferase assay was performed to analyze the influence of Sp1 transcription factor binding site deletion. Relative luciferase activity of diverse promoter constructs was compared with XYLT1 promoter activity of the wildtype promoter sequence. Each mutation of a Sp1 transcription factor binding site resulted in a decrease in relative promoter activity, as illustrated in Fig. 6. Promoter activity decreased significantly from 100% (wildtype construct) to 24.6% (SP1.1), 42.2% (SP1.3) and 54.2% (SP1.5), respectively. In summary, these results underline the relevance of Sp1 for the induction of XYLT1 expression.
Figure 5.
Complete XYLT1 promoter sequence containing the Sp1 transcription factor binding sites identified. The genomic DNA sequence of the human XYLT1 promoter region (GenBank Accession Number KM079589 and NG_015843.1) has been described recently29. Numbers on the left define the nucleotide position up- or downstream according to the translation initiation site ATG (c. +1) indicated in bold. Sp1 transcription factor binding sites identified in silico (named SP1.1 to SP1.7) are underlined, while nucleotides highlighted blue were modified by site-directed mutagenesis to delete the transcription factor binding site.
Figure 6.
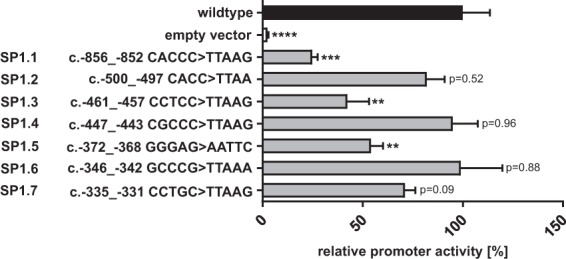
Quantification of XYLT1 promoter activity after site-directed mutagenesis of Sp1 transcription factor binding sites. The XYLT1 wildtype vector (pGL4.10 XYLT1 c. − 1639 to c. + 1complete)29 was used to delete Sp1 transcription factor binding sites by site-directed mutagenesis. Modified sequences are indicated on the left. SW1353 cells were transfected with the promoter plasmids and relative promoter activity was quantified by dual luciferase assay. Relative promoter activities are referred to the activity of the unmutated wildtype construct, which was defined as 100%. Values are means ± SEM of six biological replicates. **p < 0.01; ***p < 0.001; ****p < 0.0001 (Mann-Whitney U-test).
Sp1/Sp3 transcription factor binding sites in the XYLT1 promoter are occupied in vitro
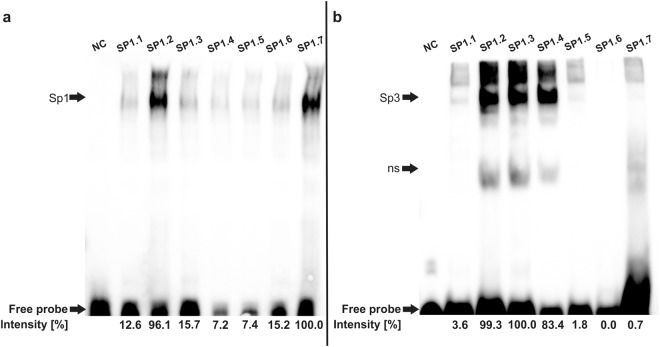
After evaluating the importance of Sp1 transcription factor binding sites for basal XYLT1 promoter activity, it was finally necessary to prove the occupation of the transcription factor binding sites by Sp1 protein. Since the Sp1 protein reveals high similarity with transcription factor Sp3, a shared occupancy of Sp1 binding sites with Sp1 and Sp3 is discussed33. An EMSA using a Sp1 or Sp3 protein was conducted to monitor not only Sp1, but also Sp3 binding to Sp1 binding sites in the XYLT1 promoter. Sp1 binds to all Sp1 binding sites of the XYLT1 promoter region, while the strongest binding was detected using an oligonucleotide probe resembling binding site SP1.2 (binding intensity 96.1%) or SP1.7 (binding intensity 100%), as depicted in Fig. 7. Contrarily, Sp3 bound strongly to binding sites SP1.2 (binding intensity 99.3%), Sp1.3 (binding intensity 100%) and Sp1.4 (binding intensity (83.4%). The remaining binding sites were not or only negligibly occupied by Sp3.
Figure 7.
Results of the electrophoretic mobility shift assay depicting Sp1 and Sp3 binding to Sp1 transcription factor binding sites identified in silico in the XYLT1 promoter region. Biotin-labeled DNA probes corresponding to Sp1 transcription factor binding sites SP1.1 to SP1.7 were incubated with recombinant Sp1 (a) or Sp3 (b) protein and analyzed on two independent gels (separated by the black line). In each line 1 (NC), an unlabeled DNA probe was used to demonstrate specificity of DNA-protein binding. Sp1 or Sp3 binding to the appropriate DNA probes was relatively quantified by using ImageJ while only a comparison between samples on one gel was performed. The strongest band in (a) or (b) represents the strongest binding of DNA to protein and was defined as 100%. ns: unspecific binding. Full-length gels (with total free probe band) are presented in Supplementary Figs S1 and S2.
Discussion
Fibrotic remodeling is characterized by ECM accumulation, tissue stiffening and progressive failure of the organs affected8,34. It has recently been investigated that fibrosis is not only associated with elevated TGF-β1 signaling, but also with aberrantly expressed miRNAs. miRNA-21, miRNA-29b and miRNA-145 are specifically suggested to play a crucial role in regulating fibrotic targets7,35.
Many studies are searching for suitable cellular points of application because no anti-fibrotic therapy has been established yet36. One of these putative targets is XT-I. Two XT isoforms, XT-I and XT-II, catalyze the rate-limiting step in PG glycosylation in humans19,20,37. Although biochemical studies revealed a strong functional similarity of both enzymes, TGF-β1 only induces XYLT1 mRNA expression38. Increased XYLT1 mRNA expression, XT activity and the resulting PG accumulation are associated with liver fibrosis23, cardiac fibrosis38, lung fibrosis39 and skin fibrosis24,25. Although many studies describe the involvement of miRNAs in pathobiochemistry, XT regulation by miRNAs has only been sparsely discussed so far. Theis et al.40 recently described an improvement of spinal cord injury recovery after miRNA-133b-mediated XYLT1 downregulation. Furthermore, altered XYLT1 expression was associated with abnormal circulating miRNA levels in patients suffering from pediatric astrocytomas41.
However, we report here on the first study dealing with miRNA-dependent regulation of XT-I in fibrosis – the most important disorder XT-I has been associated with. Only increasing knowledge about the regulation of fibrotic mediators, such as XT, might allow us to develop an anti-fibrotic treatment. Here, we focus on XT regulation via miRNA-29b, a miRNA which is aberrantly expressed in fibrosis of the kidney13, heart14, lung15 and liver26. miRNA-29b is a member of the highly conserved miRNA-29 family, which consists of three members: miRNA-29a, -29b and -29c, sharing an identical seed sequence13. Published data suggest a TGF-β1/SMAD3-mediated miRNA-29b suppression in fibrotic tissues. miRNA-29b is extensively discussed as a potential therapeutic target to heal fibrosis. Li et al.42 recently reported on the successful inhibition of endometrial fibrosis by using a miRNA-29 mimic in rats. In addition, the first clinical trial phase I (NCT02603224) using a cholesterol-conjugated miRNA-29-duplex was finished in 2017 and supports further investigation of miRNA-29b mimics as novel antifibrotic tools. We investigated for the first time whether miRNA-29b might not only be involved in fibrotic remodeling targeting collagens or fibrillin16,28, but also exert influence on XT-I expression. To examine this issue, we recently established a myofibroblast culture model, which is based on a low cellular density and supplementation with TGF-β125,43. TGF-β1 is the key mediator of fibrotic remodeling and could be demonstrated to induce XT activity during myofibroblast differentiation25. Unfortunately, several trials to inhibit TGF-β1 in an antifibrotic manner revealed that TGF-β1 is involved in too many orchestrating signal cascades and, consequently, its inhibition cannot be realized44.
Here, we describe a TGF-β1-induced downregulation of miRNA-29b expression in our cell culture model by using a PCR array. These PCR arrays represent a great tool for evaluating expression profiles, but it is controversial to quantify miRNA expression with a SYBR green-based assay due to methodical imprecision. Therefore, we confirmed our finding with a more sensitive Taqman-based assay45. TGF-β1-induced suppression of miRNA-29b expression has been reported previously46.
There are two ways in which a miRNA can exert influence on a target mRNA. On the one hand, the miRNA can bind directly to the 3′UTR, provoking mRNA decay or translational inhibition. On the other hand, indirect regulation via targeting a signaling molecule further targeting one or several mRNAs might be involved2. We could demonstrate here that XYLT1 is neither a target of miRNA-29b identified in silico nor an experimentally confirmed 3′UTR binding partner of miRNA-29b. Therefore, we can exclude a direct targeting of XYLT1 3′UTR by miRNA-29b. However, by transfecting NHDF with the miRNA-29b inhibitor resembling fibrosis, we revealed for the first time a strong induction of XYLT1 mRNA expression, which was verified by quantifying the XT activity. This important finding qualifies us to hypothesize that miRNA-29b mediates indirect XT-I regulation via one or several hitherto undescribed pathway(s) and mediator(s).
We and others have depicted that Sp1 is a direct target of miRNA-29b. Sp1 is a transcription factor expressed ubiquitously which harbors three zinc-finger DNA-binding motives and regulates the transcriptional intensity of many genes, especially housekeeping genes and genes with a TATA-less GC-rich promoter region, such as XYLT1 or XYLT232,47. Growing evidence reveals that Sp1 is not only a transcription factor which binds GC-rich motives and is involved in maintaining basal promoter activity, but also a key regulator of fibrotic remodeling48–50. Binding sites for the Sp1 family were identified in the promoter region of many ECM-coding genes and blocking of Sp1 inhibits ECM gene expression broadly in vitro and in vivo31. However, Sp1 binding can exert a repressive or inductive effect on promoter activity in response to physiologic and pathological stimuli47.
The SP1 mRNA binding score calculated was weaker than that of, for instance, COL1A1. This low target prediction score might indicate a weak binding. However, several studies have already examined and approved Sp1 regulation by miRNA-29b51,52. Müller et al.30 have pointed out previously that Sp1, in addition to the Ap1 family, drives XYLT1 expression. They quantified a decrease of XYLT1 mRNA expression after exposure of SW1353 cells to mithramycin, which prevents the binding of transcription factors to GC-rich DNA regions. These results were confirmed by Ye et al.53, who were able to reduce basal XYLT1 mRNA expression via treatment with Sp1 inhibitors, mithramycin supplementation or stable Sp1 knockdown in rat nucleus pulposus cells. These studies, therefore, underline the functional interaction of XT-I and Sp1 and give a hint regarding XT-I regulation via miRNA-29b and Sp1. We exposed NHDF to miRNA-29b or its inhibitor in parallel with TGF-β1 to verify our hypothesis of Sp1 involvement in miRNA-29b-mediated XT induction. We determined a TGF-β1-mediated XT induction, which has also been described previously25, which was significantly gradable with supplementation of a miRNA-29b inhibitor. This result approves our hypothesis of XT induction by miRNA-29b reduction. Interestingly, XYLT2 was regulated neither by TGF-β1 nor by miRNA-29b or its inhibitor. The lack of influence of TGF-β1 on XYLT2 has been described previously38.
The increase in COL1A1 mRNA expression observed after TGF-β1 treatment is in line with collagen accumulation in fibrotic tissues34 and underlines the functionality of our cell culture system. An additional cellular treatment with the miRNA-29b inhibitor resulted in a weak elevation of COL1A1 mRNA expression, while additional exposure to miRNA-29b entailed a strong diminishment of mRNA expression. Maurer et al.16 also describe a downregulation of COL1A1 mRNA expression after exposing SSc fibroblasts to miRNA-29a/b. This effect is attributed to the direct binding of miRNA-29 to the COL1A1 3′UTR. In line with our results, several published studies assess stronger effects in COL1A1 mRNA downregulation after miRNA-29 treatment than COL1A1 mRNA increase after miRNA-29b inhibitor treatment16. This might be due to the lack of a direct one-to-one stoichiometric relationship between the levels of miRNA and target gene expression, which was discussed by Li et al.46 who studied Sp1-mediated regulation of COL1A1 in the fibroblasts of a human tenon. While miRNA-29b entails mRNA degradation directly, a miRNA-29b knockdown, by using its inhibitor, only prevents mRNA degradation by miRNA-29b but does not increase the expression obviously. Therefore, independent of cell type and molecular environment, there is a multitude of other miRNAs or mediators that might exert influence on the COL1A1 mRNA expression intensity46.
Sp1 is known to be upregulated by TGF-β1. Nevertheless, apart from our observations, there are several studies which describe only a weak or no Sp1 induction after TGF-β1 treatment. Ogawa et al.51 demonstrate that miRNA-29b-induced COL1A1 suppression is mediated by its direct interaction with COL1A1 3′UTR and by its interference with Sp1 expression in human stellate cells. Notably, in this context, only a weak SP1 mRNA and protein expression reduction by miRNA-29b was detected, while TGF-β1 treatment did not exert influence on SP1 mRNA or protein expression. Contrarily, Sp1 protein is strongly increased in tubular epithelial cells after TGF-β1 supplementation13. Therefore, TGF-β1-mediated Sp1 induction seems to be generally weak and dependent on the cellular system. Vanden Oever et al.54 recently described type VII collagen downregulation in recessive dystrophic epidermolysis bullosa via miRNA-29b-mediated Sp1 repression. Comparable to the results gained here, SP1 mRNA expression was downregulated in a small but statistically significant extent by miRNA-29b transfection in dermal fibroblasts. In our study, the Sp1 protein level was not diminished by treating the cells with miRNA-29b. Contrarily, the Sp1 protein expression was significantly increased after combined TGF-β1 and miRNA-29b inhibitor treatment.
Thus, it can be hypothesized that Sp1 mediates a multitude of signaling pathways, although it is often only weakly regulated – called fine-tuning by Jiang et al.13. In summary, the results described here reveal indirect XT regulation via miRNA-29b and mention Sp1 as the putative missing link (Fig. 8). In order to determine this issue, we deleted Sp1 transcription factor binding sites and evaluated the influence on XYLT1 promoter activity, on the one hand, and proved the affinity of Sp1 protein to the binding sites identified in silico, on the other hand. We published the first successful amplification and sequencing of the complete XYLT1 promoter region in 201429 and pointed out that the XYLT1 human reference sequence had been incomplete until then. Here, we identified seven Sp1 transcription factor binding sites in the XYLT1 promoter region in silico and demonstrated in vitro that they are occupied. Deletion by site-directed mutagenesis indicated that every mutated binding site resulted in a diminished XYLT1 promoter activity. Different intensities of downregulation might possibly arise from the synergistic interaction of several transcription factors in response to the appropriate binding site. This phenomenon has already been shown concerning the XYLT2 promoter32. Khair et al.55 also performed mutation experiments of Sp1 binding sites in the XYLT1 promoter region. In contrast to our results, they only identified two binding sites and detected opposing effects of these binding sites on basal XYLT1 promoter activity. Differences in both studies might result from the reference sequence considered. Summing up, it was pointed out that Sp1 is an activating transcription factor for XT-I, which is involved in XT regulation via miRNA-29b, according to our hypothesis.
Figure 8.
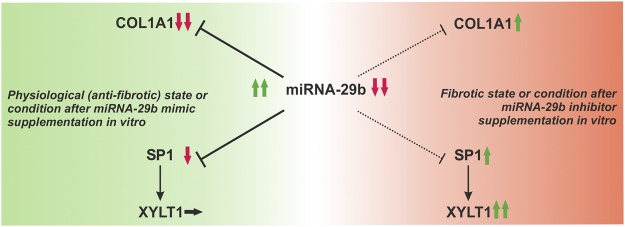
Hypothetical model of XYLT1 regulation via miRNA-29b. Physiologically, miRNA-29b suppresses the expression levels of COL1A1 and SP1, which are direct targets of the miRNA. Suppression of COL1A1 and SP1 goes along with anti-fibrotic remodeling, which is also achieved by using miRNA-29b mimics in vitro. Sp1 is known to stimulate XYLT1 mRNA expression, but the XYLT1 expression might not be affected by an SP1 decrease due to the availability of several other influencing transcription factors and mediators (left side). The miRNA-29b level is diminished in fibrosis. This state is reproducible by using miRNA-29b inhibitors in vitro. A decrease in miRNA-29b implies a minor inhibition of COL1A1 and SP1 mRNA expression, which can contribute to COL1A1 and SP1 upregulation. Sp1 upregulation provokes a XYLT1 induction (right side). Aside from XYLT1 regulation via Sp1, additional mechanisms might exist which have not yet been elucidated.
Surprisingly, it was assessed that a Sp1 knockdown did not exert influence on XYLT1 mRNA expression in SW1353 cells. We confirmed this finding because miRNA-29b-mediated Sp1 knockdown did not influence XYLT1 mRNA expression in NHDF. However, Ye et al.53 observed a strong diminishment of XYLT1 mRNA expression after stable Sp1 knockdown. Thus, Sp1-mediated XT regulation might depend on the cellular context. It is probable that a Sp1 knockdown leads to the recruitment and occupancy of Sp1 transcription factor binding sites by Sp3. Binding of Sp3 to several Sp1 binding sites, for example, in the XYLT1 promoter, has been shown here and elsewhere33. Concerning the regulatory potential of Sp1, it is expected that not only the cellular context and posttranslational Sp1 modifications, but also the molecular ratio of Sp1 to Sp3 play an important role. Additionally, Sp1 is known to interact with various other transcription factors, such as SMAD56, c-myc57 or c-jun58. Conflicting knowledge about the influence of Sp1 and Sp3 on XYLT1 promoter activity also emerges when looking at Sp3-mediated XYLT1 regulation. While Müller et al.30 and Ye et al.53 describe a decrease of XYLT1 mRNA expression indicating a positive effect on the expression level after Sp3 knockdown, Khair et al.55 propose a repressive function of Sp3 on the XYLT1 promoter in human primary chondrocytes. Hence, XYLT1 expression is influenced by Sp1 and Sp3 levels, which are, in turn, dependent on variable cellular environmental conditions.
In summary, our data suggest that miRNA-29b influences fibrotic induction of human XT-I through the Sp1 signaling pathway. This new finding does not only deepen our insights into pathobiochemical signaling in fibrosis, but also encourages us to develop targeted anti-fibrotic strategies in the future. Further studies will evaluate whether there are additional molecules, apart from Sp1, which mediate XT induction in fibrosis.
Materials and Methods
Fibroblast cultivation and treatment
Normal human dermal fibroblasts were purchased from Genlantis (San Diego, USA) and Coriell (Camden, USA). Unless otherwise stated, the NHDF were cultivated at 37 °C in a humidified atmosphere of 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, San Diego, USA) supplemented with 10% fetal calf serum (FCS; VWR, Radnor, USA), 1% antibiotic/antimycotic solution (PAN Biotech, Aidenbach, Germany) and 2% L-glutamine (PAN Biotech, Aidenbach, Germany). Cell culture medium was replaced twice a week and after reaching confluence, cells were expanded or used for experimental settings.
Cells were seeded (50 cells/mm2) and incubated for 24 h to evaluate the influence of TGF-β1 on the expression of miRNAs. Thereafter, cells were washed with phosphate-buffered saline (PBS; Thermo Fisher, San Diego, USA) and a serum withdrawal to 0.1% FCS was performed. After another 24 h, cell culture medium was replaced by medium containing 0.1% FCS and 5 ng/mL TGF-β1 (Miltenyi, Bergisch Gladbach, Germany) or PBS, respectively. Cell harvesting was executed after 48 h and cell lysates were stored at −80 °C. All treatments were performed in biological triplicates.
miRNA-29b inhibitor (UAGCACCAUUUGAAAUCAGUGUU, Thermo Fisher Scientific, San Diego, USA) or a negative control miRNA (final concentration of each miRNA: 50 nM) were transfected into NHDF using Lipofectamine 2000 (Thermo Fisher, San Diego, USA). The NHDF were seeded (50 cells/mm2) in DMEM without antibiotic/antimycotic solution in the intervening time of preparing transfection reactions. Cell culture medium was replaced 24 h after transfection. All treatments were performed in biological triplicates and transfection efficiency was determined using a FAM-labeled negative control siRNA (Thermo Fisher, San Diego, USA). Cells were harvested after 48 or 144 h and stored at −80 °C, while cell culture supernatants were stored at −20 °C.
Cells were seeded (50 cells/mm2) to analyze the effects of miRNAs under supplementation of TGF-β1. Cell culture medium was replaced with DMEM without antibiotic/antimycotic solution after 24 h. Transfection reactions (final concentration of each miRNA: 50 nM; miRNA-29b AACACUGAUUUCAAAUGGUGCUA) were prepared and added to the cells. Cell culture medium was replaced with DMEM containing PBS or TGF-β1 (5 ng/mL), respectively, 6 h post-transfection. Each treatment was seeded in biological triplicates and cells were harvested after 24 or 144 h. Cell lysates intended for gene expression analyses or Sp1 ELISA were stored at −80 °C.
Nucleic acid extraction
Cell lysates for quantification of miRNA expression via miScript miRNA HC polymerase chain reaction (PCR) array Human miFinder 384HC (Qiagen, Hilden, Germany) or TaqMan advanced miRNA assays (Thermo Fisher, San Diego, USA) were lysed in QIAzol Lysis Reagent. Biological replicates of lysates for the array were pooled. Total RNA was extracted according to the manufacturer’s description of the miRNeasy Micro Kit (Qiagen, Hilden, Germany).
Contrarily, total RNA extraction out of NHDF cell lysates to quantify mRNA expression levels via SYBR green-based real-time PCR was performed according to the manufacturer’s instructions using the NucleoSpin RNA Kit (Macherey-Nagel, Düren, Germany). Harvested cells were lysed in RA1 buffer and processed. An amount of 50 µL of the cell lysates was separated to quantify the total DNA content using the NucleoSpin Blood Kit (Macherey-Nagel, Düren, Germany). Nucleic acid concentrations were quantified using the NanoDrop 200 spectral photometer (Peqlab, Erlangen, Germany).
miRNA and mRNA expression analyses
A gene expression profile of 372 miRNAs was performed via quantitative real-time PCR using the miScript miRNA PCR Array Human miFinder 384HC (Qiagen, Hilden, Germany). After total RNA extraction, miRNAs were reverse transcribed according to the manufacturer’s instructions of the miScript II RT Kit (Qiagen, Hilden, Germany). As instructed by the manufacturer, a PCR mastermix was prepared using the miScript SYBR Green PCR Kit (Qiagen, Hilden, Germany) and added to the 384-well PCR array plate to be cycled as indicated. Data analyses of 372 target genes were calculated with a free data analysis software (miScript miRNA PCR Array Data Analysis, Qiagen, Hilden, Germany) using the delta-delta CT method.
Complementary to the PCR array, miRNA-29b expression was quantified using a Taqman probe. All reaction and incubation steps were performed according to the manufacturer’s instructions of the TaqMan Advanced miRNA cDNA Synthesis Kit (Thermo Fisher, San Diego, USA) and the TaqMan advanced miRNA Assay (Thermo Fisher, San Diego, USA). Each sample was measured in technical triplicates. miRNA-29b expression was normalized to the housekeeping gene miRNA-191 expression via the delta-delta CT method for data acquisition.
Relative quantification of mRNA expression levels with SYBR green-based real-time PCR was carried out, as has been described previously59. Briefly, 1 µg RNA was reverse transcribed to cDNA using the SuperScript II Reverse Transcriptase Kit (Thermo Fisher, San Diego, USA). Real-time PCRs were performed using SYBR Green Master Mix (Roche, Mannheim, Germany). Sequences of intron-spanning primers are listed in Table 2. Gene expression levels of XT-I (XYLT1), XT-II (XYLT2), collagen type 1 alpha 1 (COL1A1) and specificity protein 1 (SP1) were referred to a normalization factor calculated from mRNA expression levels of three housekeeping genes (β2-microglobuline (B2M), hypoxanthine phosphoribosyltransferase 1 (HPRT1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)). All samples were analyzed in technical triplicates.
Table 2.
List of PCR systems for quantitative real-time PCR.
| Gene | Protein | 5′-3′-sequence | TOA [°C] |
|---|---|---|---|
| hB2M | B2M | TGTGCTCGCGCTACTCTCTCTT CGGATGGATGAAACCCAGACA |
63.0 |
| hCOL1A1 | COL1A1 | GATGTGCCACTCTGACT GGGTTCTTGCTGATG |
63.0 |
| hGAPDH | GAPDH | AGGTCGGAGTCAACGGAT TCCTGGAAGATGGTGATG |
63.0 |
| hHPRT | HPRT | GCTGACCTGCTGGATTAC TGCGACCTTGACCATCTT |
63.0 |
| hSP1 | Sp1 | GAGCTACAGAGGCACAAACG CCTGGGCCTCCCTTCTTATTC |
63.0 |
| hXYLT1 | XT-I | GAAGCCGTGGTGAATCAG CGGTCAGCAAGGAAGTAG |
63.0 |
| hXYLT2 | XT-II | ACACAGATGACCCGCTTGTGG TTGGTGACCCGCAGGTTGTTG |
63.0 |
XT activity assay
Quantification of XT activity in cell culture supernatants has been described previously60,61. Taken together, the method relies on incorporation of the donor UDP-[14C]-xylose (Perkin Elmer, Foster City, USA) into silk fibroin protein acceptor. Quantified disintegrations per minute (dpm) reflect the XT activity, because the latter is proportional to UDP-[14C]-xylose incorporation. Each biological sample was measured in technical duplicates and referred to the total DNA content of the appropriate cell lysate.
Quantification of the SP1 protein expression by enzyme-linked immunosorbent assay
The Sp1 protein content in NHDF lysates was quantified using a commercially available ELISA kit (Cloud-Clone Corp., Katy, USA). Isolation of nuclear proteins was performed using the NE-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo Fisher, San Diego, USA), according to the manufacturer’s protocol. Absolut Sp1 protein amount was referred to total nuclear protein content. The experiment was performed twice and there was one technical replicate for each biological sample.
Verification of miRNA-29b binding to the 3′ untranslated region of XYLT1
Human chondrosarcoma SW1353 cells, purchased from ATCC (Manassas, USA), were seeded in a cell density of 180,000 cells/6-well to analyze the potential direct binding of miRNA-29b to the XYLT1 3′UTR. Unless otherwise stated, SW1353 cells were cultivated at 37 °C in a humidified atmosphere of 5% CO2 in RPMI 1640 medium containing 10% FCS and 1% antibiotic/antimycotic solution. The cell culture medium was replaced twice a week and, after reaching confluence, the cells were expanded or used for experimental settings. After 24 h, the cell culture medium was exchanged for medium without antibiotic/antimycotic solution. A negative control miRNA or miRNA-29b (final concentration 50 nM) and 50 ng of XYLT1-3′UTR coding plasmids (HmiT016805a-MT06/HmiT016805b-MT06; GeneCopoeia, Rockville, USA) was transfected into the cells using Fugene 6 (Promega, Mannheim, Germany). Cell culture medium was replaced 24 h later. The cells were lysed in lysis buffer, according to the manufacturer’s instructions of the Luc-Pair Duo-Luciferase Assay Kit 2.0 (GeneCopoeia, Rockville, USA) 48 h after transfection and stored at −80 °C. Each transfection reaction was performed in six biological replicates. Quantification of the constitutive renilla luciferase activity and XYLT1-3′UTR-regulated firefly luciferase activity was performed on a Tecan Reader infinite 200 Pro (Tecan Group, Männedorf, Swiss) and firefly luciferase activity was normalized to renilla luciferase activity.
Site-directed mutagenesis of Sp1 transcription factor binding sites in the XYLT1 promoter region
Cloning of the luciferase reporter vector pGL4.10 XYLT1 c. − 1639 to c. + 1complete containing the complete XYLT1 promoter region has been described previously29. Site-directed mutagenesis of the Sp1 transcription factor binding sites identified was performed using the QuikChange II XL Site Directed Mutagenesis Kit (Agilent, Santa Clara, USA), according to the manufacturer’s instructions. Four to six bases within an identified Sp1 transcription factor binding site were mutated to an adenine/thymine-rich DNA sequence, so that the Sp1 binding site disappeared. Successful mutations were verified by direct plasmid sequencing, as has been described previously29. Sequences of mutation and sequencing primers used are listed in Table 3.
Table 3.
List of site-directed mutagenesis (SDM) and sequencing primer sequences. TOA: optimal annealing temperature.
| Primersite | 5′-3′-sequence | TOA [°C] |
|---|---|---|
| Site-directed mutagenesis | ||
| SP1.1 | TCACCCAAAGCCTCCTCCCTTAAGCTTCAAGCCCAGGATGAA TTCATCCTGGGCTTGAAGCTTAAGGGAGGAGGCTTTGGGTGA |
60.0 |
| SP1.2 | TGTCTTCTCTGATCGCCCCTTAACCCTTCCTTTCCCTCCTCC GGAGGAGGGAAAGGAAGGGTTAAGGGGCGATCAGAGAAGACA |
60.0 |
| SP1.3 | TCCTTTTCCTCCCTCGGCCTTAAGCCCAGGCCCCGCCCTCTC GAGAGGGCGGGGCCTGGGCTTAAGGCCGAGGGAGGAAAAGG |
60.0 |
| SP1.4 | CGGCTCCTCCCCCAGGCCCTTAAGTCTCCGCCTCGGCCCGCG CGCGGGCCGAGGCGGAGACTTAAGGGCCTGGGGGAGGAGCCG |
68.0 |
| SP1.5 | CAGCGGGGACAGGGGTGTGAATTCGGGGCGCCGCGCGGGCCA TGGCCCGCGCGGCGCCCCGAATTCACACCCCTGTCCCCGCTG |
68.0 |
| SP1.6 | GGCGCCGCGCGGGCCAGGCTTAAACCCCTCCCTGCGCGCCCC GGGGCGCGCAGGGAGGGGTTTAAGCCTGGCCCGCGCGGCGCC |
68.0 |
| SP1.7 | GGCCAGGCGCCCGCCCCTCTTAAGGCGCCCCGTCCCCGAGCG CGCTCGGGGACGGGGCGCCTTAAGAGGGGCGGGCGCCTGGCC |
68.0 |
| Sequencing | ||
| XYLT1 _A | CCCTGTTTCGCGGCCCCTG | 58.8 |
| XYLT1 _B | CCCCATCCTACTCCCACTAC | 58.8 |
| XYLT1 _C | GAAGGAAAGGGAGGAGGAAA | 58.8 |
| pGL4.10 | CTTAATGTTTTTGGCATCTTCCA | 58.8 |
Quantification of XYLT1 promoter activity
As indicated above, SW1353 cells were seeded and incubated overnight. After 24 h, 1 µg of mutated firefly luciferase coding pGl4.10 plasmid and 10 ng of renilla luciferase coding pGl4.74 control plasmid were transfected into the cells using Fugene 629. Each of the mutated plasmids was amplified and, finally, transfected in six biological replicates. The medium was replaced after 24 h and cell harvesting was performed after a total of 48 h. Cell lysis and quantification of luciferase activity were conducted using the Dual Luciferase Reporter Assay System (Promega, Mannheim, Germany), as has been described previously, according to the manufacturer’s instructions29. Relative luciferase activity, resembling XYLT1 promoter activity, was calculated as the quotient of firefly/renilla luciferase activity of each sample.
Electrophoretic mobility shift assay (EMSA)
The binding of Sp1 or Sp3 to the transcription factor binding sites identified in silico was confirmed with 5′-biotinylated oligonucleotides (sequences listed in Table 4) which contained the putative Sp1 transcription factor binding sites of the XYLT1 promoter. Detection of the Sp1/3 DNA-protein complexes was performed using the LightShift chemiluminescent EMSA kit (Thermo Fisher, San Diego, USA). An amount of 1 μg recombinant Sp1 protein (OriGene, Rockville, USA) was incubated in a buffer containing 10 mM Tris, 50 mM KCl, 1 mM DTT (pH 7.5, binding buffer), 75 ng/µl poly(dI⋅dC), 0.25 mg/ml BSA and 40 fmol of the biotinylated, hybridized oligonucleotide. An amount of 1 μg of recombinant Sp3 protein (OriGene, Rockville, USA) was incubated in the same buffer to which was added 5% glycerol, 5 mM MgCl2 and 0.025% NP-40. The reaction mixture was left to incubate for 20 min at room temperature. Finally, the DNA-protein complexes were mixed with loading buffer and resolved on 6% polyacrylamide gel (Thermo Fisher, San Diego, USA) in 0.5x TBE buffer for 1.5 h at 100 V after a pre-run of the gel for 1 h at 100 V. The DNA-protein complexes were transferred to a positively charged nylon membrane (380 mA, 30 min) in 0.5x TBE and crosslinked to the membrane by ultraviolet light (312 nm) for 10 min. The following steps were performed according to the manufacturer’s protocol. Finally, DNA-protein complexes were detected with a transilluminator.
Table 4.
List of EMSA probes (5′-biotinylated).
| Probe | 5′-3′-sequence |
|---|---|
| SP1.1 | Biotin-GCCTCCTCCCCACCCCTTCAAGCCC Biotin-GGGCTTGAAGGGGTGGGGAGGAGGC |
| SP1.2 | Biotin-CTGATCGCCCCCACCCCCTTCCTTT Biotin-AAAGGAAGGGGGTGGGGGCGATCAG |
| SP1.3 | Biotin-TCCTCCCTCGGCTCCTCCCCCAGGC Biotin-GCCTGGGGGAGGAGCCGAGGGAGGA |
| SP1.4 | Biotin-CCCAGGCCCCGCCCTCTCCGCCTCG Biotin-CGAGGCGGAGAGGGCGGGGCCTGGG |
| SP1.5 | Biotin-ACAGGGGTGTGGGGAGGGGGCGCCG Biotin-CGGCGCCCCCTCCCCACACCCCTGT |
| SP1.6 | Biotin-GCCGCGCGGGCCAGGCGCCCGCCCC Biotin-GGGGCGGGCGCCTGGCCCGCGCGGC |
| SP1.7 | Biotin-CCCCTCCCTGCGCGCCCCGTCCCCG Biotin-CGGGGACGGGGCGCGCAGGGAGGGG |
In silico analyses
The Sp1 binding sites firstly had to be identified before being mutated. The Genomatix online software suite MatInspector62 was utilized for this application and to exclude the occurrence of unintended transcription factor binding sites in the mutated DNA sequences. Target prediction analysis was performed using three software tools: PicTar63, TargetScan Human64 and DIANA microT65.
Statistical analysis
All experimental data generated were processed and analyzed using the nonparametric two-tailed Mann-Whitney U Test in GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, USA). The significance level was defined as p < 0.05.
Electronic supplementary material
Acknowledgements
The authors thank Christoph Lichtenberg for his excellent technical assistance and Philip Saunders for his linguistic advice. This work was funded by the German Research Foundation (DFG, FA 1381/1-1).
Author Contributions
L.R. performed the experiments and analyzed data. B.F., T.D.L., D.H. and J.K. analyzed data and critically discussed the manuscript. C.K. and I.F. initiated and supervised the study and contributed to writing the paper. I.F. wrote the paper. All authors discussed and approved the manuscript submitted.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36217-2.
References
- 1.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 3.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 6.Jiang X, Tsitsiou E, Herrick SE, Lindsay MA. MicroRNAs and the regulation of fibrosis. FEBS J. 2010;277:2015–2021. doi: 10.1111/j.1742-4658.2010.07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel V, Noureddine L. MicroRNAs and fibrosis. Curr. Opin. Nephrol. Hypertens. 2012;21:410–416. doi: 10.1097/MNH.0b013e328354e559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wynn TA. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pakshir P, Hinz B. The big five in fibrosis: Macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol. 2018;68-69:81–93. doi: 10.1016/j.matbio.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 10.Zhou B, et al. Integration of microRNA and mRNA expression profiles in the skin of systemic sclerosis patients. Sci. Rep. 2017;7:42899. doi: 10.1038/srep42899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarjou A, Yang S, Abraham E, Agarwal A, Liu G. Identification of a microRNA signature in renal fibrosis: role of miR-21. Am. J. Physiol. Renal. Physiol. 2011;301:F793–801. doi: 10.1152/ajprenal.00273.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S, et al. miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J. 2013;27:2382–2391. doi: 10.1096/fj.12-219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L, et al. Sp1 mediates microRNA-29c-regulated type I collagen production in renal tubular epithelial cells. Exp. Cell. Res. 2013;319:2254–2265. doi: 10.1016/j.yexcr.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 14.van Rooij E, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cushing L, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am. J. Respir. Cell. Mol. Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maurer B, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010;62:1733–1743. doi: 10.1002/art.27443. [DOI] [PubMed] [Google Scholar]
- 17.Wermuth PJ, Piera-Velazquez S, Jimenez SA. Exosomes isolated from serum of systemic sclerosis patients display alterations in their content of profibrotic and antifibrotic microRNA and induce a profibrotic phenotype in cultured normal dermal fibroblasts. Clin. Exp. Rheumatol. 2017;35(Suppl 106):21–30. [PMC free article] [PubMed] [Google Scholar]
- 18.Pottier N, Cauffiez C, Perrais M, Barbry P, Mari B. FibromiRs: translating molecular discoveries into new anti-fibrotic drugs. Trends Pharmacol. Sci. 2014;35:119–126. doi: 10.1016/j.tips.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Pönighaus C, et al. Human xylosyltransferase II is involved in the biosynthesis of the uniform tetrasaccharide linkage region in chondroitin sulfate and heparan sulfate proteoglycans. J. Biol. Chem. 2007;282:5201–5206. doi: 10.1074/jbc.M611665200. [DOI] [PubMed] [Google Scholar]
- 20.Götting C, Kuhn J, Zahn R, Brinkmann T, Kleesiek K. Molecular cloning and expression of human UDP-d-Xylose:proteoglycan core protein beta-d-xylosyltransferase and its first isoform XT-II. J. Mol. Biol. 2000;304:517–528. doi: 10.1006/jmbi.2000.4261. [DOI] [PubMed] [Google Scholar]
- 21.Wilson IB. The never-ending story of peptide O-xylosyltransferase. Cell. Mol. Life Sci. 2004;61:794–809. doi: 10.1007/s00018-003-3278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu. Rev. Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn J, Gressner OA, Götting C, Gressner AM, Kleesiek K. Increased serum xylosyltransferase activity in patients with liver fibrosis. Clin. Chim. Acta. 2009;409:123–126. doi: 10.1016/j.cca.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Götting C, et al. Serum xylosyltransferase: a new biochemical marker of the sclerotic process in systemic sclerosis. J. Invest. Dermatol. 1999;112:919–924. doi: 10.1046/j.1523-1747.1999.00590.x. [DOI] [PubMed] [Google Scholar]
- 25.Faust I, et al. Human xylosyltransferase-I - a new marker for myofibroblast differentiation in skin fibrosis. Biochem. Biophys. Res. Commun. 2013;436:449–454. doi: 10.1016/j.bbrc.2013.05.125. [DOI] [PubMed] [Google Scholar]
- 26.Roderburg C, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 27.Moinzadeh P, et al. Biomarkers for skin involvement and fibrotic activity in scleroderma. J. Eur. Acad. Dermatol. Venereol. 2012;26:267–276. doi: 10.1111/j.1468-3083.2011.04206.x. [DOI] [PubMed] [Google Scholar]
- 28.Olivieri J, Smaldone S, Ramirez F. Fibrillin assemblies: extracellular determinants of tissue formation and fibrosis. Fibrogenesis Tissue Repair. 2010;3:24. doi: 10.1186/1755-1536-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faust I, et al. First description of the complete human xylosyltransferase-I promoter region. BMC Genet. 2014;15:129. doi: 10.1186/s12863-014-0129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller B, Prante C, Kleesiek K, Götting C. Identification and characterization of the human xylosyltransferase I gene promoter region. J. Biol. Chem. 2009;284:30775–30782. doi: 10.1074/jbc.M109.016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verrecchia F, Rossert J, Mauviel A. Blocking sp1 transcription factor broadly inhibits extracellular matrix gene expression in vitro and in vivo: implications for the treatment of tissue fibrosis. J. Invest. Dermatol. 2001;116:755–763. doi: 10.1046/j.1523-1747.2001.01326.x. [DOI] [PubMed] [Google Scholar]
- 32.Müller B, Prante C, Knabbe C, Kleesiek K, Götting C. First identification and functional analysis of the human xylosyltransferase II promoter. Glycoconj. J. 2013;30:237–245. doi: 10.1007/s10719-012-9439-5. [DOI] [PubMed] [Google Scholar]
- 33.Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 2002;195:27–38. doi: 10.1016/S0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 34.Hinz B, Gabbiani G. Fibrosis: recent advances in myofibroblast biology and new therapeutic perspectives. F1000 Biol. Rep. 2010;2:78. doi: 10.3410/B2-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowen T, Jenkins RH, Fraser DJ. MicroRNAs, transforming growth factor beta-1, and tissue fibrosis. J. Pathol. 2013;229:274–285. doi: 10.1002/path.4119. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh AK, Quaggin SE, Vaughan DE. Molecular basis of organ fibrosis: potential therapeutic approaches. Exp. Biol. Med. (Maywood) 2013;238:461–481. doi: 10.1177/1535370213489441. [DOI] [PubMed] [Google Scholar]
- 37.Götting C, Kuhn J, Kleesiek K. Human xylosyltransferases in health and disease. Cell. Mol. Life Sci. 2007;64:1498–1517. doi: 10.1007/s00018-007-7069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prante C, et al. Transforming growth factor beta1-regulated xylosyltransferase I activity in human cardiac fibroblasts and its impact for myocardial remodeling. J. Biol. Chem. 2007;282:26441–26449. doi: 10.1074/jbc.M702299200. [DOI] [PubMed] [Google Scholar]
- 39.Venkatesan N, et al. Glycosyltransferases and glycosaminoglycans in bleomycin and transforming growth factor-beta1-induced pulmonary fibrosis. Am. J. Respir. Cell. Mol. Biol. 2014;50:583–594. doi: 10.1165/rcmb.2012-0226OC. [DOI] [PubMed] [Google Scholar]
- 40.Theis T, et al. Lentiviral Delivery of miR-133b Improves Functional Recovery After Spinal Cord Injury in Mice. Mol. Neurobiol. 2017;54:4659–4671. doi: 10.1007/s12035-016-0007-z. [DOI] [PubMed] [Google Scholar]
- 41.Lopez-Aguilar JE, et al. Circulating microRNAs as Biomarkers for Pediatric Astrocytomas. Arch. Med. Res. 2017;48:323–332. doi: 10.1016/j.arcmed.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Li J, et al. MicroRNA-29b Inhibits Endometrial Fibrosis by Regulating the Sp1-TGF-beta1/Smad-CTGF Axis in a Rat Model. Reprod. Sci. 2016;23:386–394. doi: 10.1177/1933719115602768. [DOI] [PubMed] [Google Scholar]
- 43.Masur SK, Dewal HS, Dinh TT, Erenburg I, Petridou S. Myofibroblasts differentiate from fibroblasts when plated at low density. Proc. Natl. Acad. Sci. USA. 1996;93:4219–4223. doi: 10.1073/pnas.93.9.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varga J, Pasche B. Antitransforming growth factor-beta therapy in fibrosis: recent progress and implications for systemic sclerosis. Curr. Opin. Rheumatol. 2008;20:720–728. doi: 10.1097/BOR.0b013e32830e48e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin J, Vaud S, Zhelkovsky AM, Posfai J, McReynolds LA. Sensitive and specific miRNA detection method using SplintR Ligase. Nucleic Acids Res. 2016;44:e116. doi: 10.1093/nar/gkw399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li N, Cui J, Duan X, Chen H, Fan F. Suppression of type I collagen expression by miR-29b via PI3K, Akt, and Sp1 pathway in human Tenon’s fibroblasts. Invest. Ophthalmol. Vis. Sci. 2012;53:1670–1678. doi: 10.1167/iovs.11-8670. [DOI] [PubMed] [Google Scholar]
- 47.Tan NY, Khachigian LM. Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell. Biol. 2009;29:2483–2488. doi: 10.1128/MCB.01828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sysa P, Potter JJ, Liu X, Mezey E. Transforming growth factor-beta1 up-regulation of human alpha(1)(I) collagen is mediated by Sp1 and Smad2 transacting factors. DNA Cell. Biol. 2009;28:425–434. doi: 10.1089/dna.2009.0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park JH, et al. Antifibrotic effect through the regulation of transcription factor using ring type-Sp1 decoy oligodeoxynucleotide in carbon tetrachloride-induced liver fibrosis. J. Gene Med. 2009;11:824–833. doi: 10.1002/jgm.1355. [DOI] [PubMed] [Google Scholar]
- 50.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/S0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 51.Ogawa T, et al. Suppression of type I collagen production by microRNA-29b in cultured human stellate cells. Biochem. Biophys. Res. Commun. 2010;391:316–321. doi: 10.1016/j.bbrc.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 52.Jia LF, et al. miR-29b suppresses proliferation, migration, and invasion of tongue squamous cell carcinoma through PTEN-AKT signaling pathway by targeting Sp1. Oral Oncol. 2014;50:1062–1071. doi: 10.1016/j.oraloncology.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 53.Ye W, et al. Xylosyltransferase-1 expression is refractory to inhibition by the inflammatory cytokines tumor necrosis factor alpha and IL-1beta in nucleus pulposus cells: novel regulation by AP-1, Sp1, and Sp3. Am. J. Pathol. 2015;185:485–495. doi: 10.1016/j.ajpath.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanden Oever M, Muldoon D, Mathews W, McElmurry R, Tolar J. miR-29 Regulates Type VII Collagen in Recessive Dystrophic Epidermolysis Bullosa. J. Invest. Dermatol. 2016;136:2013–2021. doi: 10.1016/j.jid.2016.05.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khair M, et al. Regulation of xylosyltransferase I gene expression by interleukin 1beta in human primary chondrocyte cells: mechanism and impact on proteoglycan synthesis. J. Biol. Chem. 2013;288:1774–1784. doi: 10.1074/jbc.M112.419887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jungert K, et al. Smad-Sp1 complexes mediate TGFbeta-induced early transcription of oncogenic Smad7 in pancreatic cancer cells. Carcinogenesis. 2006;27:2392–2401. doi: 10.1093/carcin/bgl078. [DOI] [PubMed] [Google Scholar]
- 57.Parisi F, Wirapati P, Naef F. Identifying synergistic regulation involving c-Myc and sp1 in human tissues. Nucleic Acids Res. 2007;35:1098–1107. doi: 10.1093/nar/gkl1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McDonough PM, Hanford DS, Sprenkle AB, Mellon NR, Glembotski CC. Collaborative roles for c-Jun N-terminal kinase, c-Jun, serum response factor, and Sp1 in calcium-regulated myocardial gene expression. J. Biol. Chem. 1997;272:24046–24053. doi: 10.1074/jbc.272.38.24046. [DOI] [PubMed] [Google Scholar]
- 59.Faust I, et al. Characterization of dermal myofibroblast differentiation in pseudoxanthoma elasticum. Exp. Cell Res. 2017;360:153–162. doi: 10.1016/j.yexcr.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Campbell P, Jacobsson I, Benzing-Purdie L, Roden L, Fessler JH. Silk–a new substrate for UDP-d-xylose:proteoglycan core protein beta-D-xylosyltransferase. Anal. Biochem. 1984;137:505–516. doi: 10.1016/0003-2697(84)90119-2. [DOI] [PubMed] [Google Scholar]
- 61.Weilke C, Brinkmann T, Kleesiek K. Determination of xylosyltransferase activity in serum with recombinant human bikunin as acceptor. Clin. Chem. 1997;43:45–51. [PubMed] [Google Scholar]
- 62.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krek A, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 64.Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife4, 10.7554/eLife.05005 (2015). [DOI] [PMC free article] [PubMed]
- 65.Paraskevopoulou MD, et al. DIANA-microT web serverv5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.