Abstract
Background and Aims
Ulcerative colitis [UC] is a chronic inflammatory disease that effects the gastrointestinal tract and is considered one of the most prominent and common forms of inflammatory bowel disease [IBD]. This study aimed to define and describe the entire transcriptomic landscape in a well-stratified, treatment-naïve UC patient population compared with control patients by using next-generation technology, RNA-Seq.
Methods
Mucosal biopsies from treatment-naïve UC patients [n = 14], and healthy controls [n = 16] underwent RNA-Seq. Principal component analysis [PCA], cell deconvolution methods, and diverse statistical methods were applied to obtain and characterise a dataset of significantly differentially expressed genes [DEGs].
Results
Analyses revealed 1480 significantly DEGs in treatment-naïve UC when compared with controls. Cell populations of monocytes, T cells, neutrophils, B cells/ lymphoid cells, and myeloid cells were increased during inflammation, whereas the fraction of epithelial cells were reduced in UC, which is reflected by the DEGs; 79 DEGs were identified as IBD susceptibility genes, and 58 DEGs were expressed in a gender-specific manner. MUC5B, REG3A, DEFA5, and IL33 might be considered as colorectal cancer [CRC] risk factors following UC in males. AQP9 together with CLDN2 may have a role regulating tissue-specific physiological properties in tight junctions in UC. An additional functional role for AQP9 in the synthesis and/or the function of mucus can be implied.
Conclusions
This study reveals new potential players in UC pathogenesis in general, and provides evidence for a gender-dependent pathogenesis for UC. These results can be useful for the development of personalised treatment strategies for UC in the future.
Keywords: Gender, RNA-Seq, ulcerative colitis [UC]
1. Introduction
Ulcerative colitis [UC] is a chronic inflammatory disorder of the gastrointestinal tract, and along with Crohn’s disease [CD] comprise two of the common forms of inflammatory bowel disease [IBD]. In contrast to CD, the inflammations in UC are limited to the mucosa and submucosa of the colon, and the rectum.1 Chronic inflammations have been shown to increase the risk for the development of colorectal cancer [CRC].2 Just 10–20% of IBD cases can be explained by genetic variances,3,4 suggesting a much more complex pathogenesis, perhaps an underlying interplay between environmental factors, the intestinal microbiome, nutrition, and genetic factors. Susceptibility genes are responsible for triggering a dysregulation of the immune system and affecting the gut barrier function.5,6 Nevertheless, despite decades of investigation, the complex pathogenesis of IBD is not fully understood.
Many attempts have been made to describe transcriptional levels in UC,7–9 usually by using hybridisation-based methods like microarrays.10 These methods are restricted to predefined and often well-annotated RNA species. Next-generation sequencing [NGS] techniques [RNA-Seq] have no such restrictions. One recent report used RNA-Seq for gene expression profiling in patients with IBD including patients with different treatment strategies.11 For the present study, a clearly defined group of newly diagnosed [treatment-naïve] UC patients was used. It is believed that RNA-Seq, together with well-stratified UC patient material, can potentially provide a more comprehensive and correct transcriptomic profile of UC. The results of this study do not only reveal new potential players in treatment-naïve UC pathogenesis in general, but in addition provide evidence of a gender-dependent pathogenesis for UC. These results can be useful for the development of general and/or gender-dependent treatment strategies for UC in the future.
2. Material and Methods
2.1. Patient material
A standardised sampling method was used to collect mucosal biopsies from the colon of 14 newly diagnosed, treatment-naïve UC patients with mild to moderate disease activity and from 16 controls. Samples from subjects performing a cancer screening, with normal colonoscopy and normal colonic histological examination, served as controls. UC was diagnosed based upon established clinical endoscopic and histological criteria as defined by the ECCO guidelines.12 The grade of inflammation was assessed during colonoscopy using the UC Disease Activity Index [UCDAI] endoscopic sub-score with 3 to 10 for mild to moderate disease.13 Apart from one rectal control sample, all biopsies were taken from the sigmoid part of the colon. Tumour necrosis factor alpha [TNF-α] mRNA expression levels were measured by real-time polymerase chain reaction [PCR], thereby indicating the grade of UC activity.14 The samples were taken from an established Biobank approved by the Norwegian Board of Health. All patient characteristics are depicted in Table 1. The participants signed an informed and written consent form. The study was approved by the Regional Ethics Committee of North Norway and the Norwegian Social Science Data Services [REK Nord 2012/1349].
Table 1.
Demographic information of patient samples.
| Characteristics | Control group [n = 16] | Ulcerative colitis [n = 14] |
|---|---|---|
| Male/female | 11/5 | 9/5 |
| Age mean ± SD, years | 52.9 ± 16.9 | 39.57 ± 15.24 |
| Endo score mean ± SD | 0 | 1.93 ± 0.27 |
| Clinical score ± SD | 0 | 7.23 ± 2.45 |
| TNF-α level ± SD | 3663 ± 1973 | 15907 ± 9623 |
SD, standard deviation; TNF, tumour necrosis factor.
2.2. DNA and RNA isolation
Total RNA was isolated using the Allprep DNA/RNA Mini Kit from Qiagen [Cat. No.: 80204] and the QIAcube instrument [Qiagen], according to the manufacturer’s protocol. RNA quantity and purity were assessed by using the NanoDrop ND-1000 spectrophotometer [ThermoFisher Scientific, Wilmington, DE, USA]. The Experion Automated Electrophoresis System [Bio-Rad, Hercules, CA, USA] and the RNA StdSens Analysis Kit [Bio-Rad, cat. No.: 700–7103] was used to evaluate RNA integrity, according to the instruction manual. RNA samples were kept at −70°C until further use. All RNA samples used for analyses had a RIN value between 8.0 and 10.0.
2.3. Quantitative polymerase chain reaction
The TNF-α levels in biopsies were measured by using quantitative polymerase chain reaction [qPCR]. RNA quantity was assessed with NanoVue Plus [GE Healthcare, UK]. Synthesis of cDNA was performed using the QuantiTect Reverse Transcription Kit [Qiagen, cat. No.: 205314], and the QuantiNova Probe PCR Kit [Qiagen, cat. No.: 208256]. Beta-actin [β-actin] was used as housekeeping gene. For the detection, a CFX Connect Real Time PCR Detection System [Bio-Rad, USA] was used. The results were measured in copies/µg. Values < 7000 copies/µg protein are considered as non-inflamed tissues, and values > 7000 copies/µg protein are considered inflamed tissues.14
2.4. Library preparation & Next generation sequencing
Transcriptome libraries were prepared with the TruSeq Stranded Total RNA LT Sample Prep Kit from Illumina [Cat. No.: RS-122–2203]. The amount of input material was 1 µg of total RNA. The Bioanalyzer 2100 [Agilent Technologies, Santa Clara, USA], and the Agilent DNA 1000 kit [Cat. No.: 5067-1504] were used to assess RNA libraries quality, according to the instruction manual. The RNA libraries comprised fragments with an average size of 307 base pairs. The libraries were normalised to 10 nM and subsequently sequenced with the NextSeq 550 instrument [Illumina, USA] according to the manufacturer’s instructions. The average number of uniquely mapped reads per sequencing run was 88 million reads per sample.
2.5. Data analysis
Base calling and quality scoring were performed as a first step including quality check on the on-board computer of the NextSeq 550. The algorithm packages STAR-2.5.2b and the htseq-count were used for downstream analysis.15 Transcripts were aligned to human genome assembly GRCH38p.11 [https://www.ncbi.nlm.nih.gov/grc/human/data]. Read counts were transformed using the DESeq2-Rlog variance stabilised transform, and significantly differentially expressed transcripts were identified by including transcripts with a read count of > 30 and fold change > 2 as compared with controls. Additional annotation was added using the PANTHER classification system [http://pantherdb.org/], the Kyoto Encyclopaedia of Genes and Genomes [KEGG] [www.genome.jp/kegg/], and the human gene database GeneCards [http://www.genecards.org/]. For principal component analysis [PCA], the top 5000 most variable of the DESeq2-Rlog variance stabilised genes were used. For the estimation of specific cell populations in patient samples, all DESeq2-Rlog normalised transcripts with a log2 average mean > 5 were included. The analysis was performed using the R/Bioconductor CellMix manual [http://web.cbio.uct.ac.za/~renaud/CRAN/web/CellMix/] with the IRIS [Immune Response In Silico] weighted marker list characteristic for the different cell types.16 Epithelial markers cadherin 1 [CDH1], epithelial cell adhesion molecule [EPCAM], phosphatidylinositol glycan anchor biosynthesis class F [PIGF], L1 cell adhesion molecule [L1CAM], and laminin subunit alpha 1 [LAMA1] were added to the IRIS marker list and weighted strongly to give an estimate of epithelial presence in samples. The contrast matrix [[Nfemale - UCfemale] - [Nmale - UCmale]] was used in DESeq2 to determine differentially regulated genes between UC and controls which differed significantly between male and female. Results were limited to adjusted p-value < 0.05 and a log2 fold change > 1.0. Genes associated with the risk of IBD were downloaded from the genome-wide association studies [GWAS] catalogue using the search term IBD on 29 November 2016.17 Gene expression data for the 295 genes associated with IBD single nucleotide polymorphism [SNPs] was analysed by k-means clustering and three primary groups were identified. For interpretation, genes were further subdivided using the gene ontology according to roles in inflammatory processes. The inflammatory properties of the genes were classified with the gene ontology as part of an inflammatory response [GO: 0006954] with subcategories for negative [GO: 0050728] and positive [GO: 0050729] regulators of inflammation. Heat maps were produced for each cluster to visualise the gene expression patterns of the GWAS associated genes in conjunction with their regulatory and inflammatory roles.
3. Results
3.1. Patients
A standardised sampling method was used to collect mucosal biopsies from treatment-naïve UC patients [n = 14] and control samples [n = 16], as described above. The biopsies from UC patients showed mild to moderate disease activity [as defined by UCDAI], with clinical scores 7.23 ± standard deviation [SD] 2.45, and endo scores of 1.93 ± SD 0.27, estimated according to established clinical endoscopic and histological criteria, and as defined by the ECCO guidelines.12 The control group consisted of biopsies with normal colonoscopy, colon histology, and immunohistochemistry, and clinical and endo scores = 0. The biopsies of UC patients were taken from the sigmoid part of the colon. The gender distribution for both groups was almost equal, with nine males in the UC group and 11 males in the control group, and five females in each group. The age distribution differed between the two groups, with 39.57 ± SD 15.24 in the UC group, and 52.9 ± SD 16.9 in the control group. In order to obtain information about the inflammatory status of UC, TNF-α mRNA expression levels were measured by qPCR.14 Levels of TNF-α in control samples were estimated at 3663 ± SD 1973, and for UC samples 15907 ± SD 9623. A summary of all patient characteristics is depicted in Table 1.
3.2. Characterization of the whole transcriptome in treatment-naïve UC
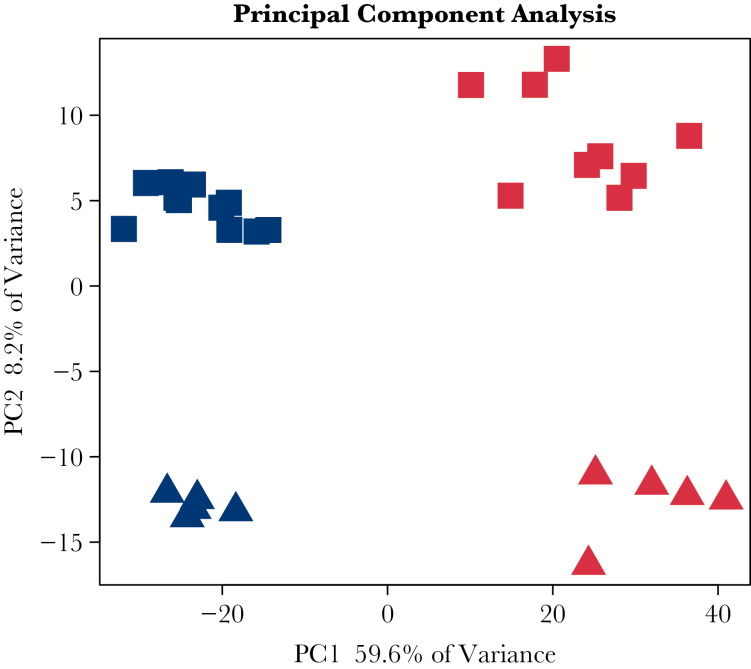
The entire transcriptome representing treatment-naïve UC was established by RNA-Seq. Pre-processing of the sequencing data revealed expression of approximately 22 000 transcripts. Initial principal component analysis [PCA] of the 5000 most variable transcripts revealed a clear distinction between UC and control samples along the first principal component with a 59.6% explained variance [Figure 1].
Figure 1.
Principal component analysis [PCA]. Unsupervised PCA analysis showing the difference between UC [red] and control [blue] as well as gender, male control [blue square], female control [blue triangle], male UC [red square] and female UC [red triangle]. There is a 59.6% variance between UC and control samples, and an 8.2% variance between male and female samples. UC, ulcerative colitis.
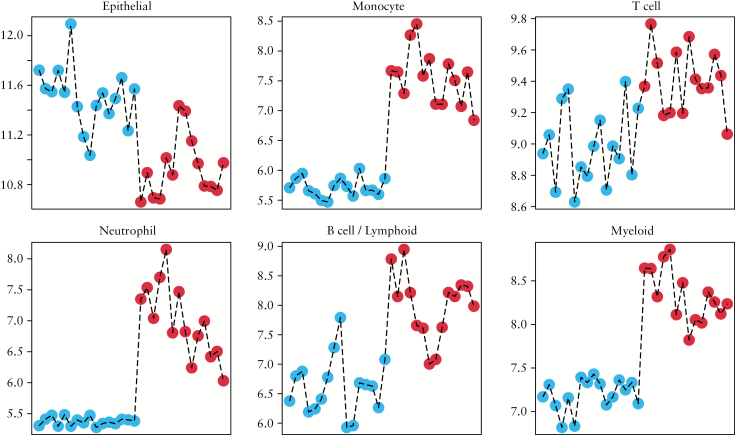
In order to estimate specific cell populations in UC and control tissue samples, a cell deconvolution method was applied as described. The deconvolutions were restricted to the following cell types: epithelial cells, monocytes, T cells, neutrophils, B cells/lymphoid cells, and myeloid cells. The results show a clear difference of cell fractions present in UC and control samples. An enrichment of monocytes, neutrophils, myeloid cells, T cells, and B/Lymphoid cells was observed in all UC samples, whereas the epithelial cell fraction was decreased in almost all UC samples when compared with control samples. The results of the deconvolution experiments are depicted in Figure 2.
Figure 2.
Estimation of cell population between samples using cell deconvolution methods using the Bioconductor CellMix package. Epithelial markers [CDH1, EPCAM, L1CAM, and LAMA1] were added to the Immune Response In Silico [IRIS] marker list and weighted heavily to help estimate epithelial contribution. The blue dots indicate control samples and red dots indicate ulcerative colitis [UC] samples, respectively.
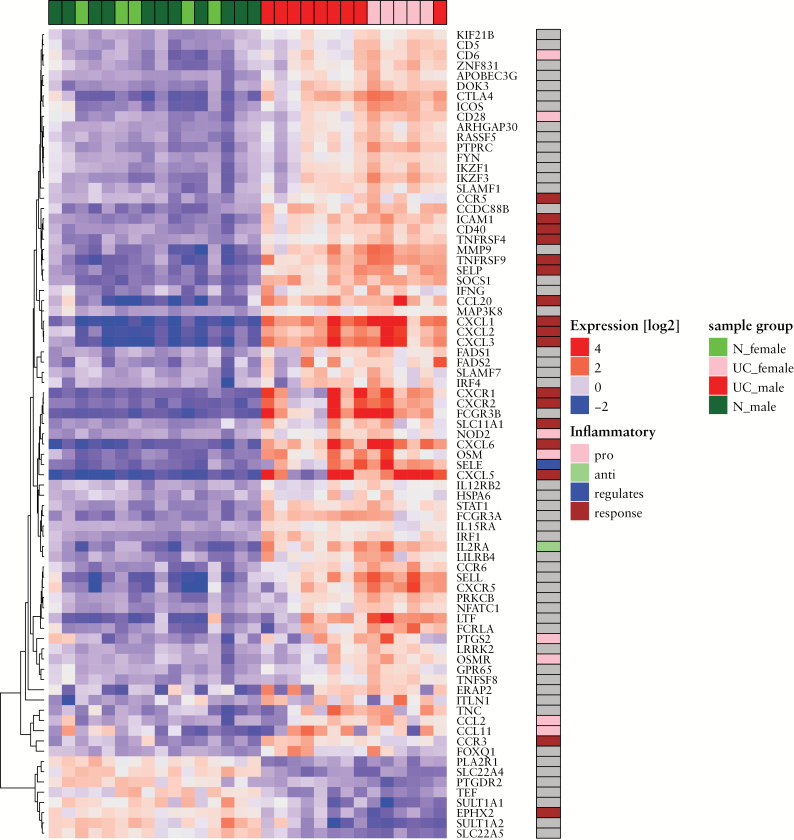
To further describe and analyse the entire transcriptome, significantly differentially expressed transcripts were adjusted to p-value < 0.05 and a cut-off of log2 fold-change > 1.0 [n = 1480] was used for downstream analyses [Supplementary Data 1, available at ECCO-JCC online] whereof the top 30 differentially expressed genes [DEGs] with log2 fold-change > 3.5 are shown in Table 2. The differentially expressed gene transcripts were related to currently known 295 IBD susceptibility genes [Supplementary Figure 1, available at ECCO-JCC online] as revealed by GWAS [see Figure 3].18,19 The identified gene transcripts have been annotated to different inflammatory processes and their transcriptional levels. Transcripts with unchanged expression are omitted in Figure 3. However, for a complete overview see Supplementary Figure 1. The data depicted in Figure 3 show 71 upregulated [24%] and eight downregulated transcripts [2.7%] linked with susceptibility to IBD. The annotation of the upregulated transcripts revealed genes involved in inflammatory responses, like chemokine receptors [CCR5, CCR3, CXCR1, CXCR2], chemokine ligands [CXCL5, CXCL1, CXCL2, CXCL3, CXCL4 [PF4], and CCL20], tumour necrosis factor receptor superfamily members (TNFRSF5 [CD40] and TNFRSF9), interleukin 19 [IL19], solute carrier family 11 member 1 [SLC11A1], intercellular adhesion molecule 1 [ICAM1], and signal transducer and activator of transcription 3 [STAT3], T cell specific antigens [CD28 and CD6], chemokine ligands [CCL2 and CCL11], oncostatin M [OSM] and its receptor oncostatin M receptor [OSMR], inflammatory bowel disease Protein 1 (NOD2 [IBD1]), and cyclooxygenase-2 (COX2 [PTGS2]). Only one of the downregulated transcripts is involved in inflammatory response, epoxide hydrolase 2 [EPHX2]. In addition, three significantly differentially expressed microRNAs [MIR155HG, MIR3936 and MIR4435-2HG] and 17 long non-coding RNAs [lncRNAs] have been identified [Supplementary Data 1].
Table 2.
Top 30 differentially expressed genes [DEGs] in treatment-naïve ulcerative colitis [UC].
| Gene symbol | Log2 FC | p-adjusted |
|---|---|---|
| ABCA12 | 4,46 | 4,98E-35 |
| AQ9 | 5,37 | 3,56E-32 |
| CHI3L1 | 5,36 | 8,62E-57 |
| CLDN2 | 4,49 | 1,82E-38 |
| CXCL1 | 4,30 | 7,06E-34 |
| CXCL5 | 4,63 | 7,17E-20 |
| CXCL6 | 4,06 | 5,46E-26 |
| CXCR1 | 4,64 | 3,93E-23 |
| DEFA5 | 4,09 | 1,1E-15 |
| DUOX2 | 5,73 | 8,62E-57 |
| DUOXA2 | 7,21 | 1,93E-77 |
| FAM83A | 4,11 | 2,94E-22 |
| FCGR3B | 4,55 | 6,03E-30 |
| FPR2 | 4,23 | 1,78E-21 |
| HCAR3 | 4,32 | 5,01E-22 |
| IL17A | 5,03 | 1,5E-33 |
| KCNJ15 | 3,95 | 1,22E-17 |
| LCN2 | 4,95 | 2,25E-53 |
| MMP10 | 4,99 | 1,44E-34 |
| MMP3 | 5,02 | 3,31E-44 |
| MMP7 | 5,32 | 2,15E-29 |
| NOS2 | 4,03 | 9,9E-40 |
| PI3 | 4,11 | 2,73E-31 |
| REG1A | 5,05 | 1,28E-22 |
| REG3A | 4,19 | 2,63E-15 |
| SAA1 | 6,40 | 5,16E-46 |
| SAA2 | 6,23 | 7,31E-43 |
| SLC6A14 | 5,66 | 1,73E-48 |
| TNIP3 | 5,60 | 3,49E-58 |
| TREM1 | 4,13 | 8,56E-26 |
Figure 3.
Gene expression pattern across controls and UC patients of differentially expressed genes that are also reported as associated with GWAS risk SNPs. Colours indicate deviation from the mean expression level for each gene. Samples are colour-coded in the top bar according to gender and diagnosis [control, UC] and genes are colour-coded in the sidebar according to their inflammation-related gene ontology annotations. Transcripts with unchanged differential expression are omitted. UC, ulcerative colitis; GWAS, genome-wide association studies; SNPs. Single nucleotide polymorphisms.
The current study was compared with microarray- based studies carried out on two different Affymetrix-based microarray platforms.20,21 Only well-characterised probes with Entrez IDs and official gene symbols were included in the comparison. Overall agreement between the studies when comparing UC patients and normal controls are shown in the Venn diagram [Supplementary Figure 2, available at ECCO-JCC online]. Only one gene, transmembrane 4 L6 family member 20 [TM4SF20], was found to be significantly [log fold-change > 1 and p < 0.05] downregulated in our study but significantly upregulated in one of the others.21
3.3. Gender-specific transcription
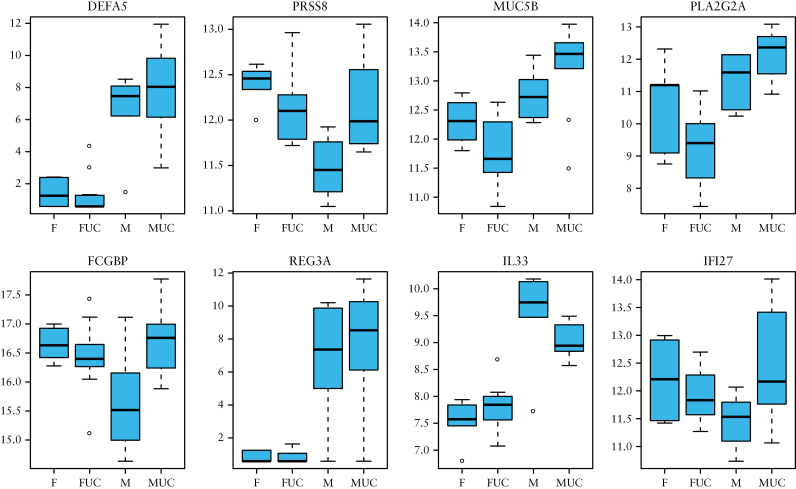
PCA analysis did not only distinguished between UC and control samples [Figure 1] but could also distinguish transcripts to a lesser extent [variance 8.2%] and in a gender- specific manner. The contrast matrix [[Nfemale - UCfemale] - [Nmale - UCmale]] was applied to determine differentially regulated genes with gender-specific effects; 58 significantly differentially expressed genes with adjusted p-value < 0.05 and log 2 FC > 1.0 were identified [see Supplementary Data 2, available at ECCO-JCC online]. A boxplot of eight selected genes show gender-specific differences [Figure 4: Paneth cell specific defensin alpha 5 [DEFA5], serine protease 8 [PRSS8], mucin 5B [MUC5B], phospholipase A2 group IIA [PLA2G2A], Fc fragment of IgG binding protein [FCGBP], regenerating family member 3 alpha [REG3A], interleukin 33 [IL33], and interferon alpha inducible protein 27 [IFI27]. The expression levels of the selected genes are higher in the UC group compared with the control group. However, the expression levels do not only show a difference between UC samples and control samples, but also show significant differences between UC samples and control samples for males and females. It is noted that thiosulphate sulphurtransferase [TST], mercaptopyruvate sulphurtransferase [MPST], and fucosyltransferase 2 [FUT2] are the only GWAS IBD susceptibility genes that are found to be expressed in a gender-specific manner [Supplementary Data 2; Figure 3].
Figure 4.
A boxplot of selected genes that show gender-specific differences within a greater subset of UC versus control significant differentially expressed genes. F and M indicate control female and male samples, FUC and MUC indicate female and male UC samples.
4. Discussion
This study provides a unique, comprehensive, and quantitative record of high-resolution gene expression in a treatment-naïve UC patient population using next-generation RNA-Seq technology. Previous gene expression studies with UC patient material mostly used hybridisation-based methodologies like microarrays,7,22–25 and only one recent study reported RNA-Seq of human IBD patient material.11 RNA-Seq technology has some advantages over microarray technology, such as the ability of impartial detection of new transcripts and the easy detection of rare and low abundance transcripts.10 In addition, RNA-Seq does not rely on pre-designed complement-sequence detection probes, and is therefore free of issues associated with probe redundancy and annotation.26 Attempts were made in order to decipher if genes found in other studies20,21 including UC patients behaved differently in the present study [Supplementary Figure 2, available at ECCO-JCC online]. However, it is difficult to assign the variability between experiments to either effects of the technical platform or different severity of disease that was mild to moderate in our dataset and involved patients resistant to standard treatment in the above-mentioned studies.
In addition, recent UC transcriptome studies were performed by using non-stratified patient populations and material including both treated and treatment-naïve UC,11 which might have resulted in biased gene expression profiles. Therefore, in this study, the UC patient material was thoroughly stratified and only treatment-naïve patients were included [see Table 1 and Material and Methods]. This approach should provide an opportunity to investigate the transcriptional profile of UC without the interference of any medications. Recent reports have shown that medication given to UC patients, such as immunosuppressant drugs, have short- and long-term side effects on the immune response.27 This treatment might introduce unwanted bias to experiments aiming to investigate the prior to medication status of UC patients. Furthermore, treatment-naïve UC transcriptomic signatures might also become important in order to decide individual treatment options for patients in the future. Our RNA-Seq revealed 1480 significantly DEGs [see Supplementary Data 1]. In the future, these DEGs could be used as a fingerprint for disease outcome and a beneficial treatment choice.
Human tissue samples are highly heterogeneous, and the amount of different cell types in a biopsy will have a certain impact on gene expression profiles. Therefore, and in order to determine the relative quantities of different immune cell types in the heterogeneous biopsies, cell deconvolution methods were applied [Figure 2]. As expected, the results clearly show and confirm that the innate and adaptive immune systems are triggered by inflammation as the fractions of monocytes, neutrophils, myeloid cells, T cells, and B/lymphoid cells are found to be increased in UC [Figure 2]. These differences could very well due to inflammatory infiltrates; as for which cells contribute the most, we will need a much larger sample size and most likely confirmatory or parallel experiments using a different technique such as single cell sequencing. One interesting aspect with these data is that one could potentially identify various patterns. For example, UC patient no. 5 from the left [Figure 2] is rather high in monocytes and neutrophils but relatively low in T cells and B/lymphoid cells, but to decipher these types of patterns would require larger study populations. In concordance with these observations, the different inflammatory responses are reflected by increased expression of genes like leucocyte immunoglobulin-like receptors, cytokines/chemokines and their respective receptors and ligands, and T cell specific antigens [see Table 2; and Supplementary Data 1]. In particular, many of the DEGs that are involved in the control of bacterial proliferation showed increased expression during mucosal inflammation, like regenerating family members [REGs] and defensins, which is in concordance with previous studies.7,28–32 It is interesting to note that the here observed increased expression of factors predictive of response to anti-TNF-α therapy, oncostatin [OSM] and its receptor [OSMR], is of particular relevance for anti-TNF-α resistant patients.33
Not surprisingly, the fraction of epithelial cells was lowered in UC [Figure 2]. Once the mucosal epithelium is compromised by inflammation, the fraction of functional epithelial cells diminishes, which then leads to a ‘leaky’ intestinal epithelium.34 The impaired ion transport and dysfunctional tight junctions in the epithelium are followed by chronic diarrhoea. In addition, increased levels of circulating TNF-α, and other cytokines lead to a rise of intestinal permeability, thereby causing oedema.35 This dysregulation of water and solute homeostasis has been suggested to play a role in UC. In concordance with our results, the expression levels of water channels like aquaporin 7 [AQP7] and aquaporin 8 [AQP8], have been shown to be reduced in the human intestinal mucosa in early stage IBD.36 Interestingly, aquaporin 9 [AQP9] was one of the most prominent expressed genes in our UC patient material, which has not been reported before [Table 2]. AQP9 plays a role in specialised leukocyte functions such as immunological response and bactericidal activity, and is located in the membranes of tight junctions in the intestine.37 In addition claudins, that are exclusively localised at tight junctions, were differentially expressed [Table 2], with claudin 2 [CLDN2] levels being increased in UC samples, which is in accordance with former results.38–40 The ‘pore-forming’CLDN2, as a component of the tight junction, forms a water channel and thus mediates para-cellular water transport across the tight junctions in impaired epithelium.41 In addition, the observed increase of CLDN2 correlates with UC severity on both protein and transcriptional levels.38 This might indicate that CLND2 and AQP9 might share similar properties regarding water transport. It is possible that like epithelial cells, other cells contribute to the elevated levels of CLDN and/or AQP9. However, no such information is available today. Cell sorting followed by single cell sequencing might shed a light on this. The increased expression of CLDN and AQP9 might be a response to inflammation, fighting the disturbances in the epithelial barrier of the colon. Taken these results together, it could be hypothesised that CLDN2 together with AQP9 regulate tissue-specific pathophysiological properties of tight junctions in UC.
Intestinal infection generally leads to depletion of goblet cells and reduction in mucin synthesis and secretion, allowing pathogens to access the underlying epithelium.42 Interestingly, AQP9 could play another role for the synthesis and/or function of mucus, as recent immuno-histological studies have shown that AQP9 is expressed in a subset of mucin-producing goblet cells in the small intestine and colon.37 However, mucin 2 [MUC2], which is the major contributor to healthy lubrication of the mucosa, was not differentially expressed, which is in concordance with previous findings.43 In contrast, several other mucins showed increased expression in UC: mucin 1 [MUC1], mucin 16 [MUC16], mucin 4 [MUC4], mucin 5Ac [MUC5AC], and mucin 5B [MUC5B] [Table 2; and Supplementary Data 1]. Elevated levels of MUC1 and MUC4 in UC have been demonstrated before.44,45 Contrary to results reported by others who have shown that expression of MUC5B is increased in colorectal cancer [CRC] but not in UC,46 we here report increased expression of MUC5B in UC. It is hereby noted that in normal colon, MUC5B has been shown to be secreted by colonic goblet cells; however it is expressed in minor quantities.47 In addition, we can report not only an increased but also gender-specific differences in the expression of MUC5B in UC, as discussed below [see also Figure 4; and Supplementary Data 2]. A cytokine-induced mucin hypersecretion has been reported for MUC5AC in an in vitro model where expression increased in a TNF-α dose-dependent manner.48 This might be also the situation in our UC material, as elevated TNF-α levels have been one criterion for patient stratification [see Materials and Methods]. A role for MUC16 in UC, which showed the most prominent elevated expression levels [log2 FC 2.16] has not been established yet but has been recently proposed to be a biomarker for epithelial ovarian cancer.49
Recently, micro RNAs [miRNAs] and long non-coding RNAs [lncRNAs] have been addressed as having a role in IBD pathogenesis.50,51 Three microRNAs have been identified in our data: MIR155HG, MIR3936, and MIR4435-2HG. It has been reported that MIR155 is involved in intestinal inflammation and immunity of UC by playing a role in the differentiation of B and T cells and dendritic cells, thereby contributing to the development of regulatory T cells.52–54 In addition, we observed 17 significantly differentially expressed lncRNAs [Supplementary Data 1]. One functional relationship between UC and a particular lncRNA has been recently demonstrated.55 However, the relationship between UC and miRNAs and/or lncRNAs still remains unclear.
In order to characterise gene expression data from RNA-Seq further, one approach was to investigate how many differentially expressed genes are associated with IBD. Until today, approximately 300 SNPs associated with IBD have been discovered through GWAS.17 Although it is expected that this RNA-Seq study cannot confirm GWAS findings, 79 of the significantly differentially expressed genes might be associated with IBD [see Table 2]. It is hereby noted that the GWAS susceptibility genes did not distinguish between UC and CD. However, although one should be cautious about assigning specificity to SNPs, since most GWAS studies are done comparing a patient population with a population of disease-free controls and not between subtypes of the disease, some SNPs are indeed more strongly related to UC than CD and vice versa.56 Among our significantly differentially expressed genes, SP140 nuclear body protein [SP140] and strawberry notch homolog 2 [SBNO2] are located close to SNPs that are more strongly related to CD risk, and both of these genes have shown upregulated gene expression in UC [Supplementary Data 1].
Recently, gender-dependent differences in IBD pathologies have been proposed.57,58 Sex-stratified analysis of long-term complications of IBD show consistently higher risk of CRC in male IBD patients.59 In addition, a recent population study reported that patients with UC are the high-risk group in incidence of CRC and that the risk is found to be higher in male than in female UC patients.60 The current available information about gender-specific differences in UC is sparse and contradictory to some extent.61 It is a common belief that understanding gender differences in any disease is important for recognising the factors contributing to the disease expression and to determine its prognosis so that clinicians can offer an appropriate medical therapy. However, molecular manifestations of a gender specificity for UC has not been established. We can for the first time show that gender differences on a molecular level occur not only between treatment-naïve UC patients [Figure 1], but also between control patients [Figure 4]. The most pronounced gender differences for UC were observed for mucin 5B [MUC5B], regenerating family member 3 alpha [REG3A], defensin A5 [DEFA5], and interleukin 33 [IL33] [Figure 4]. In UC, IL33 expression is specifically increased and has been shown to be involved in the inflammatory tumour microenvironment and to contribute to the progression of CRC.62–64 In this context, it is interesting to note that IL33 is found to be more increased in male UC than in female UC. In addition, SNPs in human DEF5A may confer susceptibility to IBD.32 Furthermore, an upregulation of REG3A in colorectal cancer cells confers proliferation and correlates with colorectal cancer risk.65 All these above-mentioned genes were more elevated in male samples than in female samples and can therefore be considered as CRC risk factors in UC. It is interesting to note that DEFA5, IL33, and REG3A showed also increased expression levels in control male samples when compared with control female samples. Taken all these observations together, a possible link between the expression of these genes and higher risk for the development of CRC following UC can be proposed for males. For some gender-specific genes, differential gene expression was observed between control male and female samples. Genes like serine protease 8 [PRSS8], Fc fragment of IgG binding protein [FCGBP], and interferon alpha-inducible protein 27 [IFAN27] showed increased expression in control female compared with control male samples. Interestingly, all these genes have been shown to convey protective properties in the mucosa: FCGBP has been shown to be involved in the maintenance of the mucosal structure as a gel-like component of the mucosa; PRSS8 preserves colonic integrity and protects against inflammation, as has been demonstrated in DSS-induced inflammation of mice; and IFN-α conveys antiviral activities.66–68 In addition, three gender-specific GWAS IBD susceptibility genes showed increased expression in female UC [Supplementary Data 2]. Two belonged to the family of sulphurtransferases, and both genes are involved in detoxification processes.69 The here observed expression of thiosulphate [TST]—and mercaptopyruvate [MPST] —sulphurtransferase is essential for sulphide detoxification in order to preserve healthy mucosa. Dysregulation of expression and/or activity of these enzymes may accompany development of UC.70 Here, fucosyltransferase 2 [FUT2] also might play a role in this regard, since this enzyme is involved in host-microbe interactions and has been shown to mediate interaction with intestinal microbiota, thereby influencing its composition.71–73
In conclusion, this study shows for the first time that the use of well-stratified treatment-naïve UC patient samples in combination with high-throughput RNA-Seq technology can reveal new molecular players that might be important in UC pathogenesis. Potentially significant might be the regulation of tissue-specific pathophysiological properties of tight junctions in the mucosa as reflected by increased expression of AQP9 and CLDN2 and the expression of different mucins, particularly MUC5B and MUC16. In addition, a gender-dependent molecular manifestation could be established. The molecular patterns of UC revealed increased expression of genes involved in preserving mucosal integrity and detoxification of microbial-derived metabolites in females. The expression of antimicrobial and cytotoxic genes in male UC patients may contribute to the higher risk for the development of CRC observed in males. These results can be useful for the development of new treatment and patient stratification strategies for UC. In addition, these expression patterns can be extremely useful if combined with UC remission data in the future.
Funding
This work was supported by the Northern Norway Regional Health Authority Helse-Nord [SPF-1209–14].
Conflict of Interest
The authors declare no conflict of interests regarding the publication of this paper.
Author Contributions
HT performed most of the experiments and wrote parts of the manuscript. CGF performed most statistical analyses and revised the manuscript. IVH performed a part of the experiments and revised the manuscript. EA performed a part of the data analysis and revised the manuscript. JF was involved in evaluating and providing clinical samples from patients and healthy controls and revised the manuscript. RHP was involved in project inception, design, supervision, and manuscript writing and revision.
Supplementary Data
Supplementary data are available at ECCO-JCC online.
Supplementary Material
Acknowledgments
The authors thank Rasmus Goll and Mona Dixon Gundersen for providing clinical evaluation of some patients, Ingrid Christiansen for the technical help performing the TNF-α levels measurements, and Odd-Sverre Moen for administrating the patient samples.
References
- 1. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Planell N, Lozano JJ, Mora-Buch R et al. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut 2013;62:967–76. [DOI] [PubMed] [Google Scholar]
- 3. Jostins L, Ripke S, Weersma RK et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;49:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu JZ, van Sommeren S, Huang H et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loddo I, Romano C. Inflammatory bowel disease: genetics, epigenetics, and pathogenesis. Front Immunol 2015;6:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007;448:427–34. [DOI] [PubMed] [Google Scholar]
- 7. Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn’s disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet 2001;10:445–56. [DOI] [PubMed] [Google Scholar]
- 8. Burczynski ME, Peterson RL, Twine NC et al. Molecular classification of Crohn’s disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells. J Mol Diagn 2006;8:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Watanabe T, Kobunai T, Yamamoto Y et al. Predicting ulcerative colitis-associated colorectal cancer using reverse-transcription polymerase chain reaction analysis. Clin Colorectal Cancer 2011;10:134–41. [DOI] [PubMed] [Google Scholar]
- 10. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holgersen K, Kutlu B, Fox B et al. High-resolution gene expression profiling using RNA sequencing in patients with inflammatory bowel disease and in mouse models of colitis. J Crohns Colitis 2015;9:492–506. [DOI] [PubMed] [Google Scholar]
- 12. Magro F, Gionchetti P, Eliakim R et al. Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 1: definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders. J Crohn’s Colitis 2017;11:649–70. [DOI] [PubMed] [Google Scholar]
- 13. Mcnally PR. Budesonide for the induction of remission for mild- moderate ulcerative colitis. Visible Hum J Endoscpy 2014;13:2–7. [Google Scholar]
- 14. Olsen T, Goll R, Cui G et al. Tissue levels of tumor necrosis factor-alpha correlate with grade of inflammation in untreated ulcerative colitis. Scand J Gastroenterol 2007;42:1312–20. [DOI] [PubMed] [Google Scholar]
- 15. Dobin A, Gingeras TR. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics 2015;51:11.14.1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abbas AR, Baldwin D, Ma Y et al. Immune response in silico [IRIS]: immune-specific genes identified from a compendium of microarray expression data. Genes Immun 2005;6:319–31. [DOI] [PubMed] [Google Scholar]
- 17. Welter D, MacArthur J, Morales J et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anderson CA, Boucher G, Lees CW et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 2011;43:246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Lange KM, Moutsianas L, Lee JC et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arijs I, De Hertogh G, Lemaire K et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One 2009;4:e7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vanhove W, Peeters PM, Staelens D et al. Strong upregulation of AIM2 and IFI16 inflammasomes in the mucosa of patients with active inflammatory bowel disease. Inflamm Bowel Dis 2015;21:2673–82. [DOI] [PubMed] [Google Scholar]
- 22. Flach CF, Eriksson A, Jennische E, Lange S, Gunnerek C, Lönnroth I. Detection of elafin as a candidate biomarker for ulcerative colitis by whole-genome microarray screening. Inflamm Bowel Dis 2006;12:837–42. [DOI] [PubMed] [Google Scholar]
- 23. Noble CL, Abbas AR, Cornelius J et al. Regional variation in gene expression in the healthy colon is dysregulated in ulcerative colitis. Gut 2008;57:1398–405. [DOI] [PubMed] [Google Scholar]
- 24. Watanabe T, Kobunai T, Toda E et al. Gene expression signature and the prediction of ulcerative colitis-associated colorectal cancer by DNA microarray. Clin Cancer Res 2007;13:415–20. [DOI] [PubMed] [Google Scholar]
- 25. Olsen J, Gerds TA, Seidelin JB et al. Diagnosis of ulcerative colitis before onset of inflammation by multivariate modeling of genome-wide gene expression data. Inflamm Bowel Dis 2009;15:1032–8. [DOI] [PubMed] [Google Scholar]
- 26. Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 2014;9:e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. D’Haens G. Risks and benefits of biologic therapy for inflammatory bowel diseases. Gut 2007;56:725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lai Y, Li D, Li C et al. The antimicrobial protein REG3A regulates keratinocyte proliferation and differentiation after skin injury. Immunity 2012;37:74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Beelen Granlund A, Østvik AE, Brenna Ø, Torp SH, Gustafsson BI, Sandvik AK. REG gene expression in inflamed and healthy colon mucosa explored by in situ hybridisation. Cell Tissue Res 2013;352:639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu F, Dassopoulos T, Cope L et al. Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis 2007;13:807–21. [DOI] [PubMed] [Google Scholar]
- 31. Ufer M, Häsler R, Jacobs G et al. Decreased sigmoidal ABCB1 [P-glycoprotein] expression in ulcerative colitis is associated with disease activity. Pharmacogenomics 2009;10:1941–53. [DOI] [PubMed] [Google Scholar]
- 32. Ferguson LR, Browning BL, Huebner C et al. Single nucleotide polymorphisms in human Paneth cell defensin A5 may confer susceptibility to inflammatory bowel disease in a New Zealand Caucasian population. Dig Liver Dis 2008;40:723–30. [DOI] [PubMed] [Google Scholar]
- 33. West NR, Hegazy AN, Owens BMJ et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mankertz J, Schulzke JD. Altered permeability in inflammatory bowel disease: pathophysiology and clinical implications. Curr Opin Gastroenterol 2007;23:379–83. [DOI] [PubMed] [Google Scholar]
- 35. He F, Peng J, Deng XL et al. Mechanisms of tumor necrosis factor-alpha-induced leaks in intestine epithelial barrier. Cytokine 2012;59:264–72. [DOI] [PubMed] [Google Scholar]
- 36. Ricanek P, Lunde LK, Frye SA et al. Reduced expression of aquaporins in human intestinal mucosa in early stage inflammatory bowel disease. Clin Exp Gastroenterol 2015;8:49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Okada S, Misaka T, Matsumoto I, Watanabe H, Abe K. Aquaporin-9 is expressed in a mucus-secreting goblet cell subset in the small intestine. FEBS Lett 2003;540:157–62. [DOI] [PubMed] [Google Scholar]
- 38. Landy J, Ronde E, English N et al. Tight junctions in inflammatory bowel diseases and inflammatory bowel disease associated colorectal cancer. World J Gastroenterol 2016;22:3117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prasad S, Mingrino R, Kaukinen K et al. Inflammatory processes have differential effects on claudins 2, 3 and 4 in colonic epithelial cells. Lab Invest 2005;85:1139–62. [DOI] [PubMed] [Google Scholar]
- 40. Oshima T, Miwa H, Joh T. Changes in the expression of claudins in active ulcerative colitis. J Gastroenterol Hepatol 2008;23[Suppl 2]:S146–50. [DOI] [PubMed] [Google Scholar]
- 41. Rosenthal R, Milatz S, Krug SM et al. Claudin-2, a component of the tight junction, forms a paracellular water channel. J Cell Sci 2010;123:1913–21. [DOI] [PubMed] [Google Scholar]
- 42. Dorofeyev AE, Vasilenko IV, Rassokhina OA, Kondratiuk RB. Mucosal barrier in ulcerative colitis and Crohn’s disease. Gastroenterol Res Pract 2013;2013:431231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Allen A, Hutton DA, Pearson JP. The MUC2 gene product: a human intestinal mucin. Int J Biochem Cell Biol 1998;30:797–801. [DOI] [PubMed] [Google Scholar]
- 44. Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep 2010;12:319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shirazi T, Longman RJ, Corfield AP, Probert CS. Mucins and inflammatory bowel disease. Postgrad Med J 2000;76:473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walsh MD, Clendenning M, Williamson E et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod Pathol 2013;26:1642–56. [DOI] [PubMed] [Google Scholar]
- 47. Einerhand AW, Renes IB, Makkink MK, van der Sluis M, Büller HA, Dekker J. Role of mucins in inflammatory bowel disease: important lessons from experimental models. Eur J Gastroenterol Hepatol 2002;14:757–65. [DOI] [PubMed] [Google Scholar]
- 48. Smirnova MG, Birchall JP, Pearson JP. TNF-alpha in the regulation of MUC5AC secretion: some aspects of cytokine-induced mucin hypersecretion on the in vitro model. Cytokine 2000;12:1732–6. [DOI] [PubMed] [Google Scholar]
- 49. Felder M, Kapur A, Gonzalez-Bosquet J et al. MUC16 [CA125]: tumor biomarker to cancer therapy, a work in progress. Mol Cancer 2014;13:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Ther Adv Gastroenterol 2015;8:4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mirza AH, Berthelsen CH, Seemann SE et al. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med 2015;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takagi T, Naito Y, Mizushima K et al. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J Gastroenterol Hepatol 2010;25[Suppl 1]:S129–33. [DOI] [PubMed] [Google Scholar]
- 53. Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol 2009;182:2578–82. [DOI] [PubMed] [Google Scholar]
- 54. O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A 2007;104:1604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Padua D, Mahurkar-Joshi S, Law IK et al. A long noncoding RNA signature for ulcerative colitis identifies IFNG-AS1 as an enhancer of inflammation. Am J Physiol Gastrointest Liver Physiol 2016;311:G446–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Peloquin JM, Goel G, Kong L et al. Characterization of candidate genes in inflammatory bowel disease-associated risk loci. JCI Insight 2016;1:e87899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Betteridge JD, Armbruster SP, Maydonovitch C, Veerappan GR. Inflammatory bowel disease prevalence by age, gender, race, and geographic location in the U.S. military health care population. Inflamm Bowel Dis 2013;19:1421–7. [DOI] [PubMed] [Google Scholar]
- 58. Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology 2011;140:1785–94. [DOI] [PubMed] [Google Scholar]
- 59. Zelinkova Z, Janneke Van Der Woude C. Gender and inflammatory bowel disease. J Clin Cell Immunol 2014;5:245–50. [Google Scholar]
- 60. Choi J, Park T. Multivariate generalized multifactor dimensionality reduction to detect gene-gene interactions. BMC Syst Biol 2013;7[Suppl 6]:S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lesuis N, Befrits R, Nyberg F, van Vollenhoven RF. Gender and the treatment of immune-mediated chronic inflammatory diseases: rheumatoid arthritis, inflammatory bowel disease and psoriasis: an observational study. BMC Med 2012;10:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kobori A, Yagi Y, Imaeda H et al. Interleukin-33 expression is specifically enhanced in inflamed mucosa of ulcerative colitis. J Gastroenterol 2010;45:999–1007. [DOI] [PubMed] [Google Scholar]
- 63. Seidelin JB, Bjerrum JT, Coskun M, Widjaya B, Vainer B, Nielsen OH. IL-33 is upregulated in colonocytes of ulcerative colitis. Immunol Lett 2010;128:80–5. [DOI] [PubMed] [Google Scholar]
- 64. Mertz KD, Mager LF, Wasmer MH et al. The IL-33/ST2 pathway contributes to intestinal tumorigenesis in humans and mice. Oncoimmunology 2016;5:e1062966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ye Y, Xiao L, Wang SJ et al. Upregulation of REG3A in colorectal cancer cells confers proliferation and correlates with colorectal cancer risk. Oncotarget 2016;7:3921–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Harada N, Iijima S, Kobayashi K et al. Human IgGFc binding protein [FcgammaBP] in colonic epithelial cells exhibits mucin-like structure. J Biol Chem 1997;272:15232–41. [DOI] [PubMed] [Google Scholar]
- 67. Keppner A, Malsure S, Nobile A, Auberson M, Bonny O, Hummler E. Altered Prostasin [CAP1/Prss8] expression favors inflammation and tissue remodeling in DSS-induced colitis. Inflamm Bowel Dis 2016;22:2824–39. [DOI] [PubMed] [Google Scholar]
- 68. Katakura K, Lee J, Rachmilewitz D, Li G, Eckmann L, Raz E. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J Clin Invest 2005;115:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Billaut-Laden I, Allorge D, Crunelle-Thibaut A et al. Evidence for a functional genetic polymorphism of the human thiosulphate sulfurtransferase [Rhodanese], a cyanide and H2S detoxification enzyme. Toxicology 2006;225:1–11. [DOI] [PubMed] [Google Scholar]
- 70. Ramasamy S, Singh S, Taniere P, Langman MJ, Eggo MC. Sulfide-detoxifying enzymes in the human colon are decreased in cancer and upregulated in differentiation. Am J Physiol Gastrointest Liver Physiol 2006;291:G288–96. [DOI] [PubMed] [Google Scholar]
- 71. Wacklin P, Mäkivuokko H, Alakulppi N et al. Secretor genotype [FUT2 gene] is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One 2011;6:e20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wacklin P, Tuimala J, Nikkilä J et al. Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PLoS One 2014;9:e94863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rausch P, Rehman A, Künzel S et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 [Secretor] genotype. Proc Natl Acad Sci U S A 2011;108:19030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.