Abstract
The processes of DNA topoisomerization and site-specific recombination are fundamentally similar: DNA cleavage by forming a phospho-protein covalent linkage, DNA topological rearrangement, and DNA ligation coupled with protein regeneration. Type IB DNA topoisomerases are structurally and mechanistically homologous to tyrosine recombinases. Both enzymes nick DNA double helices independent of metal ions, form 3´-phosphotyrosine intermediates, and rearrange the free 5´ ends relative to the uncut strands by swiveling. In contrast, serine recombinases generate 5´-phospho-serine intermediates. A 180° relative rotation of the two halves of a 100kDa terameric serine recombinase and DNA complex has been proposed as the mechanism of strand exchange. Here I propose an alternative mechanism. Interestingly, the catalytic domain of serine recombinases has structural similarity to the TOPRIM domain, conserved among all Type IA and Type II topoisomerases and responsible for metal binding and DNA cleavage. TOPRIM topoisomerases also cleave DNA to generate 5´-phosphate and 3´-OH groups. Based on the existing biochemical data and crystal structures of Topoisomerase II and serine recombinases bound to pre- and post-cleavage DNA, I suggest a strand passage mechanism for DNA recombination by serine recombinases. This mechanism is reminiscent of DNA topoisomerization and does not require subunit rotation.
Keywords: TopIB, tyrosine recombinases, Topo II, serine recombinases, DNA swivel, subunit rotation, strand passage
Introduction
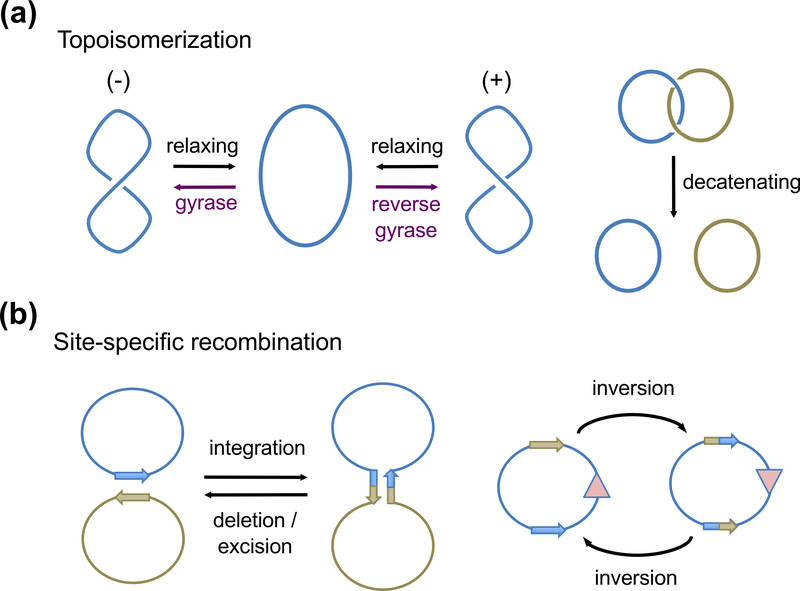
DNA topoisomerization and recombination both require breakage and rejoining of phosphodiester bonds to rearrange DNA segments. Topoisomerases cleave one or two DNA strands to change DNA linking numbers and are required during replication, transcription, recombination and repair to maintain genomic DNA integrity (Champoux, 2001; Wang, 2002; Schoeffler and Berger, 2008) (Fig. 1a). Many antibiotics and anti-cancer drugs that target topoisomerases are used to eliminate pathogens and kill tumor cells (Maxwell and Lawson, 2003; Balendiran, 2009). Conservative site-specific recombination occurs between two stringently defined DNA sequences and switches DNA connections (Fig. 1b). It is used by phages to integrate and excise from host genomes, by transposable elements to move about and spread antibiotic resistance, and by bacteria and plasmids to control plasmid copy number, regulate gene expression and separate catenated circular DNAs (Grindley et al., 2006).
Fig. 1.
Diagrams of topoisomerization and site-specific recombination. (a) DNA Topoisomerization by changing supercoils or decatenation of linked DNA circles. (b) Site-specific recombinases catalyze DNA integration, deletion (also called excision or resolution), and inversion. These reactions require two specific DNA sequences shown as arrows and negative supercols as the energy source. The color versions of all figure are available online.
Both topoisomerization and site-specific recombination form covalent protein-DNA intermediates to preserve the cleaved phosphodiester bonds. But at first glance their differences are apparent. Topoisomerization is in general non-sequence specific and requires cleavage of one strand, or at most two (Fig. 2a), whereas conservative site-specific recombination by tyrosine or serine recombinases is strictly sequence specific and requires cleavage of two double helices (four strands in total) (Fig. 2b). However, when the crystal structures of human and vaccinia topoisomerases I (type IB, TopIB) (Cheng et al., 1998; Redinbo et al., 1998) and the structures of tyrosine recombinases (YRs) of phage and bacterial origins (Guo et al., 1997; Hickman et al., 1997; Kwon et al., 1997; Subramanya et al., 1997) were determined, it was obvious that these seemingly divergent DNA enzymes share extensive sequence and structural similarity (Fig. 3). Ensuing mutagenesis and extensive biochemical and structural analyses confirm that TopIB and YRs share a conserved active site and catalytic mechanism (Gibb et al.; Perry et al.).
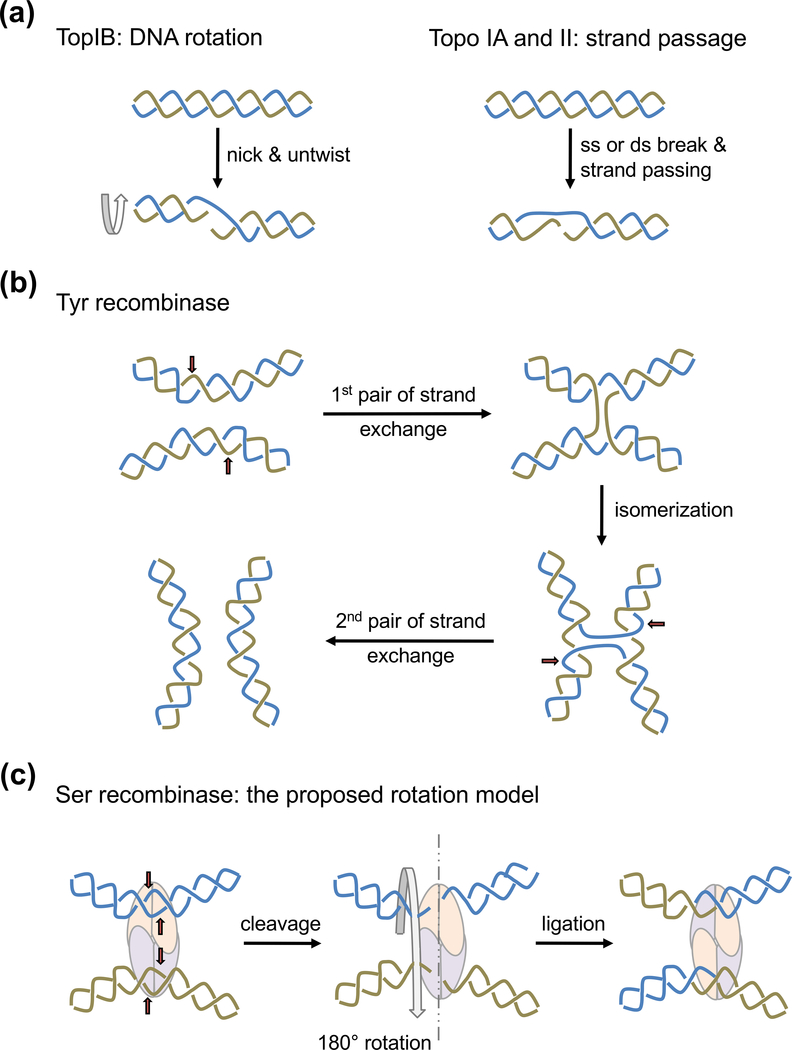
Fig. 2.
Mechanisms of topoisomerization and recombination by YRs. (a) Two mechanisms are used for topoisomerization. Type IB topoisomerases (TopIB) relax (−) or (+) supercoils by nicking one strand and allowing DNA to untwist. Topo IA and Topo II can relax as well as make supercoils by breaking one strand or duplex and passing the other through it. The intertwined blue and beige color lines represent single strands in the case of Topo IA or double helices in the case of Topo II. (b) Tyrosine recombinases (YRs) catalyze DNA recombination in two steps. One pair of strands are cleaved, exchanged and rejoined in each step (beige and blue colored lines sequentially). The red arrows point the cleavage sites. A Holliday junction is formed and isomerized between the two steps. (c) Serine recombinases make concerted double strand breaks of two recombination sites. The proposed subunit rotation mechanism requires the left half of the cleaved DNA rotate 180° relative to the right half for strand exchange.
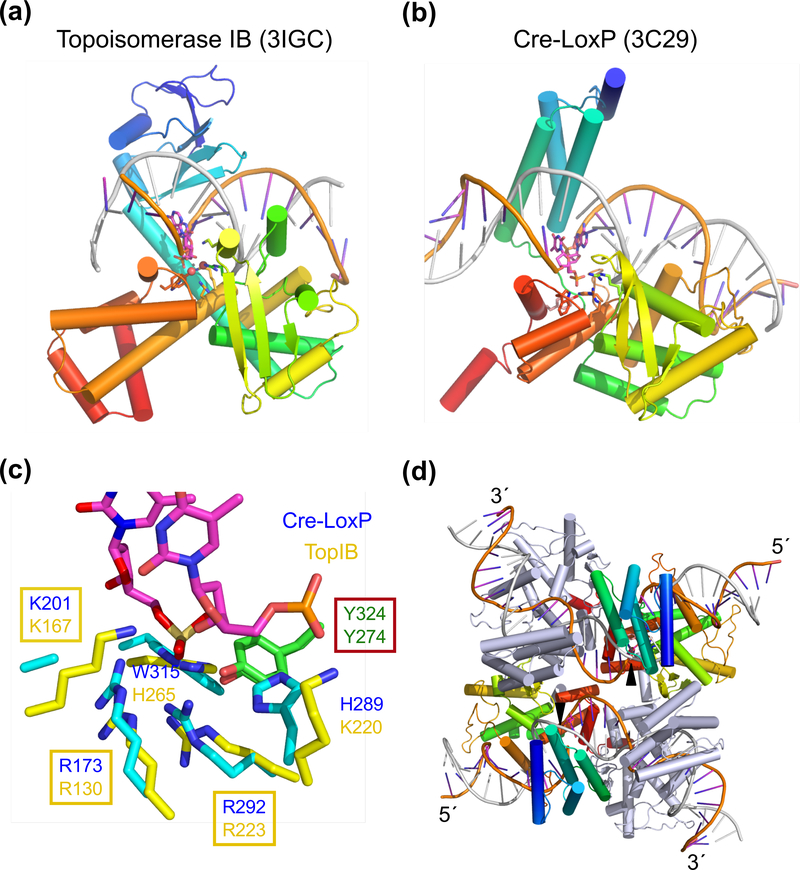
Fig. 3.
Structural comparison of TopIB and YRs. (a) The structure of smallpox viral topoisomerase complexed with DNA (PDB: 3IGC). (b) One subunit of Cre bound to one half of a loxP site (PDB: 3C29). In (a) and (b) each polypeptide chain is shown as blue (N-) to red (C-terminus) rainbow-colored ribbon diagrams. The DNA strand being nicked is colored orange and the complementary strand in silver. The active site residues are shown in sticks. (c) Superposition of the active sites of TopIB and YR. Both tyrosine nucleophiles are shown in green (carbon) and red (oxygen). Other conserved residues are shown in yellow (TopIB) and cyan (YR) with blue nitrogen. A trio of basic residues, RKR, is conserved among all TopIBs and YRs. Nearby His and Trp conserved among YRs are replaced, respectively, by Lys and His conserved in TopIBs. (d) The tetrameric Cre-loxP complex (PDB: 3C29). The view is roughly perpendicular to (b). The two Cre subunits poised to cleave DNA are shown in rainbow colors and the other two in grey. Once the orange-colored strands are cleaved (indicated by the black arrowheads), their 3´-ends are covalently linked to the Cre subunits, and the free 5´ ends reciprocally invade and rejoin with the neighboring DNAs thus forming a HJ. Isomerization of HJ activates the grey-colored Cre subunits to catalyze the exchange of silver-colored strands.
In this review, I will summarize the relationship between TopIB and YRs and focus the main discussion on the previously unnoticed similarity between type II topoisomerases (Topo II) and serine recombinases (SRs). To keep things in perspective, topoisomerization and site-specific recombination will be briefly reviewed here. For more detail, readers are referred to the following in-depth reviews on topoisomerization (Wang, 1996; Champoux, 2001; Schoeffler and Berger, 2008) and site-specific recombination (Van Duyne, 2001; Chen and Rice, 2003a; Jayaram et al., 2004; Grindley et al., 2006; Kamtekar et al., 2006).
Topoisomerization by TopIB, Topo IA and Topo II
The DNA double helix can form negative or positive supercoils (Vologodskii and Cozzarelli, 1994; Bates and Maxwell, 1997), which can lead to complex tertiary structures. DNA topology is defined by the linking number that consists of the sum of the twist and the writhe, which are the number of rotational turns within a duplex and the number of supercoils between helical segments, respectively (Crick, 1976). Twist and writhe are freely interchangeable without the breaking of phosphodiester bonds, but to change DNA topology or the linking number, at least one DNA strand must be cleaved. All topoisomerases, which cleave DNA and change its topology, use a tyrosine sidechain as the nucleophile to break a DNA phosphodiester bond and replace it with a phosphotyrosyl protein-DNA covalent bond, which is reverted back to a phosphodiester bond after the topological changes (Champoux, 1977; Wang, 1996).
Topoisomerases are divided into two types. Type I topoisomerases cleave one strand in a double helix and are often monomeric; type II topoisomerases cleave both strands and are dimeric or heterotetrameric (Wang, 1996; Forterre et al., 2007). Each type of topoisomerase can be further subdivided. The majority of topoisomerases belong to type IA, IB, IIA or IIB. These four types are segregated into two classes based on sequence conservation and cleavage mechanism (Table 1). Type IA (Topo IA) and type IB (TopIB) are unrelated in protein sequence, structure and DNA cleavage mechanism, but type IA shares a conserved catalytic domain, TOPRIM, with type IIA and IIB topoisomerases (Topo IIA and IIB, grouped together as Topo II in this review) (Forterre et al., 2007; Schoeffler and Berger, 2008).
Table 1.
Comparison of topoisomerases and site-specific recombinases
| TopIB | Tyrosine recombinases | Topo IA TopoII | Serine recombinases | |
|---|---|---|---|---|
| Nucleophile | Y | Y | Y | S |
| Catalytic residues | RKR | RKR | E-(K), DxD, R | R, DRxxR |
| Metal ion | No | No | Mg2+ | (Mg2+) |
| Cleavage product | 3´-P-Tyr, 5´-OH | 3´-P-Tyr, 5´-OH | 5´-P-Tyr, 3´-OH | 5´-P-Ser, 3´-OH |
| Multimer | monomer | tetramer | dimer, tetramer | tetramer |
| Mechanism | DNA swivel | swivel exchange | strand passage | strand passage? |
* parentheses indicate what are applicable to certain but not all members.
TopIB generates 5´-OH and 3´-phosphotyrosine cleavage products and can release both negative (−) and positive (+) supercoils by nicking double-stranded DNA (Table 1). As seen in human and several viral topoisomerases I, the active site of TopIB consists of an invariable Tyr nucleophile and positively charged Lys, Arg, and His residues (Champoux, 2001; Gibb et al.). The reaction requires no metal ions and instead relies on the positively charged sidechains to activate the phosphoryl-transfer reaction and neutralize the transition state. It doesn’t require high-energy cofactor, e.g. ATP, and uses the energy stored in the supercoils to drive DNA relaxation. Thus the efficiency is correlated with the supercoil density. Release of supercoils is achieved by rotation (untwisting) of DNA downstream from the nick around the continuous strand (Fig. 2a) (Stivers et al., 1997; Koster et al., 2005). TopIB can change the linking number by more than 1 unit in a single round of nicking and rejoining. Because of the rotational mechanism, type IB topoisomerases are also referred to as swivelases.
Topo IA and Topo II require metal ions and generate 5´-phosphate and 3´-OH cleavage products (Wang, 1996) (Table 1). Topo IA binds and cleaves a single stranded region and relaxes (−) supercoils by passing the continuous strand through the nicked one (Fig. 2a). Topo II changes DNA topology by passing a dsDNA segment (T segment) through the cleaved segment (G segment standing for gate). Therefore, the unit of linking number change per cycle is 1 by Topo IA and 2 by Topo II (Wang, 1996). Reverse gyrase, which belongs to type IA, and all Topo II enzymes are ATP dependent and contain either the SFII-helicase or GHKL ATPase domains (Confalonieri et al., 1993; Schoeffler and Berger, 2008). In the cases of gyrase (type IIA) and reverse gyrase, which introduce negative and positive supercoils to DNA, respectively, the requirement for the high-energy cofactor ATP is self-evident. The reason for the ATP-hydrolysis dependence of type II topoisomerases that relax (−) and (+) supercoils is less clear (Bates and Maxwell). ATP may play an essential regulatory role in coordinating the double-strand cleavage with DNA passage and avoiding hazardous double-strand breaks.
Site-specific recombination by YRs and SRs
Conservative site-specific recombination occurs between two DNA sites of specific sequences and, at the end of recombination there is no net loss or gain of nucleotide (Grindley et al., 2006), which differs from other types of site-specific recombination. For example, cut-and-paste recombination occurs when a donor DNA is flanked by specific sequences but the target DNA contains no specific sequence, and at the end of recombination a few basepairs of the target site are duplicated as a result of DNA synthesis (see the recent review by Hickman et al. (Hickman et al., 2010)). To date two types of recombinases have been found to catalyze conservative site-specific recombination. Based on protein sequence conservation and nucleophile for DNA cleavage (Tyr versus Ser), they are known as tyrosine recombinases (YRs) and serine recombinases (SRs) (Stark et al., 1992). YRs and SRs are unrelated in protein sequence and recombination mechanism. YRs form 3´-phosphotyrosyl intermediates, whereas SRs form 5´-phosphoseryl intermediates instead.
The archetypal YR is phage λ integrase (Nash, 1981; Campbell, 2007) (Fig 1b). This mechanism is used by other phages, e.g. HP1, for infection (Radman-Livaja et al., 2006), by bacteria for decatenation at the end of replication of circular DNAs (Sherratt et al., 2004), by bacterial transposable elements (Burrus et al., 2006; Macdonald et al., 2006) to spread antiobiotic resistance, and by fungal plasmids for copy number regulation (Jayaram et al., 2004). Cre encoded by phage P1 is a tyrosine recombinase and is a part of the Cre-loxP system that is successfully exploited for knocking out genes in higher eukaryotes. Biochemical and structural analyses of Cre-loxP, Flp and λ integrase reveal an obligatory cyclic tetramer of tyrosine recombinase subunits, which form the structural framework for recombination (Guo et al., 1997; Chen et al., 2000; Biswas et al., 2005), and DNA cleavage and strand exchange occur inside this framework. Recombination by YRs occurs in two DNA cleavage-ligation steps (Fig. 2b). The tyrosine recombinase tetramers are asymmetric with only two subunits active at any given time, so that one strand of each DNA recombination site is cleaved, exchanged and ligated in each step (Grindley et al., 2006). The two chemical steps are linked by a Holliday-junction (HJ) intermediate and, sequential activation of the recombinase subunits is accompanied by HJ isomerization (Grindley et al., 2006) (Fig. 2b).
Alternatively, various bacteriophage and transposable elements in bacteria and lower eukaryotes depend on serine recombinases (SRs) to carry out conservative site-specific recombination to infect host cells (e.g. ϕC31 and Bxb1) (Smith et al., 2010), spread antibiotic resistance (e.g. Tn21) (Liebert et al., 1999; Olorunniji and Stark, 2010) or regulate gene expression by inverting DNA segments (Van De Putte and Goosen, 1992). SRs share a highly conserved catalytic core (Simon et al., 1980) and are represented by the well-characterized γδ and Tn3 resolvases and Hin invertase (Grindley et al., 2006). Like YRs, SRs require two specific recombination signal sequences and a minimum of four SR subunits to carry out 4-strand cleavage, DNA rearrangement and ligation of new partners. Negative supercoiling is necessary to drive directional site-specific recombination by resolvases and invertases (Wasserman et al., 1985; Stark et al., 1989; Benjamin and Cozzarelli, 1990). Different from YRs, DNA cleavage by SRs is concerted and generates two double-strand breaks rather than the pairwise sequential nicking (Grindley et al., 2006). Like Topo IA and Topo II SRs cleave DNA to generate 5´-phosphate and 3´-OH groups (Table 1). SRs form no cyclic protein framework and no Holliday junction (Stark et al., 1989). The proposed architecture of SR recombination consists of a tetrameric SR core with pseudo-D2 symmetry and two DNA recombination sites bound on the outside of the protein core (Leschziner and Grindley, 2003; Dhar et al., 2004; Nollmann et al., 2004; Li et al., 2005). It has been suggested that recombination proceeds by a 180° relative rotation of the two halves of this protein-DNA complex (Fig. 2c). This rotation/swivel model fundamentally differs from that of TopIB because there is no covalent attachment between the two rotating halves.
Auxiliary DNA sites, which are bound by bacterial proteins, e.g. IHF, HU, or FIS and additional domains or recombinase subunits, often regulate the activity and directionality of site-specific recombination for their biological purposes (Nash, 1981; Johnson et al., 1986; Grindley et al., 2006; Radman-Livaja et al., 2006). Phage SR-type integrases are exceptional by catalyzing recombination with no apparent auxiliary site (Ghosh et al., 2003). In a typical case for γδ resolvase to divide a large cointegrate DNA into two catenated smaller circles, 12 resolvase subunits (6 dimers) are needed to bind to two res sites, each of which contains 3 (I, II and III) non-identical inverted repeats (Grindley et al., 2006; Olorunniji and Stark, 2010). Only site I of each res is cleaved and recombined; sites II and III are auxiliary resolvase-binding sites used to activate recombination and govern the reaction direction. When the two recombination-sites are direct repeats as in the resolvase case, the outcome is deletion (Fig. 1b, left). But if they are inverted repeats, the orientation of the genes in between becomes inverted by recombination (Fig. 1b, right). For example, salmonella uses Hin invertase and hix recombination sites to invert the promoter of flagellin genes thus achieving flagellar phase variation (Zieg et al., 1977).
To simplify the recombination system for mechanistic understanding, Tn3, γδ and Sin resolvases and Hin invertase have been genetically modified so that mutant SRs can catalyze recombination on site I (referred to as the crossover site from here on) alone without auxiliary protein and DNA (Arnold et al., 1999; Sarkis et al., 2001; Burke et al., 2004; Dhar et al., 2004; Rowland et al., 2009). But the recombination catalyzed by these “activated” enzymes has no directionality (either inversion or deletion) and may go through a different reaction pathway from wildtype recombinases (Arnold et al., 1999; Sarkis et al., 2001; Kamtekar et al., 2006; Gehman et al., 2008; Dhar et al., 2009).
Similarity between TopIB and YRs
Despite the absence of apparent similarity in protein sequences and different biological functions, crystal structures of YRs and TopIB reveal an uncanny similarity (Fig. 3a-b). Their catalytic domains are both located at the C-terminus and have the same secondary structural elements and folding topology. The catalytic residues of YRs and TopIB, including the Tyr nucleophile and a basic trio RKR that serves as the general base and acid, are completely conserved and functionally essential (Fig. 3c). The nearby H (His) and W (Trp) that are conserved in YRs are replaced by K (Lys) and H (His) in TopIBs. These residues play a role in catalysis but are not essential to YRs or TopIB (Krogh and Shuman, 2000; Yang and Champoux, 2001; Chen and Rice, 2003b; Whiteson et al., 2007; Gibb et al., 2010). In retrospect, one could have predicted that only the residues conserved in both YRs and TopIB are essential for catalysis.
In addition to the C-terminal catalytic domains, each TopIB and YR subunit contains an N-terminal DNA-binding domain. Although the structural elements are different between YRs (all α-helices) and TopIB (a central 5-stranded β-sheet) (Grindley et al., 2006), in both cases these DNA-binding domains bind the major groove (Fig. 3a-b) opposite the catalytic domain. Interestingly, even with homologous DNA-binding domains, TopIB enzymes of some viral origins have DNA sequence preferences (Shuman and Prescott, 1990; Perry et al., 2006; Minkah et al., 2007), while others, e.g. human topoisomerase I, are non-sequence specific (Redinbo et al., 1998; Champoux, 2001). With the N- and C-terminal domains together each YR and TopIB subunit covers ~13 bp of DNA substrate. Their pattern of DNA binding precludes close juxtaposition of cleavage sites on a single duplex. TopIB nicks one strand only, and the closest placement of two YRs results in cleavage 6 bp apart as observed with Cre-loxP. The spacing of 6–8 bp between the sequential nicking sites is ideal for strand exchange and formation of a Holliday junction within the framework of YR tetramers (Fig. 3d).
Some differences between TopIB and YRs may underlie the different outcomes of the two biological processes. Firstly, TopIBs are predominantly monomeric, whereas YRs may be monomeric (Kwon et al., 1997; Subramanya et al., 1997) or dimeric (Hickman et al., 1997) in the absence of DNA substrate but form functional tetramers when bound to two recombination sites (Guo et al., 1997; Chen et al., 2000; Biswas et al., 2005). Each recombination site is composed of an inverted repeat of a 13–14 bp recognition sequence separated by the 6–8 bp cleavage site. Upon association with YRs the four DNA half sites are bound by the tetramer in a pseudo P4 symmetry. The four-fold symmetry is the foundation for the square-planar Holliday junction (HJ) intermediate and also for its isomerization to activate the second pair of strand exchange (Fig. 2b). Secondly, the protein-DNA interactions play an important role in the efficiency and nature of the DNA swivel. The extent of TopIB-DNA interactions appears to influence the rate of DNA rotation due to the friction imposed by the protein-DNA interface (Koster et al., 2005). The sequence-specific interactions upstream from the scissile phosphate and small interface with the downstream DNA by the viral TopIB compared to human TopIB allows the former to relax more supercoils (5–20) than the latter, in one cleavage-religation event (Stivers et al., 1997; Koster et al., 2005). The protein-DNA interface between YRs and substrate DNA is sequence specific both upstream and downstream of the scissile phosphate. Such extensive protein-DNA interactions allow strand exchange only and no DNA rotation during recombination. Thirdly, the DNAs bound to TopIBs are more or less in a straight B form (Redinbo et al., 1998; Perry et al., 2006), whereas DNAs bound to the tetrameric YRs are inevitably bent by 70–90°, as dictated by the pseudo four-fold symmetry (Guo et al., 1997; Chen et al., 2000; Biswas et al., 2005). The bent DNA bound to YRs not only leads to asymmetric cleavage, but it probably promotes the exchange of free 5´ ends like a loaded spring.
TOPRIM domain
TOPRIM is the conserved domain found in Topo IA and Topo II as well as in bacterial DnaG-like primases, which gives rise to the name of TOPRIM (Aravind et al., 1998). In addition TOPRIM is also the catalytic domain among OLD family nucleases, including RNase M5 (Allemand et al., 2005). The tertiary structure of TOPRIM consists of four βα repeats forming a four-stranded parallel β sheet surrounded by two α helices on each face (Fig. 4). Large insertions in loops and N- and C- terminal appendages are often found in primases and topoisomerases, e.g. a large insertion after the 1st βα repeat in Topo IA, insertion in the 2nd βα repeat in Topo II (Fig. 5).
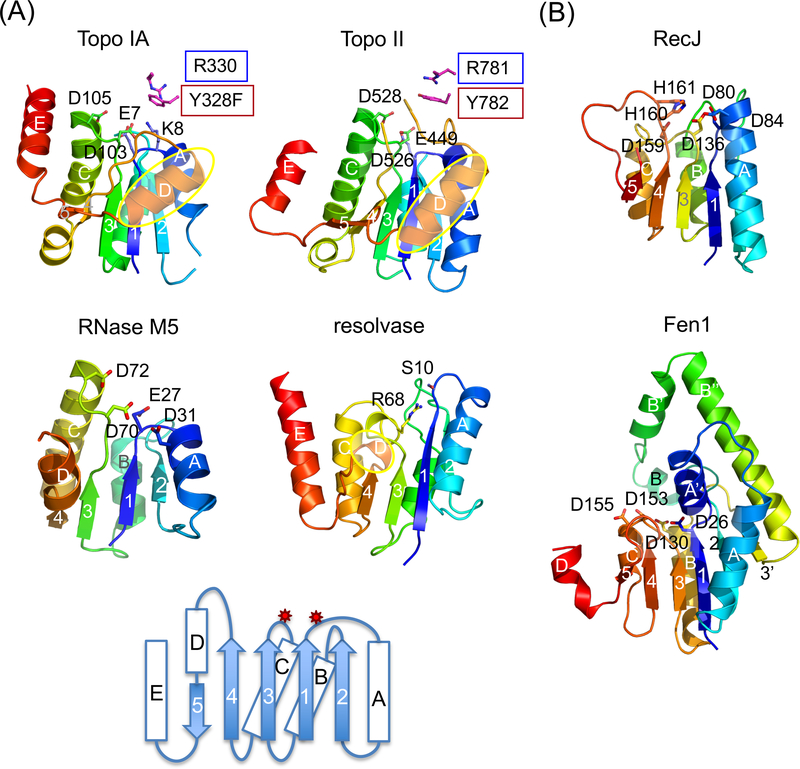
Fig. 4.
TOPRIM and SR catalytic domain are similar in the folding topology and active-site location. (a) TOPRIM domain of Topo IA (1I7D), II (2RGR) and RNase M5 (1T6T) are aligned with the catalytic domain of γδ resolvase (1GDT). Each is shown in rainbow-colored ribbon diagrams. Active site residues are highlighted as sticks. The D helices (in yellow ovals) in topoisomerases are longer than in resolvase. A topology diagram below summarizes all four examples. The catalytic residues are located on two loops indicated by the red stars. Three carboxylates (E, DxD) are conserved among all TOPRIM domains for metal-ion binding. M5 nuclease has an additional carboxylates (D31) and uses water as nucleophile. Topo IA has a catalytic Lys (K8) in the equivalent location. (b) Ribbon diagrams of the catalytic domain of RecJ (1IR6) and Fen1 (1EXN) in rainbow colors. The catalytic residues are shown and labeled. Inserted elements in Fen1 are labeled after the adjacent secondary structures with apostrophes (A’, B’, B” and 3’).
Fig. 5.
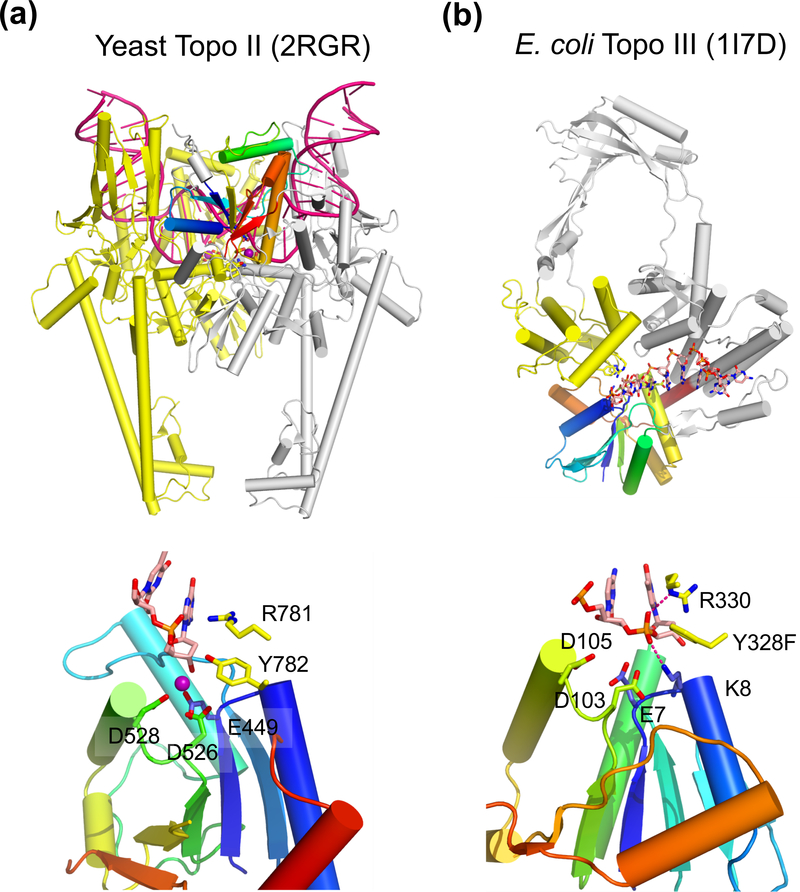
Structure of type II and type IA topoisomerase. (a) The overall structure of the dimeric yeast Topo II (type II) bound to a substrate DNA (G segment, PDB: 2RGR). One subunit is shown in silver with the TOPRIM domain in rainbow colors. The other subunit, which donates the tyrosine nucleophile to the rainbow-colored TOPRIM, is shown in yellow. The DNA substrate is severely bent and shown in dark pink. The N-terminal ATPase domain is absent in the crystal structure (not shown) and would be on top of DNA in this view. (b) E. coli Topo III (type IA, PDB: 1I7D) is monomeric. The TOPRIM is shown in rainbow colors. The tyrosine nucloephile is donated by the yellow domain. The rest of the protein is colored in silver, The ssDNA substrate is shown as multicolored sticks. The active site of Topo II and Topo III are shown below. In addition to the three carboxylates (E, DxD), the catalytically essential Arg and Tyr (nucleophile) are donated from the second subunit or domain (colored yellow) for catalysis. The Mg2+ found in Topo II is shown as a purple sphere.
Four carboxylates are conserved among the OLD-family nucleases and DnaG-like primases. They are located between the β and α elements of the first and third βα repeats. These carboxylates are likely involved in coordination of two metal ions for catalysis (Keck et al., 2000; Yang, 2010). Interestingly, only three out of these four carboxylates are conserved in the TOPRIM domains of Topo IA and Topo II (Fig. 4). The three carboxylates together with the leaving group (3´-O) of the scissile phosphate coordinate a single metal ion (at site B) (Dong and Berger, 2007; Schmidt et al., 2010) (Fig. 5a). The absence of the fourth carboxylate in the metal-binding site of the topoisomerases appears to be correlated to the one-metal-ion mechanism used by the topoisomerases versus the two-metal-ion mechanism used by the nucleases and primases. The missing metal ion in the topoisomerases is likely compensated by (1) the presence of a positively charged Lys (Topo IA) and Arg (both Topo IA and II) (Fig. 5) and (2) using Tyr as the nucleophile instead of a water molecule or 3´-OH (Table 1). The pKa of a Tyr nucleophile is much closer to neutrality than a water molecule and 3´-OH, making it a better nucleophile and less dependent on metal ions. The Tyr nucleophile is supplied in trans by a domain other than TOPRIM in Topo IA and by a second subunit in Topo II (Lima et al., 1994; Berger et al., 1996; Changela et al., 2001; Dong and Berger, 2007) (Fig. 5).
The catalytic domain of SRs is similar to TOPRIM
Crystal structures of γδ resolvase in apo form, a pre-cleavage resolvase dimer-DNA complex, and a post-cleavage resolvase tetramer-DNA complex have been determined (Sanderson et al., 1990; Yang and Steitz, 1995; Li et al., 2005; Kamtekar et al., 2006) (Fig. 6a-b). In addition, the crystal structure of a homologous Sin resolvase bound to a DNA regulatory site reveals another form of tetrameric synapse of SRs (Rowland et al., 2002; Mouw et al., 2008) (Fig. 6c). Early on mutagenesis and biochemical analyses identified residues essential for catalysis and assembly of the “resolvosome” (Newman and Grindley, 1984; Grindley, 1993; Hughes et al., 1993). The nucleophilic Ser and the nearby three Arg resides (R8, R68 and R71) and one Asp (D67) are important for DNA cleavage and rejoining (Boocock et al., 1995; Grindley et al., 2006). Residues located on the opposite side of the catalytic center, yet required for recombination, have been suggested to form protein-protein interactions and bring the auxiliary DNA sites together (Hughes et al., 1993; Rice and Steitz, 1994; Murley and Grindley, 1998). However because of the requirement for 12 γδ resolvase subunits to catalyze the site-specific recombination properly, the exact function of quite a number of conserved and essential residues is unassigned.
Fig. 6.
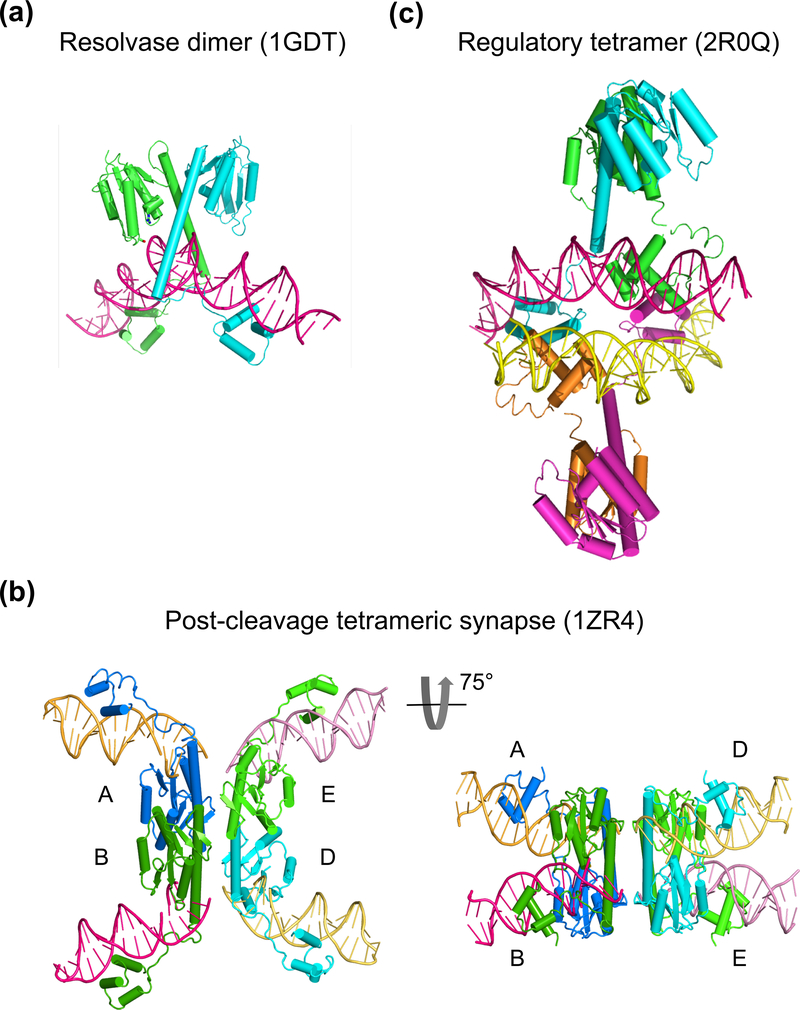
Crystal structures of resolvase-DNA complexes. (a) The pre-cleavage resolvase dimer-site I DNA complex (PDB: 1GDT). The protein subunits are shown as cyan and green ribbon diagrams and DNA in pink tube-and-ladder. (b) Two views of a post-cleavage tetrameric resolvase complex with two cleaved DNAs (PDB: 1ZR4). On the left is the conventional view with DNAs appearing to be outside of the protein tetramer. On the right is a view rotated about the horizontal axis by 75°. The diagonal subunits in the left panel are now side-by-side, and the light and dark pink as well as yellow and orange DNAs appear to be co-linear. (c) A synapse of Sin resolvase tetramer bound to two regulatory site DNAs (PDB: 2R0Q). DNAs (pink and yellow) are inside of the protein tetramer (cyan, green, orange and magenta).
Each resolvase subunit contains an N-terminal catalytic domain, a C-terminal DNA binding domain, and a long E helix connecting the two. In solution resolvases are dimers formed between the catalytic domains and mediated by E helices (Sanderson et al., 1990). The catalytic domain of SRs consists of ~ 120 residues, which form a mixed five-stranded β sheet surrounded by helices on either side. The Ser nucleophile is located at the end of the β strand of the first βα repeat. The structural similarity of resolvase with 5´ Flap endonuclease (Fen1) and RecJ has been noted previously (Artymiuk et al., 1997; Yang, 2010).
Interestingly, the catalytic domain of SRs is more similar to TOPRIM than to RecJ or Fen1 in its folding topology and active site location. SRs and TOPRIM share 4 parallel βα repeats, and both have a fifth βα repeat, with its β strand antiparallel to the first four in the mixed β sheet (Fig. 4a). The active site of SRs consists of the Ser nucleophile (S10) and conserved arginines and aspartate (Table 1). Similar to the conserved carboxylates in the TOPRIM domain, these catalytically essential residues are located in the β to α connections of the first and third βα repeats in SRs (Fig. 4a). Superposition of the catalytic core of resolvase and the TOPRIM of Topo II results in an rmsd of 2.7Å over 80 pairs of Cα atoms, which is only slightly larger than the 2.2Å rmsd over 109 pairs of Cα atoms between the TOPRIM domains of Topo IA and II. The main difference is that the D helix in resolvase is much shorter or non-existent (Fig. 4a). In contrast, the rmsd between resolvase and Flap endonuclease is 3.1Å over 73 pairs of Cα atoms. Fen1 has a βα insertion between the first and second repeat, and its catalytic residues are located in first, second and third βα repeats (Artymiuk et al., 1997) (Fig. 4b). Although the rmsd between RecJ and resolvase is relatively low, 2.3Å over 68 pairs of Cα atoms, RecJ doesn’t have the fifth βα repeat and its catalytic residues are located in first, third and fourth βα repeats instead (Yang, 2010) (Fig. 4b).
Metal dependence of TOPRIM enzymes and SRs
Whether TOPRIM topoisomerases require metal ions to cleave DNA was unclear and debated for a long time. DNA cleavage by Topo IA could occur in the presence of EDTA (Tse-Dinh et al., 1983; Domanico and Tse-Dinh, 1991) and metal ions were absent in crystal structures of Topo IA and Topo IA-DNA complexes (Lima et al., 1994; Mondragon and Digate, 1999; Changela et al., 2001). The hallmark of TOPRIM is the conserved carboxylates, however, and their role normally is to coordinate metal ions (Aravind et al., 1998; Keck et al., 2000; Kato et al., 2003; Rezacova et al., 2008). Mutations of the conserved carboxylates in Topo IA do diminish the DNA cleavage activity (Zhu and Tse-Dinh, 2000). The recently determined crystal structures of Topo II and Topo IV (type II) complexed with DNA substrate and cleaved intermediate consistently show a metal ion coordinated by the conserved carboxylates in TOPRIM domain (Laponogov et al.; Dong and Berger, 2007; Schmidt et al., 2010). Structural superposition of the TOPRIM domains of Topo IA and Topo II suggests that the metal-ion binding site coordinated by the conserved carboxylates and the leaving group of the scissile phosphate is present in Topo IA and both Topo IA and II likely use the one-metal-ion mechanism to cleave and/or ligate DNA (Dong and Berger, 2007; Yang, 2010). The absence of metal ion in the crystal structures of Topo IA and ssDNA complex is likely due to the greater than 1 M salt concentration in the crystallization buffer (Changela et al., 2001). The Lys conserved among type IA topoisomerases (K8 in Fig. 5) may alleviate the metal dependence in the cleavage step, but Topo IA requires metal ions for the ligation step (the second chemical reaction) (Tse-Dinh et al., 1983; Domanico and Tse-Dinh, 1991). According to its location observed in the Topo II structures (Dong and Berger, 2007; Schmidt et al., 2010), the metal ion would be in position to coordinate and activate the nucleophile 3´-OH in the ligation step.
Early on, γδ resolvase was shown to cleave DNA without Mg2+ but require Mg2+ for strand rejoining (Reed and Grindley, 1981). The metal ion dependence of γδ resolvase is thus similar to Topo IA and Topo II. Interestingly, ensuing studies of other resolvases and phage integrase show that metal ion is not essential for recombination (Castell et al., 1986; Stark et al., 1989; Ghosh et al., 2003). Phage Bxb1 integrase has additional 350 residues of unknown structure and function after the conserved SR core and can catalyze DNA recombination between linear DNAs (Kim et al., 2003). Nevertheless Mg2+ is usually included in the reaction buffer of resolvasaes and invertase to increase recombination rate and efficiency (Leschziner and Grindley, 2003; Dhar et al., 2009; Rowland et al., 2009). Structural comparison of resolvase with Fen1 and RecJ suggest that for the strand cleavage reaction, positively charged sidechains (e.g. R68 of γδ resolvase) may replace the metal ion that is coordinated by the 3´ leaving group. During the strand rejoining reaction, the Ser (in phospho-serine form) becomes the leaving group, and a Mg2+ ion may facilitate the activation of nucleophile 3´-OH.
Strand passage mechanism by Topo II and subunit rotation by SRs
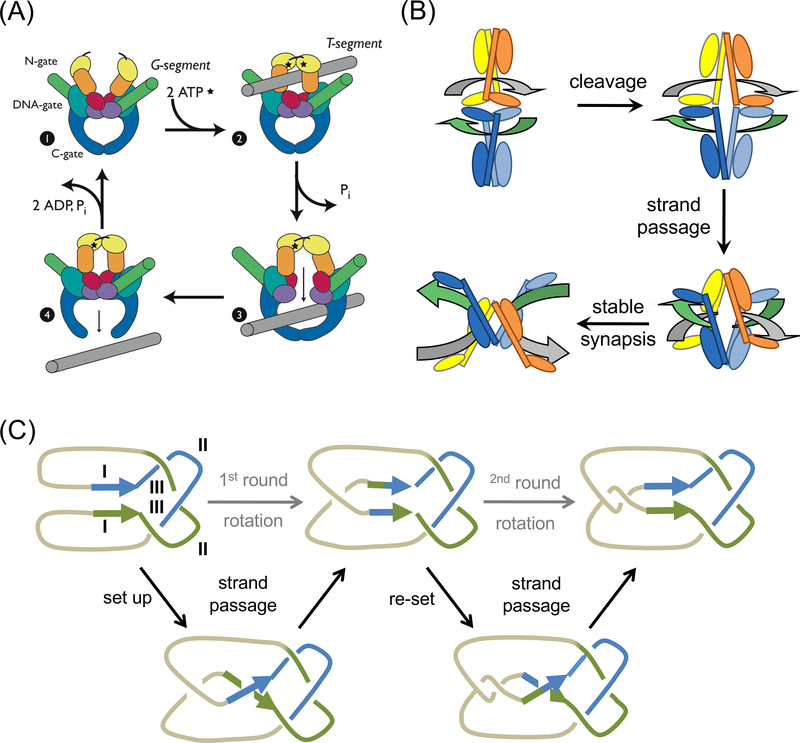
Topo IA and Topo II alter DNA linking number by a “strand passage” mechanism, which is fundamentally different from the swivel mechanism used by TopIB (Fig. 2a). After breaking a single strand, Topo IA allows the continuous (uncut) strand to pass through the gap of the cleaved DNA before re-ligation, thus changing linker number by 1. Similarly Topo II allows the passage of a double-stranded DNA segment (T) through the cleaved DNA (G), thus changing the linker number by 2. Ample biochemical, mutagenic and structural data support the strand passage mechanism (Kreuzer and Cozzarelli, 1980; Liu et al., 1980; Wang, 1996; Schoeffler and Berger, 2008). The DNA passage is blocked by several protein- and DNA-gates (Fig. 8a), which open and close concertedly in a DNA-binding and ATP-hydrolysis dependent manner.
Fig. 8.
Strand passage mechanism. (a) A diagram of topoisomerization by a type II enzyme. The dimeric Topo II protein itself has two gates (N and C), which can open and close in an ATP hydrolysis and DNA dependent manner. DNA T segment (shown in grey) is transported through the open protein N gate, the cleaved DNA gate (G segment, shown in green, and eventually out of the protein C gate. This figure is a reproduction of Schoeffler et al. (Schoeffler and Berger, 2008) with permission. (b) A diagram of SRs recombining DNAs by the strand passage mechanism. Each SR subunit contains a large N-terminal catalytic domain and a small C-terminal DNA binding domain linked by the long E helix. Two SR dimers (in distinct colors) enclose the two DNA recombination sites (grey and green). Polarity of res DNA is indicated by the arrowhead. DNA cleavage requires some conformational changes at the dimer interface. After DNA cleavage, the two SR dimer-cleaved DNA complexes approach each other and DNAs cross and pass each other through the cleaved DNA and the open C-termini of E helices. The two DNAs are recombined from being horizontally connected at the beginning to vertically connected, which is reminiscent of recombination by YRs as diagramed in Fig. 2b. (c) A topological diagram of DNA resolution by Tn21-family resolvases. The two res sites (direct repeats) are shown in green and blue and the intervening DNAs shown in light brown. Each res site consists of site I, II and III as labeled. The recombination synapse traps 3 negative supercoils as experimentally determined. Additional local + and - supercoil occur but cancel one another. After strand cleavage, crossing and partner switching, the ligation product two singly linked catenanes.
In contrast, the recombination mechanism of SRs has been proposed to involve a subunit rotation of 180° (Fig. 2c). In this model, SRs use the molecular surface distal to the catalytic centers to form a tetramer and the two DNA crossover sites are bound on the outside of the tetramer and do no interact (Grindley et al., 2006). The DNA outside model was derived from the elegant biochemical and small-angle X-ray scattering studies of a hyperactive SR tetramer complexed with two non-cleavable crossover sites (Sarkis et al., 2001; Nollmann et al., 2004). It was later supported by the crystal structure of the post-cleavage tetrameric DNA complex (Li et al., 2005). For strand exchange, one half of the protein tetramer-DNA complex would have to rotate 180° relative to the other half, before DNA rejoining (Li et al., 2005; Grindley et al., 2006; Dhar et al., 2009). Although this is occasionally referred to as a “swivel”, there is no covalent linkage between the two rotating parts, quite unlike the case of TopIB. Protein-protein crosslinking experiments appear to support the protein-DNA arrangement and the subunit rotation model (Dhar et al., 2004; Li et al., 2005; Dhar et al., 2009). In the absence of direct evidence, however, one questions the probability of a 180° rotation of two halves of a ~100 kDa protein-DNA complex sharing a 700 to 1200 Å2 protein-protein interface, all supposed to occur with no energy input (Li et al., 2005).
A strand passage model for DNA exchange by SRs
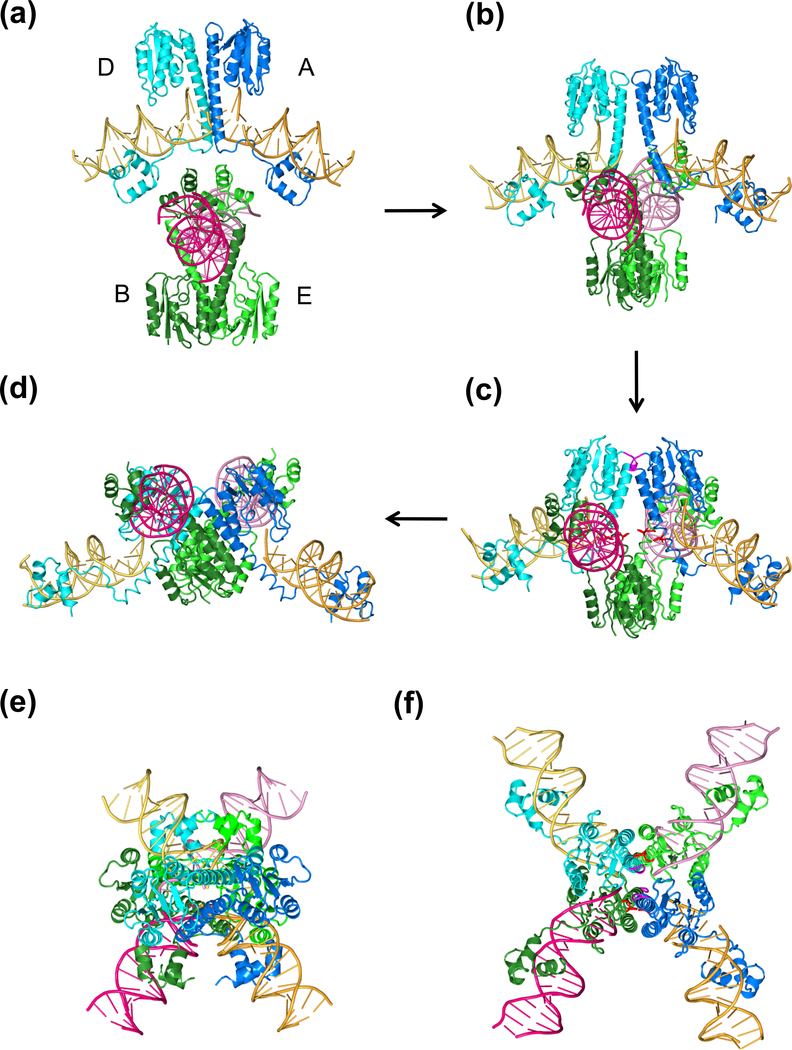
I wish to propose a different model for SR recombination. Examination of the crystal structures of the post-cleavage SR-DNA synaptic tetramer (PDB: 1ZR4 and 2GM4) reveals that strand passage, analogous to that used by TOPRIM topoisomerases, can account for DNA exchange without the 180° rotation (Fig. 7, 8b). This alternative mechanism involves the following steps: (1) two DNA crossover sites face each other and the two associated SR dimers are outside (the parental dimer is based on PDB: 1GDT) (Fig. 7a); (2) DNA cleavage occurs after the protein conformational changes necessary to unblock Ser10 (Fig. 7b); (3) the two dimer-cleaved DNA complexes approach each other, and the two DNAs cross and pass one another through the cleaved gaps, ending with the synapse as observed in the crystal structure (1ZR4) (Fig. 7c-d); and (4) the left and right halves of each initial crossover site are aligned with new partners and ready to be rejoined. The structures shown in Fig. 6b and 7d are identical, but appear different because of different viewing angles.
Fig. 7.
Four intermediates of the proposed strand passage model. (a) Two pre-cleavage SR dimers are facing each other at ~90° right-handed crossing angle. (b) During or after DNA cleavage, the E helices within each dimer become uncrossed and start to open at the C-termini. Two opposite dimers approach each other. (c) The E helices continue to open and the two cleaved crossover-site DNAs are about to cross each other. The conserved E124 residues from the four SR subunits (highlighted in red) are in close proximity with each other. (d) The final state of the strand passage. The view is related to the right panel in Fig. 6b by a 60° rotation around the vertical axis. (e) and (f) Recombination intermediates in the (a) and (c) states are viewed in the same orientation as in the left panel of Fig. 6b.
The fundamental difference between the subunit rotation model and this strand passage model is whether the side-by-side dimers (A and E, B and D) or the diagonal dimers (A and D, B and E) in the post-cleavage tetrameric synapse (Fig. 6b) correspond to the parental dimer before cleavage and recombination. According to the subunit rotation model, the side-by-side dimers correspond to the parental SR-DNA complex, and DNA substrates are bound on the outside of the recombinase tetramer throughout the reaction process. According to the strand passage model, DNA substrates are bound inside before DNA cleavage and move to the “outside” during strand passage, leading to strand exchange without subunit rotation. Both the DNA “inside” and “outside” models have the potential to make a tetrameric synapse according to the structure of dimeric γδ resolvase complexed with the site I DNA (Yang and Steitz, 1995). The subunit rotation model implies that the crystal structure of the tetrameric synapse represents the state before DNA exchange, whereas the strand passage model suggests that in the observed tetrameric synapse two halves of crossover-site DNA’s are already exchanged and waiting to be rejoined.
Support for the strand passage model
Three observations provide critical support for the strand passage model. Firstly, when viewed at a 75° angle to the canonical view of the post-cleavage resolvase-DNA complex (Fig. 6b), the diagonal dimers in the tetrameric synapse (labeled as A/D and B/E in Fig. 6b) are as likely to be the pre-cleavage parental dimers as the side-by-side dimers (A/E and B/D), if DNAs are allowed to pass through each other. To transform the pre-cleavage dimer (PDB: 1GDT) to the post-cleavage side-by-side dimer (PDB: 1ZR4), the E helices at the dimer interface need to splay open at the N-termini and increase the crossing angle from ~30° to 90° (Supplementary Movie 1). Additional movements of the catalytic core domain also accompany the changes between E helices. In comparison, transformation to the diagonal dimers requires uncrossing of the E helices using their N-terminal contact as a hinge (Supplementary Movie 2). Experimentally it was reported more than a decade ago that γδ resolvase and Hin dimers disulfide crosslinked at the N-termini of E helices (M101C or M106C) support the synapse of two recombination sites and DNA nicking (Hughes et al., 1993; Merickel et al., 1998). Because the crosslink prevents the N-termini of E helices from splaying, the pro-nicking synaptic complexes of M101C-Hin and M106-resolvase have to be different from the synaptic tetramers observed with the hyperactive mutants.
The triple mutation (G101S, E102Y and M103I) that make Tn3 and γδ resolvases hyperactive and able to carry out recombination without sites II and III are located at the N-terminus of the E helix (Arnold et al., 1999; Sarkis et al., 2001; Li et al., 2005). The increased hydrophobicity and rigidity caused by these mutations appears to be consistent with their role as a linchpin to maintain the N-terminal contact of E helices and thus keep the cleaved two halves of the resolvase dimer-DNA complex together during the strand crossing and passage. For the subunit rotation model, these mutations are suggested to promote tetramerization by creating a hydrophobic interface. This implies that the E helices spontaneously open at the N-termini without binding to DNA. Indeed tetramerization of hyperactive SRs was observed independent of DNA binding or cleavage (Nollmann et al., 2004; Kamtekar et al., 2006; Gehman et al., 2008). This suggests that the hyperactive SRs and ready-made tetramers may use unorthodox routes to form the tetrameric SR-DNA synapse. Consistent with the prediction that the synapse thus made is different from the normal, recombination pathways catalyzed by wildtype and hyperactive mutant Hin differ significantly (Dhar et al., 2009).
Secondly, E124 is highly conserved among SRs. Initially E124 was thought to play a role in DNA cleavage because of its proximity to the scissile phosphate and S10 nucleophile (Yang and Steitz, 1995). But mutation of E124 to Q only activates rather than diminishes the recombinase activity and makes recombination independent of negative supercoiling (Arnold et al., 1999). E124 was thus proposed to sequester the active residues from cleaving DNA. With Gln being both a hydrogen bond donor and acceptor, however, E124Q can maintain the hydrogen bond with S10 and R68. Neither the location of E124 in the crystal structures of pre or post-cleavage resolvase-DNA complex nor the subunit rotation model explains its regulatory role in DNA topology dependence. In contrast, restriction of recombination by E124 and the permissive nature of E124Q mutation are readily explained by the strand passage model. Four E124’s of the four SR subunits have to approach and cross each other during DNA strand passage (Fig. 7c). The conserved negative charges in the vicinity of the crossing DNAs likely serve as a barrier to prevent unwanted DNA recombination, e.g. on linear DNAs. This charge barrier, however, can be overcome by negative supercoiling.
Thirdly, the proposed strand passage model suggests that before strand passage, DNAs are inside of the resolvase subunits and in close proximity with one another. The DNA inside configuration has been observed in the crystal structure of Sin resolvase (a homologue of γδ and Tn3 resolvases) bound to a regulatory DNA (site III equivalent), which is not subject to cleavage (Mouw et al., 2008). Two Sin resolvase dimer-DNA complexes form a synapse in the crystal lattice via interactions between the C-terminal DNA-binding domains and sandwich the regulatory DNAs inside (Fig. 6c). Mutagenic studies confirm the importance of the protein-protein interactions observed in the crystal structure (Mouw et al., 2008). The DNA-inside configuration before cleavage is consistent with the strand passage model and contrary to the subunit rotation model (Fig. 2c).
Finally, the strand passage model is analogous to the DNA rearrangement catalyzed by Topo IA and Topo II. Topoisomerization and recombination both use DNA supercoiling as the energy source. All recombinases bend DNA substrates, as does yeast topoisomerase II (Dong and Berger, 2007). The supercoils and torsional strains on the double helix likely facilitate DNA movement relative to protein in all these cases. The difference is that two DNA duplexes are cleaved by SRs instead of a single strand or a single duplex by the topoisomerases. The strand passage and rearrangement as shown in Fig. 8 account for the linking number change of +3, which is one fewer than experimentally determined for serine recombinases (Benjamin and Cozzarelli, 1988; Kanaar et al., 1988; Stark et al., 1989; Merickel and Johnson, 2004). DNA untwisting of the cleaved crossover sites, for example, the four free 3´-ends, can potentially make up for the addition +1 linking number change. A topological diagram of recombination by strand passage, which produces catenated and knotted recombination products in consecutive cycles same as by the subunit rotation model (Wasserman et al., 1985; Stark et al., 1989; Benjamin and Cozzarelli, 1990), is shown in Fig. 7C.
The strand passage model of SRs is compatible with the crosslinking data
A number of Cys-based protein disulfide crosslinking experiments have been employed to test and verify the subunit rotation model (Mcilwraith et al., 1996; Sanders and Johnson, 2004; Li et al., 2005; Dhar et al., 2009). Interesting, the crosslinking data are equally supportive of the strand passage model because the post-cleavage tetrameric complex is the reference frame in both proposals. For example, each SR subunit associates with the same DNA half-site throughout the strand passage in agreement with the C-terminal domain crosslinking data (Mcilwraith et al., 1996). Crosslinking in the middle of the E helices (residue 114 in Hin) allows DNA cleavage to occur but not strand exchange (Dhar et al., 2004), indicating that cleavage occurs before any dramatic protein conformational change. It is thought to support the subunit rotation model because the crosslink prevents the 180° rotation between the subunits of a parental dimer (Fig. 2c). However it is equally consistent with the strand passage model, where opening of the E helices by pivoting about the N-termini is also necessary for the pre-cleavage dimer to transform to the diagonal dimer and for DNA strands to cross and pass each other after cleavage (supplementary Movies 2, 3).
Interestingly other crosslinking mutations are more consistent with the DNA passage model than the subunit rotation model. When Lys residues at the C-terminus of E helix were replaced by Cys (K136C in γδ resolvase or K134C in Hin invertase), the mutant recombinases supported recombination (Dhar et al., 2004; Li et al., 2005). The Cys residues are far apart from each other in both the pre-cleavage dimeric or post-cleavage tetrameric complex, but they are efficiently crosslinked in the cleaved intermediate. It is suggested that the Cys residues are crosslinked in the middle of the 180° rotation (at ~75°) of the two halves of the tetrameric synapse (Li et al., 2005). It is rather mysterious that the proposed 180° subunit rotation would take place continuously in the cleaved recombination intermediate and pause frequently in the middle for the disulfide bond to form. In contrast, in the DNA passage model the C-termini of the E helices of the opposite dimers are adjacent to each other during the strand passage process, thus allowing the crosslinking to occur efficiently as observed (Dhar et al., 2009). In the tetrameric synapse trapped after DNA cleavage and before rejoining, the two cleaved SR dimer-DNA complexes may be in an equilibrium of crossing and uncrossing each other, thus allowing crosslinking along the entire E helices to take place with various efficiencies (Sanders and Johnson, 2004; Dhar et al., 2009).
Crosslinking of residues preceding the E helix, e.g. S94C of Hin (Sanders and Johnson, 2004), can also be explained by the strand passage model without invoking subunit rotation. The equivalent of S94 in γδ resolvase is G96. Although the G96 residues are more than 15Å from each other both in the pre-cleavage dimer and post-cleavage tetramer structures, the Cα atoms of G96 belonging to the diagonal dimers are within 8Å in the activated apo-protein core tetramer (PDB: 2GM5). The protein core tetramer is a slight variation of the post-cleavage synapse without significant subunit rotation (Kamtekar et al., 2006).
Future experiments to test the strand exchange mechanism
The experimental data available to date do not critically distinguish whether recombination occurs by the subunit rotation or strand passage mechanism. Given the dramatic conformational changes in the recombination process and the few structures available at the atomic resolution along the reaction pathway, it is not surprising that the biochemical data support more than one potential mechanism. The hyperactive recombinases generated by the protein-interface mutations are helpful in simplifying the reaction components, but their spontaneous tetramerization also reduces the stringency of recombination, thus possibly creating unorthodox reaction paths and forming SR-DNA synapses through a “backdoor”. In light of the proposed strand passage model, the existing data may be revisited and reinterpreted. To distinguish the different models proposed for SRs, new experiments are also necessary.
The DNA passage and subunit rotation mechanism have several distinct biochemical and biophysical properties. Firstly, the strand passage model predicts that each set up of the recombination assembly (resolvosome or invertasome, including the cleavage and the regulatory DNA sites) allows one strand exchange event only. Multiple rounds of recombination require multiple rounds of assembly. The subunit rotation model, however, predicts that multiple recombination events can occur with one single setup, and changes of supercoiling occur outside of recombination synaptosomes as diagrammed originally (Wasserman et al., 1985). Based on energetic consideration and the involvement of regulatory factors, a single assembly capable of multiple cycles of recombination seems implausible. Time-resolved analyses of Hin recombination indicate that multiple rounds of strand swapping don’t occur (Dhar et al., 2009). Similar time resolved experiments are needed for γδ and Tn3 resolvases to determine whether a resolvasome re-assembles after each cycle of strand exchange (Mcilwraith et al., 1997). Secondly, the strand passage model implies that the two crossover-site DNAs are at a ~90° right-handed crossing angle (Fig. 7e-f). In contrast, the subunit rotation model suggests the two sites are near parallel. Experimental data that support the right-handed local super-twist between two crossover-site DNAs, however, also suggest the DNAs being outside (Leschziner and Grindley, 2003). These data may be re-interpreted in consideration of the strand passage model. Thirdly, the DNA passage model predicts that the two recombination sites are close to each other before cleavage when assembled inside of the tetrameric SR (like the Sin regulatory assembly, 2R0Q) and become further apart after strand crossing and formation of the tetrameric synapse (PDB: 1ZR4 and 2GM5). In contrast, the subunit rotation model predicts the distance remains the same and rather far apart throughout recombination (Li et al., 2005; Dhar et al., 2009). Single-molecule fluorescence energy transfer experiments with fluorescence donor and acceptor appropriately attached to each of recombination site have the potential to distinguish the two mechanisms. If recombination takes place by strand passage, the FRET signal would change from strong to weak from the beginning to the end. But if it happens by subunit rotation, the FRET signal would remain weak throughout.
Concluding remarks
It is rather intriguing and satisfying that topoisomerization and site-specific recombination share similarity and homology at multiple levels. TopIB and tyrosine recombinases are highly homologous. The different multimerization states of these proteins lead to the different outcomes. The lesson learned from their relationship is that homologies in structures, catalytic residues and reaction products imply an identical catalytic mechanism. DNA swiveling catalyzed by TopIB and strand exchange catalyzed by tyrosine recombinases are two alternative modes for releasing topological strain. A partially functional site-specific recombinase can readily become a topoisomerase. The similarities between TOPRIM topoisomerases and serine recombinases are less obvious, but the related folding topology, catalytic site location, metal ion dependence and reaction products confers plausibility to the proposal of a similar strand passage mechanism (Fig. 8).
Supplementary Material
Animation Animation of two pre-cleavage γδ resolvase- crossover-site DNA complexes (PDB: 1GDT) transformed to the post-cleavage tetrameric synapse (PDB: 1ZR4). Two pre-cleavage resolvase dimer complexes face each with DNAs at a ~90° right-handed crossing angle. One dimer is colored blue and cyan and binds the yellow and orange DNA. The other dimer is colored dark and light green and binds the dark and light pink DNA. They are placed without any contact for visual clarity. In reality, the two complexes probably interact with each other and also with the proteins bound to the auxiliary sites. These interactions are likely necessary to activate the cleavage reaction. The conformational changes needed to cleave DNAs are built in the morphing from the pre-cleavage dimer to post-cleavage tetramer. Associated with the opening of the E helices, the cleaved two halves of DNA separate. The two SR dimers and their covalently linked cleaved DNA advance towards each other in a straight line, and the DNAs cross each other without any steric clash. The C-termini of the four E helices meet first and then pass each other. The opening of the E helices is concurrent with the penetrating of the two resolvase dimers into one another and forming the final tetrameric synapse. The recombination intermediate is the tetrameric crystal structure (1ZR4). To bring it to the conventional viewpoint, it is rotated 60° around the horizontal axis and 75° around the vertical axis. Recombination is completed when the half-site DNAs, pink with orange, yellow with dark pink, are rejoined.
Acknowledgement
I thank D. Leahy and M. Gellert for critical reading of the manuscript, and for K. Mizuuchi and F. Dyda for the stimulating discussions. I also deeply appreciate Drs. N. Grindley, M. Stark and M. Boocock for extensive comments and corrections.
Footnotes
Declaration of Interest
The research was funded by the intramural research program of NIDDK, NIH.
Declaration of no interest
The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
References
- Allemand F, Mathy N, Brechemier-Baey D & Condon C. 2005. The 5S rRNA maturase, ribonuclease M5, is a Toprim domain family member. Nucleic Acids Res 33: 4368–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Leipe DD & Koonin EV. 1998. Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res 26: 4205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold PH, Blake DG, Grindley ND, Boocock MR & Stark WM. 1999. Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. EMBO J 18: 1407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artymiuk PJ, Ceska TA, Suck D & Sayers JR. 1997. Prokaryotic 5’−3’ exonucleases share a common core structure with gamma-delta resolvase. Nucleic Acids Res 25: 4224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balendiran GK. 2009. Fibrates in the chemical action of daunorubicin. Curr Cancer Drug Targets 9: 366–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates AD & Maxwell A. 1997. DNA topology: topoisomerases keep it simple. Curr Biol 7: R778–81. [DOI] [PubMed] [Google Scholar]
- Bates AD & Maxwell A. 2010. The role of ATP in the reactions of type II DNA topoisomerases. Biochem Soc Trans 38: 438–42. [DOI] [PubMed] [Google Scholar]
- Benjamin HW & Cozzarelli NR. 1988. Isolation and characterization of the Tn3 resolvase synaptic intermediate. EMBO J 7: 1897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin HW & Cozzarelli NR. 1990. Geometric arrangements of Tn3 resolvase sites. J Biol Chem 265: 6441–7. [PubMed] [Google Scholar]
- Berger JM, Gamblin SJ, Harrison SC & Wang JC. 1996. Structure and mechanism of DNA topoisomerase II. Nature 379: 225–32. [DOI] [PubMed] [Google Scholar]
- Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A & Ellenberger T. 2005. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature 435: 1059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boocock MR, Zhu X & Grindley ND. 1995. Catalytic residues of gamma delta resolvase act in cis. EMBO J 14: 5129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke ME, Arnold PH, He J, Wenwieser SV, Rowland SJ, Boocock MR & Stark WM. 2004. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol Microbiol 51: 937–48. [DOI] [PubMed] [Google Scholar]
- Burrus V, Marrero J & Waldor MK. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55: 173–83. [DOI] [PubMed] [Google Scholar]
- Campbell A 2007. Phage integration and chromosome structure. A personal history. Annu Rev Genet 41: 1–11. [DOI] [PubMed] [Google Scholar]
- Castell SE, Jordan SL & Halford SE. 1986. Site-specific recombination and topoisomerization by Tn21 resolvase: role of metal ions. Nucleic Acids Res 14: 7213–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ. 1977. Strand breakage by the DNA untwisting enzyme results in covalent attachment of the enzyme to DNA. Proc Natl Acad Sci U S A 74: 3800–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ. 2001. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70: 369–413. [DOI] [PubMed] [Google Scholar]
- Changela A, DiGate RJ & Mondragon A. 2001. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature 411: 1077–81. [DOI] [PubMed] [Google Scholar]
- Chen Y, Narendra U, Iype LE, Cox MM & Rice PA. 2000. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol Cell 6: 885–97. [PubMed] [Google Scholar]
- Chen Y & Rice PA. 2003a. New insight into site-specific recombination from Flp recombinase-DNA structures. Annu Rev Biophys Biomol Struct 32: 135–59. [DOI] [PubMed] [Google Scholar]
- Chen Y & Rice PA. 2003b. The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis. J Biol Chem 278: 24800–7. [DOI] [PubMed] [Google Scholar]
- Cheng C, Kussie P, Pavletich N & Shuman S. 1998. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92: 841–50. [DOI] [PubMed] [Google Scholar]
- Confalonieri F, Elie C, Nadal M, de La Tour C, Forterre P & Duguet M. 1993. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc Natl Acad Sci U S A 90: 4753–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH. 1976. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A 73: 2639–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar G, Heiss JK & Johnson RC. 2009. Mechanical constraints on Hin subunit rotation imposed by the Fis/enhancer system and DNA supercoiling during site-specific recombination. Mol Cell 34: 746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar G, Sanders ER & Johnson RC. 2004. Architecture of the hin synaptic complex during recombination: the recombinase subunits translocate with the DNA strands. Cell 119: 33–45. [DOI] [PubMed] [Google Scholar]
- Domanico PL & Tse-Dinh YC. 1991. Mechanistic studies on E. coli DNA topoisomerase I: divalent ion effects. J Inorg Biochem 42: 87–96. [DOI] [PubMed] [Google Scholar]
- Dong KC & Berger JM. 2007. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature 450: 1201–5. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S, Gadelle D & Serre MC. 2007. Origin and evolution of DNA topoisomerases. Biochimie 89: 427–46. [DOI] [PubMed] [Google Scholar]
- Gehman JD, Cocco MJ & Grindley ND. 2008. Chemical shift mapping of gamma delta resolvase dimer and activated tetramer: mechanistic implications for DNA strand exchange. Biochim Biophys Acta 1784: 2086–92. [DOI] [PubMed] [Google Scholar]
- Ghosh P, Kim AI & Hatfull GF. 2003. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol Cell 12: 1101–11. [DOI] [PubMed] [Google Scholar]
- Gibb B, Gupta K, Ghosh K, Sharp R, Chen J & Van Duyne GD. 2010. Requirements for catalysis in the Cre recombinase active site. Nucleic Acids Res 38: 5817–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley ND. 1993. Analysis of a nucleoprotein complex: the synaptosome of gamma delta resolvase. Science 262: 738–40. [DOI] [PubMed] [Google Scholar]
- Grindley ND, Whiteson KL & Rice PA. 2006. Mechanisms of site-specific recombination. Annu Rev Biochem 75: 567–605. [DOI] [PubMed] [Google Scholar]
- Guo F, Gopaul DN & van Duyne GD. 1997. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389: 40–6. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Chandler M & Dyda F. 2010. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit Rev Biochem Mol Biol 45: 50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman AB, Waninger S, Scocca JJ & Dyda F. 1997. Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution. Cell 89: 227–37. [DOI] [PubMed] [Google Scholar]
- Hughes RE, Rice PA, Steitz TA & Grindley ND. 1993. Protein-protein interactions directing resolvase site-specific recombination: a structure-function analysis. EMBO J 12: 1447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram M, Mehta S, Uzri D, Voziyanov Y & Velmurugan S. 2004. Site-specific recombination and partitioning systems in the stable high copy propagation of the 2-micron yeast plasmid. Prog Nucleic Acid Res Mol Biol 77: 127–72. [DOI] [PubMed] [Google Scholar]
- Johnson RC, Bruist MF & Simon MI. 1986. Host protein requirements for in vitro site-specific DNA inversion. Cell 46: 531–9. [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Ho RS, Cocco MJ, Li W, Wenwieser SV, Boocock MR, Grindley ND & Steitz TA. 2006. Implications of structures of synaptic tetramers of gamma delta resolvase for the mechanism of recombination. Proc Natl Acad Sci U S A 103: 10642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, van de Putte P & Cozzarelli NR. 1988. Gin-mediated DNA inversion: product structure and the mechanism of strand exchange. Proc Natl Acad Sci U S A 85: 752–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Ito T, Wagner G, Richardson CC & Ellenberger T. 2003. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol Cell 11: 1349–60. [DOI] [PubMed] [Google Scholar]
- Keck JL, Roche DD, Lynch AS & Berger JM. 2000. Structure of the RNA polymerase domain of E. coli primase. Science 287: 2482–6. [DOI] [PubMed] [Google Scholar]
- Kim AI, Ghosh P, Aaron MA, Bibb LA, Jain S & Hatfull GF. 2003. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol Microbiol 50: 463–73. [DOI] [PubMed] [Google Scholar]
- Koster DA, Croquette V, Dekker C, Shuman S & Dekker NH. 2005. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434: 671–4. [DOI] [PubMed] [Google Scholar]
- Kreuzer KN & Cozzarelli NR. 1980. Formation and resolution of DNA catenanes by DNA gyrase. Cell 20: 245–54. [DOI] [PubMed] [Google Scholar]
- Krogh BO & Shuman S. 2000. Catalytic mechanism of DNA topoisomerase IB. Mol Cell 5: 1035–41. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Tirumalai R, Landy A & Ellenberger T. 1997. Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science 276: 126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM & Sanderson MR. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One 5: e11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leschziner AE & Grindley ND. 2003. The architecture of the gamma delta resolvase crossover site synaptic complex revealed by using constrained DNA substrates. Mol Cell 12: 775–81. [DOI] [PubMed] [Google Scholar]
- Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley ND & Steitz TA. 2005. Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs. Science 309: 1210–5. [DOI] [PubMed] [Google Scholar]
- Liebert CA, Hall RM & Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 63: 507–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima CD, Wang JC & Mondragon A. 1994. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature 367: 138–46. [DOI] [PubMed] [Google Scholar]
- Liu LF, Liu CC & Alberts BM. 1980. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell 19: 697–707. [DOI] [PubMed] [Google Scholar]
- MacDonald D, Demarre G, Bouvier M, Mazel D & Gopaul DN. 2006. Structural basis for broad DNA-specificity in integron recombination. Nature 440: 1157–62. [DOI] [PubMed] [Google Scholar]
- Maxwell A & Lawson DM. 2003. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr Top Med Chem 3: 283–303. [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, Boocock MR & Stark WM. 1996. Site-specific recombination by Tn3 resolvase, photocrosslinked to its supercoiled DNA substrate. J Mol Biol 260: 299–303. [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, Boocock MR & Stark WM. 1997. Tn3 resolvase catalyses multiple recombination events without intermediate rejoining of DNA ends. J Mol Biol 266: 108–21. [DOI] [PubMed] [Google Scholar]
- Merickel SK, Haykinson MJ & Johnson RC. 1998. Communication between Hin recombinase and Fis regulatory subunits during coordinate activation of Hin-catalyzed site-specific DNA inversion. Genes Dev 12: 2803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merickel SK & Johnson RC. 2004. Topological analysis of Hin-catalysed DNA recombination in vivo and in vitro. Mol Microbiol 51: 1143–54. [DOI] [PubMed] [Google Scholar]
- Minkah N, Hwang Y, Perry K, Van Duyne GD, Hendrickson R, Lefkowitz EJ, Hannenhalli S & Bushman FD. 2007. Variola virus topoisomerase: DNA cleavage specificity and distribution of sites in Poxvirus genomes. Virology 365: 60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon A & DiGate R. 1999. The structure of Escherichia coli DNA topoisomerase III. Structure 7: 1373–83. [DOI] [PubMed] [Google Scholar]
- Mouw KW, Rowland SJ, Gajjar MM, Boocock MR, Stark WM & Rice PA. 2008. Architecture of a serine recombinase-DNA regulatory complex. Mol Cell 30: 145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley LL & Grindley ND. 1998. Architecture of the gamma delta resolvase synaptosome: oriented heterodimers identity interactions essential for synapsis and recombination. Cell 95: 553–62. [DOI] [PubMed] [Google Scholar]
- Nash HA. 1981. Integration and excision of bacteriophage lambda: the mechanism of conservation site specific recombination. Annu Rev Genet 15: 143–67. [DOI] [PubMed] [Google Scholar]
- Newman BJ & Grindley ND. 1984. Mutants of the gamma delta resolvase: a genetic analysis of the recombination function. Cell 38: 463–9. [DOI] [PubMed] [Google Scholar]
- Nollmann M, He J, Byron O & Stark WM. 2004. Solution structure of the Tn3 resolvase-crossover site synaptic complex. Mol Cell 16: 127–37. [DOI] [PubMed] [Google Scholar]
- Olorunniji FJ & Stark WM. 2010. Catalysis of site-specific recombination by Tn3 resolvase. Biochem Soc Trans 38: 417–21. [DOI] [PubMed] [Google Scholar]
- Perry K, Hwang Y, Bushman FD & Van Duyne GD. 2006. Structural basis for specificity in the poxvirus topoisomerase. Mol Cell 23: 343–54. [DOI] [PubMed] [Google Scholar]
- Perry K, Hwang Y, Bushman FD & Van Duyne GD. 2010. Insights from the structure of a smallpox virus topoisomerase-DNA transition state mimic. Structure 18: 127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Biswas T, Ellenberger T, Landy A & Aihara H. 2006. DNA arms do the legwork to ensure the directionality of lambda site-specific recombination. Curr Opin Struct Biol 16: 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redinbo MR, Stewart L, Kuhn P, Champoux JJ & Hol WG. 1998. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 279: 1504–13. [DOI] [PubMed] [Google Scholar]
- Reed RR & Grindley ND. 1981. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell 25: 721–8. [DOI] [PubMed] [Google Scholar]
- Rezacova P, Borek D, Moy SF, Joachimiak A & Otwinowski Z. 2008. Crystal structure and putative function of small Toprim domain-containing protein from Bacillus stearothermophilus. Proteins 70: 311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA & Steitz TA. 1994. Model for a DNA-mediated synaptic complex suggested by crystal packing of gamma delta resolvase subunits. EMBO J 13: 1514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland SJ, Boocock MR, McPherson AL, Mouw KW, Rice PA & Stark WM. 2009. Regulatory mutations in Sin recombinase support a structure-based model of the synaptosome. Mol Microbiol 74: 282–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland SJ, Stark WM & Boocock MR. 2002. Sin recombinase from Staphylococcus aureus: synaptic complex architecture and transposon targeting. Mol Microbiol 44: 607–19. [DOI] [PubMed] [Google Scholar]
- Sanders ER & Johnson RC. 2004. Stepwise dissection of the Hin-catalyzed recombination reaction from synapsis to resolution. J Mol Biol 340: 753–66. [DOI] [PubMed] [Google Scholar]
- Sanderson MR, Freemont PS, Rice PA, Goldman A, Hatfull GF, Grindley ND & Steitz TA. 1990. The crystal structure of the catalytic domain of the site-specific recombination enzyme gamma delta resolvase at 2.7 A resolution. Cell 63: 1323–9. [DOI] [PubMed] [Google Scholar]
- Sarkis GJ, Murley LL, Leschziner AE, Boocock MR, Stark WM & Grindley ND. 2001. A model for the gamma delta resolvase synaptic complex. Mol Cell 8: 623–31. [DOI] [PubMed] [Google Scholar]
- Schmidt BH, Burgin AB, Deweese JE, Osheroff N & Berger JM. 2010. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature 465: 641–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeffler AJ & Berger JM. 2008. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys 41: 41–101. [DOI] [PubMed] [Google Scholar]
- Sherratt DJ, Soballe B, Barre FX, Filipe S, Lau I, Massey T & Yates J. 2004. Recombination and chromosome segregation. Philos Trans R Soc Lond B Biol Sci 359: 61–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S & Prescott J. 1990. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J Biol Chem 265: 17826–36. [PubMed] [Google Scholar]
- Simon M, Zieg J, Silverman M, Mandel G & Doolittle R. 1980. Phase variation: evolution of a controlling element. Science 209: 1370–4. [DOI] [PubMed] [Google Scholar]
- Smith MC, Brown WR, McEwan AR & Rowley PA. 2010. Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem Soc Trans 38: 388–94. [DOI] [PubMed] [Google Scholar]
- Stark WM, Boocock MR & Sherratt DJ. 1992. Catalysis by site-specific recombinases. Trends Genet 8: 432–9. [PubMed] [Google Scholar]
- Stark WM, Sherratt DJ & Boocock MR. 1989. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell 58: 779–90. [DOI] [PubMed] [Google Scholar]
- Stivers JT, Harris TK & Mildvan AS. 1997. Vaccinia DNA topoisomerase I: evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry 36: 5212–22. [DOI] [PubMed] [Google Scholar]
- Subramanya HS, Arciszewska LK, Baker RA, Bird LE, Sherratt DJ & Wigley DB. 1997. Crystal structure of the site-specific recombinase, XerD. EMBO J 16: 5178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse-Dinh YC, McCarron BG, Arentzen R & Chowdhry V. 1983. Mechanistic study of E. coli DNA topoisomerase I: cleavage of oligonucleotides. Nucleic Acids Res 11: 8691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Putte P & Goosen N. 1992. DNA inversions in phages and bacteria. Trends Genet 8: 457–62. [DOI] [PubMed] [Google Scholar]
- Van Duyne GD. 2001. A structural view of cre-loxp site-specific recombination. Annu Rev Biophys Biomol Struct 30: 87–104. [DOI] [PubMed] [Google Scholar]
- Vologodskii AV & Cozzarelli NR. 1994. Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct 23: 609–43. [DOI] [PubMed] [Google Scholar]
- Wang JC. 1996. DNA topoisomerases. Annu Rev Biochem 65: 635–92. [DOI] [PubMed] [Google Scholar]
- Wang JC. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3: 430–40. [DOI] [PubMed] [Google Scholar]
- Wasserman SA, Dungan JM & Cozzarelli NR. 1985. Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science 229: 171–4. [DOI] [PubMed] [Google Scholar]
- Whiteson KL, Chen Y, Chopra N, Raymond AC & Rice PA. 2007. Identification of a potential general acid/base in the reversible phosphoryl transfer reactions catalyzed by tyrosine recombinases: Flp H305. Chem Biol 14: 121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W 2010. Nucleases: diversity of structrue, function and mechanism. Quarterly Review of Biophysics: in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W & Steitz TA. 1995. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell 82: 193–207. [DOI] [PubMed] [Google Scholar]
- Yang Z & Champoux JJ. 2001. The role of histidine 632 in catalysis by human topoisomerase I. J Biol Chem 276: 677–85. [DOI] [PubMed] [Google Scholar]
- Zhu CX & Tse-Dinh YC. 2000. The acidic triad conserved in type IA DNA topoisomerases is required for binding of Mg(II) and subsequent conformational change. J Biol Chem 275: 5318–22. [DOI] [PubMed] [Google Scholar]
- Zieg J, Silverman M, Hilmen M & Simon M. 1977. Recombinational switch for gene expression. Science 196: 170–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animation Animation of two pre-cleavage γδ resolvase- crossover-site DNA complexes (PDB: 1GDT) transformed to the post-cleavage tetrameric synapse (PDB: 1ZR4). Two pre-cleavage resolvase dimer complexes face each with DNAs at a ~90° right-handed crossing angle. One dimer is colored blue and cyan and binds the yellow and orange DNA. The other dimer is colored dark and light green and binds the dark and light pink DNA. They are placed without any contact for visual clarity. In reality, the two complexes probably interact with each other and also with the proteins bound to the auxiliary sites. These interactions are likely necessary to activate the cleavage reaction. The conformational changes needed to cleave DNAs are built in the morphing from the pre-cleavage dimer to post-cleavage tetramer. Associated with the opening of the E helices, the cleaved two halves of DNA separate. The two SR dimers and their covalently linked cleaved DNA advance towards each other in a straight line, and the DNAs cross each other without any steric clash. The C-termini of the four E helices meet first and then pass each other. The opening of the E helices is concurrent with the penetrating of the two resolvase dimers into one another and forming the final tetrameric synapse. The recombination intermediate is the tetrameric crystal structure (1ZR4). To bring it to the conventional viewpoint, it is rotated 60° around the horizontal axis and 75° around the vertical axis. Recombination is completed when the half-site DNAs, pink with orange, yellow with dark pink, are rejoined.