Abstract
Rationale: Hematopoietic cell transplant (HCT) is a common treatment for hematological neoplasms and autoimmune disorders. Among HCT recipients, pulmonary complications are common, morbid, and/or lethal, and they have recently been associated with gut dysbiosis. The role of lung microbiota in post-HCT pulmonary complications is unknown.
Objectives: To investigate the role of lung microbiota in post-HCT pulmonary complications using animal modeling and human BAL fluid.
Methods: For animal modeling, we used an established murine model of HCT with and without postengraftment herpes virus infection. For human studies, we characterized lung microbiota in BAL fluid from 43 HCT recipients. Lung bacteria were characterized using 16S ribosomal RNA gene sequencing and were compared with lung histology (murine) and with alveolar inflammation and pulmonary function testing (human).
Measurements and Main Results: Both HCT and viral infection independently altered the composition of murine lung microbiota, but they had no effect on lung microbial diversity. By contrast, combined HCT and viral infection profoundly altered lung microbiota, decreasing community diversity with an associated pneumonitis. Among human HCT recipients, increased relative abundance of the Proteobacteria phylum was associated with impaired pulmonary function, and lung microbiota were significantly associated with alveolar concentrations of inflammatory cytokines.
Conclusions: In animal models and human subjects, lung dysbiosis is a prominent feature of HCT. Lung dysbiosis is correlated with histologic, immunologic, and physiologic features of post-HCT pulmonary complications. Our findings suggest the lung microbiome may be an unappreciated target for the prevention and treatment of post-HCT pulmonary complications.
Keywords: hematopoietic stem cell transplantation, pulmonary complications, microbiome, microbiota, murine herpes virus
At a Glance Commentary
Scientific Knowledge on the Subject
Pulmonary complications of hematopoietic cell transplant (HCT) are heterogeneous, common, and cause considerable morbidity and mortality. Recent work has reported associations between gut dysbiosis and rates of bacteremia, mortality, and pulmonary complications in HCT recipients. Yet, the presence and significance of pulmonary dysbiosis after HCT is unknown.
What This Study Adds to the Field
To our knowledge, this study is the first report of pulmonary dysbiosis after HCT, both in animal models and in human subjects. We found significant correlations between pulmonary dysbiosis and key histologic, immunologic, and physiologic features of post-HCT pulmonary complications. Our results provide both experimental and observational human plausibility to the hypothesis that lung microbiota perpetuate the alveolar host response after HCT, promoting acute and chronic pulmonary inflammation and fibrosis.
Hematopoietic cell transplant (HCT) is a potentially curative treatment for a variety of malignancies, inherited disorders, and autoimmune diseases (1). However, HCT therapy is commonly complicated by the development of acute and chronic lung diseases that are associated with significant morbidity and mortality (2–4). Lung injury is heterogeneous in HCT patients and can include both infectious and noninfectious pathologies. Bronchiolitis obliterans syndrome (BOS) and idiopathic pneumonia syndrome (IPS) are poorly understood HCT complications associated with high mortality and irreversible pulmonary dysfunction (5). Recent evidence supports the case for occult herpes viral infection promoting disease pathogenesis in IPS (6). In addition, clinical observations in pediatric HCT recipients support an association between early post-HCT viral infection and the risk of developing IPS and BOS (7). We have previously demonstrated the potential for virus exposure (murine gammaherpesvirus 68 [MHV or γ-HV-68]) in murine models to promote a nonresolving pneumonitis and fibrosis that shares several features of pulmonary disorders that complicate HCT (8, 9).
In a recent study published in the Journal, the risk of pulmonary complications after HCT was associated with features of gut dysbiosis (10). The integrity of the gut mucosal barrier is impaired by HCT conditioning regimens. Physiologic homeostasis is altered with loss of microbial diversity and bacterial invasion of the subepithelial space leading to bacteremia (11). This loss of diversity at engraftment is an independent predictor of mortality after HCT (12). Reductions in bacterial diversity, characterized by intestinal domination by Enterococcus spp. and the Proteobacteria phylum, predict several-fold increases in rates of bacteremia (11). Gut dysbiosis is important in HCT complications, yet the role of lung dysbiosis is unknown; however, dysbiosis of lung microbiota has been associated with lung pathology and altered pulmonary immunity (13, 14). Interestingly, HCT patients have impaired pulmonary immunity and demonstrate reduced pathogen clearance through altered cytokine and immune cell functions, with an increased risk of bacterial pulmonary infections (9). Therefore, the impact of disordered lung microbiota on local alveolar immunity in HCT pulmonary complications requires study.
In this article, we report, for the first time to our knowledge, that pulmonary complications in HCT are associated with dysbiosis of lung microbiota, both in animal models and in human patients. We report associations between lung dysbiosis and histologic, immunologic, and physiologic features of post-HCT pulmonary complications. This work provides biological plausibility for the hypothesis that lung microbiota perpetuate the alveolar host response after HCT, promoting pulmonary inflammation and pathology. Some of the results of these studies were reported previously in the form of an abstract (15).
Methods
Ethics Statement
All animal studies reported in this paper were approved by the institutional animal care and use committee at the University of Michigan. Laboratory animal care policies at the University of Michigan follow the Public Health Service policy on Humane Care and Use of Laboratory Animals. All clinical study components described in this paper were conducted under the principles of the Declaration of Helsinki. The human study protocol and biological sample repository were approved by the institutional review board at the University of Michigan. Informed consent was provided by all participating patients or legal surrogates.
Human Study Population
All patients enrolled in this study were seen at the University of Michigan Medical Center between 2010 and 2016. All patients presented with new-onset respiratory symptoms and/or fever with new radiological infiltrates. See the online supplement for further details.
BAL Cytokine Measures
BAL fluid cytokine concentrations were measured by using a human cytokine/chemokine magnetic bead panel (EMD Millipore). See Table E4 and the methods section in the online supplement for further details.
Syngeneic Bone Marrow Transplant and MHV Infection
Syngeneic bone marrow transplant between C57BL/6 recipient and donor mice followed by MHV infection was performed as previously described (8).
Bacterial DNA Isolation
Genomic DNA was extracted from mouse tissue and human HCT patient BAL supernatants using a modified protocol previously demonstrated to isolate bacterial DNA (16).
16S Ribosomal RNA Gene Sequencing
The V4 region of the 16S ribosomal RNA (rRNA) gene was amplified using primers as previously published (17, 18). Sequencing was performed using the MiSeq platform (Illumina) and a previously described dual-indexing sequencing strategy (17, 18) using a MiSeq Reagent Kit V2 (500 cycles) according to the manufacturer’s instructions with previously published modifications (19, 20). PCR cycling conditions were as previously published (21). All bacterial sequences, as well as operational taxonomic unit (OTU), taxonomy, and metadata tables, are available for download from: https://github.com/dicksonlunglab/HCT_lung_microbiome.
Statistical Analyses
Sequence data processing and analysis were completed as previously published (19–22). In murine experiments, cage number was recorded and incorporated into multivariable statistical analysis. In human studies, associations between lung microbiota and measures of pulmonary function (percent predicted) were performed using linear regression modeling and multivariable regression modeling adjusted with the following variables: age at BAL, type of HCT (autologous vs. allogenic), source of HCT, conditioning regimen (myeloablative vs. reduced toxicity), presence or absence of chronic graft-versus-host disease, time from pulmonary function test to BAL, time from HCT to BAL, and the use of antimicrobials at treatment doses using a weighted score (23). See the online supplement for further details.
Linear regressions between relative abundances of microbiota and cytokine concentrations were performed using log10-transformed cytokine data and adjusted using the same multivariable model applied for pulmonary function with the addition of variables, FEV1, FVC, and DlCO (percent predicted). Cytokine data were logarithmically transformed to improve normality assumptions for statistical models. To account for multiple comparisons, we performed false discovery rate calculations using the Benjamini-Hochberg procedure with a FDR level of 0.1. We performed all analyses in R (R Core Team, 2017) and Prism version 7 software (GraphPad Software). In analysis of murine data, means were compared using the paired t test for parametric data and paired ANOVA with Tukey’s multiple comparisons test for nonparametric data.
Identification of Procedural Contaminants
To identify potential sources of contamination, we collected multiple procedural controls as per our previously published protocol (21). See the online methods section for further details.
Results
Experimental HCT Alters Community Composition, but Not Diversity, of Lung Microbiota
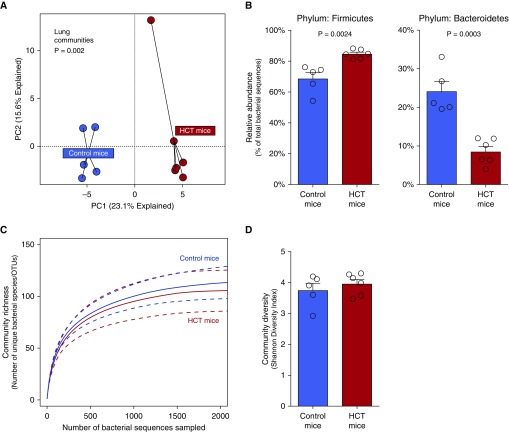
To determine whether syngeneic HCT alters the microbiota of the lung, we used a well-established experimental model in which recipient C57BL/6 mice are irradiated followed by syngeneic HCT (8, 9, 24) (Figure E1). We characterized the microbiota of lungs, feces, and tongues of transplanted and control (nontransplanted) mice using 16s rRNA gene sequencing. Compared with the lungs of nontransplanted mice, lungs of HCT-treated mice contained distinct bacterial communities (P = 0.002 via permutational multivariate ANOVA) (Figure 1A). Post-HCT lungs were enriched with the Firmicutes phylum (P = 0.002) (Figure 1B) with relative loss of Bacteroidetes phylum members (P = 0.003) (Figure 1B). This change in lung microbiota was driven by enrichment with members of the Lachnospiraceae family (P = 0.005), a gut-associated taxonomic group that comprised 22.6% of bacterial sequences within mouse fecal specimens. Bacterial taxa detected in the lungs of both experimental arms are compared in Figure E2, with significant differences demonstrated in Table E1 and Figure E3. By contrast, we found no significant change in diversity of the lung microbiota as measured by community richness (Figure 1C) or the Shannon diversity index (which reflects both richness and evenness) (Figure 1D). As shown in Figure E4, HCT had no effect on β-diversity of lung, oral, or fecal bacteria; within-group similarity of communities was comparable in HCT and control mice. We concluded that experimental HCT alters lung microbial communities without influencing lung microbial diversity. Changes in upper airway/tongue and gut microbiota after HCT are reported in Figure E5.
Figure 1.
Hematopoietic cell transplant (HCT) changes the community composition of the lung microbiota but does not alter community diversity. (A) Principal component (PC) analysis of untreated control mice and HCT mice demonstrates changes in lung community composition (P = 0.002, permutational multivariate ANOVA). (B) This change in community composition was driven by enrichment with the Firmicutes phylum (P = 0.0024) and relative loss of the Bacteroidetes phylum (P = 0.0003). (C) Yet, rarefaction analysis revealed that community richness—the number of unique species per specimen—was not changed by HCT (P > 0.05). (D) Similarly, the lungs of HCT-treated mice did not differ from those of untreated mice in the Shannon diversity index (P > 0.05). OTU = operational taxonomic unit.
MHV Exposure Alters Community Composition, but Not Diversity, of Lung Microbiota
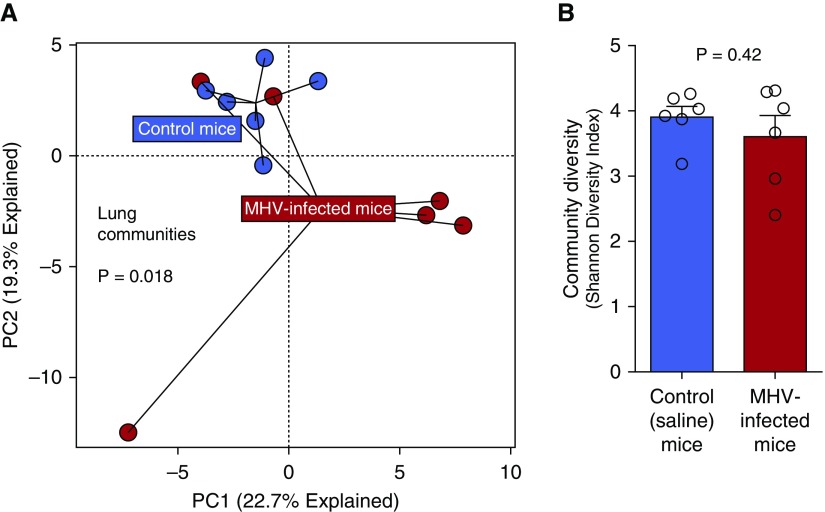
Herpesvirus exposure in humans is associated with an increased risk of post-HCT lung injury (6). MHV is a member of the subfamily Gammaherpesvirinae in the family of Herpesviridae and related to Epstein-Barr virus and herpesvirus saimiri (25). MHV has been studied extensively as a mediator of pulmonary inflammation and subsequent pulmonary fibrosis in preclinical models (8, 9, 24, 26–29). The exposure of otherwise untreated (non-HCT) control mice to MHV by intranasal inoculation generates a resolving pneumonitis. Among nontransplanted mice, MHV infection had a significant but modest effect on lung community composition (P = 0.018) (Figure 2A). Yet, MHV infection had no effect on diversity of the lung communities measured by the Shannon diversity index (P > 0.05) (Figure 2B). As shown in Figure E4, MHV had no effect on β-diversity of lung, oral, or fecal bacteria; within-group similarity of communities was comparable in MHV and control mice. We thus concluded that MHV infection causes a self-limiting pneumonitis associated with modest changes in pulmonary microbiota composition with no detectable effect on lung microbial diversity. Changes in upper airway/tongue and gut microbiota after viral infection are reported in Figure E5.
Figure 2.
Murine herpesvirus (MHV) infection alters the community composition of lung microbiota but has no effect on community diversity. (A) MHV infection results in a resolving pneumonitis that is associated with significant changes in pulmonary microbial communities (P = 0.018, permutational multivariate ANOVA). (B) Despite the differences in community composition, lung bacterial diversity (as measured by the Shannon diversity index) was not significantly altered by viral infection (P > 0.05). PC = principal component.
Viral Infection in the Post-HCT Lung Alters Both Community Composition and Diversity of Lung Microbiota with Associated Fibrosis
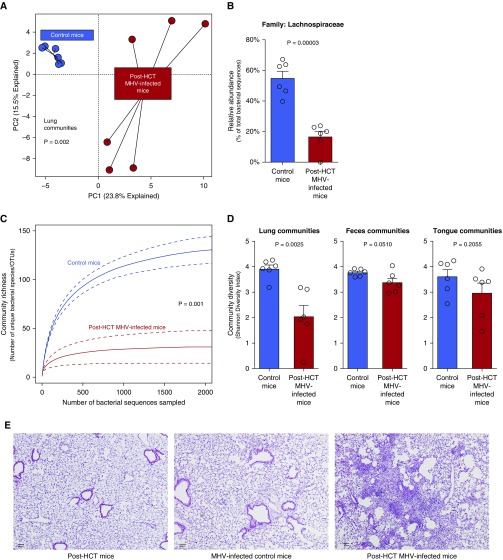
Having demonstrated that HCT and viral infection both alter lung microbiota without affecting lung diversity, we then asked what effect combined HCT and viral infection have on lung microbiota. As previously demonstrated, MHV infection in post-HCT mice initiates a pneumonitis with persistent pulmonary inflammation, lung dysfunction, and fibrosis at 21 days postinfection (8). We found that viral infection profoundly altered microbial lung communities of post-HCT mice compared with uninfected post-HCT mice (P = 0.002) (Figure 3A). This shift in community structure was driven by a relative decrease in the Lachnospiraceae family (Figure 3B). Changes in upper airway/tongue and gut community composition after combined HCT and viral infection are reported in Figure E5. Combined viral infection and HCT resulted in a comparably profound loss of lung microbial diversity, reflected in both decreased community richness (P = 0.001) (Figure 3C) and the Shannon diversity index (P = 0.002) (Figure 3D). This change in diversity was restricted to lung bacteria; no change was observed in the α-diversity of tongue and gut communities (P < 0.05 for both) (Figure 3D). As shown in Figure E4, combined viral infection and HCT significantly increased the β-diversity of lung and fecal bacteria; within-group similarity of communities was lower in the lungs and feces of post-HCT virus-infected mice (P < 0.0001 and P = 0.0119, respectively). Consistent with prior reports, combined viral infection and HCT resulted in pneumonitis (Figure 3E) and collagen deposition consistent with pulmonary fibrosis.
Figure 3.
Combined hematopoietic cell transplant (HCT) and viral infection results in altered lung microbiota, profound loss of lung microbial diversity, and persistent pulmonary pathology. (A) Combined HCT and viral infection profoundly altered the community composition of lung microbiota (P = 0.002, permutational multivariate ANOVA). (B) This change in community composition was driven by a profound decrease in the Lachnospiraceae family. (C) The community richness of lung microbiota was profoundly decreased by combined HCT and viral infection. (D) Lung microbial diversity was decreased by combined HCT and viral infection. This difference was restricted to lung communities; no effect on fecal or oral communities was detected. (E) Compared with those of control mice, lungs of virus-infected mice exhibited transient inflammation with no persistent fibrosis. Combined HCT and viral infection resulted in persistent pneumonitis (hematoxylin and eosin stain) and pulmonary fibrosis (see Reference 8). Scale bars, 100 μm. MHV = murine herpesvirus; OTU = operational taxonomic unit; PC = principal component.
Lung Dysbiosis Is Associated with Pulmonary Dysfunction in Human HCT Recipients
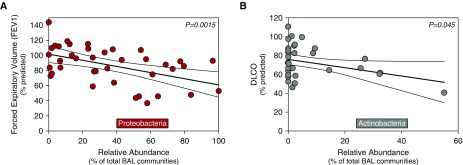
We next characterized microbiota in BAL fluid from 43 HCT recipients with post-HCT pulmonary complications in a novel exploratory study. All patients underwent bronchoscopy for clinical indications, including cough, dyspnea, or fever, with infiltrates on radiological imaging. Clinical characteristics of this cohort of patients are reported in Table 1. Comparison of HCT BAL pulmonary microbiota with taxa detected in negative (procedural and reagent) sequencing controls is reported in Figure E6. Further details, including BAL descriptors and antimicrobial use, are provided in Table E2, and clinical characteristics of HCT recipients with pulmonary complications not enrolled in the study are reported in Table E3. Study HCT recipients had significantly increased relative abundance of Proteobacteria in BAL (see Figure E7). Examples of the most abundant taxa included OTU0012 (Prevotella, Bacteroidetes), OTU0002 (Haemophilus, Proteobacteria), OTU0006 (Bacteroides, Bacteroidetes), and OTU0003 (Veillonella, Firmicutes). We found significant associations between lung microbiota and pulmonary function. Relative abundance of the Proteobacteria phylum, which has previously been associated with pulmonary inflammation (21, 30–32), was negatively associated with FEV1 (P = 0.0015) (Figure 4A). Significance was maintained after adjusting for several potential confounding variables and multiple comparisons (P = 0.0073) (Table E4).
Table 1.
Clinical Characteristics and Demographics of Hematopoietic Cell Transplant Study Population
| Characteristics | Data |
|---|---|
| Total patients, n | 43 |
| Age, yr, mean (SD) | 47.8 (18.7) |
| Male sex, n (%) | 23 (60.5) |
| Primary diagnosis, n (%)* | |
| NHL/Hodgkin’s lymphoma | 9 (20.9) |
| AML/MDS/MF | 22 (51.1) |
| ALL | 4 (9.3) |
| Myeloma | 1 (2.3) |
| Other | 7 (16.4) |
| Type, n (%) | |
| Allogenic | 36 (83.7) |
| Autologous | 7 (16.3) |
| Source, n (%) | |
| PB | 33 (76.7) |
| BM | 4 (9.3) |
| UCB | 6 (14.0) |
| Conditioning, n (%) | |
| Myeloablative | 32 (74.4) |
| Reduced toxicity | 11 (25.6) |
| CGVHD, n (%) | |
| Yes | 26 (60.5) |
| No | 17 (39.5) |
| Pulmonary complications, n | |
| BOS | 12 |
| IPS | 5 |
| RLD | 6 |
| BAL pathogen positive, n (%)† | 14 (32.5) |
| BAL pathogen negative, n (%)‡ | 29 (67.5) |
Definition of abbreviations: ALL = acute lymphoblastic leukemia; AML = acute myeloid leukemia; BM = bone marrow; BOS = bronchiolitis obliterans syndrome; CGVHD = presence of chronic graft-versus-host disease; IPS = idiopathic pneumonia syndrome; MDS = myelodysplastic syndrome; MF = marrow failure; NHL = non-Hodgkin’s lymphoma; PB = peripheral blood; RLD = restrictive lung disease after hematopoietic cell transplant; UCB = umbilical cord and double cord.
Pretransplant diagnosis
BAL culture positive.
BAL culture negative.
Figure 4.
Pulmonary microbiota correlate with measures of pulmonary dysfunction after hematopoietic cell transplant. 16S ribosomal RNA gene community sequencing of BAL samples from 43 hematopoietic cell transplant patients with new pulmonary infiltrates undergoing diagnostic bronchoscopy is reported. (A) The relative abundance of the Proteobacteria phylum was significantly and negatively associated with patients’ FEV1 (adjusted P value = 0.0073). (B) Similarly, the relative abundance of the Actinobacteria phylum was significantly and negatively associated with patients’ gas exchange (DlCO) (adjusted P value = 0.012). Results depicted are from univariate linear regression modeling of logarithmically transformed alveolar cytokine data with regression line and confidence intervals.
For every 10% increase in the relative abundance of BAL Proteobacteria, we found an associated decrease of 4.25% in FEV1 percent predicted. We found a negative association between the relative abundance of the Actinobacteria phylum and DlCO (P = 0.045) (Figure 4B), a significant correlation that was strengthened after multivariable analysis (P = 0.012) (Table E4). For every 10% increase in the relative abundance of BAL Actinobacteria, we found an associated 6.2% decrease in DlCO (percent predicted). The relative abundance of Proteobacteria in BAL was also negatively associated with FVC and of marginal significance (P = 0.052), but it was strengthened after adjustment for covariates (P = 0.048) (Table E4). However, significance was not maintained after accounting for multiple comparisons. We did not find a significant association using linear regression between relative abundance of Proteobacteria and airway obstruction (FEV1) in our limited number of patients with BOS (n = 12). Results of univariate and multivariable linear regression models are reported in Table E4. We concluded that pulmonary dysfunction after HCT is associated with prominent features of the lung microbiome.
Lung Microbiota Are Associated with Alveolar Inflammation in Human HCT Recipients
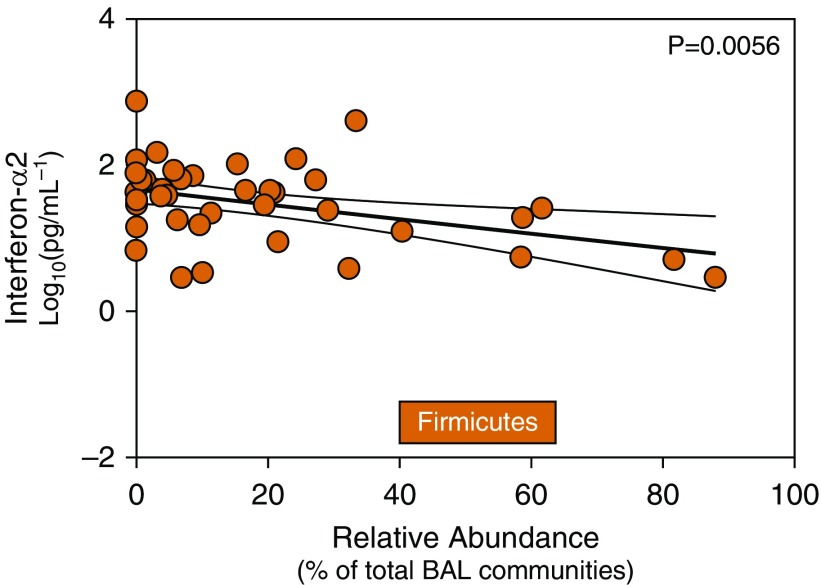
Previous studies in humans have identified associations with gut microbiota and inflammatory cytokine production (33). We thus asked if differences in lung microbiota are associated with indices of alveolar inflammation from human HCT recipient BAL fluid. We found associations between pulmonary microbiota and concentrations of alveolar inflammatory cytokines in this exploratory analysis. Cytokines measured in HCT recipient BAL fluid are reported in Table E5. Innate antiviral cytokines such as IFN-α2 are proinflammatory cytokines, often responsible for modulating the host response to intracellular pathogens and promoting autoimmune responses. In post-HCT patients, concentrations of alveolar IFN-α2 were negatively associated with the relative abundance of the Firmicutes phylum (P = 0.006) on univariate analysis (Figure 5). We found an 11% relative fold decrease in BAL concentrations of IFN-α2 for every 10% increase in the relative abundance of the Firmicutes phylum. This association maintained its significance after adjustment for confounding variables (P = 0.0002) and accounting for multiple comparisons (Table E6). We also report significant negative associations after multivariable analysis between Firmicutes and IL-13 (P = 0.013), epithelial growth factor (P = 0.017), vascular endothelial growth factor (P = 0.031), granulocyte-macrophage colony-stimulating factor (P = 0.032), eotaxin (P = 0.036), tumor necrosis factor (TNF)-β (P = 0.039), TNF-α (P = 0.045), and IL-10 (P = 0.048). The relative abundance of BAL Bacteroidetes was associated with IL-4 (P = 0.03) and IL-13 (P = 0.03) (Table E6). However, these were not considered significant after accounting for multiple comparisons and are presented for hypothesis generation only. Interestingly, we found no significant associations between BAL Proteobacteria and alveolar cytokines. Results of all univariate and multivariable linear regression models examining associations between relative abundance and diversity of pulmonary microbiota and alveolar cytokine expression are reported in Table E6. We conclude that alveolar inflammation after HCT is associated with features of the lung microbiome.
Figure 5.
Pulmonary microbiota correlate with changes in alveolar IFN-α2. To examine possible associations between alveolar inflammation and pulmonary microbiota, we analyzed microbiota at a phylum level with alveolar T-helper cell type 1 cytokines by linear regression. We found significant associations between alveolar cytokines and lung microbiota in hematopoietic cell transplant recipients. Relative abundance of the Firmicutes phylum was negatively associated with alveolar concentrations of IFN-α2 (adjusted P value = 0.0073). Correlations are made using linear regression of relative abundance microbiota data and logarithmically transformed cytokine data. Results depicted are from univariate linear regression modeling with regression line and confidence intervals.
Discussion
The core finding of our study is that after HCT, lung dysbiosis is associated with histologic, immunologic, and physiologic features of pulmonary complications. In an animal model of HCT, developed to recapitulate features of pulmonary complications, we found significant dysbiosis of pulmonary microbiota and biodiversity that correlated with parenchymal changes of inflammation and organ fibrosis. In an exploratory study of human HCT recipients, features of the lung microbiome are associated with lung dysfunction and alveolar inflammation. Our results build on previously reported associations between gut microbiota and HCT outcomes, including bacteremia (11), pulmonary complications (10), and mortality (12). Our findings, which, to our knowledge, represent the first study of lung microbiota after HCT, provide biological plausibility for the hypothesis that altered lung microbiota perpetuate the alveolar host response after HCT, promoting acute and chronic pulmonary inflammation and pathology.
The use of preclinical models has provided crucial insight into the pathogenesis of human disease. However, although animal models of gut microbiota–host interactions have been extensively reported, the study of pulmonary host–microbiota interactions in preclinical models is limited to date (34). In this study, we sought to explore changes within lung microbiota that may contribute to disease pathology in HCT using animal modeling. Human observational studies of chronic lung disease have identified associations between lung dysbiosis, patient survival, host defense, and gene expression of innate immune mediators and antimicrobial peptides (35–38). However, studies of host–microbiota interactions in human lungs are difficult and limited by confounding, potential for contamination, and access to appropriate serial samples. Animal models of host and pulmonary microbiota interactions provide an important evolving tool for these investigations (34, 39). We have recently demonstrated that the murine lung microbiome is variable and strongly correlated with pulmonary immune tone, even in health (34). In murine models of emphysema, the absence of pulmonary microbiota–derived products led to attenuated pulmonary IL-17A production, and microbiota transplant into microbiota-depleted mice enhanced IL-17A expression (40). In our present study, we demonstrate that both HCT and virus-mediated lung injury are associated with significant changes in pulmonary microbial community composition in animal models. This report, in conjunction with previous studies, demonstrates the feasibility of preclinical models in the investigation of pulmonary host–microbiota interactions and generates further support for their continued use and refinement.
Recent studies have reported observations that support a potential relationship between gut and lung microbiota. This purported “gut–lung axis” may serve a key role in early host defense responses (41). The gut harbors a relatively vast biomass of microbiota that has established roles in systemic and local mucosal immunity, inflammation, and metabolism, among others (42, 43). Gut microbiota composition has an immunomodulatory effect in HCT, and preengraftment antibiotic therapy that targets anaerobic species in the gut has recently been observed to increase the risk of colonic graft-versus-host disease in HCT recipients (44). We have recently reported that the lung microbiome is enriched with gut-associated bacteria, both in experimental sepsis and in humans with acute respiratory distress syndrome (ARDS) (21). Lung dysbiosis in ARDS was correlated with key features of both systemic and alveolar inflammation. Importantly, ARDS is clinically indistinguishable from IPS in post-HCT patients. In the present study, the post-HCT lung microbiome in our animal model was significantly enriched with gut-associated bacteria. The gut microbiome in HCT recipients is altered by various ecological pressures, including T cell–mediated changes and conditioning regimens. These changes include dysbiosis and domination by specific species, which predict bacteremia and mortality (11, 12). Recently, Harris and colleagues identified gut Proteobacteria domination as a major predictor for the development of pulmonary complications in HCT patients (10). Interestingly, in our study of post-HCT lung microbiota in humans, increased relative abundance of Proteobacteria in the lung was correlated with impaired lung function (decreased FEV1). This taxonomic parallel across studies may reflect a disordered gut–lung axis underlying post-HCT pulmonary complications and should be investigated further using paired gut and lung human microbiome specimens. In our post-HCT cohort, Proteobacteria domination could not be explained solely by the presence of pneumonia, because only 3 of 43 BAL specimens grew Proteobacteria spp. by culture. The well-described mucosal injury observed in the gut of post-HCT patients may also occur at other mucosal sites, including the respiratory tract, favoring the local outgrowth of Proteobacteria. Impaired gut wall integrity may permit translocation of gut bacteria to the lungs, provoking alveolar inflammation and pulmonary dysfunction.
We report, for the first time to our knowledge, significant correlations in HCT recipients between pulmonary microbiota, indices of alveolar inflammation, and pulmonary dysfunction. These observations, though exploratory, provide further evidence of a connection between local alveolar inflammation and pulmonary microbiota. A major cause of pulmonary morbidity and worsening airflow obstruction in HCT is the development of BOS. Lung transplant may also be complicated by BOS, and pulmonary dysbiosis in lung transplant recipients is enriched with Proteobacteria and associated with an “inflammatory” alveolar macrophage phenotype (45). Like BOS, asthma and COPD are characterized by airflow obstruction and are both similarly characterized by increased relative abundance of airway Proteobacteria (30–32, 46, 47). Relative enrichment with Proteobacteria may be either a cause or effect of airway inflammation, and it may propel the small airway remodeling of BOS.
Although altered community composition is a consistent feature of respiratory dysbiosis commonly reported in acute and chronic lung disease (13), the role and significance of lung microbial diversity are less clear. Certainly, acute infections in the lower respiratory tract are characterized by diminished lung diversity (48); the clinical, microbiological, and immunological significance of differences in diversity outside of infections is undetermined. Increases in diversity within the lung microbiome can reflect either increased immigration of species (e.g., gut–lung translocation or increased pharyngeal aspiration) or decreased elimination of species (e.g., due to impaired host defenses) (14). We have previously shown that HCT results in impaired pulmonary clearance of bacterial pathogens as a result of altered cytokine and immune cell function (9). It is also biologically plausible, given our data and previous work, that the loss of gut mucosal integrity observed in HCT may result in increased immigration of bacteria to the lung. By contrast, within our animal model, dual exposure to HCT and herpesvirus resulted in a catastrophic loss of lung diversity concurrent with the development of lung fibrosis. The ecologic mechanism and pathophysiologic significance of this collapse in diversity remain undetermined.
There are several limitations within our work. No consensus method exists for determining statistical power for microbiome comparisons, and it is possible that the lack of effect of isolated HCT and MHV infection on lung diversity represents a type II (false-negative) error. Our murine microbiome data were derived from single representative time points, though surely the effects of HCT and viral infection on lung microbiota are dynamic and time dependent. Our human microbiome data were generated from acellular BAL, which differs in its microbial content from whole BAL (14). This taxonomic bias should be taken into consideration when comparing our results with those of other studies. In our attempt to control for the confounding of antibiotic exposure in our human cohort, we chose to employ a previously published model derived from sputum samples collected from patients with cystic fibrosis (23). The generalizability of this model to human BAL specimens in a population without cystic fibrosis is unestablished. Procedural contamination control specimens (e.g., bronchoscope rinse specimens) were not collected at the time of sample collection in our human cohort, and should be a part of the design of subsequent studies. Our human cohort of HCT patients lacks a non-HCT control group, and the modest size of this human cohort limits further subgroup analysis. Despite our cohort size, we uncovered novel and plausible correlations between lung microbiota, airway obstruction, and inflammatory cytokine expression, all congruent with published observations from non-HCT populations. The issue of multiple comparisons is intrinsic to high-dimensional analyses such as our comparison of lung microbiota with inflammatory and physiologic indices. We addressed this source of type I error using an FDR approach, and we sought to determine biological plausibility of our findings by comparing them with prior non-HCT lung microbiome studies. We plan further validation of our reported associations.
In summary, in animal models and human subjects, lung dysbiosis is prominent after HCT and is correlated with the histologic, immunologic, and physiologic features of post-HCT pulmonary complications. Further study is required to explore potential causal associations or pathological contributions of pulmonary dysbiosis and lung injury in HCT. Our findings suggest the lung microbiome may be an unappreciated target for the prevention and treatment of post-HCT pulmonary complications.
Acknowledgments
Acknowledgment
The authors acknowledge all patients who participated in this study, our study coordinator Connie Varner, and the Host Microbiome Initiative at the University of Michigan.
Footnotes
Supported by NIH grants K99HL139996 (D.N.O’D.), AI117229 (B.B.M.), HL127805 (B.B.M.), and K23HL130641 (R.P.D.), and the Host Microbiome Initiative Explorer Program at the University of Michigan. This research was also supported by the National Cancer Institute under award number P30 CA046592 by the use of the Cancer Center Shared Resource: Experimental Irradiation Core.
Author Contributions: D.N.O’D., B.B.M., J.R.E.-D., and R.P.D.: designed study; acquired, analyzed, and interpreted data; and drafted the manuscript; C.A.W., N.R.F., K.C.N., K.B.A., and X.Z.: acquired data; M.X. and S.M.: analyzed and interpreted data and contributed to the manuscript; G.A.Y.: obtained patient samples; and G.A.Y. and G.B.H.: revised the manuscript. All authors approved the final version of the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201712-2456OC on June 7, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Gooley TA, Chien JW, Pergam SA, Hingorani S, Sorror ML, Boeckh M, et al. Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363:2091–2101. doi: 10.1056/NEJMoa1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucena CM, Torres A, Rovira M, Marcos MA, de la Bellacasa JP, Sánchez M, et al. Pulmonary complications in hematopoietic SCT: a prospective study. Bone Marrow Transplant. 2014;49:1293–1299. doi: 10.1038/bmt.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afessa B, Peters SG. Major complications following hematopoietic stem cell transplantation. Semin Respir Crit Care Med. 2006;27:297–309. doi: 10.1055/s-2006-945530. [DOI] [PubMed] [Google Scholar]
- 4.Wingard JR, Hsu J, Hiemenz JW. Hematopoietic stem cell transplantation: an overview of infection risks and epidemiology. Infect Dis Clin North Am. 2010;24:257–272. doi: 10.1016/j.idc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Yoshihara S, Yanik G, Cooke KR, Mineishi S. Bronchiolitis obliterans syndrome (BOS), bronchiolitis obliterans organizing pneumonia (BOOP), and other late-onset noninfectious pulmonary complications following allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2007;13:749–759. doi: 10.1016/j.bbmt.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Seo S, Renaud C, Kuypers JM, Chiu CY, Huang ML, Samayoa E, et al. Idiopathic pneumonia syndrome after hematopoietic cell transplantation: evidence of occult infectious etiologies. Blood. 2015;125:3789–3797. doi: 10.1182/blood-2014-12-617035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Versluys AB, Rossen JW, van Ewijk B, Schuurman R, Bierings MB, Boelens JJ. Strong association between respiratory viral infection early after hematopoietic stem cell transplantation and the development of life-threatening acute and chronic alloimmune lung syndromes. Biol Blood Marrow Transplant. 2010;16:782–791. doi: 10.1016/j.bbmt.2009.12.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Loomis-King H, Gurczynski SJ, Wilke CA, Konopka KE, Ptaschinski C, et al. Bone marrow transplantation alters lung antigen-presenting cells to promote TH17 response and the development of pneumonitis and fibrosis following gammaherpesvirus infection. Mucosal Immunol. 2016;9:610–620. doi: 10.1038/mi.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coomes SM, Hubbard LL, Moore BB. Impaired pulmonary immunity post-bone marrow transplant. Immunol Res. 2011;50:78–86. doi: 10.1007/s12026-010-8200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris B, Morjaria SM, Littmann ER, Geyer AI, Stover DE, Barker JN, et al. Gut microbiota predict pulmonary infiltrates after allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med. 2016;194:450–463. doi: 10.1164/rccm.201507-1491OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Dwyer DN, Dickson RP, Moore BB. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J Immunol. 2016;196:4839–4847. doi: 10.4049/jimmunol.1600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384:691–702. doi: 10.1016/S0140-6736(14)61136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Dwyer DN, Wilke CA, Zhou X, Falkowski NR, Yanick GA, Moore BB, et al. Lung microbiome disruption in a murine model of post-bone marrow transplant pulmonary fibrosis [abstract] Am J Respir Crit Care Med. 2017;195:A7293. [Google Scholar]
- 16.Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, et al. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun. 2012;80:3371–3380. doi: 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schloss PD.454 SOP – mothur 2015 [accessed 2017 Sept 25]. Available from: http://www.mothur.org/wiki/454_SOP
- 20.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Murray S, Lipuma JJ. Modeling the impact of antibiotic exposure on human microbiota. Sci Rep. 2014;4:4345. doi: 10.1038/srep04345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coomes SM, Wilke CA, Moore TA, Moore BB. Induction of TGF-β1, not regulatory T cells, impairs antiviral immunity in the lung following bone marrow transplant. J Immunol. 2010;184:5130–5140. doi: 10.4049/jimmunol.0901871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efstathiou S, Ho YM, Hall S, Styles CJ, Scott SD, Gompels UA. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J Gen Virol. 1990;71:1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- 26.McMillan TR, Moore BB, Weinberg JB, Vannella KM, Fields WB, Christensen PJ, et al. Exacerbation of established pulmonary fibrosis in a murine model by gammaherpesvirus. Am J Respir Crit Care Med. 2008;177:771–780. doi: 10.1164/rccm.200708-1184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vannella KM, Luckhardt TR, Wilke CA, van Dyk LF, Toews GB, Moore BB. Latent herpesvirus infection augments experimental pulmonary fibrosis. Am J Respir Crit Care Med. 2010;181:465–477. doi: 10.1164/rccm.200905-0798OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashley SL, Jegal Y, Moore TA, van Dyk LF, Laouar Y, Moore BB. γ-Herpes virus-68, but not Pseudomonas aeruginosa or influenza A (H1N1), exacerbates established murine lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2014;307:L219–L230. doi: 10.1152/ajplung.00300.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurczynski SJ, Procario MC, O’Dwyer DN, Wilke CA, Moore BB. Loss of CCR2 signaling alters leukocyte recruitment and exacerbates γ-herpesvirus-induced pneumonitis and fibrosis following bone marrow transplantation. Am J Physiol Lung Cell Mol Physiol. 2016;311:L611–L627. doi: 10.1152/ajplung.00193.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. National Heart, Lung, and Blood Institute’s Asthma Clinical Research Network. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381.e3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sze MA, Dimitriu PA, Suzuki M, McDonough JE, Campbell JD, Brothers JF, et al. Host response to the lung microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;192:438–445. doi: 10.1164/rccm.201502-0223OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167:1125–1136.e8. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dickson RP, Erb-Downward JR, Falkowski NR, Hunter EM, Ashley SL, Huffnagle GB. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am J Respir Crit Care Med. 2018;198:497–508. doi: 10.1164/rccm.201711-2180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molyneaux PL, Willis-Owen SAG, Cox MJ, James P, Cowman S, Loebinger M, et al. Host-microbial interactions in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;195:1640–1650. doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y, Ma SF, Espindola MS, Vij R, Oldham JM, Huffnagle GB, et al. COMET-IPF Investigators. Microbes are associated with host innate immune response in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;196:208–219. doi: 10.1164/rccm.201607-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han MK, Zhou Y, Murray S, Tayob N, Noth I, Lama VN, et al. COMET Investigators. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med. 2014;2:548–556. doi: 10.1016/S2213-2600(14)70069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190:906–913. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. The role of the lung microbiome in health and disease: a National Heart, Lung, and Blood Institute workshop report. Am J Respir Crit Care Med. 2013;187:1382–1387. doi: 10.1164/rccm.201303-0488WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadava K, Pattaroni C, Sichelstiel AK, Trompette A, Gollwitzer ES, Salami O, et al. Microbiota promotes chronic pulmonary inflammation by enhancing IL-17A and autoantibodies. Am J Respir Crit Care Med. 2016;193:975–987. doi: 10.1164/rccm.201504-0779OC. [DOI] [PubMed] [Google Scholar]
- 41.Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 42.Blacher E, Levy M, Tatirovsky E, Elinav E. Microbiome-modulated metabolites at the interface of host immunity. J Immunol. 2017;198:572–580. doi: 10.4049/jimmunol.1601247. [DOI] [PubMed] [Google Scholar]
- 43.Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol. 2017;35:8–15. doi: 10.1016/j.mib.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med. 2016;8:339ra71. doi: 10.1126/scitranslmed.aaf2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernasconi E, Pattaroni C, Koutsokera A, Pison C, Kessler R, Benden C, et al. SysCLAD Consortium. Airway microbiota determines innate cell inflammatory or tissue remodeling profiles in lung transplantation. Am J Respir Crit Care Med. 2016;194:1252–1263. doi: 10.1164/rccm.201512-2424OC. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Q, Cox M, Liang Z, Brinkmann F, Cardenas PA, Duff R, et al. Airway microbiota in severe asthma and relationship to asthma severity and phenotypes. PLoS One. 2016;11:e0152724. doi: 10.1371/journal.pone.0152724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marsland BJ, Gollwitzer ES. Host-microorganism interactions in lung diseases. Nat Rev Immunol. 2014;14:827–835. doi: 10.1038/nri3769. [DOI] [PubMed] [Google Scholar]
- 48.Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, et al. Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol. 2014;52:3605–3613. doi: 10.1128/JCM.01028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]