Abstract
Data analytics play vital roles in diagnosis and treatment in the health care sector. To enable practitioner decision-making, huge volumes of data should be processed with machine learning techniques to produce tools for prediction and classification. Diseases like breast cancer can be classified based on the nature of the tumor. Finding an effective algorithm for classification should help resolve the challenges present in analyzing large volume of data. The objective with this paper was to present a report on the performance of Bayes classifiers like Tree Augmented Naive Bayes (TAN), Boosted Augmented Naive Bayes (BAN) and Bayes Belief Network (BBN). Among the three approaches, TAN produced the best performance regarding classification and accuracy. The results obtained provide clear evidence for benefits of TAN usage in breast cancer classification. Applications of various machine learning algorithms could clearly assist breast cancer control efforts for identification, prediction, prevention and health care planning.
Keywords: Tree Augmented Naive Bayes (TAN), Boosted Augmented Naive Bayes (BAN)
Introduction
The risk of developing breast cancer rises throughout a woman’s lifespan, and the disease is relatively rare in very young women. Breast cancer remains the most common cancer among women in the United States and its association with increasing age is consistent. Breast cancer is normally identified either during a screening check, perhaps before symptoms have developed, or after noticing a lump (Anuranjeeta et al., 2017). Most masses seen on a mammogram and most breast lumps turn out to be benign that do not grow uncontrollably or spread, and are not life-threatening (Stojadinovic et al., 2010). To suspect cancer health care practitioners use microscopic analysis of breast tissue in order determine the extent of spread and the type of the disease (Torosian, 2002). At the age of informatics era usage of computational methods helps the practitioners in classifying the characteristics of the cancer (Abdel-Zaher and Eldeib, 2016).One such effective tool for analysis and decision making is machine learning algorithms (Bishop, 2006).
Machine learning is a data analytics method that uses computational methods to “discover” information directly from data without depending on encoded equation. The algorithms rapidly improve their performance as the number of samples available for learning increases (Basu et al., 2018). With the rise in volume of data, machine learning has become a solution for solving problems in areas like health care sector (Araujo et al., 2017). Machine learning algorithms are used in various applications for classification, regression, estimation and novelty detection (Somla and Vishwanathan, 2008).
In the work herein described, analysis of bayes classifiers are executed by SAS -EM (Statistical Analytical Software Enterprise Miner). SAS-EM rationalizes the data to create highly accurate predictive and descriptive models (Hall et al., 2014). The models are based on investigation of vast amount of data from across an enterprise or user defined data set. All analyses begin with a data set. SAS-EM is used primarily to examine large, complex data sets with tens of thousands to millions of records and hundreds to thousands of variables. SAS-EM uses SEMMH (Sample, Explore, Modify, Model and High performance Data mining) model development process for finding patterns and relationships in the data and thereby determines whether the discovered patterns are valid. The basic data preprocessing can be done in SAS-EM. After preprocessing partitioning of the data can be designed (Baxter and Huddleston, 2015).
Related Works
Kharya et al., (2014) used Naive Bayes (NB) algorithm for breast cancer detection and demonstrated the accuracy results as 93%. However evaluation and improvement measures for NB algorithm has not been proposed by the researchers. Chaurasia et al., (2018) compared algorithms like NB, Radial Basis Function Network and J48 for breast cancer prediction and proved the performance of NB algorithm. Stojadinovic et al., (2010) applied NB algorithm for breast cancer risk stratification. Mandal et al., (2017) analyzed the performance of NB, Logistic Regression and Decision Tree for breast cancer detection and proved the performance of NB classifier. Huang et al., (2017) compared Support Vector Machines (SVM) and SVM based ensemble method. The researchers proved the performance of SVM based ensemble and suggested the usage of boosting method with machine learning techniques for better performance and accuracy. Jing et al., (2008) proposed Boosted Bayesian Network classifier for breast cancer classification. Most of the researchers suggested the usage of NB based classifiers for breast cancer prediction (Ren et al., 2015). Researchers investigated Genetic Programming (GP), SVM, Multilayered Perceptrons (MLP) and Random Forest for classifying cancer patients into risk classes and suggested to use GP. However the time taken for the convergence has not been discussed (Vanneschi et al., 2011). Asria et al., (2016) investigated four different classifiers like Decision Tree (C4.5), K-Nearest Neighbor (k-NN), NB, SVM and suggested SVM for breast cancer prediction. However the algorithmic complexity and high memory requirements has not been addressed by the researchers for prediction.
Materials and Methods
Wisconsin Diagnostic Breast Cancer (WDBC) data set is used for the analysis. Total number of instances present in the dataset is about 569 instances with 32 attributes. Among the attributes diagnosis is used as classification and the following ten attributes like radius (mean of distances from center to points on the perimeter), texture, perimeter, area, smoothness, compactness, concavity concave points, symmetry and fractal dimension are used for the research work (Aalaei et al.,2016). These attributes are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They define the characteristics of the cell nuclei present in the image (Sountharrajan et al., 2017). Hence the major attributes are extracted for the comparison.
There are five types of BN classifiers. They are Naive-Bayes, Tree augmented Naïve-Bayes (TANs), Bayesian network augmented Naive-Bayes (BANs), Bayesian multi-nets and general Bayesian networks (GBNs). The preface of investigations involves a study of evolutionary methods to analyze the challenges present in bayes classifier. Naive Bayes is one among the statistical classifier used to predict class membership probability (Zaidi et al., 2013). It detects the class membership based on the maximum probability obtained for the given tuple to a particular class. It assumes all variables take part in classification to be sovereign and provides the outcome for prediction. The algorithm leads to a simple prediction framework which yields good results in many cases as proved by the researchers. But the algorithm treats discrete and continuous variables in different way (Soria et al., 2008). In recent years, researchers focused on improving Naive-Bayesian classifiers.
TAN classifiers extend Naive-Bayes by allowing the attributes to construct a tree for classification. BAN classifiers extend TAN classifiers by permitting the attributes to form an arbitrary graph, rather than building a tree (Jiang et al., 2005). A Bayesian network is an annotated directed graph that translates the probabilistic associations among variables of interest. However all these classifiers produce minimal accuracy. In order to improve the accuracy, all the classifiers are combined with Gradient Boosting (GB) technique. The objective of GB algorithm is to minimize the loss function defined as mean squared error (MSE) given in Formula 1.
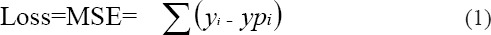
Where yi is the target value, ypi ith prediction.
GB algorithm strengthens the model with weak accuracy by evaluating average squared error (ASE) in restricted number of iterations. Based on the ASE value obtained in the iteration, the new model is calculated by finding the difference across target value and ASE. Procedure for finding new predicted value will be repeated until the loss function becomes a constant value for the remaining iterations.
The model is designed in SAS-EM 14.3, depicted in Figure 1, consist of series of nodes. SAS-EM 14.3 has been accessed through SAS OnDemand for Academics portal (SAS 2018). The data downloaded from WDBC is loaded in the file import node and the attributes are selected for the model. Among the attributes diagnosis is considered as the target attribute. Then the data set is partitioned as 70:30 for training and testing. Further the nodes are connected with High performance Data Mining (HPDM) nodes and the algorithms like BAN, TAN and Bayes Network are selected. HPDM offers In-Memory processing and thereby the data set is manipulated in a computer’s RAM.As a result the amount of time required for computation will be reduced. Finally the nodes are connected to GB node by assigning the loss function as mean squared error.For evaluating the classifiers the models are connected to model comparison node.
Figure 1.
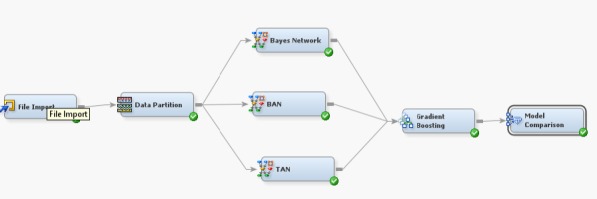
SAS-EM Design for Comparative Analysis of Classifiers
Results
The results of the comparison analysis using bayes classifiers like bayes network, BAN and TAN presented here are based on two parameters: benign and malignant cancer patients. TAN classifier outperforms from other classifiers in terms of performance analysis parameters like accuracy, sensitivity and specificity. All the three classifiers produced almost similar accuracy 90.1% before applying GB process. However the results of all the classifiers are enhanced with GB and the accuracy, specificity and sensitivity results are shown in the Figure 2.
Figure 2.
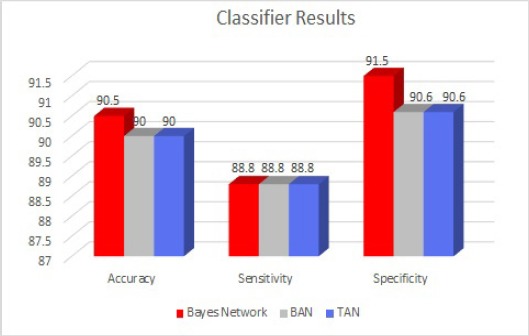
Comparative Results of Classifiers by Accuracy, Specificity and Sensitivity
Figure 3.

Comparative Results of Classifiers with Gradient Boosting by Accuracy, Specificity and Sensitivity
The results shows that TAN classifier performs well by GB technique by fine-tuning the MSE loss function. Although the standard TAN algorithm is stable, the accuracy level can be improved by boosting technique. The average squared error has been reduced to .04% and the misclassification rate is .05% for TAN with GB and subsequently as .07% and .09% for BAN, whereas for Bayes Network as .07% and .1%. Overall the error rate is reduced when GB is applied for TAN classifier.
Discussion
In this paper a comparative study on different bayes classification techniques are investigated along with boosting method in terms of accuracy percentage, sensitivity and specificity. The study reveals that Tree Augmented Naive Bayes Classifier along with Gradient Boosting delivers the maximum accuracy with reduced Mean Squared Error when compared to bayes network and BAN. This work can be further be enhanced by identification of dynamic class labels for the prediction of breast cancer with various attributes.
References
- 1.Aalaei S, Shahraki H, Rowhanimanesh A, Eslami S. Feature selection using genetic algorithm for breast cancer diagnosis: experiment on three different datasets. Iran J Basic Med Sci. 2016;19:476–82. [PMC free article] [PubMed] [Google Scholar]
- 2.Abdel-Zaher AM, Eldeib AM. Breast cancer classification using deep belief networks. Expert Syst Appl. 2016;46:139–44. [Google Scholar]
- 3.Anuranjeeta A, Shukla KK, Tiwari A, Sharma S. Classification of histopathological images of breast cancerous and non cancerous cells based on morphological features, biomed. Pharmacol J. 2017;10:353–66. [Google Scholar]
- 4.Araujo T, Aresta G, Castro E, et al. Classification of breast cancer histology images using Convolutional Neural Networks. PLoS One. 2017;12:1–14. doi: 10.1371/journal.pone.0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asria H, Mousannif H, Moatassime HA, Noel T. Using machine learning algorithms for breast cancer risk prediction and diagnosis. Procedia Comput Sci. 2016;83:1064–9. [Google Scholar]
- 6.Basu A, Roy R, Savitha N. Performance analysis of regression and classification models in the prediction of breast cancer. Indian J Sci Technol. 2018;11:1–6. [Google Scholar]
- 7.Baxter A, Huddleston ED. Getting started with SAS®Enterprise Miner™14.1: High-performance procedures. NC, USA: SAS institute Inc, Cary; 2015. pp. 1–44. [Google Scholar]
- 8.Bishop C. Pattern recognition and machine learning. New York, USA: Springer science and business media; 2006. pp. 359–70. [Google Scholar]
- 9.Chaurasia V, Pal S, Tiwari BB. Prediction of benign and malignant breast cancer using data mining techniques. J Algorithm Comput Technol. 2018;12:119–26. [Google Scholar]
- 10.Hall P, Dean J, Kabul IK, Silva J. An overview of machine learning with SAS®Enterprise Miner. SAS Institute Inc, Cary, NC, USA, pp. 2014:1–24. [Google Scholar]
- 11.Huang MW, Chen CW, Lin WC, Ki SW, Tsai CF. SVM and SVM ensembles in breast cancer prediction. PLoS One. 2017;12:1–14. doi: 10.1371/journal.pone.0161501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Zhang H, Cai Z, Su J. Learning tree augmented naive bayes for ranking DASFAA. LNCS. 2005;3453:688–98. [Google Scholar]
- 13.Jing Y, Pavlovi V, Rehg JM. Boosted bayesian network classifiers. Machine Learning. 2008;73:155–84. [Google Scholar]
- 14.Kharya S, Agrawal S, Soni S. Naive Bayes classifiers: A probabilistic detection model for breast cancer. Int J Comput Appl. 2014;92:26–31. [Google Scholar]
- 15.Mandal SK. Performance analysis of data mining algorithms for breast cancer cell detection using Naive Bayes, logistic regression and decision. Int J Eng Comput Sci. 2017;6:20388–91. [Google Scholar]
- 16.Ren P. A tree augmented naive Bayesian network experiment for breast cancer prediction. ArXiv e-prints. 2015:1–24. [Google Scholar]
- 17.SAS OnDemand for Academics. Resources/Sas Products and Solutions. 2018. [[Accessed on: Apr/2018]]. Available from: http://support.sas.com/software/products/ondemand-academics/
- 18.Somla A, Vishwanathan SVN. Introduction to machine learning. UK: Cambridge University Press; 2008. pp. 16–32. [Google Scholar]
- 19.Soria D, Garibaldi JM, Biganzoli E, Ellis IO. A comparison of three different methods for classification of breast cancer data. Proceedings –ICMLA. 2008:619–24. [Google Scholar]
- 20.Sountharrajan S, Karthiga M, Suganya E, Rajan C. Automatic classification on bio medical prognosis of invasive breast cancer. Asian Pac J Cancer Prev. 2017;18:2541–4. doi: 10.22034/APJCP.2017.18.9.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stojadinovic A, Eberhard C, Henry L. Development of a Bayesian classifier for breast cancer risk stratification: A feasibility study. Eplasty. 2010;10:203–16. [PMC free article] [PubMed] [Google Scholar]
- 22.Torosian MH. Curr Oncol. otowa, NJ: Humana Press; 2002. Breast cancer: a guide to detection and multidisciplinary therapy; pp. 81–3. [Google Scholar]
- 23.Vanneschi L, Farinaccio A, Mauri G, et al. A comparison of machine learning techniques for survival prediction in breast cancer. Biodata Min. 2011;4:12–25. doi: 10.1186/1756-0381-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaidi NA, Cerquides J, Carman MJ, Webb GI. Alleviating Naive Bayes attribute independence assumption by attribute weighting. J Mach Learn Res. 2013;14:1947–88. [Google Scholar]


