Abstract
Peroxisome proliferator-activated receptor-γ (PPARγ) plays an important role in lipid and glucose metabolism. In this study, the function of PPARγ on lung development was investigated. Lung-specific Pparg conditional knockout mice (PpargΔLuEpC) were developed using Cre-Lox system. PpargΔLuEpC mice showed abnormal lung development with enlarged airspaces and followed by increase of apoptotic cells at E14.5 to E18.5. Gene analysis revealed that expression of Pmaip1, a gene related to apoptosis, was significantly increased while expression of Retnla, a gene related to anti-apoptosis, was dramatically decreased in the fetal lung (E14.5) of PpargΔLuEpC mice. In addition, expression of Pthlh, a gene pheno-typically expressed in the congenital cystic adenomatoid malformation (CCAM), was increased at E14.5 to E18.5 in the lung of PpargΔLuEpC mice. Cell culture studies revealed that PPARγ could bind to promoter region of Pthlh gene as a repressor in the immortalized mouse lung epithelial cell line MLE-15. Surprisingly, phenotypic changes in MLE-15-shPparg cells, stably transfected with shPparg plasmid, were similar to the PpargΔLuEpC mice model. In addition, MLE-15-shPparg cells were easily detached from the cultured plate when cold phosphate buffered saline was applied. Furthermore, expression of Cdh1, a gene related to cell adhesion, was significantly reduced in the MLE-15-shPparg cells. Taken together, PPARγ may play an important role in fetal lung development via alveolar cell-to-cell adhesion system.
Keywords: PPARγ, Cystic adenomatoid malformation
1. Introduction
Peroxisome proliferator-activated receptor-γ (PPARγ, NPIX3), along with PPARα (NRIC1) and PPARβ/δ (NRIC2), are ligand-activated transcription factor in the nuclear receptor superfamily [1,2]. PPARγ is highly expressed in white and brown adipose tissue and to a lesser extent in macrophage, liver, colon, and kidney [3]. Together with the retinoid X receptor (RXR), PPARγ binds to PPAR responsive-regulatory elements (PPRE) of genes involved in adipogenesis, lipid metabolism, inflammation, and glucose homeostasis [3,4]. PPARγ is activated by endogenous metabolites such as 15-deoxy-delta 12, 14 prostaglandin J2 [5] and PPARγ agonists including thiazolidinediones pioglitazone and rosiglitazone have been used for the treatment of metabolic disorders such as hyperlipidemia and diabetes mellitus type II [6].
Because PPARγ is mostly distributed in adipose tissues, its roles in different tissues or cells have not received much attention. Lung tissue is composed of different cell types such as alveolar type I and type II epithelial cells, and club cells and ciliated cells in airway epithelia. Alveolar type II epithelial cells and club cells express PPARγ. One study suggested a role for PPARγ in lung development. Airway epithelial cell PPARγ-targeted mice display enlarged airspaces resulting from insufficient postnatal lung maturation [7]. More recently, it was reported that PPARγ activation enhances lung maturation in a neonatal rat model [8]. These findings suggest that PPARγ is necessary for normal lung structure during developmental stage. In this study, the function of PPARγ on fetal lung development was investigated using lung-specific Pparg-null mice.
2. Materials and methods
2.1. Mouse
Pparg-floxed (Pparg fl/fl) mice containing loxP sites flanking exon 2 [9] were crossed with mice harboring the Cre recombinase under control of the surfactant protein C promoter [10] to generate PpargΔLuEC mice. Mice were interbred for over six generations to produce littermates with the same mixed genetic background (FVB). Mice, housed in temperature- and light-controlled rooms, were given water and pelleted chow ad libitum. All animal studies were carried out in accordance with Institute of Laboratory Animal Resources guidelines and approved by the National Cancer Institute Animal Care and Use Committee.
2.2. Isolation of lung epithelial cells
Lung epithelial cells were isolated from PpargΔLuEC and Ppargfl/fl fetuses at E18.5 as described previously [11] with minor modifications. Briefly, minced lung was incubated in Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Cellgro, Herndon, VA) containing 10% heat-inactivated fetal bovine serum (FBS, Gemini Bio-Products, Woodland, CA), 1 U/ml Dispase® (Invitrogen, Grand Island, NY) and 100 U/ml collagenase type I (Invitrogen) at 37 °C for 30 min. Cells were washed three times in DMEM/F12 containing 10% fetal bovine serum. Cells were seeded and incubated at 37 °C for 20 min to separate epithelial cells from mesenchymal cells. Epithelial cells that remained in the media were transferred to a 12-well plate and incubated at 37 °C for 24 h in 5% CO2 incubator.
2.3. Cell culture and transfection
Immortalized mouse lung epithelial MLE-15 cells (kindly provided by Dr. Jeffrey Whitsett, Cincinnati Children’s Hospital [12]) were maintained at 37 °C in a humidified atmosphere of 5% CO2/95% air in DMEM supplemented with 10% heat-inactivated FBS and 50 U/ml of penicillin/streptomycin mixture (Invitrogen). Cells were grown to 60e80% confluence and trypsinized with 0.05% trypsin containing 2 mM EDTA. For silencing of PPARγ in the MLE-15 cells, cells were transfected with 3 μg of the pGFP-V-RS-GI56785 construct (Origene) using JetPEI transfection reagent (Polyplus transfection) or transfected with mouse siPparg (Santa Cruz biotechnology) using Lipofectamine RNAiMAX reagent (Thermofisher).
2.4. Reporter gene assays
For Pthlh-promoter luciferase assays, MLE-15 cells were transfected with different sizes of pGL3-pthlh constructs (−7648/+ 442, −6055/+442, −3767/+442, and −1876/+442) in the presence of pSG5-Pparg and pSG5-RXRα plasmids or rosiglitazone. The assay was analyzed using luciferase assay kit (Promega).
2.5. DNA-protein pulldown assay
For protein-DNA pulldown assay, MLE-15 cells were plated in 100-mm culture dishes at 70% confluence and cultured overnight. Then, the cells were transfected with pEGFP-Pparg plasmid (3 μg) for 24 h and nuclear protein was isolated using NE-PER kit (Thermo Fisher Scientific). The nuclear proteins (50 μg) were incubated with biotinylated Pthlh promoter DNA fragments (1 μg) in the incubation buffer (10 mM Trise‒HCl, pH 7.5; 150 mM NaCl; 1 mM DTT; 1 mM EDTA, 0.1% NP-40, 0.1 mg/ml sonicated salmon sperm DNA) and the protein-DNA complexes were pull-downed with 50 μl of streptavidin-labelled dynabead (Thermo Fisher Scientific). After washing the beads three times with same buffer, samples were subjected to Western blotting. To analyze the PPARγ binding sites in the Pthlh promoter, chromatin immunoprecipitation (ChIP) assay was executed as previously described [13]. Briefly, cultured MLE-15 cells were fixed with 1% formaldehyde for 10 min at room temperature and the reaction was stopped by 500 μl of 1.25 M glycine. The cells were then scraped and lysed with SDS lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8.1) and protease inhibitor cocktail], the samples were sonicated using the Bioruper system (Cosmo Bio, Tokyo, Japan) for 15 min (30 s ON/30 s OFF mode) in an ice-water bath and centrifuged at 12,000 r.p.m. for 5 min at 4 °C. Each sample was mixed with ChIP dilution buffer(0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris‒HCl, pH8.1, 167 mM NaCl and protease inhibitor cocktail) in the presence of anti-PPARγ antibody (Santa Cruz Biotechnology). After washing and reverse cross-linking, eluted DNA was subjected to PCR reaction using the following primers for the two different regions of PPREs and one negative control: A region (89 bp): 5’- GAGACAGGGTTTCTCTGTATAGCCC-3’ and 5’-CAGAGGCAGGTGGATTTCTGAGTT-3’; B region (105 bp): 5’e TACACTGAAGTAGCACAGTGGGCA-3’ and 5’-CCACTCGGCTGTATTTCCCATGGAT-3’; negative control (99 bp): 5’-ATGGCAGAACACCTGCAGTTCCAA-3’ and 5’- AGGGTCTCTCAATGAGGTCTAGGA-3’. PCR was performed using KOD Hot start DNA polymerase (EMD Milipore) under the following reaction conditions: an initial activation step at 95 °C for 2 min, 32 cycles of 95 °C for 20 s, 60 °C for 20 s and 72 °C for 20 s, and a final extension at 72 °C for 5 min. PCR amplicons were visualized by running the gel electrophoresis.
2.6. Western blotting
MLE-15 cell lysates were subjected to Western blotting as previously described [13]. Briefly, 30 μg of whole cell extracts were run on the SDS PAGE gels and transferred to nitrocellulose membranes. The membranes were blocked with 5% fat-free dry milk in phosphate-buffered saline containing 0.1% Tween-20 (PBST) for 1 h, and incubated with primary antibodies and horseradish peroxidase-conjugated secondary antibodies using standard western blotting procedures. Proteins were visualized using the Femto signal chemiluminescent substrate (Thermo Fisher Scientific) under an image analyzer (Bio-Rad).
2.7. Southern blot analysis
Southern blotting was performed as described previously [9].
2.8. RNA analysis
Total RNA was extracted from MLE-15 cells or lung of PpargΔLuEC and Ppargfl/fl mice using TRIzol reagent (Invitrogen). Quantitative real-time PCR (qPCR) was carried out using cDNA generated from 1 μg total RNA with SuperScript III Reverse Transcriptase Kit (Invitrogen). qPCR was performed using an Applied Biosystems Prism 7900HT Sequence Detection System (Foster City, CA). All reactions were performed in a 10 μl volume consisting of 25 ng of cDNA, 300 nM of each primer and 5 μl of SYBR Green PCR Master Mix (Applied Biosystems) at 95 °C for 10 min and 40 cycles of 95 °C for 3 s and 60 °C for 30 s. Expression levels of mRNA were normalized to Actb mRNA. Primers for the qPCR were designed using qPrimer Depot (http://mouseprimerdepot.nci.nih.gov).
2.9. Lung histology
The lungs of PpargΔLuEC and Ppargfl/fl fetal mice were fixed in 10% neutral phosphate-buffered formalin (Fisher, Pittsburg, PA) overnight. The tissues were processed through conventional paraffin embedding. Four-mm sections were cut, deparaffinized with xylene, and hydrated in an ethanol gradient. The sections were stained with hematoxylin-eosin. Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay was carried out with a DeadEnd™ Fluorometric TUNEL System (Promega) according to the manufacturer’s instructions.
2.10. Data analysis
All data are expressed as the mean ± S.D. Statistical significance was determined by the Student’s t-test for unpaired samples.
3. Results and discussion
3.1. PPARγ deficiency causes cystic adenomatoid malformations in mouse fetal lung
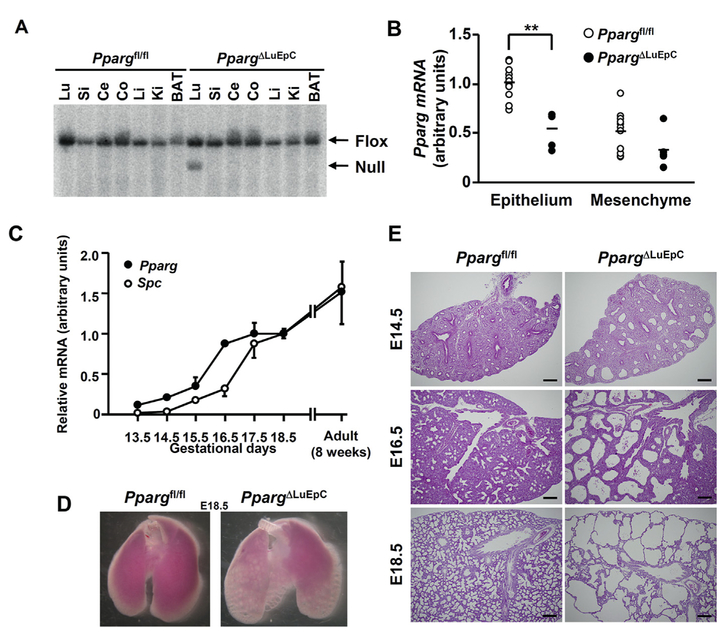
Since PPARγ expression in mouse fetal lung has hitherto not been reported, developmental expression of Pparg in the fetal lung was examined. A lung-specific PPARγ knockout mice was developed using the Cre-lox system (Fig. 1A) and the level of Pparg mRNA was measured in both fetal lung epithelial cells and mesenchymal cells (Fig. 1B). Only epithelial cells expressed the Pparg mRNA. Pparg mRNA increased in a time-dependent manner and reached a plateau at E16.5. Expression of Spc mRNA, which is a marker of alveolar type II epithelial cell and its progenitor cells, progressively increased and reached a plateau at E17.5 (Fig. 1C). As the expression of Pparg and Spc appeared in different developmental stages of the fetus, the phenotypic change at E18.5 in PpargΔLuEC fetal lung was monitored. Enlarged alveolar sacs were noted grossly in the PpargΔLuEC fetal lung, but not in the Ppargfl/fl fetal lung (Fig. 1D). This abnormal feature of the fetal lung was confirmed by H&E staining (Fig. 1E). Based on these results, epithelial PPARγ may play an important role in fetal lung development.
Fig. 1.
Ablation of Pparg gene resulted in abnormal development of mouse fetal lung. (A) Southern blotting of the recombination of Pparg allele in lung (Lu), small intestine (Si), caecum (Ce), colon (Co), liver (Li), kidney (Ki) and brown adipose tissue (BAT) genomic DNA isolated from Ppargfl/fl and PpargΔLuEC mice. (B) qPCR analysis of Pparg mRNA in epithelial and mesenchymal cells isolated from Ppargfl/fl (open circles, n = 11) and PpargΔLuEC (closed circles, n = 5) lungs at E18.5. Data points and bars represent individual values and the means, respectively. (C) qPCR quantitation of Pparg (closed circles) and Spc (open circles) mRNAs using whole lungs from normal mice at gestational days of 13.5e18.5 and postnatal weeks of 8. Individual values represent as mean ± S.D. of three lungs. (D) Fetal lung morphology is shown at gestational days of 18.5 in Ppargfl/fl and PpargΔLuEC mice. (E) Fetal lung histology at different gestational days is shown in Ppargfl/fl and PpargΔLuEC mice. Scale bars: 200 μm **, p < 0.001.
3.2. Apoptotic cell death in the alveolar sac is increased in PpargΔLuEC fetal lung
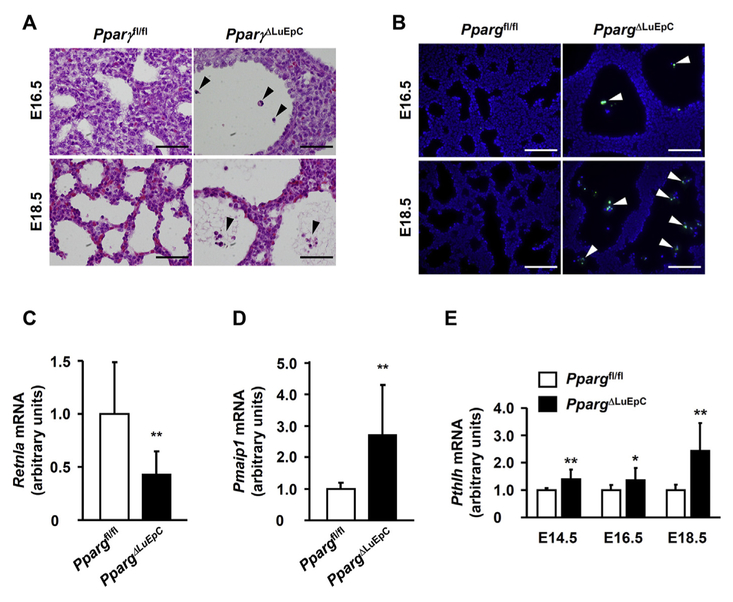
Based on the data above, the abnormal fetal alveolar formation appears to be derived from the lack of PPARγ in lung epithelial cells. However, mechanism of alveolar sac enlargement is not clear. Some cells at E18.5 were found in the alveolar sac of PpargΔLuEC fetal lung (Fig. 2A). To investigate the phenomena, apoptotic cell death was examined in fetal lung tissues by fluorometric TUNEL staining, which revealed that cells in the lumen were positive (Fig. 2B). Expression of Retnla (resistin like beta), a gene related to anti-apoptosis, was dramatically decreased (Fig. 2C), while Pmaip1 (phorbol-12-myristate-13-acetate-induced protein 1), a gene related to apoptosis, was significantly increased (Fig. 2D) at E14.5 of PpargΔLuEC fetal lung.
Fig. 2.
Ablation of lung Pparg gene shows signature of apoptotic cell death. (A) Sections of Ppargfl/fl and PpargΔLuEC lungs at E16.5 and E18.5 were subjected to H&E staining. Note the presence of cellular debris (arrowheads). Scale bars: 50 mm. (B) Immunofluorescence for TUNEL and DAPI (nuclei). Note the presence of apoptotic cells (arrowheads). Scale bars: 100 μm. Expression of anti-apoptotic Retnla mRNA (C) and apoptotic Pmaip1 mRNA (D) were measured by qPCR at E14.5. (E) Pthlh mRNA measured from Ppargfl/fl and PpargΔLuEC fetus lung at E14.5 (n = 6 and 10, respectively), E16.5 (n = 13 and 10, respectively) and E18.5 (n = 6 and 10, respectively). Individual values represent as mean ± S.D. *p < 0.05 and **p < 0.01 vs Ppargfl/fl at the corresponding days.
The abnormal phenotype observed in PpargΔLuEC fetus resembles congenital cystic adenomatoid malformation (CCAM), which is a relatively rare lesion characterized by an increase in terminal respiratory structures forming varying-sized cysts. It was previously reported that localized overexpression of Fgf10 (fibro-blast growth factor 10), Bmp4 (bone morphogenetic protein 4), and Pthlh (parathyroid hormone like hormone) in fetal lungs induces a phenotype of CCAM [14,15]. Thus, expression of these genes was investigated in whole lungs from PpargΔLuEC fetuses. Interestingly, like other genes such as Fgf10 and Bmp4 (data not shown), Pthlh was significantly increased during E14.5 to E18.5 in PpargΔLuEC fetal lung (Fig. 2E). Based on the fetal lung histology and its gene analysis, CCAM may be characterized by apoptotic cell death in alveolar epithelial cells in the absence of PPARγ.
3.3. PPARγ binds to Pthlh promoter regions as repressor in MLE-15 cells
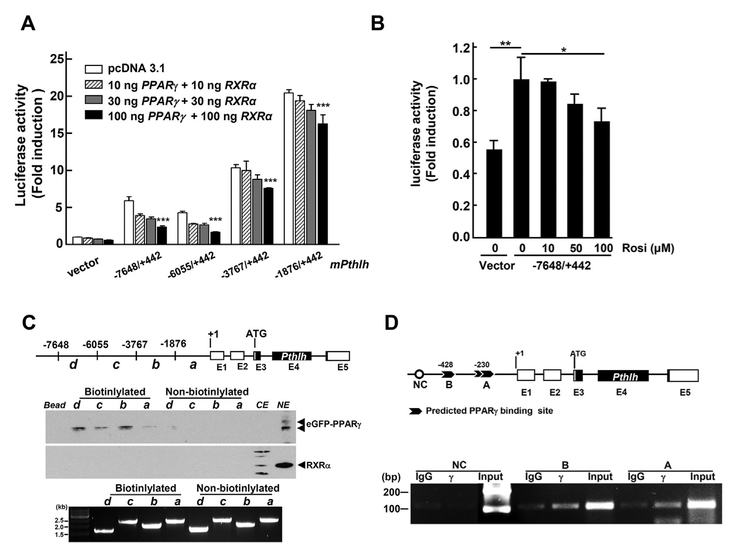
To investigate whether PPARγ could bind to Pthlh promoter, the possible PPARγ binding site was predicted using Genomatix software. MLE-15 cells, immortalized mouse lung epithelial cells [12], were transfected with various Pthlh-luciferase constructs having different promoter sequences between −7648 and + 442 bp relative to the transcription start site in the presence of both PPARγ and RXRα expression plasmids or treated with rosiglitazone after transfection with Pthlh-luciferase constructs (Fig. 3). Pthlh-lucif-erase activity was dramatically decreased in all sizes of Pthlh-luciferase constructs by PPARγ/RXRα in dose dependent manner. In addition, the longer size of 5’ flanking region of Pthlh-promoter-luciferase resulted in a significant decrease of the luciferase activity (Fig. 3A). Rosiglitazone, a PPARγ agonist, showed an inhibitory effect on Pthlh-luciferase activity (Fig. 3B). Furthermore, the binding study showed that PPARγ could bind to the Pthlh promoter region from the 5’ flanking region (Fig. 3C) and possible PPARγ binding sites were found in the Pthlh promoter by ChIP assay (Fig. 3D).
Fig. 3.
PPARγ binds to Pthlh promoter regions as repressor. (A) Pthlh-luciferase activity analyzed after transfection of different sizes of pGL3-Pthlh-promoter constructs with plasmid expressing PPARγ or RXRα proteins in the MLE-15 cells. (B) Pthlh-luciferase activity analyzed after transfection with pGL3-Pthlh-promoter constructs (−7648/+442) with different concentrations of rosiglitazone, a PPARg agonist in the MLE-15 cells. (C) MLE-15 cells were transfected with plasmids expressing eGFP-PPARγ or RXRα protein and then, nuclear proteins were incubated with DNA fragments from the Pthlh promoter and subjected to Western blotting. The bottom gel shows PCR amplicons for the study. a, b, c, d: DNA fragment covering different areas used in pull down assays. CE: cytosolic extract, NE: nuclear extract. (D) PPRE sites were monitored by ChIP assay. Predicted PPRE sites were analyzed using Genomatix software (www.genomatix.de). γ, PPARγ binding site; NC, negative control. ***, p < 0.0001.
Previously, it was suggested that parathyroid hormone-related protein (PTHrP/PTHLH), known as a paraneoplastic protein, may play an important role in the non-small lung carcinomas (NSCLC) and its receptor is frequently present in lung cancers. Also the protein has been implicated in cell growth, apoptosis and invasion [16–19]. Thus, increased Pthlh by ablation of Pparg may change the fetal lung architecture.
3.4. PPARγ may contribute cell-cell adhesion in MLE-15 cells
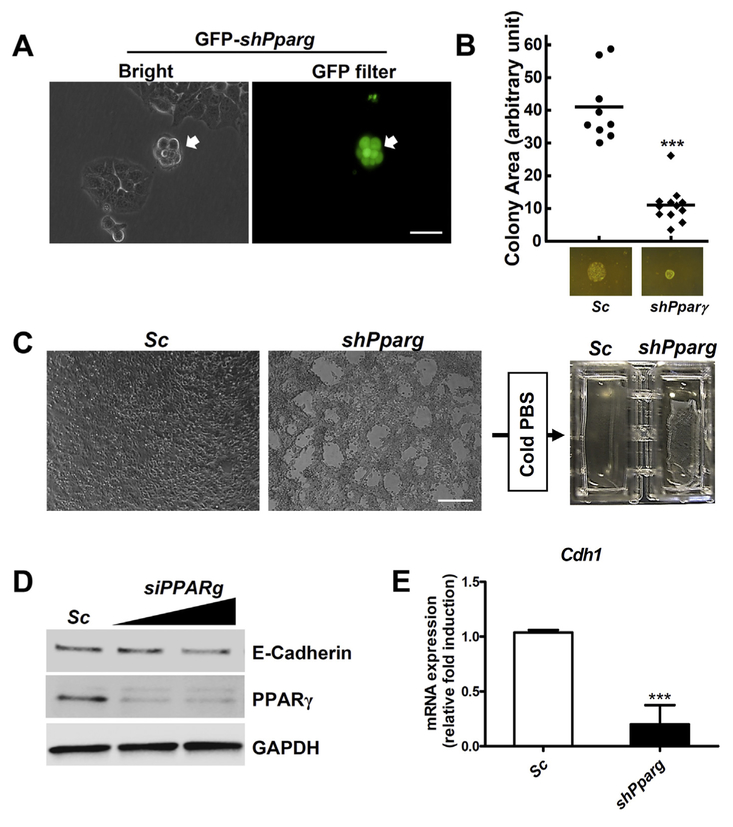
Due to the limitation of molecular studies using fetal lung epithelial cells, immortalized mouse lung epithelial cells (MLE-15) was employed. When MLE-15 cells were transfected with a Pparg silencing construct MLE-15-shPparg, Pparg silencing in MLE-15 cells resulted in a a pattern of colonization growth (Fig. 4A). Colony size from single cell culture was significantly reduced in the MLE-15-shPparg stable cells (Fig. 4B). Interestingly, monolayer growth pattern of MLE-15-shPparg cells was similar to the abnormal phenotype of PpargΔLuEC fetal lung. Furthermore, when the MLE-15-shPparg cells were rinsed with cold PBS, the cell layer was easily lifted compared to the parent cells (Fig. 4C). These results revealed that silencing of Pparg can inhibit cell-cell or cell-matrix interactions and cause abnormal fetal lung growth similar to CCAM. To support this conclusion, E-cadherin protein levels were measured after transfection with the siPparg. E-cadherin and its mRNA were significantly reduced by siPparg or shPparg in MLE-15 cells, respectively (Fig. 4D–E). Because cell adhesion molecules may play a critical role not only in maintaining tissue architecture or morphogenesis [20] but also in cancer cell invasion [21], if the adhesion system in the fetal epithelium is weakened, it can lead to abnormal fetal lung development. Previously, it was demonstrated that E-cadherin can be regulated by PPARγ and inhibit growth and invasion of prostate cancer or MCF-7 cells [22,23]. Based on these results, Pparg silencing may lose the epithelial architecture of fetal lung, resulting in CCAM.
Fig. 4.
Cellular adhesion of MLE-15 cells was reduced by knock-down of Pparg gene. (A) Colony shape of MLE15 cells is shown after transfection with shPparg plasmid for 4 days. Transfected cells were visualized by turbo GFP (tGFP) protein. Scale bars: 100 μm. (B) Colony areas from stable MLE-15-shPparg and parental cells. (C) MLE-15-shPparg and parental cells were grown to 100% confluence and challenged with cold PBS. Lifted cells are shown in MLE-15-shPparg cells. Scale bars: 300 μm. (D) MLE-15 cells were transfected with siPparg or scramble RNA and total protein extracts were subjected to Western blotting. (E) Expression of Cdh1 in MLE-15-shPparg cells was measured by qRT-pCR. Gapdh mRNA was used as internal control. ***, p < 0.0001.
Acknowledgments
We thank Yatrik M. Shah for technical advice. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (NRF-2015R1A5A2008833), Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2015R1C1A1A02036595), and National Cancer Institute Intramural Research Program.
Abbreviations:
- PPARγ
peroxisome proliferator-activated receptor gamma
- Retnla
Resistin-like alpha
- Pthlh
parathyroid hormone-like hormone
- Cdh1
Cadherin-1
Footnotes
Conflict of interest
The authors have no financial conflicts of interest.
References
- [1].Barish GD, Narkar VA, Evans RM, PPAR delta: a dagger in the heart of the metabolic syndrome, J. Clin. Invest 116 (2006) 590e597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Evans RM, Barish GD, Wang YX, PPARs and the complex journey to obesity, Nat. Med 10 (2004) 355e361. [DOI] [PubMed] [Google Scholar]
- [3].Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, Vidal-Puig A, Flier J, Briggs MR, Staels B, Vidal H, Auwerx J, The organization, promoter analysis, and expression of the human PPARgamma gene, J. Biol. Chem 272 (1997) 18779e18789. [DOI] [PubMed] [Google Scholar]
- [4].Wahli W, Braissant O, Desvergne B, Peroxisome proliferator activated receptors: transcriptional regulators of adipogenesis, lipid metabolism and more, Chem. Biol 2 (1995) 261e266. [DOI] [PubMed] [Google Scholar]
- [5].Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM, 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma, Cell 83 (1995) 803e812. [DOI] [PubMed] [Google Scholar]
- [6].Li Y, Qi Y, Huang TH, Yamahara J, Roufogalis BD, Pomegranate flower: a unique traditional antidiabetic medicine with dual PPAR-alpha/-gamma activator properties, Diabetes Obes. Metab 10 (2008) 10e17. [DOI] [PubMed] [Google Scholar]
- [7].Simon DM, Tsai LW, Ingenito EP, Starcher BC, Mariani TJ, PPARgamma deficiency results in reduced lung elastic recoil and abnormalities in airspace distribution, Respir. Res 11 (2010) 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang Y, Santos J, Sakurai R, Shin E, Cerny L, Torday JS, Rehan VK, Peroxisome proliferator-activated receptor gamma agonists enhance lung maturation in a neonatal rat model, Pediatr. Res 65 (2009) 150e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Akiyama TE, Sakai S, Lambert G, Nicol CJ, Matsusue K, Pimprale S, Lee YH, Ricote M, Glass CK, Brewer HB Jr., Gonzalez FJ, Conditional disruption of the peroxisome proliferator-activated receptor gamma gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux, Mol. Cell Biol 22 (2002) 2607e2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Okubo T, Knoepfler PS, Eisenman RN, Hogan BL, Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation, Development 132 (2005) 1363e1374. [DOI] [PubMed] [Google Scholar]
- [11].Lebeche D, Malpel S, Cardoso WV, Fibroblast growth factor interactions in the developing lung, Mech. Dev 86 (1999) 125e136. [DOI] [PubMed] [Google Scholar]
- [12].Wikenheiser KA, Vorbroker DK, Rice WR, Clark JC, Bachurski CJ, Oie HK, Whitsett JA, Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice, Proc. Natl. Acad. Sci. U. S. A 90 (1993) 11029e11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim JH, Yu S, Chen JD, Kong AN, The nuclear cofactor RAC3/AIB1/SRC-3 enhances Nrf2 signaling by interacting with transactivation domains, Oncogene 32 (2013) 514e527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gonzaga S, Henriques-Coelho T, Davey M, Zoltick PW, Leite-Moreira AF, Correia-Pinto J, Flake AW, Cystic adenomatoid malformations are induced by localized FGF10 overexpression in fetal rat lung, Am. J. Respir. Cell Mol. Biol 39 (2008) 346e355. [DOI] [PubMed] [Google Scholar]
- [15].Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL, Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis, Development 122 (1996) 1693e1702. [DOI] [PubMed] [Google Scholar]
- [16].Hastings RH, Burton DW, Nefzi A, Montgrain PR, Quintana R, Deftos LJ, Combinatorial library discovery of small molecule inhibitors of lung cancer proliferation and parathyroid hormone-related protein expression, Cancer Biol. Ther 10 (2010) 1067e1075. [DOI] [PubMed] [Google Scholar]
- [17].Hastings RH, Montgrain PR, Quintana R, Rascon Y, Deftos LJ, Healy E, Cell cycle actions of parathyroid hormone-related protein in non-small cell lung carcinoma, Am. J. Physiol. Lung Cell Mol. Physiol 297 (2009) L578eL585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hastings RH, Araiza F, Burton DW, Deftos LJ, Role of parathyroid hormone-related protein in lung cancer cell survival, Chest 125 (2004) 150S. [DOI] [PubMed] [Google Scholar]
- [19].Hastings RH, Araiza F, Burton DW, Zhang L, Bedley M, Deftos LJ, Parathyroid hormone-related protein ameliorates death receptor-mediated apoptosis in lung cancer cells, Am. J. Physiol. Cell Physiol 285 (2003) C1429eC1436. [DOI] [PubMed] [Google Scholar]
- [20].Gumbiner BM, Cell adhesion: the molecular basis of tissue architecture and morphogenesis, Cell 84 (1996) 345e357. [DOI] [PubMed] [Google Scholar]
- [21].Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Lochner D, Birchmeier W, E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells, J. Cell Biol 113 (1991) 173e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bocca C, Bozzo F, Francica S, Colombatto S, Miglietta A, Involvement of PPAR gamma and E-cadherin/beta-catenin pathway in the antiproliferative effect of conjugated linoleic acid in MCF-7 cells, Int. J. Cancer 121 (2007) 248e256. [DOI] [PubMed] [Google Scholar]
- [23].Annicotte JS, Iankova I, Miard S, Fritz V, Sarruf D, Abella A, Berthe ML, Noel D, Pillon A, Iborra F, Dubus P, Maudelonde T, Culine S, Fajas L, Peroxisome proliferator-activated receptor gamma regulates E-cadherin expression and inhibits growth and invasion of prostate cancer, Mol. Cell Biol 26 (2006) 7561e7574. [DOI] [PMC free article] [PubMed] [Google Scholar]