Abstract
The last three decades have witnessed an explosion in mechanistic details on how model bacterial organisms such as Escherichia coli, Bacillus subtilis, and Caulobacter crescentus undergo binary fission. These advances were possible by not only advances in microscopy that allowed cell biological questions to be answered, but also by the clever use of genetic manipulations in these systems in which specific hypothesis could be directly and easily tested. More recently, research using traditionally understudied organisms, or “non-model” systems, has revealed several alternate mechanistic strategies that bacteria use to divide and propagate. In this review, we will highlight these new findings and compare these strategies to cell division mechanisms elucidated in well-established model organisms.
Keywords: Min system, FtsZ, DivIVA, nucleoid occlusion, Noc
INTRODUCTION
The field of bacterial cell division has relied heavily on model organisms such as the Gram-negative Escherichia coli and Gram-positive Bacillus subtilis, largely because of the abundance of available genetic tools in these organisms. Since E. coli and B. subtilis are both rod-shaped cells that divide symmetrically along the short axis of the cell, it has been relatively straightforward to compare and contrast the mechanisms involved in regulating cell division in these two species, leading to tremendous progress over the last three decades in understanding the fundamentals of bacterial cell division. To gain a deeper appreciation of how bacterial cells divide, several labs have begun to examine differently shaped organisms that may undergo more complex cell cycles and occupy a variety of ecological niches, where many of the lessons learned from studying model organisms appear not to apply. Certainly, our understanding of molecular details in these systems is still in its infancy compared to what is known in model systems, but a wide array of interesting cell division mechanisms is already being reported (Fig. 1). Therefore, this review will highlight new research in traditionally understudied systems and compare these systems to cell division mechanisms elucidated in well-studied model organisms.
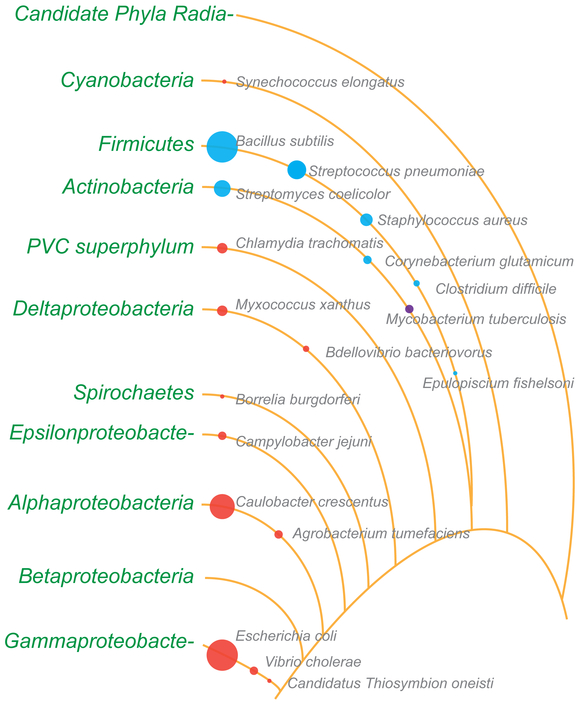
Figure 1.
Representation of the relative number of reports describing cell division in various bacterial species. The diameters of the circles roughly indicate the number of cell division publications available for organisms highlighted in this review. Note: The diameter of the circles for E. coli and B. subtilis are capped at an arbitrary number so that other circles are visible. Red circles, Gram-negative; blue circles, Gram-positive; violet, M. tuberculosis. Lines depict phyla and lineages loosely based on the bacterial branch of the tree of life (PMID: 27572647). Several phyla and lineages were omitted for clarity. PVC superphylum comprises of Planctomycetes, Verrucomicrobia, and Chlamydiae. The branch length, spaces between them, and the order in which some organisms are listed are not based on phylogeny.
The bacterial tubulin homolog, FtsZ, is almost universally conserved in different bacterial species. FtsZ assembles as a ring (termed the “Z-ring”) and marks the site for division by subsequently recruiting components of the divisome to initiate cytokinesis (58). A central question has been to understand how the correct placement of the Z-ring initially occurs. In E. coli two negative regulatory systems influence Z-ring assembly and localization: nucleoid occlusion (NO), mediated by the SlmA protein which prevents cell division atop the nucleoid, and the Min system, composed of three proteins in E. coli, which prevents cell division near the polar regions of the cell (131) (Fig. 2A). B. subtilis also harbors a NO system, mediated by the Noc protein which is not homologous to the E. coli SlmA protein and also functions in a different fashion (131). In E. coli, the Min system oscillates from one cell pole to another thereby creating a low time-averaged concentration at mid-cell, permitting Z-ring assembly to take place only near mid-cell (89) (Fig. 2A). Although B. subtilis harbors components of the Min system, it functions more to mediate the fidelity of cell division via the cell division protein DivIVA, rather than the actual placement of the Z-ring (45, 56, 136) (Fig. 2B). Curiously, both well-studied systems are somewhat dispensable for correct Z-ring placement, suggesting the presence of other, heretofore undiscovered, division factors that is the major focus of current research (7, 116). The notion that negative regulation can determine Z-ring positioning was also observed in another model organism, C. crescentus, that lacks both Min and NO systems, but instead employs a protein termed MipZ to prevent Z-ring assembly near the cell poles (discussed in detail below; Fig. 2C) (53, 129).
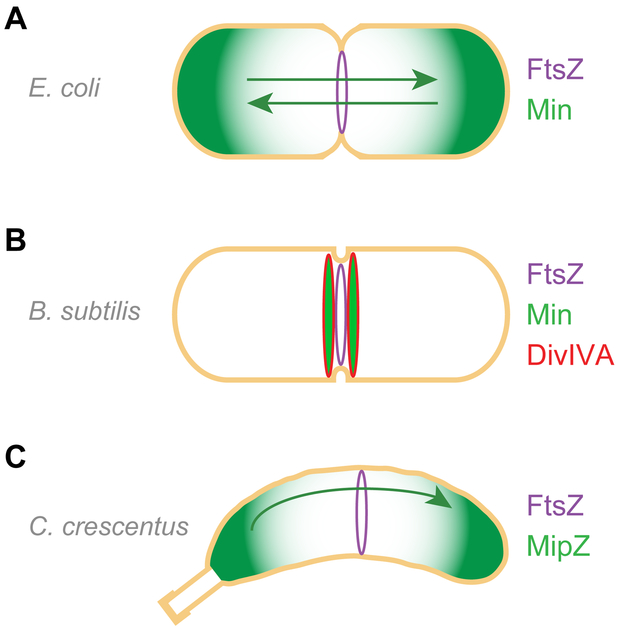
Figure 2.
Regulation of cell division in model organisms is achieved predominantly by negative regulators. (A) In E. coli, proteins that comprise the Min system (green), which prevent FtsZ (purple) ring assembly, oscillate between the poles and inhibit cell division close to cell poles. (B) In B. subtilis, the Min system (green) is recruited to sites adjacent to newly forming septa by DivIVA (red); it does not mediate division site selection, but maintains cell division fidelity by preventing aberrant septation from occurring at mid-cell adjacent to a newly formed septum. (C) In C. crescentus, MipZ (green)interacts with chromosome-bound ParB and co-migrates to the stalk-less pole and displaces polar-localized FtsZ (not shown), to permit FtsZ ring assembly at mid-cell. The negative regulators of FtsZ are shown in green. Nucleoid occlusion system is not depicted for clarity.
The model systems set up a central notion that placement of the division septum is largely the result of negative regulation, but recent results have also indicated that Z-ring placement in multiple species may be positively influenced, thereby setting up an entirely new paradigm for bacterial cell division. We will also highlight studies in several systems that are less well established but point to other novel mechanisms for cell division regulation, including those found in pathogens and symbionts. Finally, we will also review cell division behaviors of bacteria that break fundamental “rules” learned from model systems more dramatically: dividing along alternate axes of the cell and not utilizing the nearly universally conserved FtsZ protein at all.
Cell division in polar flagellates
Cell division in the monotrichous (one polar flagellum per cell) dimorphic prosthecate alphaproteobacterium C. crescentus, has been extensively studied. This bacterium lacks both MinCD and NO systems to regulate the placement of the FtsZ ring. Instead it employs a ParA-like ATPase MipZ (Midcell positioning of FtsZ) to regulate the assembly site of the Z-ring (34, 111, 129). MipZ forms a gradient by interacting directly with origin-proximal DNA-bound ParB-parS complexes at the flagellated (stalked) pole prior to cell division and translocating with the newly replicated origin to the non-flagellated pole (Fig. 2C). At both poles, the presence of the MipZ gradient displaces polar-localized FtsZ through direct interaction, thereby creating an FtsZ polymerization-permissive zone near mid-cell where FtsZ is allowed to assemble into a Z-ring and form the division septum (72, 129). The formation of minicells has been observed in this bacterium dating back to 1978 (107) and, not surprisingly, cells in which MipZ is depleted produce minicells, due to the mis-regulated assembly of FtsZ at non-permissive subcellular regions (129). Similarly, the multi-functional polar-localized protein PopZ (Pole-Organizing Protein that affects FtsZ) undergoes transition from being unipolar to bipolar and captures the ParB-parS complex at the non-flagellated pole. Cells lacking popZ were unable to produce stalks, formed minicells and appeared elongated due to erroneous cell division (14, 38). These phenotypes were due to a malfunction of chromosome segregation and subsequent incorrect MipZ localization, linking stalk formation with cell division. TipN (Tip of New pole) is another protein involved in marking the new pole (the site of flagellar assembly) after cell division. Interestingly, overproduction of TipN resulted in the formation of both minicells and elongated cells(64, 79, 81). Absence of TipN together with TipF, a protein essential for flagellar assembly, results in cell elongation and filamentation (64). In this manner, a mechanism that coordinates cell division with flagellar assembly in this fresh water organism may provide a dispersal mechanism for progeny cells.
Campylobacter species exploit the formation of amphitrichous flagella (one flagellum per pole on both poles) to regulate FtsZ placement. These organisms require the correct number of flagella on each pole to be present to exhibit a behavior termed darting motility and for successful host colonization (119, 126). Campylobacter species lack a MinCD system and instead utilize a MinD/ParA-like ATPase protein FlhG (FleN), a known regulator of flagellar number (Fig. 3A). In Campylobacter jejuni, cells lacking flhG often produce more than one flagellum per pole, but intriguingly also forms minicells, suggesting a role in cell division for FlhG (8). Consistent with a MinD-like role for FlhG in cell division regulation, cells of an FlhG mutant lacking ATPase activity exhibit cell length elongation – a phenotype that was suppressed by increasing the levels of FtsZ (8, 70). By making use of the amphitrichous flagellation as a mechanism to regulate cell division, Campylobacter species ensure that cell division occurs away from the pole to increase the chances of daughter cell survival by confirming the formation of a flagellum and preventing minicell formation. Interestingly, flhG of Helicobacter pylori, which is a lophotrichous organism containing several flagella at one pole is able to complement the defect of a flhG null mutant of C. jejuni indicating that the usage of flagellar assembly to regulate cell division may not be unique to Campylobacter species (8).
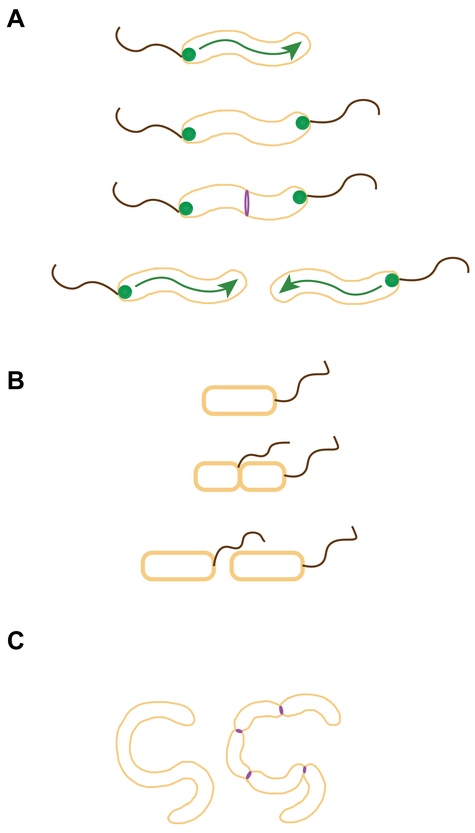
Figure 3.
Cell division regulation in polar flagellates. (A) In C. jejuni, FlhG (green circles), a MinD/ParA-like ATPase that regulates flagellar copy number, also participates in negatively regulates FtsZ (purple) assembly near poles. (B) In a magnetotactic Gammaproteobacterium, flagella arise from the site of cell division (suggesting positive cell division regulation), perhaps to ensure that the flagella of daughter cells are oriented in the same direction as the parental cells along earth’s the magnetic dipole. (C) Cells of B. bacteriovorus grow as filaments inside the periplasm of Gram-negative bacteria. Just prior to host cell lysis, the filamentous cell undergoes synchronous septation (reminiscent of sporulating S. coelicolor cells) to liberate motile daughter cells. Possible sites of FtsZ ring assembly (purple) are shown; flagella arising from the site of septation are not depicted.
In contrast, in a monotrichous magnetotactic bacterium of the Gammaproteobacteria class, the site of the cell division septum appears to dictate the site for the construction of the new flagellum: specifically, at the side of the septum facing the daughter cell that has no flagellum (83) (Fig. 3B). It is hypothesized that this mechanism permits offspring to align their cells along the same polarity of magnetic dipole as the parental cell and position the flagellated pole accordingly (83).
Together, these set of data indicate that regulation of flagella or stalk formation may be tightly intertwined with cell division. Further research in these organisms will shed light on the precise molecular mechanisms by which cell division is regulated via flagellar assembly.
The monotrichous Gram-negative bacterium, Bdellovibrio bacteriovorus, belongs to the class of deltaproteobacteria that parasitize other Gram-negative bacteria. In this organism, the non-flagellated pole is utilized to recognize and penetrate the outer membrane of the prey (121). Once inside the periplasm of prey such as E. coli, B. bacteriovorus undergoes growth in the form of filamentation, by undergoing unipolar growth (40), and feeds off of the nutrients provided by the prey. Just prior to inducing host lysis, B. bacteriovorus produces multiple motile daughter cells (18, 46) (Fig. 3C). The manner in which this bacterium spatially and temporally regulates this remarkable cell division event and the identity of cell division factors required for this remains to be elucidated. Although this organism encodes FtsZ that is speculated to be involved in cell division (33, 114), the Min system is completely absent (114). Unlike most bacteria, B. bacteriovorus encodes two copies of MreB (111), a protein that regulates peptidoglycan synthesis, and at least one MreB homolog appears to play a role in regulating cell division and nucleoid organization (20, 47). In this organism, concurrent septation has been reported (Fig. 3C), which appears similar to the event that occurs during Streptomyces sporulation (see below), and it was also observed that flagellation occurs before daughter cell separation (18), similar to the magnetotactic bacterium describe above. At this time, there is no knowledge of any positive regulatory mechanism similar to the one present in Streptomyces (discussed under positive regulation) or if flagellation has a role in regulating cell division in B. bacteriovorus. Being a small bacterium (1 µm in length and 0.25 µm in width, which is about 1/3rd size of E. coli or B. subtilis), light and fluorescence microscopy studies have been limited (46), but perhaps the increased use of super-resolution microscopy techniques will help trigger a renewed interest in understanding the cell division process in B. bacteriovorus.
Cell division in bacteria that exhibit polar growth
Unlike E. coli and B. subtilis which grow by incorporating peptidoglycan near mid-cell, the rod-shaped, Gram-negative, alphaproteobacterium, Agrobacterium tumefaciens undergoes growth by adding peptidoglycan specifically at the new poles (22, 63) (Fig. 4A). The non-growing pole, usually the old pole, is reserved for the secretion of an adhesive molecule to facilitate attachment to various surfaces. This organism possesses three copies of ftsZ. One copy encodes FtsZ that is equivalent to that of E. coli, where the gene is present within an operon coding for ftsQ and ftsA. The other two ftsZ copies exhibit varying levels of truncation and are encoded elsewhere on the genome. A recent study has elegantly shown that after septation, which occurs near mid-cell in A. tumefaciens, FtsA and FtsZ stay at the newly formed growth pole and presumably facilitate polar growth. Subsequent to cell enlargement, FtsA and FtsZ relocate to mid-cell to form an FtsZ ring which, upon completion of septum formation, mark the new pole for polar growth (15, 55, 146) (Fig. 4A). Two proteins (PopZ and PodJ) were identified and reported to mark the growth pole and old pole, respectively (55). An interesting feature that is present in both A. tumefaciens and C. crescentus, also an alphaproteobacterium, is the pole-localized intermediate population of FtsZ (129, 146), although the significance of this in C. crescentus is not clear. How the division site is determined in A. tumefaciens and if there is a MipZ-like factor regulating FtsZ placement also remains to be determined.
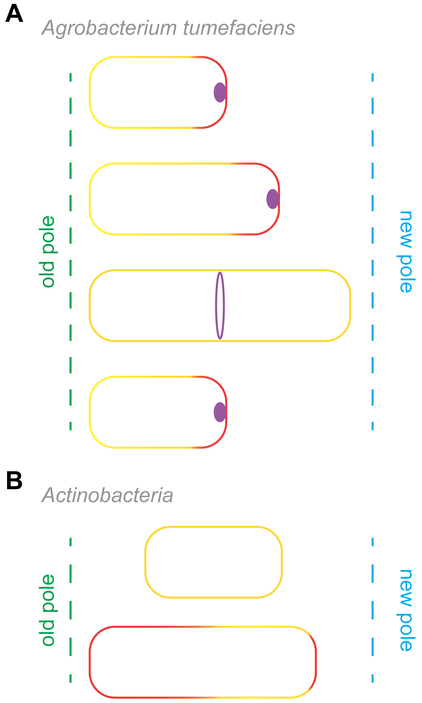
Figure 4.
Cell division in bacteria that display polar growth. (A) FtsZ (purple foci) in A. tumefaciens remain localized at the pole after cell division and likely facilitate polar elongation specifically at the new pole (red line). Subsequently FtsZ relocates to the mid-cell site (purple ring) to assemble the divisome and mediate cell division. (B) Cells of both C. glutamicum and M. tuberculosis (labeled “Actinobacteria”) undergo uneven growth at cells poles with older poles exhibiting faster growth than the new pole.
Another group of bacteria that display polar growth is predominantly in the Actinobacteria phylum. Some examples include Mycobacterium tuberculosis, Streptomyces coelicolor (see positive regulation section below), and Corynebacterium glutamicum. The bacterium C. glutamicum is reported to undergo uneven polar growth in which the old poles appear to grow faster than the newly-formed poles (120) (Fig. 4B). This rod-shaped bacterium lacks both Min and NO systems and the mid-cell division site is not precisely chosen, unlike its well-studied rod-shaped counterparts B. subtilis and E. coli. As a result, cell division often results in grossly unequal-sized daughter cells (31). C. glutamicum contains two ParA proteins, one encoded by the parAB operon and the other ParA encoded elsewhere as a standalone gene (32). The orphan ParA-like protein, referred to as PldP, appears to play a role in regulating cell division. Deletion of pldP results in a minicell phenotype while overexpression of pldP results in increased cell length, consistent with its role in regulating cell division. In further support of this idea, PldP localizes to the nascent division sites (31).
In M. tuberculosis, which also undergoes polar growth, cell division produces two unequal-sized daughter cells and it appears that the old poles elongate faster than the new pole (73) (Fig. 4B). The well-studied FtsZ anchoring proteins such as FtsA are curiously absent in this organism (25). However, M. tuberculosis harbors other FtsZ anchoring proteins such as FhaB (also referred to as FipA), FtsW, and CrgA (26, 73, 106, 125). Mycobacteria lack both a Noc-like and complete Min system (60). However, a protein homologous to MinD, termed septum site determining protein Ssd (Rv3660c), plays a critical role in determining the site of FtsZ ring assembly and appears to link cell division with environmental adaptation (41). The AAA+ family chaperone ClpX that is known to regulate and repurpose FtsZ in E. coli (21, 50, 124) and B. subtilis (57, 139), also plays a role in negatively regulating cell division in M. tuberculosis (37). It was also observed that in this organism a cell wall hydrolase, ChiZ, helps regulate FtsZ localization (134). Additionally, it was noted that a Ser/Thr Phosphatase (PstP) is important for cell division regulation (118), and that the GTPase activity of FtsZ and interaction with other divisome partners of FtsZ are negatively affected via phosphorylation by a Ser/Thr kinase PknA (125, 128). Several studies reported that the transcription factor-like WhiB homologs may be involved in the non-transcriptional regulation of cell division in Mycobacteria by either directly interacting with FtsZ or acting as a chaperone, depending on the species (11, 54, 77). Furthermore, there is evidence that ftsZ levels could be up- or down-regulated for adapting to various extracellular environments (74). Regulation of cell division in non-pathogenic Mycobacterium smegmatis may harbor a largely similar cell division regulation system as M. tuberculosis, since M. smegmatis ftsZ can complement the deletion of ftsZ in M. tuberculosis (36). Taken together, it appears that for cell division in bacteria that undergo polar growth, the use of either the Min or NO systems may be eschewed and several other components are used to couple cell division to cell growth.
Examples of positive regulation
For many years, a common mechanistic theme in bacterial cell division was that, as exemplified by the Min and NO systems, placement of the cell division machinery may be primarily mediated by negative regulation, whereby the Z-ring would assemble at subcellular locations where negative regulators were largely absent. However, several recent examples in Streptomyces coelicolor, Myxococcus xanthus, and Streptococcus pneumoniae, which all lack both Min- and NO-like systems (105, 117), suggest that positive regulatory elements in some species may pre-localize to the future site of cell division to position the cell division machinery (Fig. 5).
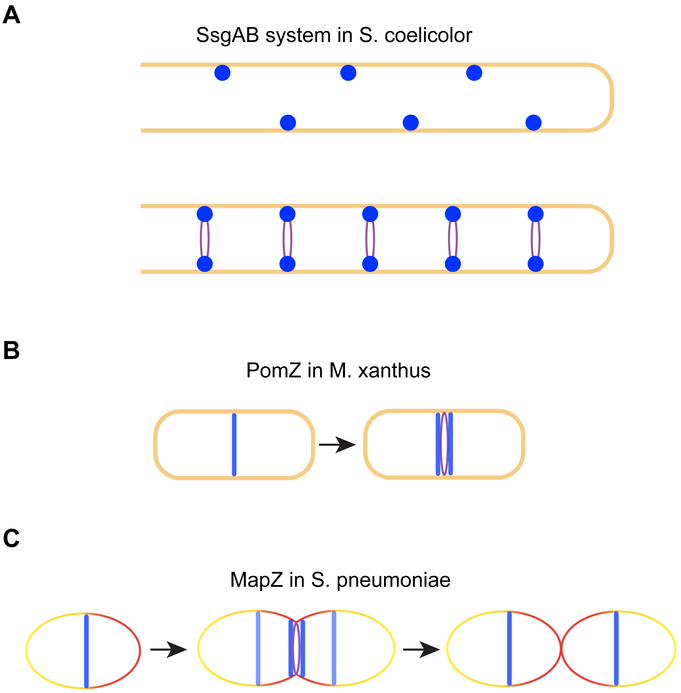
Figure 5.
Examples of positive regulation of cell division. (A) During sporulation in S. coelicolor, SsgB interacts with SsgA (complex depicted in dark blue) to mark potential division sites and recruit FtsZ (purple). (B) Cells of M. xanthus uses PomZ (blue) to indicate the site for FtsZ localization and assembly. (C) In S. pneumoniae, MapZ (LocZ; depicted in blue) localizes to the mid-cell division site facilitate the process of FtsZ ring assembly. Just prior to septum formation, a subpopulation of MapZ (light blue) binds to the nascent peptidoglycan during the lateral cell wall synthesis (shown as red line) and hitchhikes to mark the future division sites.
In S. coelicolor, ftsZ is a non-essential gene that is likely only required for sporulation (93). Although it does contain a negative FtsZ regulator, CrgA, that plays a poorly defined function (27), it harbors another system mediated by SsgAB that positively affects FtsZ assembly by actively recruiting FtsZ to the division sites (140) (Fig. 5A). SsgB, the protein that brings FtsZ to the division sites during sporulation, mislocalizes in the absence SepG, a transmembrane protein that may be involved in nucleoid compaction (144), suggesting that chromosome organization and cell division may be coordinately regulated during sporulation in S. coelicolor. Consistent with this notion, the chromosome segregation proteins ParAB in this organism is able to regulate cell tip elongation and FtsZ assembly (30, 76), and the broadly conserved multi-functional cell division protein DivIVA interacts with ParA at least indirectly via a cytoskeletal protein Scy (29, 62). Finally, it is worth noting that, although most of the cell division research in S. coelicolor has been focused on sporulation in this organism due to the dispensability of ftsZ during normal growth, new observations suggest that this naturally filamentous bacterium does indeed routinely separate its cytosol and achieve compartmentalization by formation of FtsZ-independent “cross-membranes” that represent a form of cell division whose molecular details of formation and regulation await further study (23, 143).
Another example of positive regulation is observed in the deltaproteobacterium M. xanthus. In this organism, the ParA-like protein PomZ localizes to mid-cell division sites, which immediately precedes localization of FtsZ, and does so in an FtsZ-independent manner (130) (Fig. 5B). However, cells lacking pomZ formed filamentous cells and nucleoid-less minicells, suggesting that PomZ plays a central, currently undefined, role in dictating where FtsZ localizes and polymerizes within the cell to form Z-rings.
A third case of positive regulation is reported in the ovoid bacterium S. pneumoniae, where a recently identified protein MapZ (LocZ) localizes to the site of cell division prior to FtsZ and the FtsZ-anchoring protein FtsA, and recruits downstream divisome proteins beginning with FtsZ (48, 61) (Fig. 5C). Deletion of mapZ resulted in suboptimal septum placement and production of nucleoid-less minicells. MapZ is conserved only within Streptococcaceae and Enterococcaceae families among Firmicutes (52). MapZ is capable of binding nascent peptidoglycan and due to cell elongation, which slightly precedes septation during cell division process, it has been proposed that a MapZ subpopulation splits from the mid-cell associated population to migrate and subsequently mark new cell division sites (52). MapZ is under post-translational regulation via phosphorylation by a Ser/Thr kinase StkP, which also localizes to the division site and acts as a critical switch coordinating peptidoglycan synthesis during elongation and septation (48, 49). Depending on the strain background, the ΔmapZ phenotype may be less dramatic, indicating that there could be other redundant factors that may perform a similar role to that of MapZ (12). Another report shows that, FtsA, an essential protein in S. pneumoniae (80), is required for proper localization of FtsZ and may play a role in regulating peptidoglycan synthesis during both septation as well as lateral cell elongation (98).
Cell division in bacteria that contain multiple chromosomes
A gammaproteobacterial cousin of E. coli, Vibrio cholerae, contains two chromosomes. As there are many bacterial species with multiple chromosomes that are being discovered, there is an accumulating interest in understanding how the faithful segregation of chromosomes is coordinated with cell division, and V. cholerae has emerged as a model to address these questions (39, 66, 108). Chromosome 1, the larger chromosome, communicates with chromosome 2 and signals the proper time to initiate DNA replication while undergoing its own replication (6, 109, 135). A recent report has provided evidence that mutants that lack both Min and NO systems are viable (51). However, the Min system becomes critical in maintaining cell division fidelity when the chromosome organization is compromised. Similar to C. crescentus, this bacterium also displays pole-localized FtsZ. The absence of SlmA, which mediates NO in this organism, results in the untimely accumulation of FtsZ at midcell, suggesting that chromosome segregation in V. cholerae is linked to the proper timing of cell division. Specifically, it has been proposed that the SlmA binding sites, especially on chromosome 2, may act as a cell division timing device that allows FtsZ assembly based on the status of nucleoid segregation (51).
Cell division in cocci
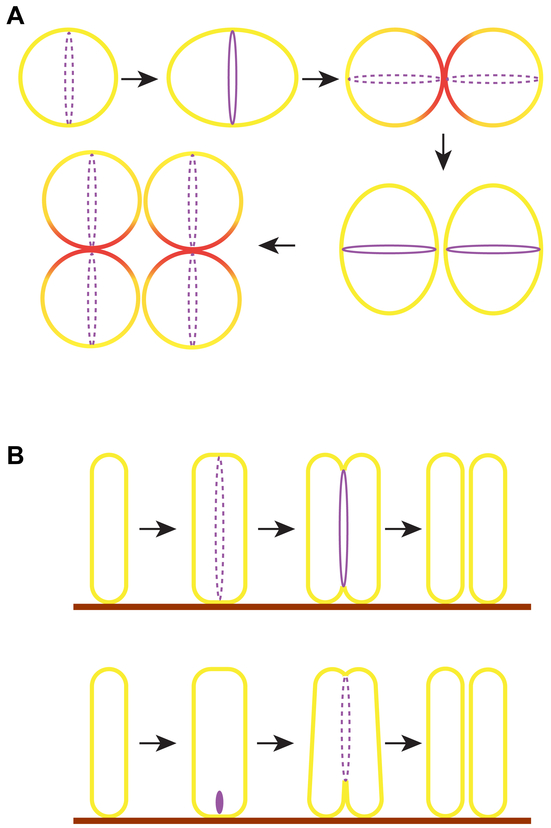
In the Gram-positive spherical bacterium, Staphylococcus aureus, new cell division planes arise orthogonal to the two previous division planes, which results in the formation of its signature grape-like clusters (78, 133) (Fig. 6A). In recent years this organism has become an increasingly-studied model for interrogating cell division in spherical bacteria, where the lack of distinctly symmetrical planes imposes an additional challenge in establishing the plane of cell division (105). S. aureus lacks a Min system and possess a NO system that may link nucleoid segregation and cell division (137). The gene coding for DivIVA, which spatially regulates the Min system in B. subtilis (45), is present in S. aureus but deletion of divIVA does not obviously affect cell division or chromosome segregation (104). The cell shape-determining protein MreB that is widely conserved in rod-shaped bacteria is absent in S. aureus. Interestingly, the shape-determining proteins MreC and MreD proteins are present in this organism and localize to division sites. However, deletion studies indicated that these proteins are not major factors in regulating septal or peripheral peptidoglycan synthesis (127). The homolog of a negative regulator of FtsZ, EzrA, which is conserved among several Gram-positive bacteria (86), directly interacts with FtsZ in S. aureus (122), and localizes to division sites and participates in regulating cell size in S. aureus by controlling peptidoglycan synthesis and cell division (68, 123). Recent reports have shown that, subsequent to cell division, daughter cell separation happens very quickly, within a span of two milliseconds, indicating a mechanism that uses physical forces rather than enzymes alone for cell separation (96, 145). A traditional view of Staphylococcal cell division was that the biosynthesis of peptidoglycan is restricted to division sites. This view also suggested a mechanism for the identification of sequential orthogonal mid-cell division plane selection that invoked the presence of peptidoglycan-based scar-like marks that appear after each round of divisions (132). In fact, DivIB, which was shown to bind peptidoglycan was speculated to be a pointer for previously used sites for division, since depletion of DivIB resulted in incorrect placement of septa (13). However, newer measurements, generated with advanced microscopy and newly developed cell wall labeling techniques, indicated that septal cell wall material accounted for less than half of the cell wall material in a newly separated cell (96, 145) (Fig. 6), which contradicted the previously held model of restricted deposition of peptidoglycan (103, 132). The newer measurements also indicated that peptidoglycan synthesis occurs along the entire circumference of S. aureus cells, mediated by at least one of the four penicillin-binding proteins (96, 145). Thus, given new observations that revealed the non-restricted insertion of peptidoglycan, a model based on a peptidoglycan-based division marker appears less likely and the question of how cell division plane selection occurs in S. aureus, remains an open question.
Figure 6.
Other modes of bacterial cell division. (A) Subsequent generations of the spherical S. aureus divide in a plane orthogonal to the two previous generations. Although cell wall synthesis was speculated to occur only during septum synthesis, new evidence suggests that circumferential cell wall synthesis also occurs in this organism. Subsequent to cell division, the septum-derived cell wall material (red) contributes to less than half of the newly formed daughter cell. The division plane of the next generation lies orthogonal to that of the preceding generation. (B) Top: The marine gammaproteobacterium Candidatus Thiosymbion oneisti attaches via one pole to the surface (brown lines) of the nematode Laxus oneistus. This bacterium grows wider along the short axis prior to FtsZ (purple) assembly, which occurs at mid-cell parallel to the long axis of the cell, presumably to permit both daughter cells to remain adhered to the host. Bottom: In another gammaproteobacterial relative of this organism (yet to be named), that lives on the surface of a different nematode Robbea hypermnestra, FtsZ localizes closer to the host-proximal pole and trigger septum formation. Subsequently, several fragmented FtsZ foci continue to mediate septum synthesis towards the distal pole. The factors involved in the regulation of longitudinal division remains to be elucidated.
Cell division in bacteria that lack FtsZ
Since FtsZ is apparently the central bacterial cell division factor, the discovery of a growing number of organisms that do not encode ftsZ posed the question of how cell division occurs in these FtsZ-less bacteria (10, 43, 67, 115). One such FtsZ-less organism is an obligate intracellular pathogen, Chlamydia trachomatis. Adding to this anomaly of the absence of FtsZ was the longstanding conundrum that, although these organisms possess the genes encoding peptidoglycan biosynthesis machinery, peptidoglycan production could not be measured directly. Nonetheless Chlamydial cells were sensitive to antibiotics that target peptidoglycan synthesis, indicating that cell wall material was likely present (97). With the aid of fluorescent D-amino acid labeling techniques that measure the nascent peptidoglycan incorporation, it was recently shown that peptidoglycan is indeed present in C. trachomatis and other related organisms (88, 100, 102). However, in C. trachomatis, what likely precluded earlier attempts at detecting peptidoglycan was the observation that peptidoglycan synthesis primarily occurred transiently during cell division and predominantly at the division site. This led to the model that peptidoglycan synthesis may drive cytokinesis in this organism (88). Indeed, in C. trachomatis and a related organism, MreB and RodZ (9), which are involved in spatially regulating peptidoglycan incorporation, localize to cell division sites (65, 71, 87). Interestingly, recent observations have suggested that, even in E. coli, FtsZ may not provide the force for membrane invagination and constriction during cell division and that peptidoglycan assembly may instead be the rate-limiting step that deforms the membrane (142), suggesting that cell wall-driven cytokinesis may be a widely conserved mechanism driving bacterial cell division. Recent work has shown that C. trachomatis can also exhibit asymmetric budding-like cell division, which is documented in another FtsZ-less evolutionary relative Gemmata obscuriglobus, a member of the Planctomycetes phylum (1, 82, 87) and in L-forms of B. subtilis that do not produce FtsZ or peptidoglycan (44). Interestingly the C. trachomatis FtsQ ortholog, similar to the E.coli divisome protein FtsQ (99), localizes to the site where budding occurs and stays at the asymmetric division septum (1), but it is not yet clear what regulates the choice between symmetric or budding division in this organism (87).
Bacteria that undergo extra-ordinary modes of cell division
Beyond molecular anomalies, such as the absence of certain conserved factors (Min and NO systems, or FtsZ, for example) that are increasingly apparent in the course of studying cell division in diverse bacterial species, more fundamental physical differences have emerged in certain organisms that violate long-held central premises of bacterial cell division. Chief among these exceptional situations include violating the notion that bacteria must produce exactly two daughter cells by binary fission, and that cell division must occur along the short axis of a rod-shaped cell.
A bacterial endosymbiont that belongs to Epulopiscium spp., which hosts the largest known bacterium measuring more than 600 × 80 microns (5), inhabits the intestines of surgeon fish and “gives birth” to multiple live offspring (3). In these organisms, it has been shown that FtsZ assembles into polar rings and a minimum of two (and sometimes up to twelve) live offspring emerge from each pole and grow side by side which are then “birthed” subsequent to programmed mother cell lysis (4, 138), in a manner reminiscent of endospore formation in Firmicutes. Thus, in Epulopiscium spp., asymmetric division in the progenitor cell gives rise to multiple progeny and the lysis of the progenitor, not as a stress response, but as part of the normal cell cycle.
A longstanding challenge in bacterial cell biology has been to determine how FtsZ assembles at mid-cell, but septum placement has been assumed to lie along the short axis of a rod-shaped cell - not from pole to pole. A stunning mode of cell division has been observed recently in some marine gammaproteobacteria ectosymbionts and are in the process of receiving a proper binomial name (17, 101). A rod-shaped bacterium now referred to as Candidatus Thiosymbion oneisti grows as a monolayer, where each bacterium is attached at a bacterial pole on the surface of a marine nematode Laxus oneistus. In this organism, fragmented FtsZ rings form parallel to the long axis of the cell, instead of perpendicular to the long axis like in commonly studied model organisms (85) (Fig. 6B). This organism possesses an operon that encodes for a Min system, but it is not yet clear if Z-ring placement is regulated by the Min system. In another ectosymbiont of same family that also undergoes longitudinal division, it was recently shown that FtsZ accumulation occurs first at a pole proximal to the host (84) (Fig. 6C). Interestingly, instead of initiating cell division from both poles at the same time, this organism initiates the process at the host-proximal pole and drives the division and daughter cell separation towards the other pole making use of several FtsZ foci aligned at the division plane, instead of an FtsZ ring. One model posits that division along the long axis permits the continued association of daughter cells with the host surface on which the monolayer of bacteria is formed. Division along the long axis has also been reported for a helix-shaped, Drosophila melanogaster endosymbiont, Spiroplasma poulsonii, in which longitudinal branching occurs and FtsZ localizes to the branching sites (110).
Concluding thoughts
In recent years, increasingly new observations have been made in non-model organisms. For example, a new operon containing mid-cell associated probable division genes has been reported that is specific for obligate anaerobe Clostridium difficile (113). It was also reported that the C. difficile Min system, which includes an E. coli-like MinE protein in addition to the Firmicutes-like DivIVA, is capable of oscillation when produced in B. subtilis, suggesting that C. difficile may exhibit features that integrate both systems (91). These advances were aided by new cell biological tools that now permit the use of fluorescence microscopy probes in anaerobes such as C. difficile (16, 112). Further variations on existing themes are also now emerging by studying non-model organisms. In a twist on positive regulation of cell division site selection in certain spirochetes such as in the etiological agent of Lyme disease, Borrelia burgdorferi, peptidoglycan growth was observed to occur at precise sub-regions within a growing cell, so that the daughter cell division site is actually marked one generation in advance (69). Even the well-studied Min system displays variety, in that Min oscillation can occur in the cyanobacterium Synechococcus elongatus even in the presence of physical barriers set-up by thylakoid membranes (90). Similar to C. difficile, S. elongates also harbors both MinE and DivIVA-like proteins to regulate cell division in this organism (90).
Even in model organisms, new discoveries are continuing to be made such as the one that indicates that MinD in E. coli and B. subtilis may have an additional role in DNA segregation (28, 75). In B. subtilis, for example, the NO protein Noc does not directly interact with FtsZ in the way that the NO protein in E. coli (SlmA) does (24, 35, 141). Instead, it was recently proposed that Noc anchors the chromosome to the membrane and physically prevents regions of the cell that are conducive for FtsZ ring assembly during the process of nucleoid segregation (2). Another example of linking chromosome replication to cell division involves a nucleoid replication region organization factor in E. coli (MatP) which, together with other divisome proteins, positively facilitates Z-ring assembly and its constriction (7, 19, 92). There is also evidence suggesting the existence of hitherto undiscovered FtsZ regulators, since division site selection may still occur in the absence of the Min and NO systems in E. coli and B. subtilis (7, 116), and new evidence continues to emerge that links nutrition status to cell division (59, 95). Finally, FtsZ, the well-studied protein central to cell division in most bacteria, is still under active investigation, as new evidence has questioned if FtsZ is the predominant force generator that drives membrane invagination during cytokinesis (42, 94, 142). Instead, an emerging model proposes that assembled FtsZ is a platform to recruit peptidoglycan synthetic enzymes to provide directionality for peptidoglycan synthesis, and that it is peptidoglycan synthesis per se that directly contributes the force for septum formation (142).
The well-established genetics afforded by model systems has provided detailed molecular insights into how these species grow and give rise to progeny. Moving forward, the increasing availability of genome sequences even for unculturable bacteria, combined with the advent of molecular tools that increase the ease of genetic manipulation of these organisms, promises to open avenues of research into traditionally understudied systems to reveal the full repertoire of prokaryotic cell division strategies.
ACKNOWLEDGEMENTS
This work was funded by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (K.S.R) and by a start-up grant from the University of South Florida (P.J.E.).
REFERENCES
- 1.Abdelrahman Y, Ouellette SP, Belland RJ, Cox JV. 2016. Polarized Cell Division of Chlamydia trachomatis. PLoS Pathog 12: e1005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams DW, Wu LJ, Errington J. 2015. Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J 34: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angert ER. 2005. Alternatives to binary fission in bacteria. Nat Rev Microbiol 3: 214–24 [DOI] [PubMed] [Google Scholar]
- 4.Angert ER, Clements KD. 2004. Initiation of intracellular offspring in Epulopiscium. Mol Microbiol 51: 827–35 [DOI] [PubMed] [Google Scholar]
- 5.Angert ER, Clements KD, Pace NR. 1993. The largest bacterium. Nature 362: 239–41 [DOI] [PubMed] [Google Scholar]
- 6.Baek JH, Chattoraj DK. 2014. Chromosome I controls chromosome II replication in Vibrio cholerae. PLoS Genet 10: e1004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Mannik J. 2014. Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet 10: e1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balaban M, Hendrixson DR. 2011. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog 7: e1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernander R, Ettema TJ. 2010. FtsZ-less cell division in archaea and bacteria. Curr Opin Microbiol 13: 747–52 [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharya D, Kumar A, Panda D. 2017. WhmD promotes the assembly of Mycobacterium smegmatis FtsZ: A possible role of WhmD in bacterial cell division. Int J Biol Macromol 95: 582–91 [DOI] [PubMed] [Google Scholar]
- 12.Boersma MJ, Kuru E, Rittichier JT, VanNieuwenhze MS, Brun YV, Winkler ME. 2015. Minimal Peptidoglycan (PG) Turnover in Wild-Type and PG Hydrolase and Cell Division Mutants of Streptococcus pneumoniae D39 Growing Planktonically and in Host-Relevant Biofilms. J Bacteriol 197: 3472–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottomley AL, Kabli AF, Hurd AF, Turner RD, Garcia-Lara J, Foster SJ. 2014. Staphylococcus aureus DivIB is a peptidoglycan-binding protein that is required for a morphological checkpoint in cell division. Mol Microbiol [DOI] [PubMed] [Google Scholar]
- 14.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, et al. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134: 945–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, et al. 2012. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci U S A 109: 1697–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buckley AM, Jukes C, Candlish D, Irvine JJ, Spencer J, et al. 2016. Lighting Up Clostridium Difficile: Reporting Gene Expression Using Fluorescent Lov Domains. Sci Rep 6: 23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulgheresi S 2016. Bacterial cell biology outside the streetlight. Environ Microbiol 18: 2305–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burnham JC, Hashimoto T, Conti SF. 1970. Ultrastructure and cell division of a facultatively parasitic strain of Bdellovibrio bacteriovorus. J Bacteriol 101: 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buss J, Coltharp C, Shtengel G, Yang X, Hess H, Xiao J. 2015. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet 11: e1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butan C, Hartnell LM, Fenton AK, Bliss D, Sockett RE, et al. 2011. Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus. J Bacteriol 193: 1341–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camberg JL, Hoskins JR, Wickner S. 2009. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci U S A 106: 10614–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cameron TA, Zupan JR, Zambryski PC. 2015. The essential features and modes of bacterial polar growth. Trends Microbiol 23: 347–53 [DOI] [PubMed] [Google Scholar]
- 23.Celler K, Koning RI, Willemse J, Koster AJ, van Wezel GP. 2016. Cross-membranes orchestrate compartmentalization and morphogenesis in Streptomyces. Nat Commun 7: ncomms11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho H, Bernhardt TG. 2013. Identification of the SlmA active site responsible for blocking bacterial cytokinetic ring assembly over the chromosome. PLoS Genet 9: e1003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das N, Dai J, Hung I, Rajagopalan MR, Zhou HX, Cross TA. 2015. Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis, in lipid bilayers. Proc Natl Acad Sci U S A 112: E119–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datta P, Dasgupta A, Singh AK, Mukherjee P, Kundu M, Basu J. 2006. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol Microbiol 62: 1655–73 [DOI] [PubMed] [Google Scholar]
- 27.Del Sol R, Mullins JG, Grantcharova N, Flardh K, Dyson P. 2006. Influence of CrgA on assembly of the cell division protein FtsZ during development of Streptomyces coelicolor. J Bacteriol 188: 1540–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Ventura B, Knecht B, Andreas H, Godinez WJ, Fritsche M, et al. 2013. Chromosome segregation by the Escherichia coli Min system. Mol Syst Biol 9: 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ditkowski B, Holmes N, Rydzak J, Donczew M, Bezulska M, et al. 2013. Dynamic interplay of ParA with the polarity protein, Scy, coordinates the growth with chromosome segregation in Streptomyces coelicolor. Open Biol 3: 130006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donczew M, Mackiewicz P, Wrobel A, Flardh K, Zakrzewska-Czerwinska J, Jakimowicz D. 2016. ParA and ParB coordinate chromosome segregation with cell elongation and division during Streptomyces sporulation. Open Biol 6: 150263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donovan C, Schauss A, Kramer R, Bramkamp M. 2013. Chromosome segregation impacts on cell growth and division site selection in Corynebacterium glutamicum. PLoS One 8: e55078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donovan C, Schwaiger A, Kramer R, Bramkamp M. 2010. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum. J Bacteriol 192: 3441–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dori-Bachash M, Dassa B, Pietrokovski S, Jurkevitch E. 2008. Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl Environ Microbiol 74: 7152–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du S, Lutkenhaus J. 2012. MipZ: one for the pole, two for the DNA. Mol Cell 46: 239–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du S, Lutkenhaus J. 2014. SlmA antagonism of FtsZ assembly employs a two-pronged mechanism like MinCD. PLoS Genet 10: e1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dziadek J, Rutherford SA, Madiraju MV, Atkinson MA, Rajagopalan M. 2003. Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology 149: 1593–603 [DOI] [PubMed] [Google Scholar]
- 37.Dziedzic R, Kiran M, Plocinski P, Ziolkiewicz M, Brzostek A, et al. 2010. Mycobacterium tuberculosis ClpX interacts with FtsZ and interferes with FtsZ assembly. PLoS One 5: e11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134: 956–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egan ES, Fogel MA, Waldor MK. 2005. Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol Microbiol 56: 1129–38 [DOI] [PubMed] [Google Scholar]
- 40.Eksztejn M, Varon M. 1977. Elongation and cell division in Bdellovibrio bacteriovorus. Arch Microbiol 114: 175–81 [DOI] [PubMed] [Google Scholar]
- 41.England K, Crew R, Slayden RA. 2011. Mycobacterium tuberculosis septum site determining protein, Ssd encoded by rv3660c, promotes filamentation and elicits an alternative metabolic and dormancy stress response. BMC Microbiol 11: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74: 504–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erickson HP, Osawa M. 2010. Cell division without FtsZ--a variety of redundant mechanisms. Mol Microbiol 78: 267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Errington J 2013. L-form bacteria, cell walls and the origins of life. Open Biol 3: 120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eswaramoorthy P, Erb ML, Gregory JA, Silverman J, Pogliano K, et al. 2011. Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis. MBio 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fenton AK, Kanna M, Woods RD, Aizawa SI, Sockett RE. 2010. Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J Bacteriol 192: 6329–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fenton AK, Lambert C, Wagstaff PC, Sockett RE. 2010. Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J Bacteriol 192: 1299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleurie A, Lesterlin C, Manuse S, Zhao C, Cluzel C, et al. 2014. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516: 259–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleurie A, Manuse S, Zhao C, Campo N, Cluzel C, et al. 2014. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet 10: e1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11: 671–83 [DOI] [PubMed] [Google Scholar]
- 51.Galli E, Poidevin M, Le Bars R, Desfontaines JM, Muresan L, et al. 2016. Cell division licensing in the multi-chromosomal Vibrio cholerae bacterium. Nat Microbiol 1: 16094. [DOI] [PubMed] [Google Scholar]
- 52.Garcia PS, Simorre JP, Brochier-Armanet C, Grangeasse C. 2016. Cell division of Streptococcus pneumoniae: think positive! Curr Opin Microbiol 34: 18–23 [DOI] [PubMed] [Google Scholar]
- 53.Goley ED, Iniesta AA, Shapiro L. 2007. Cell cycle regulation in Caulobacter: location, location, location. J Cell Sci 120: 3501–7 [DOI] [PubMed] [Google Scholar]
- 54.Gomez JE, Bishai WR. 2000. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc Natl Acad Sci U S A 97: 8554–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grangeon R, Zupan JR, Anderson-Furgeson J, Zambryski PC. 2015. PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 112: 11666–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gregory JA, Becker EC, Pogliano K. 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev 22: 3475–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haeusser DP, Lee AH, Weart RB, Levin PA. 2009. ClpX inhibits FtsZ assembly in a manner that does not require its ATP hydrolysis-dependent chaperone activity. J Bacteriol 191: 1986–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haeusser DP, Margolin W. 2016. Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol 14: 305–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, et al. 2008. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321: 1673–5 [DOI] [PubMed] [Google Scholar]
- 60.Hett EC, Rubin EJ. 2008. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev 72: 126–56, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holeckova N, Doubravova L, Massidda O, Molle V, Buriankova K, et al. 2014. LocZ is a new cell division protein involved in proper septum placement in Streptococcus pneumoniae. MBio 6: e01700–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holmes NA, Walshaw J, Leggett RM, Thibessard A, Dalton KA, et al. 2013. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc Natl Acad Sci U S A 110: E397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Howell M, Brown PJ. 2016. Building the bacterial cell wall at the pole. Curr Opin Microbiol 34: 53–59 [DOI] [PubMed] [Google Scholar]
- 64.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124: 1025–37 [DOI] [PubMed] [Google Scholar]
- 65.Jacquier N, Frandi A, Pillonel T, Viollier PH, Greub G. 2014. Cell wall precursors are required to organize the chlamydial division septum. Nat Commun 5: 3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jha JK, Baek JH, Venkova-Canova T, Chattoraj DK. 2012. Chromosome dynamics in multichromosome bacteria. Biochim Biophys Acta 1819: 826–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang C, Caccamo PD, Brun YV. 2015. Mechanisms of bacterial morphogenesis: evolutionary cell biology approaches provide new insights. Bioessays 37: 413–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jorge AM, Hoiczyk E, Gomes JP, Pinho MG. 2011. EzrA contributes to the regulation of cell size in Staphylococcus aureus. PLoS One 6: e27542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jutras BL, Scott M, Parry B, Biboy J, Gray J, et al. 2016. Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. Proc Natl Acad Sci U S A 113: 9162–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kazmierczak BI, Hendrixson DR. 2013. Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol Microbiol 88: 655–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kemege KE, Hickey JM, Barta ML, Wickstrum J, Balwalli N, et al. 2015. Chlamydia trachomatis protein CT009 is a structural and functional homolog to the key morphogenesis component RodZ and interacts with division septal plane localized MreB. Mol Microbiol 95: 365–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kiekebusch D, Michie KA, Essen LO, Lowe J, Thanbichler M. 2012. Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol Cell 46: 245–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kieser KJ, Rubin EJ. 2014. How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol 12: 550–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kiran M, Maloney E, Lofton H, Chauhan A, Jensen R, et al. 2009. Mycobacterium tuberculosis ftsZ expression and minimal promoter activity. Tuberculosis (Edinb) 89 Suppl 1: S60–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kloosterman TG, Lenarcic R, Willis CR, Roberts DM, Hamoen LW, et al. 2016. Complex polar machinery required for proper chromosome segregation in vegetative and sporulating cells of Bacillus subtilis. Mol Microbiol 101: 333–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kois-Ostrowska A, Strzalka A, Lipietta N, Tilley E, Zakrzewska-Czerwinska J, et al. 2016. Unique Function of the Bacterial Chromosome Segregation Machinery in Apically Growing Streptomyces - Targeting the Chromosome to New Hyphal Tubes and its Anchorage at the Tips. PLoS Genet 12: e1006488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Konar M, Alam MS, Arora C, Agrawal P. 2012. WhiB2/Rv3260c, a cell division-associated protein of Mycobacterium tuberculosis H37Rv, has properties of a chaperone. FEBS J 279: 2781–92 [DOI] [PubMed] [Google Scholar]
- 78.Koyama T, Yamada M, Matsuhashi M. 1977. Formation of regular packets of Staphylococcus aureus cells. J Bacteriol 129: 1518–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lam H, Schofield WB, Jacobs-Wagner C. 2006. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124: 1011–23 [DOI] [PubMed] [Google Scholar]
- 80.Lara B, Rico AI, Petruzzelli S, Santona A, Dumas J, et al. 2005. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol Microbiol 55: 699–711 [DOI] [PubMed] [Google Scholar]
- 81.Lasker K, Mann TH, Shapiro L. 2016. An intracellular compass spatially coordinates cell cycle modules in Caulobacter crescentus. Curr Opin Microbiol 33: 131–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee KC, Webb RI, Fuerst JA. 2009. The cell cycle of the planctomycete Gemmata obscuriglobus with respect to cell compartmentalization. BMC Cell Biol 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lefevre CT, Bennet M, Klumpp S, Faivre D. 2015. Positioning the flagellum at the center of a dividing cell to combine bacterial division with magnetic polarity. MBio 6: e02286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leisch N, Pende N, Weber PM, Gruber-Vodicka HR, Verheul J, et al. 2016. Asynchronous division by non-ring FtsZ in the gammaproteobacterial symbiont of Robbea hypermnestra. Nat Microbiol 2: 16182. [DOI] [PubMed] [Google Scholar]
- 85.Leisch N, Verheul J, Heindl NR, Gruber-Vodicka HR, Pende N, et al. 2012. Growth in width and FtsZ ring longitudinal positioning in a gammaproteobacterial symbiont. Curr Biol 22: R831–2 [DOI] [PubMed] [Google Scholar]
- 86.Levin PA, Kurtser IG, Grossman AD. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci U S A 96: 9642–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liechti G, Kuru E, Packiam M, Hsu YP, Tekkam S, et al. 2016. Pathogenic Chlamydia Lack a Classical Sacculus but Synthesize a Narrow, Mid-cell Peptidoglycan Ring, Regulated by MreB, for Cell Division. PLoS Pathog 12: e1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, et al. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506: 507–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lutkenhaus J, Pichoff S, Du S. 2012. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton (Hoboken) 69: 778–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.MacCready JS, Schossau J, Osteryoung KW, Ducat DC. 2016. Robust Min-system oscillation in the presence of internal photosynthetic membranes in cyanobacteria. Mol Microbiol [DOI] [PubMed] [Google Scholar]
- 91.Makroczyova J, Jamroskovic J, Krascsenitsova E, Labajova N, Barak I. 2016. Oscillating behavior of Clostridium difficile Min proteins in Bacillus subtilis. Microbiologyopen 5: 387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mannik J, Castillo DE, Yang D, Siopsis G, Mannik J. 2016. The role of MatP, ZapA and ZapB in chromosomal organization and dynamics in Escherichia coli. Nucleic Acids Res 44: 1216–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McCormick JR, Su EP, Driks A, Losick R. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol 14: 243–54 [DOI] [PubMed] [Google Scholar]
- 94.Meier EL, Goley ED. 2014. Form and function of the bacterial cytokinetic ring. Curr Opin Cell Biol 26: 19–27 [DOI] [PubMed] [Google Scholar]
- 95.Monahan LG, Harry EJ. 2016. You Are What You Eat: Metabolic Control of Bacterial Division. Trends Microbiol 24: 181–9 [DOI] [PubMed] [Google Scholar]
- 96.Monteiro JM, Fernandes PB, Vaz F, Pereira AR, Tavares AC, et al. 2015. Cell shape dynamics during the staphylococcal cell cycle. Nat Commun 6: 8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moulder JW. 1993. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect Agents Dis 2: 87–99 [PubMed] [Google Scholar]
- 98.Mura A, Fadda D, Perez AJ, Danforth ML, Musu D, et al. 2016. Roles of the essential protein FtsA in cell growth and division in Streptococcus pneumoniae. J Bacteriol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ouellette SP, Rueden KJ, AbdelRahman YM, Cox JV, Belland RJ. 2015. Identification and Partial Characterization of Potential FtsL and FtsQ Homologs of Chlamydia. Front Microbiol 6: 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Packiam M, Weinrick B, Jacobs WR Jr., Maurelli AT. 2015. Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly”. Proc Natl Acad Sci U S A 112: 11660–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Petersen JM, Kemper A, Gruber-Vodicka H, Cardini U, van der Geest M, et al. 2016. Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat Microbiol 2: 16195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, et al. 2013. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun 4: 2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pinho MG, Errington J. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol Microbiol 50: 871–81 [DOI] [PubMed] [Google Scholar]
- 104.Pinho MG, Errington J. 2004. A divIVA null mutant of Staphylococcus aureus undergoes normal cell division. FEMS Microbiol Lett 240: 145–9 [DOI] [PubMed] [Google Scholar]
- 105.Pinho MG, Kjos M, Veening JW. 2013. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol 11: 601–14 [DOI] [PubMed] [Google Scholar]
- 106.Plocinski P, Ziolkiewicz M, Kiran M, Vadrevu SI, Nguyen HB, et al. 2011. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J Bacteriol 193: 3246–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Poindexter JS. 1978. Selection for nonbuoyant morphological mutants of Caulobacter crescentus. J Bacteriol 135: 1141–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ramachandran R, Jha J, Chattoraj DK. 2014. Chromosome segregation in Vibrio cholerae. J Mol Microbiol Biotechnol 24: 360–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ramachandran R, Jha J, Paulsson J, Chattoraj D. 2017. Random versus Cell Cycle-Regulated Replication Initiation in Bacteria: Insights from Studying Vibrio cholerae Chromosome 2. Microbiol Mol Biol Rev 81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramond E, Maclachlan C, Clerc-Rosset S, Knott GW, Lemaitre B. 2016. Cell Division by Longitudinal Scission in the Insect Endosymbiont Spiroplasma poulsonii. MBio 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Randich AM, Brun YV. 2015. Molecular mechanisms for the evolution of bacterial morphologies and growth modes. Front Microbiol 6: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ransom EM, Ellermeier CD, Weiss DS. 2015. Use of mCherry Red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl Environ Microbiol 81: 1652–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ransom EM, Williams KB, Weiss DS, Ellermeier CD. 2014. Identification and characterization of a gene cluster required for proper rod shape, cell division, and pathogenesis in Clostridium difficile. J Bacteriol 196: 2290–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, et al. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303: 689–92 [DOI] [PubMed] [Google Scholar]
- 115.Rivas-Marin E, Canosa I, Devos DP. 2016. Evolutionary Cell Biology of Division Mode in the Bacterial Planctomycetes-Verrucomicrobia- Chlamydiae Superphylum. Front Microbiol 7: 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rodrigues CD, Harry EJ. 2012. The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet 8: e1002561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rowlett VW, Margolin W. 2015. The Min system and other nucleoid-independent regulators of Z ring positioning. Front Microbiol 6: 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sharma AK, Arora D, Singh LK, Gangwal A, Sajid A, et al. 2016. Serine/Threonine Protein Phosphatase PstP of Mycobacterium tuberculosis Is Necessary for Accurate Cell Division and Survival of Pathogen. J Biol Chem 291: 24215–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shigematsu M, Umeda A, Fujimoto S, Amako K. 1998. Spirochaete-like swimming mode of Campylobacter jejuni in a viscous environment. J Med Microbiol 47: 521–6 [DOI] [PubMed] [Google Scholar]
- 120.Sieger B, Schubert K, Donovan C, Bramkamp M. 2013. The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Mol Microbiol 90: 966–82 [DOI] [PubMed] [Google Scholar]
- 121.Sockett RE. 2009. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol 63: 523–39 [DOI] [PubMed] [Google Scholar]
- 122.Son SH, Lee HH. 2013. The N-terminal domain of EzrA binds to the C terminus of FtsZ to inhibit Staphylococcus aureus FtsZ polymerization. Biochem Biophys Res Commun 433: 108–14 [DOI] [PubMed] [Google Scholar]
- 123.Steele VR, Bottomley AL, Garcia-Lara J, Kasturiarachchi J, Foster SJ. 2011. Multiple essential roles for EzrA in cell division of Staphylococcus aureus. Mol Microbiol 80: 542–55 [DOI] [PubMed] [Google Scholar]
- 124.Sugimoto S, Yamanaka K, Nishikori S, Miyagi A, Ando T, Ogura T. 2010. AAA+ chaperone ClpX regulates dynamics of prokaryotic cytoskeletal protein FtsZ. J Biol Chem 285: 6648–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sureka K, Hossain T, Mukherjee P, Chatterjee P, Datta P, et al. 2010. Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS One 5: e8590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 126.Szymanski CM, King M, Haardt M, Armstrong GD. 1995. Campylobacter jejuni motility and invasion of Caco-2 cells. Infect Immun 63: 4295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tavares AC, Fernandes PB, Carballido-Lopez R, Pinho MG. 2015. MreC and MreD Proteins Are Not Required for Growth of Staphylococcus aureus. PLoS One 10: e0140523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thakur M, Chakraborti PK. 2006. GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J Biol Chem 281: 40107–13 [DOI] [PubMed] [Google Scholar]
- 129.Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147–62 [DOI] [PubMed] [Google Scholar]
- 130.Treuner-Lange A, Aguiluz K, van der Does C, Gomez-Santos N, Harms A, et al. 2013. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol Microbiol 87: 235–53 [DOI] [PubMed] [Google Scholar]
- 131.Tsang MJ, Bernhardt TG. 2015. Guiding divisome assembly and controlling its activity. Curr Opin Microbiol 24: 60–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Turner RD, Ratcliffe EC, Wheeler R, Golestanian R, Hobbs JK, Foster SJ. 2010. Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nat Commun 1: 26. [DOI] [PubMed] [Google Scholar]
- 133.Tzagoloff H, Novick R. 1977. Geometry of cell division in Staphylococcus aureus. J Bacteriol 129: 343–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vadrevu IS, Lofton H, Sarva K, Blasczyk E, Plocinska R, et al. 2011. ChiZ levels modulate cell division process in mycobacteria. Tuberculosis (Edinb) 91 Suppl 1: S128–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Val ME, Marbouty M, de Lemos Martins F, Kennedy SP, Kemble H, et al. 2016. A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae. Sci Adv 2: e1501914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van Baarle S, Bramkamp M. 2010. The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly. PLoS One 5: e9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Veiga H, Jorge AM, Pinho MG. 2011. Absence of nucleoid occlusion effector Noc impairs formation of orthogonal FtsZ rings during Staphylococcus aureus cell division. Mol Microbiol 80: 1366–80 [DOI] [PubMed] [Google Scholar]
- 138.Ward RJ, Clements KD, Choat JH, Angert ER. 2009. Cytology of terminally differentiated Epulopiscium mother cells. DNA Cell Biol 28: 57–64 [DOI] [PubMed] [Google Scholar]
- 139.Weart RB, Nakano S, Lane BE, Zuber P, Levin PA. 2005. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol Microbiol 57: 238–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev 25: 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu LJ, Errington J. 2011. Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol 10: 8–12 [DOI] [PubMed] [Google Scholar]
- 142.Xiao J, Goley ED. 2016. Redefining the roles of the FtsZ-ring in bacterial cytokinesis. Curr Opin Microbiol 34: 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yague P, Willemse J, Koning RI, Rioseras B, Lopez-Garcia MT, et al. 2016. Subcompartmentalization by cross-membranes during early growth of Streptomyces hyphae. Nat Commun 7: 12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang L, Willemse J, Claessen D, van Wezel GP. 2016. SepG coordinates sporulation-specific cell division and nucleoid organization in Streptomyces coelicolor. Open Biol 6: 150164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou X, Halladin DK, Rojas ER, Koslover EF, Lee TK, et al. 2015. Bacterial division. Mechanical crack propagation drives millisecond daughter cell separation in Staphylococcus aureus. Science 348: 574–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zupan JR, Cameron TA, Anderson-Furgeson J, Zambryski PC. 2013. Dynamic FtsA and FtsZ localization and outer membrane alterations during polar growth and cell division in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 110: 9060–5 [DOI] [PMC free article] [PubMed] [Google Scholar]