Abstract
An increasing number of antibody-based therapies are being considered for controlling bacterial infections, including Clostridium difficile by targeting toxins A and B. In an effort to develop novel C. difficile immunotherapeutics, we previously isolated several single-domain antibodies (VHHs) capable of toxin A neutralization through recognition of the extreme C-terminal combined repetitive oligopeptides (CROPs) domain, but failed at identifying neutralizing VHHs that bound a similar region on toxin B. Here we report the isolation of a panel of 29 VHHs targeting at least seven unique epitopes on a toxin B immunogen composed of a portion of the central delivery domain and the entire CROPs domain. Despite monovalent affinities as high as KD = 70 pM, none of the VHHs tested were capable of toxin B neutralization; however, modest toxin B inhibition was observed with VHH-VHH dimers and to a much greater extent with VHH-Fc fusions, reaching the neutralizing potency of the recently approved anti-toxin B monoclonal antibody bezlotoxumab in in vitro assays. Epitope binning revealed that several VHH-Fcs bound toxin B at sites distinct from the region recognized by bezlotoxumab, while other VHH-Fcs partially competed with bezlotoxumab for toxin binding. Therefore, the VHHs described here are effective at toxin B neutralization when formatted as bivalent VHH-Fc fusions by targeting toxin B at regions both similar and distinct from the bezlotoxumab binding site.
Introduction
Clostridium difficile is a Gram-positive spore-forming bacterium that continues to be a problematic nosocomial pathogen. The symptoms of gastrointestinal C. difficile infections can range from mild diarrhea to pseudomembrane colitis and death. The spore-forming nature of the pathogen coupled with an ability to rapidly colonize patients on broad-spectrum antibiotics presents a significant challenge to infection control in healthcare settings. Healthcare associated costs of managing C. difficile infection were estimated to exceed a staggering $4.5 billion annually in the US alone [1]. Despite the introduction of therapies that include new antibiotic modalities, fecal transplantation and antibody-based immunotherapy, efficacy limitations remain which necessitate the continuous search for more potent therapeutic agents [2,3,4].
C. difficile secretes two large toxins (TcdA and TcdB) that act upon the epithelial cells lining the gastrointestinal tract by first internalizing and then inactivating Rho/Ras proteins, which leads to cell-cytoskeleton disruption and ultimately a loss of epithelial barrier function, severe inflammation and apoptosis [5,6]. TcdA and TcdB are each four-domain proteins composed of an N-terminal glucosyltransferase domain, a central autoprotease cutting domain, a neighboring central delivery/translocation domain and a C-terminal region referred to as the combined repetitive oligopeptides (CROPs) domain. These two toxins, along with transferase toxin (CDT), are considered the primary virulence factors of C. difficile and have been targeted by toxin-binding polymers, vaccines and antibodies as strategies to control C. difficile-associated disease [5]. With respect to antibodies a number of anti-toxin formats have been explored, including intravenous immunoglobulin therapy, IgG, IgA, IgY, single-chain fragment variable (scFv) and single-domain antibodies (sdAbs) [2,7]. Recently, the anti-TcdB monoclonal antibody (mAb) bezlotoxumab was FDA-approved for the treatment of recurrent C. difficile infection [8,9].
In an effort to develop novel antibody-based therapeutics for C. difficile infection, our group and others have explored the use of sdAbs as potent anti-toxin neutralizing agents [10,11,12,13,14,15,16,17,18,19]. Camelid-sourced sdAbs (VHHs or Nanobodies) are recombinant antibody fragments that offer the benefits of full-sized mAbs–high target affinity and specificity–with unique properties, most notably their amenability to tandem formatting in various geometries such that multi-specificities can be attained within a single molecule [20,21,22]. We previously isolated several moderate-affinity llama VHHs that targeted the C-terminal CROPs domain (also referred to as the receptor binding domain or RBD) of TcdA (aa 2304–2710) that were neutralizers of TcdA as single VHHs or in combination [10]. At the same time, immunization with a small C-terminal fragment of the TcdB CROPs domain (aa 2286–2366) failed to produce neutralizing VHHs.
Here, we immunized a llama with a recombinant TcdB fragment (aa 1751–2366) that encompasses a portion of the central delivery/translocation domain (aa 1751–1833) and the entire CROPs domain (aa 1834–2366). We hypothesized that extending our previous immunogen design to include a portion of the delivery/translocation domain and the full-length CROPs domain may lead to neutralizing antibodies. This is supported by recent reports showing TcdB binding the frizzled (FZD2) family of Wnt receptors [23,24] and the poliovirus receptor-like 3 (PVRL3) receptor [25] through the central delivery/translocation domain, and TcdB binding the chondroitin sulfate proteoglycan 4 (CSPG4) receptor [26] through a region adjacent to the CROPs domain. In addition, several earlier studies showed the isolation of neutralizing antibodies to the CROPs region of TcdB [15,27,28]. Despite isolating numerous VHHs with affinities as high as KD = 70 pM, none of the monomeric antibodies were capable of preventing TcdB-induced cytotoxicity in cell-based assays. However, when reformatted as Fc-fusions, several VHH-Fcs were transformed into potent TcdB neutralizers on par with the neutralizing efficacy of the recently approved anti-TcdB mAb bezlotoxumab. Thus, TcdB-binding VHHs targeting a fragment of the delivery domain and the complete CROPs domain are effective neutralizers when presented in the context of larger bivalent VHH-Fc fusions, presumably due to steric and/or avidity effects not afforded to monomeric VHHs.
Materials and methods
Llama immunization, serum fractionation and serology
Recombinant TcdB (aa 1751–2366; hereafter referred to as TcdB1751-2366) was a generous gift from Dr. Kenneth Ng (University of Calgary, Calgary, AB, Canada). A llama was immunized with 100 μg of TcdB1751-2366 per injection, following a similar immunization schedule and adjuvanted as previously reported [29]. Serum from blood drawn 42 days post immunization was tested for binding to TcdB1751-2366 by ELISA essentially as described [10]. Serum was fractionated according to established protocols with protein G and protein A affinity columns [30] and conventional and heavy-chain IgG fractions tested for TcdB1751-2366 binding by ELISA [10]. The ability of polyclonal fractions to neutralize TcdB VPI 10463 (List Biological Laboratories) was examined by Vero cell toxin inhibition assays, essentially as described for TcdA [19] with minor modifications. Vero cell monolayers were incubated with a final TcdB concentration of 10 pM (2.7 ng/mL) or 30 pM (8.1 pg/mL) and 1 μM of day 42 fractionated serum for 72 h at 37°C and 5% CO2 before addition of WST-1 cytotoxicity reagent (Roche, Mississauga, ON, Canada) for 30 min and subsequent absorbance measurement at 450 nm.
Research involving animals
All procedures involving llamas and their care were approved by the National Research Council Canada Animal Care Committee and by the Animal Care Committee of Cedarlane Laboratories who is licensed by the Ontario Ministry of Agriculture, Food and Rural Affairs.
Library construction, VHH isolation and expression
Lymphocytes obtained from serum drawn 42 days post immunization served as a starting point for phagemid library construction. RNA extraction, cDNA synthesis, two rounds of PCR, restriction digestion, ligation into the pMED1 phagemid vector, transformation of electrocompetent E. coli and preparation of library phage were all performed as described [10,30]. VHHs were then selected by two approaches. In the first approach, TcdB1751-2366 was coated directly onto microtiter plate wells, plates were blocked with 5% non-fat skimmed milk in PBS-T (PBS + 0.5% (v/v) Tween 20), and library phage applied, washed and eluted with 0.1 M triethylamine, all essentially as reported [10], for three rounds. In the second approach, TcdB1751-2366 was first biotinylated (TcdB1751-2366-Biotin) using a commercial EZ-LinkTM Sulfo-NHS-Biotinylation Kit (ThermoFisher, Ottawa, ON, Canada), according to the manufacturer’s instructions, and confirmed by Western blotting by probing with streptavidin (SA) conjugated with AP (ThermoFisher). Next, library phage were incubated with TcdB1751-2366-Biotin (5 nM) in a 1.5 mL Eppendorf tube for 10 min before addition of non-biotinylated TcdB1751-2366 competitor (2.5 μM) for 10 min. The mixture was then applied to streptavidin coated microtiter plates (ThermoFisher) for 5 min before a series of washes with PBS and PBS-T and elution with 0.1 M triethylamine. In each subsequent round the concentration of TcdB1751-2366-Biotin target was reduced and the incubation time with TcdB1751-2366 competitor increased, except for the fourth round in which the incubation time was held at 60 min. Eluted phage clones displaying VHHs from both isolation methods were tested for binding to immobilized TcdB1751-2366 in monoclonal phage ELISA as described [10] and those clones producing the highest absorbance (450 nm) were sequenced, subcloned into the pSJF2H expression vector [10], and expressed and purified from E. coli by IMAC [10,30]. Purified VHHs were probed by Western blotting using an α-His tag specific IgG conjugated with AP (ThemoFisher).
VHH characterization
The aggregation state of VHHs was assessed by size-exclusion chromatography (SEC) using a SuperdexTM 75 Increase column (GE Healthcare, Mississauga, ON, Canada) as described [10,31]. VHH thermal unfolding and refolding were examined by circular dichroism spectroscopy, essentially as reported [31], with the exception of data collection every 0.2°C and the VHHs were allowed to cool at 25°C for 3 h before the second thermal melt was performed. The monovalent binding affinities of VHHs were determined using a Biacore 3000 surface plasmon resonance (SPR) instrument (GE Healthcare) and the Biotin CAPture Kit (GE Healthcare). Approximately 900 resonance units (RUs) of TcdB1751-2366-Biotin were captured before flowing a two-fold dilution series of SEC-purified VHHs (concentrations ranging from as low as 0.13–2 nM to as high as 31.3–500 nM) over the TcdB1751-2366-Biotin surface using single-cycle kinetic analysis. All experiments were performed in HBS-EP buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% (v/v) Surfactant P20; GE Healthcare) at 25°C at a flow rate of 30 μL/min, with a contact time of 2 min and dissociation time of 10 min. Surfaces were regenerated according to the Biotin CAPture Kit instructions (Regeneration stock 1 and 2, at 10 μL/min). Reference flow cell subtracted sensorgrams were fit to a 1:1 binding model using the BIAevaluation 4.1 software (GE Healthcare). VHH epitope binning was also performed by SPR on TcdB1751-2366-Biotin captured surfaces by injection of the first VHH at 10× KD concentration for 2 min followed immediately by injection of a mixture of the first VHH + a second VHH at 10× KD concentration for 2 min, all at a flow rate of 30 μL/min. Binning experiments were also performed in the reverse format (i.e., VHH2 followed by VHH2 + VHH1). The ability of VHHs to neutralize TcdB VPI 10463 was examined by Vero cell toxin inhibition assays, essentially as described above for fractionated serum. Vero cell monolayers were incubated with a final TcdB concentration of 1 pM (270 pg/mL) and 1 μM of VHH for 72 h at 37°C and 5% CO2 before addition of WST-1 cytotoxicity reagent (Roche) for 30 min and subsequent absorbance measurement at 450 nm. Before performing TcdB inhibition assays that involved VHHs, dimers and Fc-fusions, dose-response experiments were always conducted to identify a working TcdB concentration that provided ~90% toxicity to account for batch-to-batch variability in TcdB potency.
Generation and characterization of VHH-VHH dimers
Using the three highest affinity VHHs targeting unique epitopes (B39, B74 and B167) all VHH-VHH dimer formats were constructed by splice overlap extension PCR, essentially as described [31], before ligation into pSJF2H expression vector. Each dimer was separated by a 25 amino acid linker (Gly4Ser)5 and contained a C-terminal His6 tag for purification. Dimers were expressed in 1 L E. coli cultures, extracted from the periplasm by osmotic shock, purified by IMAC and assessed for purity and aggregation by SDS-PAGE, Western blot and SEC with a Superdex 200 Increase column (GE Healthcare), according to standard methods. When VHH-VHHs showed higher order aggregates or signs of degradation by SEC, the non-aggregating monomer peaks were collected and used for subsequent assays. The thermal unfolding temperature of each dimer was determined as described above for monomer VHHs. The dissociation rate of each VHH-VHH dimer was determined by SPR using conditions described above for binding to TcdB1751-2366-Biotin captured surfaces. Briefly, dimers and control monomers were injected at 1 nM for 5 min before 30 min of dissociation, all at 25°C with a flow rate of 30 μL/min and in HBS-EP buffer (GE Healthcare). Dissociation phases were fit to a 1:1 dissociation model using the BIAevaluation 4.1 software (GE Healthcare) in order to calculate kds (s-1). The ability of VHH-VHH dimers to neutralize TcdB (List Biological Laboratories) was examined by Vero cell toxin inhibition assays as described above for fractionated serum and VHH monomers. A final TcdB concentration of 3 pM (810 pg/mL), based on dose-response experiments, and a dimer concentration of 1 μM were used. All other assay conditions remained the same. An irrelevant dimer T5/TC9 [32] was used at 1 μM as a negative control while the TcdB-binding mAb bezlotoxumab (MDX-1388) was used at 250 nM as a positive control.
Generation and characterization of VHH-Fc fusions
VHHs fused to the N-termini of human IgG1 Fcs were synthesized and subcloned into the pTT5 vector and expressed through transient transfection of mammalian HEK293-6E cells before protein A purification, as described [33,34]. Each VHH was separated from the IgG1 Fc region by either the human IgG1 hinge or a 35-residue camel/llama γ2a hinge [35]. The aggregation profiles of VHH-Fcs were assessed by SEC using a Superdex 200 column (GE Healthcare). SPR was used to determine the dissociation rates of VHH-Fcs by flowing 1 nM of SEC-purified antibody over TcdB1751-2366-Biotin surfaces for 2 min, followed by a dissociation time of 30 min, all at 25°C with a 30 μL/min flow rate in HBS-EP buffer (GE Healthcare). SPR-based binning experiments between VHH-Fcs and MDX-1388 [36] were performed essentially as described above for VHH monomers. The ability of VHH-Fcs to neutralize TcdB (List Biological Laboratories) was examined by Vero cell toxin inhibition assays as described above for VHH monomers and dimers. A final TcdB concentration of 500 fM (135 pg/mL), based on dose-response experiments, and a VHH-Fc fusion concentration of 250 nM were used. All other assay conditions remained the same. When two VHH-Fc fusions were used in combination for TcdB inhibition, 125 nM of each VHH-Fc was added. TcdB and VHH-Fcs were not pre-incubated before the addition to Vero cells.
Results
Llama immunization, serum fractionation and serology
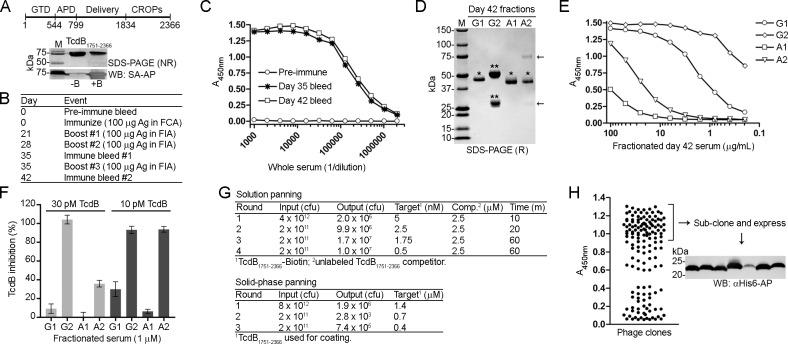
In an attempt to generate TcdB neutralizing VHHs, we started by immunizing a llama with recombinant TcdB1751-2366 which consists of a portion of the central delivery/translocation domain and entire CROPs domain (Fig 1A and 1B). Serum drawn 35 days and 42 days post immunization showed a clear response to TcdB1751-2366 by ELISA (Fig 1C). Serum drawn on day 42 was separated by fractionation into conventional IgG (cIgG; G2 fraction) and heavy-chain IgG (hcIgG; G1 fraction–a long hinge isotype based on SDS-PAGE, and A1 and A2 fractions–a short hinge isotype(s) based on SDS-PAGE, Fig 1D) and showed a typical reactivity pattern by TcdB1751-2366 binding in ELISA (Fig 1E). The G2 fraction (cIgG) showed the strongest reactivity towards TcdB1751-2366 with an EC50 ~ 0.1 μg/mL compared to the G1 fraction (hcIgG) with an EC50 ~ 2 μg/mL. A1 and A2 fractions (hcIgG) possessed considerably lower TcdB binding titers. We next examined if the fractionated polyclonal sera could neutralize TcdB in Vero cell cytotoxicity assays. TcdB (10 or 30 pM, final) was added to Vero cells alone or in combination with fractionated serum (1 μM, final) for 72 h before addition of WST-1 reagent (Fig 1F). At 30 pM of TcdB the G2 fraction fully inhibited TcdB, while approximately 30% was inhibited by the A2 fraction, 10% by the G1 fraction and no inhibition with A1. A similar pattern was seen when 3-fold less TcdB was used: near complete inhibition with G2 and A2, approximately 30% by G1 and less than 10% by A1. The inhibition pattern largely matches the TcdB binding data (Fig 1E), although the A2 heavy-chain fraction showed greater inhibition than G1, presumably due to the presence of minor IgM contaminants (Fig 1D) that would efficiently neutralize TcdB due to size and valency. We next proceeded with phage display library construction and created a phage display library with a size of ~ 3 x 107 independent transformants.
Fig 1. Isolation of high-affinity TcdB-binding VHHs.
(A, top) Schematic of TcdB. GTD, glucosyltransferase domain; APD, autoprotease domain, Delivery, delivery/receptor binding domain, CROPs, combined repetitive oligopeptides domain. (A, bottom) A 71.6 kDa fragment of C. difficile toxin B (TcdB1751-2366) was purified, biotinylated and analyzed by non-reducing (NR) SDS-PAGE and Western blot (WB) probed with streptavidin-AP (SA-AP). Approximately 1 μg of protein was loaded per lane. M, protein molecular weight marker; -B, unlabeled TcdB1751-2366; +B, biotinylated TcdB1751-2366. (B) TcdB1751-2366 was used as an antigen (Ag) for llama immunization with the schedule shown. FCA, Freund’s complete adjuvant; FIA, Freund’s incomplete adjuvant. (C) ELISA showing the binding response from pre-immune and immune llama sera collected on days 35 and 42 post immunization to coated TcdB1751-2366. (D) Serum from day 42 post immunization was fractionated using protein G and protein A affinity columns and analyzed by SDS-PAGE under reducing (R) conditions. G1, A1 and A2 fractions contain heavy-chain IgG (hcIgG, *) while the G2 fraction contains conventional IgG (cIgG, **). The arrows denote IgM heavy and light chains in the A2 fraction. (E) ELISA showing the binding response of fractionated day 42 serum to coated TcdB1751-2366. (F) Vero cell cytotoxicity assay demonstrating the effect of fractionated day 42 serum on TcdB inhibition, 72 h post addition of TcdB and polyclonal antibodies to Vero cell monolayers. TcdB was used at 10 and 30 pM and polyclonal fractionated sera at 1 μM. (G, top) Summary of solution-phase library panning titers using off-rate selection. In each round, the amount of target (biotinylated TcdB1751-2366) was reduced, the amount of competitor (“Comp”; unlabeled TcdB1751-2366) was held constant, and the incubation time of phage + target + competitor was increased. After incubation, the complex of phage and biotinylated TcdB1751-2366 was captured on streptavidin coated microtiter plates, washed and eluted. (G, bottom) Summary of solid-phase library panning titers using coated TcdB1751-2366. (H) Phage ELISA showing binding of phage displayed VHHs to coated TcdB1751-2366. VHHs on phage producing the highest ELISA signals were expressed, purified and characterized. The Western blot (WB) shows detection of purified VHHs with an α-His6-AP secondary antibody.
VHH isolation and expression
For the isolation of VHHs, two different selection strategies were used: solution panning (off-rate based selection) and solid-phase panning (Fig 1G). In off-rate based selections 5 nM of biotinylated TcdB1751-2366 (Fig 1A) was incubated with the phage-displayed VHH library followed by addition of a 500-fold molar excess of non-biotinylated TcdB1751-2366 (2.5 μM) for 10 min before capture on streptavidin coated wells, washing and elution in round 1 (Fig 1G). In each subsequent round the amount of biotinylated TcdB1751-2366 was decreased and the incubation time with non-biotinylated TcdB1751-2366 competitor increased. For solid-phase selection, TcdB1751-2366 was coated directly on standard microtiter plate wells, incubated with library phage and eluted with high pH. In each round of panning the amount of TcdB1751-2366 coated was reduced. Monoclonal phage ELISA was performed on clones derived from both methods after three or four rounds of panning and those producing the highest ELISA signals were sequenced and sub-cloned for expression in E. coli (Fig 1H). A total of 29 unique VHH sequences (all differing in CDR3; S1 Table) were expressed and purified by immobilized metal-ion affinity chromatography (IMAC) with purification yields ranging from 3.1–41.3 mg/L before extensive biophysical characterization.
Biophysical characterization of VHHs
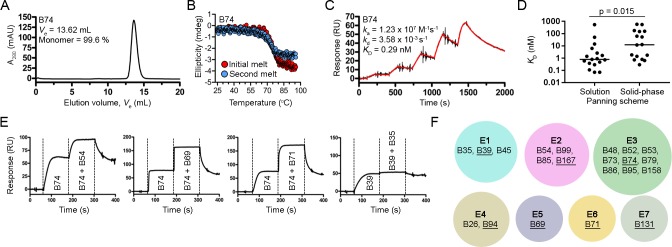
Size-exclusion chromatography (SEC) profiles of the VHHs showed predominantly single monodispersed peaks devoid of higher order aggregates as expected (Table 1, Fig 2A and S1 Fig). Circular dichroism spectroscopy was used to determine VHH melting temperatures (Tms) and to assess VHH refolding (Table 1, Fig 2B and S2 Fig). The Tms ranged from 57.6 to 87.2°C (median Tm = 73.4°C) and most VHHs could refold, although the completeness of refolding was sequence dependent. VHH binding affinities and kinetics were determined by SPR single-cycle kinetic analysis (Table 1, Fig 2C and S3 Fig). VHH affinities (KDs) ranged from 70 pM to 576 nM and, when classified by selection method (Fig 2D), the 18 VHHs isolated from solution panning were of statistically higher affinity than the 15 VHHs obtained by solid-phase panning (median KDs of 0.79 nM and 12.3 nM, respectively, and note that the four VHHs found by both selection methods were counted in each). Next, VHHs with KDs of ~50 nM and stronger were subjected to epitope binning by SPR-based co-injection (VHH 1 followed by VHH 1 + VHH 2) experiments (Table 1, Fig 2E and S4 Fig). From the 21 VHHs binned a total of seven non-overlapping TcdB epitopes were found (Fig 2F). The nine VHHs residing in epitope bin E3 were clonally related (Fig 2F, S1 Table) while the other six bins contained predominantly unique, unrelated VHHs. Finally, using VHHs with the highest affinities and/or slowest off-rates (kds) in each epitope bin (B39, B69, B71, B74, B94, B131 and B167), we examined the TcdB neutralization capacity in Vero cell assays. At the highest VHH concentration tested (1 μM) none of the antibodies were capable of inhibiting the cytotoxic effects of TcdB on cells at 1 pM.
Table 1. Biophysical properties of anti-TcdB VHHs.
| VHH | Panning source | Ve (mL) | SEC (%)a | Tm (°C)b | ka (M-1 s-1) | kd (s-1) | KD (nM) | Rmax (RU)c | Epitope bin |
|---|---|---|---|---|---|---|---|---|---|
| B26 | solution | 12.97 | 99.8 | 68.3±0.3 | 5.25×106 | 3.45×10−2 | 6.3 | 149 | 4 |
| B35 | solution | 14.07 | 99.8 | 76.2±0.4 | 1.16×106 | 1.05×10−3 | 0.9 | 50 | 1 |
| B39 | solution | 16.05 | 99.8 | 87.2±0.2 | 3.44×107 | 2.56×10−3 | 0.07 | 43 | 1 |
| B45 | solution | 14.84 | 99.4 | 79.5±0.4 | 3.52×106 | 3.84×10−4 | 0.1 | 55 | 1 |
| B46 | solid phase | 13.18 | 99.3 | 61.0±1.4 | 1.53×105 | 9.43×10−3 | 62 | 51 | n.d. |
| B48 | both | 13.07 | 99.7 | 82.5±0.3 | 4.11×106 | 5.71×10−3 | 1.4 | 73 | 3 |
| B52 | both | 12.77 | 99.9 | 65.3±0.7 | 1.29×107 | 1.01×10−2 | 0.8 | 63 | 3 |
| B53 | solid phase | 12.97 | 99.6 | 77.1±0.0 | 4.38×106 | 7.98×10−3 | 1.8 | 72 | 3 |
| B54 | solution | 12.85 | 95.3 | 81.7±0.3 | 3.86×107 | 1.47×10−2 | 0.4 | 65 | 2 |
| B55 | solid phase | 13.35 | 99.8 | 66.6±1.1 | 3.34×105 | 4.93×10−2 | 147 | 142 | n.d. |
| B56 | solid phase | 14.00 | 86.7 | 57.6±0.3 | 5.46×104 | 3.15×10−2 | 576 | 18 | n.d. |
| B65 | solid phase | 13.26 | 96.3 | 84.4±0.2 | 1.32×104 | 2.33×10−3 | 176 | 17 | n.d. |
| B69 | solid phase | 13.11 | 82.1 | 80.9±0.5 | 1.17×107 | 1.50×10−1 | 12.8 | 102 | 5 |
| B71 | solid phase | 12.88 | 85.9 | 63.3±0.3 | 7.76×106 | 9.18×10−2 | 11.8 | 61 | 6 |
| B73 | both | 13.41 | 99.7 | 80.2±0.0 | 1.27×107 | 9.19×10−3 | 0.7 | 63 | 3 |
| B74 | both | 13.62 | 99.6 | 75.0±0.2 | 1.23×107 | 3.58×10−3 | 0.3 | 66 | 3 |
| B76 | solid phase | 12.48 | 99.4 | 84.2±1.1 | 1.38×106 | 2.17×10−1 | 157 | 74 | n.d. |
| B79 | solid phase | 13.48 | 99.1 | 66.9±0.1 | 5.59×106 | 9.72×10−3 | 1.7 | 75 | 3 |
| B85 | solution | 12.94 | 99.4 | 69.7±0.1 | 1.93×107 | 1.14×10−2 | 0.6 | 73 | 2 |
| B86 | solution | 13.03 | 99.6 | 72.9±0.1 | 8.25×106 | 3.98×10−3 | 0.5 | 71 | 3 |
| B92 | solid phase | 12.85 | 98.9 | 78.0±0.1 | 1.51×106 | 8.28×10−1 | 548 | 72 | n.d. |
| B94 | solution | 12.58 | 96.7 | 69.7±0.4 | 6.19×105 | 8.77×10−3 | 14.2 | 385 | 4 |
| B95 | solid phase | 13.97 | 95.1 | 80.8±0.6 | 1.12×107 | 7.05×10−3 | 0.6 | 72 | 3 |
| B96 | solution | 13.17 | 99.7 | 70.5±0.2 | 1.37×106 | 7.33×10−2 | 53.6 | 104 | n.d. |
| B99 | solution | 13.05 | 98.7 | 69.8±2.2 | 6.33×107 | 1.04×10−1 | 1.6 | 76 | 2 |
| B131 | solution | 13.01 | 100 | 69.3±2.3 | 5.47×107 | 1.40×10−1 | 2.6 | 73 | 7 |
| B149 | solution | 13.45 | 97.4 | 83.9±0.0 | 4.12×104 | 2.20×10−2 | 534 | 133 | n.d. |
| B158 | solution | 13.44 | 99.4 | 73.4±0.8 | 3.44×104 | 1.74×10−3 | 50.5 | 67 | 3 |
| B167 | solution | 13.14 | 99.6 | 72.1±0.4 | 1.53×107 | 1.10×10−3 | 0.07 | 76 | 2 |
a% monomer determined by area under the curve
b(mean ± SD)
cObserved Rmax was obtained from a TcdB surface with a theoretical Rmax of 200 RUs; n.d., not determined.
Fig 2. Biophysical characterization of VHHs.
Representative SEC profile (A) and thermal unfolding curve from initial and refolded thermal melts (B). (C) Representative SPR single-cycle kinetics sensorgram showed high-affinity binding of VHHs to biotinylated TcdB1751-2366 immobilized on a CAP sensor chip. (D) Plot comparing KDs of VHHs isolated from solution and solid-phase panning schemes. The four VHHs isolated from both panning schemes were included in the analyses. Bars represent the median KD and a P-value < 0.05 was considered significant (Mann Whitney two-tailed unpaired t-test). (E) Representative sensorgrams demonstrating SPR-based epitope binning. All VHHs were injected at 10x KD concentrations. (F) Summary of the TcdB1751-2366 epitope bins identified in this study by the pool of VHHs tested. The VHHs in each epitope (E) bin are noted and the underlined VHHs represent the highest affinity and/or slowest dissociating antibodies in each bin.
Reformatting VHHs as dimeric molecules
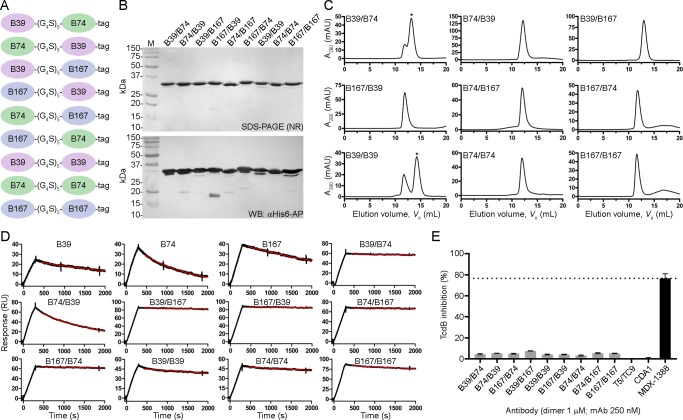
We next generated VHH-VHH dimers using the three highest affinity VHHs that targeted unique epitopes (B39, B74 and B167) to determine if biparatopic designs could impart a measureable level of TcdB neutralization not seen with VHH monomers. All possible combinations of the three antibodies were created including homodimers (Fig 3A), with a standard 25 amino acid linker separating each VHH. Dimers were expressed in E. coli, purified by IMAC with yields ranging from 2.0 to 12.0 mg/L (Fig 3B) and assessed by SEC (Fig 3C) which revealed a predominantly single monodispersed species with the exception of B39/B74 and B39/B39 dimers that showed higher order aggregates. VHH-VHH dimer Tms ranged from 64.4 to 73.1°C. SPR off-rate analysis demonstrated nearly irreversible bivalent binding to TcdB1751-2366 surfaces for many of the dimers, approaching the instrument limit of detection (Table 2, Fig 3D). Vero cell neutralization assays using the dimers showed minor TcdB inhibition with all nine formats tested ranging from 3.2% (B39/B39) to 7.5% (B167/B39) maximum inhibition (Table 2, Fig 3E). The negative control T5/TC9 dimer did not inhibit TcdB and the benchmark control mAb MDX-1388 reached a maximum inhibition of 76.6%.
Fig 3. Generation and characterization of VHH-VHH dimers.
(A) Cartoon diagram of VHH-VHH dimers separated by a 25 amino acid linker (G4S)5 and containing a C-terminal His6 “tag”. (B) Dimers were expressed, purified and analyzed by non-reducing (NR) SDS-PAGE and probed by α-His6-AP in Western blot (WB). M, protein molecular mass marker. (C) SEC profiles of dimers. Asterisks denote the peaks that were selected for SPR analysis when aggregates were present. (D) SPR sensorgrams showing dissociation phases of VHH-VHHs and parent monomers. (E) TcdB neutralization with VHH-VHH dimers. The final TcdB concentration of 3–10 pM and VHH-VHH concentration of 1 μM was co-incubated with Vero cell monolayers for 72 h before addition of WST-1 cytotoxicity reagent. MDX-1388 was used at 250 nM. Neutralization values are presented as mean ± SD from n = 3 independent experiments.
Table 2. Biophysical properties of anti-TcdB VHH-VHH dimers.
| VHH- VHH | Yield (mg/ L)a | Ve (mL)b | Tm (°C)c | kd (s-1) | Maximum TcdB neutralization (%)c,d |
|---|---|---|---|---|---|
| B39/B74 | 9.2 | 13.2 | 71.6 ± 0.2 | 2.2 × 10−5 | 4.4 ± 0.4 |
| B74/B39 | 3.6 | 12.2 | 70.9 ± 1.6 | 5.9 × 10−4 | 5.3 ± 0.1 |
| B39/B167 | 9.0 | 13.0 | 73.1 ± 1.7 | 2.4 × 10−5 | 4.9 ± 0.3 |
| B167/B39 | 2.0 | 11.9 | 67.7 ± 1.4 | 1.7 × 10−5 | 7.5 ± 0.1 |
| B74/B167 | 5.8 | 12.0 | 64.4 ± 2.1 | 1.4 × 10−5 | 4.2 ± 0.2 |
| B167/B74 | 2.4 | 11.7 | 66.5 ± 0.6 | 2.4 × 10−5 | 4.1 ± 0.2 |
| B39/B39 | 12.0 | 14.2 | 79.5 ± 3.4 | 9.7 × 10−5 | 3.2 ± 0.3 |
| B74/B74 | 2.2 | 12.0 | 70.4 ± 3.3 | 6.3 × 10−5 | 5.4 ± 0.3 |
| B167/B167 | 6.8 | 11.6 | 67.5 ± 2.3 | 6.0 × 10−5 | 5.1 ± 0.2 |
| MDX-1388 | - | n.d. | n.d. | n.d. | 76.6 ± 4.4e |
aPurification yield from 1 L E. coli culture
bVe from Superdex 200
c(mean ± SD)
d1 μM or
e250 nM antibody + 3 pM TcdB vero cell cytotoxicity (72 h); n.d., not determined.
Reformatting VHHs as Fc fusions
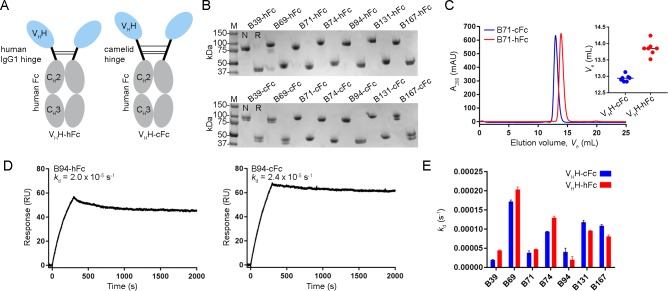
Given the modest level of TcdB inhibition seen with the VHH-VHH dimers, we next explored if construction of larger molecules may lead to greater TcdB neutralizing potency. VHH-Fc fusions (Fig 4A) were constructed using each of the seven high-affinity VHHs from distinct epitope bins (B39, B69, B71, B74, B94, B131 and B167). The designs consisted of a set of VHH-Fcs with a 15-residue human IgG1 hinge (EPKSCDKTHTCPPCP) and another set of complementary molecules with a 35 residue camel/llama γ2a hinge (EPKIPQPQPKPQPQPQPQPKPQPKPEPECTCPKCP), to explore the possible advantages a longer, more flexible hinge may have on TcdB inhibition. The VHH-Fcs, denoted “VHH-hFc” for molecules containing the human hinge and “VHH-cFc” for molecules containing the camel hinge, were expressed in mammalian cells and purified by protein A with yields ranging from 2.5 to 21.6 mg/100 mL of culture (Table 3, Fig 4B). SEC analyses of VHH-Fcs revealed single, monodispersed peaks and consistently showed VHHs with camel hinges eluting earlier, most likely due to their larger hydrodynamic radius (Table 3, Fig 4C and S5A Fig). SPR-determined off-rates demonstrated nearly irreversible bivalent binding to TcdB1751-2366 surfaces (Table 3, Fig 4D and S5B Fig). Comparison of the off-rates (kds) of VHH-Fcs with human or camel hinges did not reveal significant differences in their ability to bind TcdB1751-2366 (Fig 4E).
Fig 4. Generation and characterization of VHH-Fcs with human or camelid hinges.
(A) Cartoon diagram illustrating the two VHH-Fc formats used with varying hinge length and composition. (B) Purified VHH-Fcs were analyzed by SDS-PAGE under non-reducing (N) and reducing (R) conditions. (C) Representative VHH-Fc SEC elution profile from a Superdex 200 column. Inset, plot of SEC elution volumes for all VHH-cFcs and VHH-hFcs. (D) Representative SPR sensorgrams showing 30 min (1800 s) dissociations of VHH-Fc fusions flowing over immobilized TcdB1751-2366 surfaces. (E) Dissociation rate comparison of VHH-Fcs with human or camelid hinges.
Table 3. Biophysical properties of anti-TcdB VHH-Fc fusions.
| VHH-Fc/mAb | Hinge | Yield (mg/ 100 mL)a | Ve (mL)b | kd (s-1) | Maximum TcdB neutralization (%)c,d |
|---|---|---|---|---|---|
| B39-hFc | human | 5.7 | 17.64 | 4.5 × 10−5 | 0.0 |
| B69-hFc | human | 13 | 14.24 | 2.0 × 10−4 | 18.0 ± 10.2 |
| B71-hFc | human | 6 | 13.91 | 4.7 × 10−5 | 11.0 ± 7.0 |
| B74-hFc | human | 4.4 | 13.82 | 1.3 × 10−4 | 26.3 ± 4.5 |
| B94-hFc | human | 12.2 | 13.52 | 2.0 × 10−5 | 55.9 ± 22.4 |
| B131-hFc | human | 14.6 | 13.70 | 9.6 × 10−5 | 18.0 ± 13.4 |
| B167-hFc | human | 2.5 | 13.85 | 8.1 × 10−5 | 37.5 ± 2.7 |
| B39-cFc | camelid | 7.9 | 15.07 | 2.0 × 10−5 | 0.1 ± 0.1 |
| B69-cFc | camelid | 9.1 | 13.13 | 2.0 × 10−4 | 20.2 ± 5.7 |
| B71-cFc | camelid | 11.3 | 12.98 | 4.7 × 10−5 | 8.9 ± 5.5 |
| B74-cFc | camelid | 6.8 | 12.95 | 9.4 × 10−5 | 22.3 ± 4.2 |
| B94-cFc | camelid | 11.8 | 12.84 | 2.4 × 10−5 | 61.2 ± 1.3 |
| B131-cFc | camelid | 21.6 | 12.84 | 9.6 × 10−5 | 18.7 ± 12.3 |
| B167-cFc | camelid | 9.2 | 12.90 | 8.1 × 10−5 | 22.8 ± 2.9 |
| MDX-1388 | human | - | n.d. | n.d. | 70.7 ± 13.9 |
aPurification yield from 100 mL HEK293 culture
bVe from Superdex 200
c(mean ± SD)
d250 nM antibody + 500 fM TcdB vero cell cytotoxicity (72 h); n.d., not determined.
TcdB neutralization with VHH-Fcs
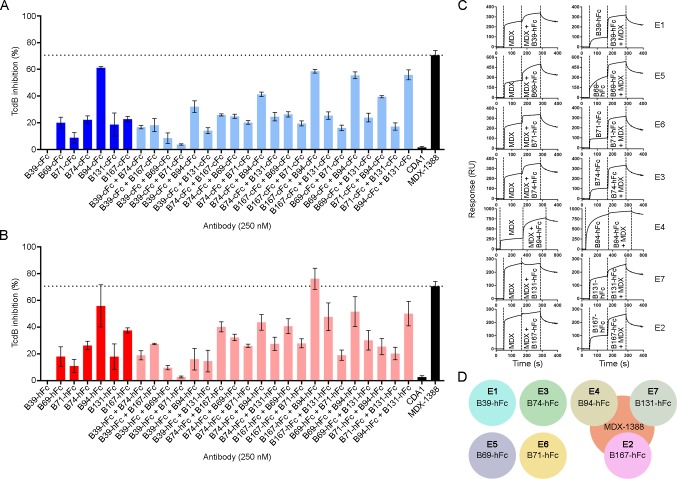
The neutralization potencies of VHH-Fcs were compared to the benchmark mAb bezlotoxumab (MDX-1388) in TcdB inhibition assays using 500 fM TcdB and 250 nM antibody (Table 3, Fig 5A and 5B). MDX-1388 showed a maximum TcdB inhibition of 70.7% compared to VHH-Fcs that ranged from 8.9% (B71-cFc) to 61.2% (B94-cFc). There were essentially no differences between the neutralizing capacities of VHH-Fcs containing the short IgG1 hinge or the longer camel hinge, mirroring the near identical off-rates determined by SPR. Neither format of B39-Fc neutralized TcdB. CDA1 (an anti-TcdA mAb) [36] was included as a negative control and did not neutralize TcdB as expected. To examine possible synergistic effects from combinations of VHH-Fcs on TcdB inhibition, pairs of VHH-Fcs (125 nM + 125 nM) were examined in neutralization assays (S2 Table, Fig 5A and 5B). The VHH-Fc pair of B94-hFc + B167-hFc achieved a maximum TcdB inhibition of 76.2%, slightly exceeding the neutralization potency of each VHH-Fc alone and that of MDX-1388. In pairs containing B39-Fc, the non-neutralizing antibody, overall neutralization was reduced by approximately 50% of the maximum inhibiting potency of the second antibody partner, reflecting the fact that 50% less inhibitory antibody was present (Fig 5A and 5B).
Fig 5. TcdB neutralization assays with VHH-Fcs.
(A) TcdB neutralization with VHH-Fcs containing a camelid hinge. (B) TcdB neutralization with VHH-Fcs containing a human hinge. In (A) and (B), the final TcdB concentration of 500 fM and final antibody concentration of 250 nM (single antibody) or 125 + 125 nM (pairs) were co-incubated with Vero cell monolayers for 72 h before addition of WST-1 cytotoxicity reagent. Neutralization values are presented as mean ± SD from n = 4 independent experiments. (C) Sensorgrams showing SPR-based epitope binning of MDX-1388 mAb with each VHH-hFc, in both orientations, with antibodies injected at 25–50 × KD concentrations over TcdB1751-2366 surfaces. Epitope bins corresponding to each VHH-hFc are noted. (D) Summary of VHH-Fc reactivity to TcdB1751-2366 illustrating that several VHH-Fcs bind TcdB at sites distinct from MDX-1388, while others bind TcdB at regions that partially overlap with MDX-1388.
VHH-Fc competition with bezlotoxumab
To determine if our panel of neutralizing VHH-Fcs recognized similar or unique TcdB epitopes from that of bezlotoxumab we performed SPR co-injection experiments (Fig 5C and 5D). B39-hFc, B69-hFc, B71-hFc and B74-hFc did not compete with MDX-1388 in either injection sequence, indicating that the four VHH-Fcs bind sites on TcdB completely independent of the MDX-1388 binding site. This is unsurprising for B39-hFc given the inability of this antibody to neutralize TcdB. The other three VHHs were previously shown to bind unique epitopes as monomers (Fig 2F) suggesting B69-hFc, B71-hFc and B74-hFc recognize three novel TcdB epitopes that support neutralizing antibodies. The results of B94-hFc, B131-hFc and B167-hFc binning with MDX-1388 revealed significant overlap in TcdB binding patterns. When MDX-1388 was injected first, B94-hFc was partially blocked by pre-bound MDX-1388 and B131-hFc and B167-hFc were completely blocked by pre-bound MDX-1388. In the opposite orientation, MDX-1388 binding was completed blocked by pre-bound B94-hFc, and partially blocked by pre-bound B131-hFc or B167-hFc. Collectively this data suggests the most potent TcdB neutralizing VHH-Fcs (B94-hFc, B167-cFc) bind TcdB at sites that partially overlap with the MDX-1388 binding site at the N-terminal end of the CROPs domain [27].
Discussion
In this work we set out to identify high-affinity VHHs capable of neutralizing C. difficile TcdB. We previously failed to identify monomeric VHH neutralizers when immunizing with a small C-terminal fragment of the CROPs domain [10]. Here our expanded immunogen design containing a portion of the central delivery domain and the entire CROPs domain yielded a number of VHHs that were capable of TcdB inhibition when formatted as dimers and more so as Fc fusions. It should be noted that our previous TcdB-binding VHHs [10] were not tested as VHH-Fc fusions and may have been capable of TcdB inhibition, although their affinities were considerably weaker than the VHHs isolated in this work. Once again the monomeric VHHs did not inhibit TcdB, suggesting a steric element that is required for TcdB inhibition when targeting the CROPs domain. Consistent with our findings, Yang et al isolated several TcdB-binding VHHs and found five high-affinity VHHs targeting the C-terminal CROPs domain that failed to inhibit TcdB cytotoxicity while those targeting the N-terminal GTD domain were potent neutralizers [13].
Our work is not the first to report TcdB-inhibiting VHHs binding the CROPs domain. Andersen et al isolated several inhibitory monomeric VHHs binding this domain, indicating that TcdB neutralization is possible with monomeric antibodies [15]. It is not clear how they successfully identified VHHs that recognize critical epitope(s) for TcdB function/cell binding, that were not identified here, in our early work [10] or by [13], while employing a similar immunization strategy. Interestingly two of their non-neutralizing CROPs-binding VHHs were converted into inhibitory antibodies when displayed on the surfaces of lactobacilli [15], again pointing to a large steric element in imparting TcdB inhibition with antibodies binding this region of the toxin.
Beyond VHHs there are several potent TcdB-neutralizing mAbs that have been well studied. The most advanced anti-TcdB mAb is bezlotoxumab which received FDA approval for recurrent C. difficile infection in 2016. This antibody was originally isolated in a study reported by [36] and demonstrated to bind the C-terminal receptor binding domain (CROPs domain). Structural studies performed by [27] revealed the precise binding site of the mAb lies within the first two of four CROP repeat domains, with each CROP domain consisting of three short repeating units (SRs) followed by one long repeating unit (LR) and two more SRs. SPR binding data showed bezlotoxumab bound two distinct epitopes which was later supported by X-ray crystal structures and homology modeling showing two Fab fragments binding adjacent to each other in the N-terminal half of the CROPs domain (aa 1834–2101). Given bezlotoxumab neutralizes TcdB by preventing binding to mammalian cells [27,37], one can assume that some of our most potent VHH-Fc fusions (B94-Fc, B131-Fc and B167-Fc) neutralize TcdB in a similar manner given that they partially overlap with bezlotoxumab for TcdB binding. Whether the exact mechanism of inhibition is due to blocking putative carbohydrate binding site interactions with a host-cell receptor, or by steric effects precluding the central/delivery domain from making contacts with FZD2, PVRL3 or CSPG4 receptors is unknown. It is interesting to note that the observed Rmax of B94 in SPR experiments was considerably higher than other antibodies as both a VHH monomer (Table 1) and as a VHH-Fc in binning experiments (Fig 5C), suggesting that B94 may bind to repeating TcdB CROPs domain epitopes. Supporting this idea is the fact that B94-Fc was the only VHH-Fc to completely block MDX-1388 binding when B94-Fc was bound to TcdB first. If a repeating epitope is bound by B94-Fc, it would suggest a similar mechanism of inhibition to that of MDX-1388 and that affinity improvements may lead to greater neutralizing potency since the monovalent affinity of B94 is relatively weak (KD = 14 nM) compared to a Fab fragment from MDX-1388 (KD = 19 pM or 370 pM; depending on the TcdB epitope) [27].
For the other three neutralizing VHH-Fcs described here (B69-Fc, B71-Fc and B74-Fc) their location for toxin binding and subsequently their mechanism for toxin inhibition also remain unknown. B39, which failed to neutralize TcdB as an Fc fusion and did not overlap with bezlotoxumab as expected, was previously [12] co-crystalized with a C-terminal fragment of the TcdB CROPs domain (aa 2248–2367 of TcdB from strain 10463) and definitively showed only recognition of a single, non-repeating TcdB epitope. This may suggest that antibodies binding at a distance from the central delivery/translocation domain have minimal effects on TcdB inhibition compared to antibodies binding nearby in the N-terminal half of the CROPs domain. Elsewhere, [28] have successfully identified four inhibitory mAbs targeted to the CROPs domain of TcdB that were neutralizers alone and to a greater extent when combined in pairs. Additionally, TcdB-neutralizing mAbs recognizing the C-terminal GT domain of TcdB have proven to be both potent inhibitors and effective in in vivo protection assays [38,39,40].
There were several other interesting observations of note from this work. The conventional IgG (cIgG) fraction obtained from llama serum post immunization with TcdB1751-2366 was capable of inhibiting TcdB in cytotoxicity assays more efficiently than the three heavy-chain IgG (hcIgG) fractions. While the cIgG binding titer for TcdB was higher than the best hcIgG fraction by approximately 10-fold, which is a typical binding pattern we have observed in several other immunization campaigns, there were dramatic differences in the inhibition pattern seen between G1 (hcIgG) and G2 (cIgG) fractions. It is possible that a greater agglutination mechanism with camelid cIgGs compared to camelid hcIgGs is at play here and could be due to the physical distance separating the binding arms of each antibody format. We hypothesize that the more compact hcIgG footprint may bias the polyclonal pool toward intra-toxin binding events and that the greater distance afforded to cIgGs promotes both intra-toxin and inter-toxin binding events, leading to increased agglutination and ultimately greater TcdB inhibition. Supporting this is the fact the A2 (hcIgG) fraction, which showed a much weaker TcdB binding titer than the G1 fraction, possessed considerably greater neutralizing potency likely driven by IgM antibodies found as contaminants in the fractionated serum.
We performed panning experiments in solution using off-rate based selection and this method produced statistically higher affinity VHHs compared to panning on immobilized antigen. This method of selection was also the source of the VHH-Fcs with the highest neutralizing potency, even though two of the three most potent TcdB inhibitors did not possess the highest overall monovalent VHH affinities. It is probable that the solution panning scheme presented the randomly biotinylated TcdB in a more native-like conformation and made more areas of the protein available for VHH binding compared to selection of VHHs on TcdB-coated wells. This may explain the higher average binding affinity and greater neutralization potency of VHHs isolated by the solution panning approach. We also examined the use of a longer camel hinge in place of the human IgG1 hinge to present the VHH in a more natural context when tethered to the Fc domain. While SEC profiles clearly showed that VHH-Fcs with camel hinges eluted earlier and thereby suggest a larger hydrodynamic volume, similar dissociation rate constants and TcdB neutralizing potency demonstrated there was no clear benefit to using the longer hinge. We do not completely understand why this was the case but speculate that the camel hinge only moderately expands the binding distance between VHH arms, still less than the footprint of a cIgG, and that neutralization is dependent on combination of factors including the location, geometry and accessibility of TcdB epitopes.
In summary, the VHHs described here were potent TcdB neutralizing antibodies on par with bezlotoxumab when formatted as Fc fusions. We envision creating even more effective TcdB-neutralizing agents through optimization of affinities and binding geometries, such as through structure-guided biparatopic designs. Combining anti-TcdB biparatopic VHH-VHH designs with TcdA-neutralizing VHHs onto a human Fc scaffold would allow for the generation of ultra-potent toxin inhibitors in a single antibody format, similar to approaches described previously [13,17], while maintaining the steric requirements for TcdB neutralization and long serum half-life. The in vivo efficacy previously demonstrated with multivalent designs that include linking anti-TcdA and anti-TcdB VHHs suggests the inclusion of GTD-targeting anti-TcdB antibodies is critical [13,17,18]. Our results make a case for targeting the central delivery and CROPs domains with VHHs. Whether including CROPs-targeting VHHs in these designs will be as potent as those targeting the GTD region remains to be seen. In addition, finer epitope mapping of the VHH-Fcs that bound TcdB at regions distinct from bezlotoxumab will reveal if these antibodies recognize the central delivery domain or the CROPs domain and the nature of these unique inhibitory epitopes. Finally, the VHHs described here will serve as useful additions to the reagent toolkit for further refinement of the mechanism of TcdB-host cell interactions.
Supporting information
VHHs were passed over a Superdex 75 column at a flow rate of 0.5 mL/min in HBS-EP buffer. Molecular mass standards are shown. The percent monomer was calculated from the peak area under the curve. Ve, elution volume.
(TIF)
(A) Thermal unfolding of VHHs (50 μg/mL, 3.2 μM) was performed in phosphate buffer and measured in a 5 mm cuvette at 215 nm. VHHs were allowed to cool at 25°C for 3 h before the second thermal unfolding (refolded melting curve) was performed. (B) Voltage comparing two VHHs (B54 and B99) that aggregate as a consequence of unfolding versus one VHH that does not (B53).
(TIF)
Black lines represent raw data and red lines represent 1:1 binding model fits. Rate constants and affinities are shown for each antibody. The irrelevant VHH served as a negative control. For experimental conditions see Materials and methods.
(TIF)
The first VHH was injected at 10× KD concentration followed immediately by injection of a mixture of the first VHH + second VHH at 10× KD concentration. For experimental conditions see Materials and methods.
(TIF)
(A) SEC chromatograms of VHH-Fcs on a Superdex 200 column at a flow rate of 0.5 mL/min in HBS-EP buffer. (B) SPR sensorgrams illustrating the dissociation of 1 nM VHH-Fcs from TcdB1751-2366 surfaces. For experimental conditions see Materials and methods.
(TIF)
(PDF)
(PDF)
Acknowledgments
We thank Kenneth Ng (University of Calgary) for providing the recombinant TcdB fragment, Shreya Jain (NRC) for assisting in protein expression and Yves Durocher and Denis L’Abbé (NRC) for producing the CDA1 and MDX-1388 control mAbs.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was funded by the National Research Council Canada. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dubberke ER, Olsen MA (2012) Burden of Clostridium difficile on the healthcare system. Clin Infect Dis 55 Suppl 2: S88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussack G, Tanha J (2016) An update on antibody-based immunotherapies for Clostridium difficile infection. Clin Exp Gastroenterol 9: 209–224. 10.2147/CEG.S84017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Napolitano LM, Edmiston CE Jr. (2017) Clostridium difficile disease: Diagnosis, pathogenesis, and treatment update. Surgery 162: 325–348. 10.1016/j.surg.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 4.Hopkins RJ, Wilson RB (2018) Treatment of recurrent Clostridium difficile colitis: a narrative review. Gastroenterol Rep 6: 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aktories K, Schwan C, Jank T (2017) Clostridium difficile toxin biology. Ann Rev Microbiol 71: 281–307. [DOI] [PubMed] [Google Scholar]
- 6.Chandrasekaran R, Lacy DB (2017) The role of toxins in Clostridium difficile infection. FEMS Microbiol Rev 41: 723–750. 10.1093/femsre/fux048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussack G, Tanha J (2010) Toxin-specific antibodies for the treatment of Clostridium difficile: current status and future perspectives. Toxins 2: 998–1018. 10.3390/toxins2050998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullard A (2016) FDA approves antitoxin antibody. Nat Rev Drug Discov 15: 811. [DOI] [PubMed] [Google Scholar]
- 9.Navalkele BD, Chopra T (2018) Bezlotoxumab: an emerging monoclonal antibody therapy for prevention of recurrent Clostridium difficile infection. Biologics 12: 11–21. 10.2147/BTT.S127099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussack G, Arbabi-Ghahroudi M, van Faassen H, Songer JG, Ng KK, MacKenzie R, et al. (2011) Neutralization of Clostridium difficile toxin A with single-domain antibodies targeting the cell receptor binding domain. J Biol Chem 286: 8961–8976. 10.1074/jbc.M110.198754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussack G, Keklikian A, Alsughayyir J, Hanifi-Moghaddam P, Arbabi-Ghahroudi M, van Faassen H, et al. (2012) A V(L) single-domain antibody library shows a high-propensity to yield non-aggregating binders. Protein Eng Des Sel 25: 313–318. 10.1093/protein/gzs014 [DOI] [PubMed] [Google Scholar]
- 12.Murase T, Eugenio L, Schorr M, Hussack G, Tanha J, Kitova EN, et al. (2013) Structural basis for antibody Recognition in the receptor-binding domains of toxins A and B from Clostridium difficile. J Biol Chem 289: 2331–2343. 10.1074/jbc.M113.505917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Schmidt D, Liu W, Li S, Shi L, Sheng J, et al. (2014) A novel multivalent, single-domain antibody targeting TcdA and TcdB prevents fulminant Clostridium difficile infection in mice. J Infect Dis 210: 964–972. 10.1093/infdis/jiu196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shkoporov AN, Khokhlova EV, Savochkin KA, Kafarskaia LI, Efimov BA (2015) Production of biologically active scFv and VHH antibody fragments in Bifidobacterium longum. FEMS Microbiol Lett 362: fnv083. [DOI] [PubMed] [Google Scholar]
- 15.Andersen KK, Strokappe NM, Hultberg A, Truusalu K, Smidt I, Mikelsaar RH, et al. (2015) Neutralization of Clostridium difficile toxin B mediated by engineered lactobacilli producing single domain antibodies. Infect Immun 84: 395–406. 10.1128/IAI.00870-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unger M, Eichhoff AM, Schumacher L, Strysio M, Menzel S, Schwan C, et al. (2015) Selection of nanobodies that block the enzymatic and cytotoxic activities of the binary Clostridium difficile toxin CDT. Sci Rep 5: 7850 10.1038/srep07850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt DJ, Beamer G, Tremblay JM, Steele JA, Kim HB, Wang Y, et al. (2016) A tetraspecific VHH-based neutralizing antibody modifies disease outcome in three animal models of Clostridium difficile infection. Clin Vaccine Immunol 23: 774–784. 10.1128/CVI.00730-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Shi L, Yu H, Zhang Y, Chen K, Saint Fleur A, et al. (2016) Intravenous adenovirus expressing a multi-specific, single-domain antibody neutralizing TcdA and TcdB protects mice from Clostridium difficile infection. Pathog Dis 74: ftw078 10.1093/femspd/ftw078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulea T, Hussack G, Ryan S, Tanha J, Purisima EO (2018) Application of assisted design of antibody and protein therapeutics (ADAPT) improves efficacy of a Clostridium difficile toxin A single-domain antibody. Sci Rep 8: 2260 10.1038/s41598-018-20599-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frère JM, Kinne J, et al. (2001) Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother 45: 2807–2812. 10.1128/AAC.45.10.2807-2812.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muyldermans S (2013) Nanobodies: natural single-domain antibodies. Ann Rev Biochem 82: 775–797. 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- 22.Iezzi ME, Policastro L, Werbajh S, Podhajcer O, Canziani GA (2018) Single-domain antibodies and the promise of modular targeting in cancer imaging and treatment. Front Immunol 9: 273 10.3389/fimmu.2018.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen P, Tao L, Wang T, Zhang J, He A, Lam KH, et al. (2018) Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B. Science 360: 664–669. 10.1126/science.aar1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao L, Zhang J, Meraner P, Tovaglieri A, Wu X, Gerhard R, et al. (2016) Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538: 350–355. 10.1038/nature19799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaFrance ME, Farrow MA, Chandrasekaran R, Sheng J, Rubin DH, Lacy DB (2015) Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc Natl Acad Sci U S A 112: 7073–7078. 10.1073/pnas.1500791112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan P, Zhang H, Cai C, Zhu S, Zhou Y, Yang X, et al. (2015) Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res 25: 157–168. 10.1038/cr.2014.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orth P, Xiao L, Hernandez LD, Reichert P, Sheth PR, Beaumont M, et al. (2014) Mechanism of action and epitopes of Clostridium difficile toxin B-neutralizing antibody bezlotoxumab revealed by X-ray crystallography. J Biol Chem 289: 18008–18021. 10.1074/jbc.M114.560748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies NL, Compson JE, Mackenzie B, O'Dowd VL, Oxbrow AK, Heads JT, et al. (2013) A mixture of functionally oligoclonal humanized monoclonal antibodies that neutralize Clostridium difficile TcdA and TcdB with high levels of in vitro potency shows in vivo protection in a hamster infection model. Clin Vaccine Immunol 20: 377–390. 10.1128/CVI.00625-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry KA, Tanha J, Hussack G (2015) Identification of cross-reactive single-domain antibodies against serum albumin using next-generation DNA sequencing. Protein Eng Des Sel 28: 379–383. 10.1093/protein/gzv039 [DOI] [PubMed] [Google Scholar]
- 30.Baral TN, MacKenzie R, Arbabi Ghahroudi M (2013) Single-domain antibodies and their utility. Curr Protoc Immunol 103: Unit 2 17. [DOI] [PubMed] [Google Scholar]
- 31.Hussack G, Hirama T, Ding W, Mackenzie R, Tanha J (2011) Engineered single-domain antibodies with high protease resistance and thermal stability. PLoS One 6: e28218 10.1371/journal.pone.0028218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossotti MA, Gonzalez-Techera A, Guarnaschelli J, Yim L, Camacho X, Fernández M, et al. (2015) Increasing the potency of neutralizing single-domain antibodies by functionalization with a CD11b/CD18 binding domain. MAbs 7: 820–828. 10.1080/19420862.2015.1068491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durocher Y, Perret S, Kamen A (2002) High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res 30: E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henry KA, Kandalaft H, Lowden MJ, Rossotti MA, van Faassen H, Hussack G, et al. (2017) A disulfide-stabilized human VL single-domain antibody library is a source of soluble and highly thermostable binders. Mol Immunol 90: 190–196. 10.1016/j.molimm.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 35.Conrath KE, Wernery U, Muyldermans S, Nguyen VK (2003) Emergence and evolution of functional heavy-chain antibodies in Camelidae. Dev Comp Immunol 27: 87–103. [DOI] [PubMed] [Google Scholar]
- 36.Babcock GJ, Broering TJ, Hernandez HJ, Mandell RB, Donahue K, Boatright N, et al. (2006) Human monoclonal antibodies directed against toxins A and B prevent Clostridium difficile-induced mortality in hamsters. Infect Immun 74: 6339–6347. 10.1128/IAI.00982-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z, Ramsey J, Hamza T, Zhang Y, Li S, Yfantis HG, et al. (2015) Mechanisms of protection against Clostridium difficile infection by the monoclonal antitoxin antibodies actoxumab and bezlotoxumab. Infect Immun 83: 822–831. 10.1128/IAI.02897-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marozsan AJ, Ma D, Nagashima KA, Kennedy BJ, Kang YK, Arrigale RR, et al. (2012) Protection against Clostridium difficile infection with broadly neutralizing antitoxin monoclonal antibodies. J Infect Dis 206: 706–713. 10.1093/infdis/jis416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anosova NG, Cole LE, Li L, Zhang J, Brown AM, Mundle S, et al. (2015) A combination of three fully human toxin A- and toxin B-specific monoclonal antibodies protects against challenge with highly virulent epidemic strains of Clostridium difficile in the hamster model. Clin Vaccine Immunol 22: 711–725. 10.1128/CVI.00763-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kroh HK, Chandrasekaran R, Rosenthal K, Woods R, Jin X, Ohi MD, et al. (2017) Use of a neutralizing antibody helps identify structural features critical for binding of Clostridium difficile toxin TcdA to the host cell surface. J Biol Chem 292: 14401–14412. 10.1074/jbc.M117.781112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
VHHs were passed over a Superdex 75 column at a flow rate of 0.5 mL/min in HBS-EP buffer. Molecular mass standards are shown. The percent monomer was calculated from the peak area under the curve. Ve, elution volume.
(TIF)
(A) Thermal unfolding of VHHs (50 μg/mL, 3.2 μM) was performed in phosphate buffer and measured in a 5 mm cuvette at 215 nm. VHHs were allowed to cool at 25°C for 3 h before the second thermal unfolding (refolded melting curve) was performed. (B) Voltage comparing two VHHs (B54 and B99) that aggregate as a consequence of unfolding versus one VHH that does not (B53).
(TIF)
Black lines represent raw data and red lines represent 1:1 binding model fits. Rate constants and affinities are shown for each antibody. The irrelevant VHH served as a negative control. For experimental conditions see Materials and methods.
(TIF)
The first VHH was injected at 10× KD concentration followed immediately by injection of a mixture of the first VHH + second VHH at 10× KD concentration. For experimental conditions see Materials and methods.
(TIF)
(A) SEC chromatograms of VHH-Fcs on a Superdex 200 column at a flow rate of 0.5 mL/min in HBS-EP buffer. (B) SPR sensorgrams illustrating the dissociation of 1 nM VHH-Fcs from TcdB1751-2366 surfaces. For experimental conditions see Materials and methods.
(TIF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.