Summary
mRNA interferases are sequence-specific endoribonucleases encoded by toxin-antitoxin (TA) systems in bacterial genomes. Previously, we demonstrated that Mycobacterium tuberculosis contains at least seven genes encoding MazF homologues (MazF-mt1 to –mt7) and determined cleavage specificities for MazF-mt1 and MazF-mt6. Here we have developed a new general method for the determination of recognition sequences longer than three bases for mRNA interferases with the use of phage MS2 RNA as a substrate and CspA, an RNA chaperone, which prevents the formation of secondary structures in the RNA substrate. Using this method, we determined that MazF-mt3 cleaves RNA at UUˇCCU or CUˇCCU and MazF-mt7 at UˇCGCU (ˇindicates the cleavage site). Since pentad sequence recognition is more specific than those of previously characterized mRNA interferases, bioinformatics analysis was carried out to identify M. tuberculosis mRNAs that may be resistant to MazF-mt3 and MazF-mt7 cleavage. The pentad sequence was found to be significantly underrepresented in several genes, including members of the PE and PPE families, large families of proteins that play a role in tuberculosis immunity and pathogenesis. These data suggest that MazF-mt3 and MazF-mt7 or other mRNA interferases that target longer RNA sequences may alter protein expression though differential mRNA degradation, a regulatory mechanism that may allow adaptation to environmental conditions, including those encountered by pathogens such as M. tuberculosis during infection.
Introduction
mRNA interferases are sequence-specific endoribonucleases encoded by toxin-antitoxin systems that are present in a wide range of bacterial genomes (Zhang et al., 2003). These endoribonucleases usually have specific cleavage sites within single-strand RNA; for example, E. coli MazF specifically cleaves at ACA sequences (Zhang et al., 2005), ChpBK at ACY sequences (Y is U, A, or G) (Zhang et al., 2005), PemK from plasmid R100 at UAH sequences (where H is C, A, or U) (Zhang et al., 2004), and MazF-mt1 and MazF-mt6 from M. tuberculosis at UAC and U rich regions, respectively (Zhu et al., 2006). Recently, a MazF homologue from Staphylococcus aureus was reported to cleave at VUUV’ (where V and V’ are A, C, or G and may or may not be identical) (Fu et al., 2007).
The overproduction of MazF in E. coli causes complete cell growth arrest, resulting in a quasi-dormant state in which the cells still retain full capacity for protein synthesis (Suzuki et al., 2005). The genes for MazF homologues are usually co-expressed with their cognate antitoxin genes, encoding MazE, and appear to function primarily to regulate cell growth under stress conditions. Interestingly, a subset of genes encoding MazF homologues, are not co-expressed with their cognate antitoxins; for example, Myxococcus xanthus, a developmental soil-dwelling bacterium, contains a solitary mazF gene, whose developmental expression is essential for obligatory lysis of cells during differentiation to form fruiting bodies (Nariya et al., 2008). Some bacteria contain more than one MazF homologue. As we recently demonstrated there are 60 TA systems encoded by the Mycobacterium tuberculosis genome, of which at least seven are MazF homologues, suggesting that these MazF homologues may have individual roles in the physiology of this pathogen (Zhu et al., 2006). Interestingly in contrast to M. tuberculosis, the non-pathogenic mycobacterium, M. smegmatis has only two TA systems (mazEF and phd/doc) in its genome (Pandey and Gerdes, 2005).
Our previous attempts to determine the cleavage specificities of some of the MazF homologues of M. tuberculosis failed. One possible reason for this could be a low frequency of toxin-specific target sequences, which could result from differences in GC content and/or more stringent (longer) sequence requirements for these toxins. For this reason, in the present paper, we developed a general method to determine the cleavage-site specificity for mRNA interferases using bacteriophage MS2 RNA. This commercially available RNA is 3569 bases in length, and sufficiently complex to allow determination of the cleavage-site specificity for sites that are longer than three-bases. However, MS2 RNA contains extensive secondary structures (more than 70%; (Chon et al., 2002)) and has a relatively high melting temperature (58°C). Therefore, this RNA is highly resistant to endoribonucleases or mRNA interferases, which cleave only single-stranded regions of RNA. In order to make the entire sequence of the RNA susceptible to mRNA interferases, we sought to facilitate melting of secondary structures in the RNA. For this purpose, we used CspA, the major cold-shock protein of E. coli, which functions as an RNA chaperone (Jiang et al., 1997).
Using this method, we identified the cleavage sites for MazF-mt3 and MazF-mt7 to be CUˇCCU /UUˇCCU and UˇCGCU (ˇ indicates the cleavage site), respectively. This approach enabled us to detect almost all the cleavage sites for MazF-mt3 as well as MazF-mt7, demonstrating the sensitivity and general applicability of the method for identifying cleavage sites targeted by sequence-specific endoribonucleases.
Results
Development of a general method to determine the cleavage specificity of mRNA interferases using CspA, an RNA chaperone —
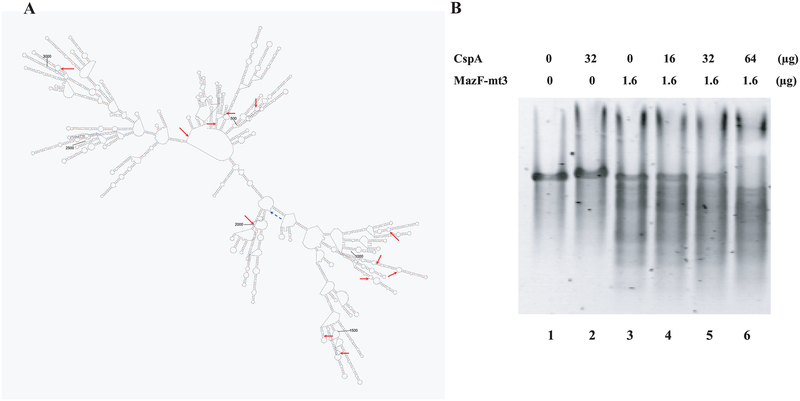
MazF-mt3 is a M. tuberculosis homologue of E. coli MazF. Previously we demonstrated that induction of MazF-mt3 causes cell growth inhibition in E. coli (Zhu et al., 2006). We attributed this toxic effect of MazF-mt3 to its mRNA interferase activity; however we were not able to determine its cleavage specificity, possibly due to its highly specific cleavage sequences. Particularly for the determination of the cleavage specificity of mRNA interferases that recognize a specific sequence longer than three bases, it is essential to use an RNA substrate that is long enough to cover all possible target sequences; for example an RNA substrate should be longer than 1024 bases (= 45) to contain all possible five base cleavage sites, assuming that the RNA contains an equal number of each base. For this reason, we chose bacteriophage MS2 RNA, consisting of 3569 bases and having a fairly even base content (26% G, 23% A, 26% C and 25% U) as a substrate for the determination of the cleavage specificity of MazF-mt3. A major problem in using MS2 RNA (and any long RNA) as a substrate, however, is that although it is a single-stranded RNA, it forms extensive stable secondary structures (see Figure 1A; (Chon et al., 2002)). Therefore, in order to use this RNA as a substrate for endoribonucleases, its secondary structures have to be unfolded. Since the major cold-shock protein of E. coli, CspA is known to function as an RNA chaperone and prevent the formation of secondary structures in RNAs (Jiang et al., 1997), we first attempted to develop a general method for determination of cleavage specificity of endoribonucleases using MS2 RNA in the presence of CspA.
Figure 1. Secondary structure of MS2 RNA and enhancement of MazF-mt3 endoribonucleases activity of CspA.
A. Predicted secondary structure of MS2ssRNA by MFOLD (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). Minimum Folding Energy at 37oC is −1460.69 kcal/mole. The red arrows indicate cleavage sites of MazF-mt3 identified by primer extension analysis and the blue dotted arrow indicates the predicted cleavage sites of MazF-mt3 that were not detected by primer extension analysis.
B. Enhancement of endoribonuclease activity by RNA chaperone CspA.
The full-length MS2 mRNAs were partially digested at 37oC with (lanes 3,4,5 and 6) or without (lanes 1 and 2) purified N-terminal His-tagged MazF-mt3, with (lanes 2 and 5, 32μg; lane 4, 16μg; lane 6, 64μg) or without (lanes 1 and 3) purified CspA protein. The digestion reaction mixture (10 μl) consisted of 1.6 μg of MS2 RNA substrate, 1.6 μg of MazF-mt3(His)6, 16, 32 or 64 μg CspA and 0.5 μl of RNase inhibitor (Roche) in 10 mM Tris-HCl (pH 7.8). The reaction products were run on a 1.2% (1x TBE) agarose gel.
We purified (His)6MazF-mt3 protein expressed from pET-28a-MazF-mt3 as previously described for MazF-mt1 and –mt6, using Ni-nitrilotriacetic acid resin (Zhu et al., 2006). When purified MazF-mt3 was incubated with MS2 RNA in the absence of CspA, partial cleavage of the RNA was observed (Figure 1B lane 3). As increasing amounts of CspA protein were added to the reaction mixture, the cleavage of the MS2 RNA substrate was enhanced (lanes 4 to 6). At the highest concentration of CspA no full-sized MS2 RNA was detected (lane 6). This concentration (0.86 mM) is 32 fold higher that the Kd value of CspA for RNA binding, indicating that the MS2 RNA was almost completely saturated with CspA, which binds every 6 bases of RNA (Jiang et al., 1997). These results also demonstrate that MazF-mt3 is an mRNA interferase capable of cleaving single-stranded RNAs that are generated when CspA unfolds MS2 secondary structure.
MazF-mt3 specifically cleaves RNA at CUCCU and UUCCU —
Next, we carried out primer extension experiments to determine the cleavage sites of MazF-mt3 in the MS2 RNA using the reaction mixture in the presence of 0.43 mM CspA. In order to cover the entire MS2 RNA sequence, a total of 22 primers were synthesized for the primer extension experiments. As shown in Figure 2A to H, in most cases the addition of CspA, significantly enhanced RNA cleavage. Notably, the RNA cleavage was detectable only in the presence of CspA for the cleavage site shown in Figure 2F. The cleavage of the RNA at base 1028 (Figure 2B) appears to be reduced in the presence of CspA, which is due to enhanced cleavage of a site at base 1078 immediately downstream of the cleavage site at base 1028, located between the primer binding site and the upstream cleavage site. This highly enhanced cleavage site at base 1078 in the presence of CspA is shown in Figure 2C. Through these experiments, eight cleavage sites were identified, as listed in Table 2. The consensus sequence from these cleavage sites is CUCCU, where MazF-mt3 cleaves between the U residue in the second position and the C residue in the third position. Although we identified eight MazF-mt3 cleavage sites, there are nine CUCCU sequences in the MS2 RNA according to the published MS2 sequence (Refseq: NC_001417 from NCBI website). We therefore re-sequenced the region containing the ninth CUCCU sequence and found that there is no CUCCU sequence in the region. The MS2 RNA used in these experiments (Roche) contained CCUCU (base 2158 to base 2162) instead of CUCCU. Therefore, all the CUCCU sequences in MS2 RNA were cleaved by MazF-mt3.
Figure 2. MazF-mt3 cleaves specifically at UUCCU sites.
A-H, In vitro cleavage of the MS2 RNA with (His)6MazF-mt3. Lane 1 represents a control reaction in which no proteins were added; lane 2, a control reaction in which only CspA protein was added; lane 3, MS2 RNA only with (His)6MazF-mt3; lane 4, MS2 RNA incubated with (His)6MazF-mt3 and CspA protein. Cleavage sites are indicated by red arrows on the RNA sequence and were determined using the RNA ladder shown on the right. In Fig 2H the RNA ladder was compressed between the C (base 3390) and G (base 3394) residues, but we were able to elucidate the sequence though comparison of the upstream and downstream regions with the standard sequence.
Table 2.
Cleavage sites of MazF-mt3 in MS2 RNA

|
We also detected four other cleavage sites which are different from those found in Figure 2, as shown in Figure 3A to D. The consensus sequence from these primer extension experiments is UUCCU, where MazF-mt3 cleaves between the U residue in the second position and the C residue in the third position. There are five UUCCU sequence in the MS2 RNA all of which were confirmed by RNA sequence. At present it is not clear why the fifth sequence (from base 1804 to base 1808; see Table 2) cannot be cleaved by MazF-mt3. Under the conditions used, the region containing the sequence might not be fully unfolded even in the presence of CspA. All the cleavage sites for MazF-mt3 are shown by red arrows in Figure 1a.
Figure 3. MazF-mt3 cleaves specifically at CUCCU sites.
A-D, In vitro cleavage of the MS2 RNA with (His)6MazF-mt3. Lane 1, control reaction in which no proteins were added; lane 2, control reaction in which only CspA protein was added; lane 3, MS2 RNA incubated only with (His)6MazF-mt3; lane 4, MS2 RNA incubated with (His)6MazF-mt3 and CspA protein. Cleavage sites are indicated by red arrows (strong cleavage sites) or pink arrows (weak cleavage sites) on the RNA sequence and were determined using the RNA ladder shown on the right.
The target pentad sequence for MazF-mt3 is underrepresented in PE and PPE family proteins in M. tuberculosis–
It is tempting to speculate that the pentad sequences identified as the cleavage sites for MazF-mt3 (CUCCU and UUCCU) might allow more stringent control of specific protein synthesis under distinct conditions, either by protecting mRNAs from being cleaved or by being more sensitive to MazF-mt3 endoribunuclease activity. Therefore, we examined the entire M. tuberculosis H37Rv genome to search the open reading frames (ORFs) that contain the pentad sequences at a much lower frequency than expected or conversely at a much higher frequency than expected, calculated from the frequency of each pentad sequence in the gene based on the nucleotide compositions of each ORF.
Interestingly, we found that there are certain genes that have the MazF-mt3 pentad sequences at a much lower frequency than expected. Using a cutoff of p<0.01 we identified ten such genes in the genome. Interestingly, four of these ten genes were identified as members of the PPE gene family, which has been proposed to play a role in the immunopathogenicity of M. tuberculosis (Brennan and Delogu, 2002) (Table 3). Three of these four genes, PPE54, PPE55 and PPE56, are in a single locus in the chromosome of M. tuberculosis though they do not appear to be part of a single operon. All four of these PPE genes encode members of the major polymorphic tandem repeat subfamily of PPE proteins (PPE_MPTR) (Gey van Pittius et al., 2006). Substituting n = 10 and m = 4 in Equation (2), this over-representation of the PPE family has a p-value less than 0.0006. This result suggests that the mRNAs from these genes may be more resistant to MazF-mt3 cleavage. Extending this analysis to genes with a lower than expected number of cleavage sites with p<0.02, we found a total of 20 genes ((including the ten with a probability lower than 1%). Seven out of these 20 genes (35%) were found to belong to the PE or PPE families (see Supplementary Data 1). Substituting n = 20 and m = 7 in Equation (2), this over-representation of the PE and PPE proteins has a p-value less than 0.00002. This analysis indicates that the MazF-mt3-recognizing pentad sequences are underrepresented in specific members of the PE and PPE family suggesting that the mRNAs for these genes are relatively resistant to MazF-mt3 compared to most mRNA’s encoded in the M. tuberculosis genome.
Table 3.
The top 10 MazF-mt3-resistant genes in the M. tuberculosis genome
| CDS position | length | expected motif counts | actual motif counts | probability | locus | gene |
|---|---|---|---|---|---|---|
| complement(3755952..3767102) | 11151 | 20.39 | 6 | 0.00019 | Rv3350c | PPE56 |
| complement(2294531..2336986) | 12456 | 18.71 | 6 | 0.00063 | Rv2048c | pks12 |
| complement(3743711..3753184) | 9474 | 18.25 | 6 | 0.00087 | Rv3347c | FPE55 |
| complement(510702..515321) | 4620 | 6.37 | 0 | 0.00170 | Rv0425c | ctpH |
| 2886373..2889795 | 3423 | 5.88 | 0 | 0.00277 | Rv2566 | |
| 2917871..2921377 | 3507 | 5.75 | 0 | 0.00315 | Pv2590 | fadD9 |
| complement(3729364..3736935) | 7572 | 17.75 | 7 | 0.00337 | Rv3343c | PPE54 |
| complement(2854938..2857781) | 2844 | 5.17 | 0 | 0.00568 | Rv2531c | |
| complements(721729..726013) | 3285 | 4.73 | 0 | 0.00882 | Rv0630c | recB |
| complement(372820..375711) | 2892 | 4.65 | 0 | 0.00956 | Rv0305c | PFE6 |
MazF-mt7 is also a sequence-specific mRNA interferase. —
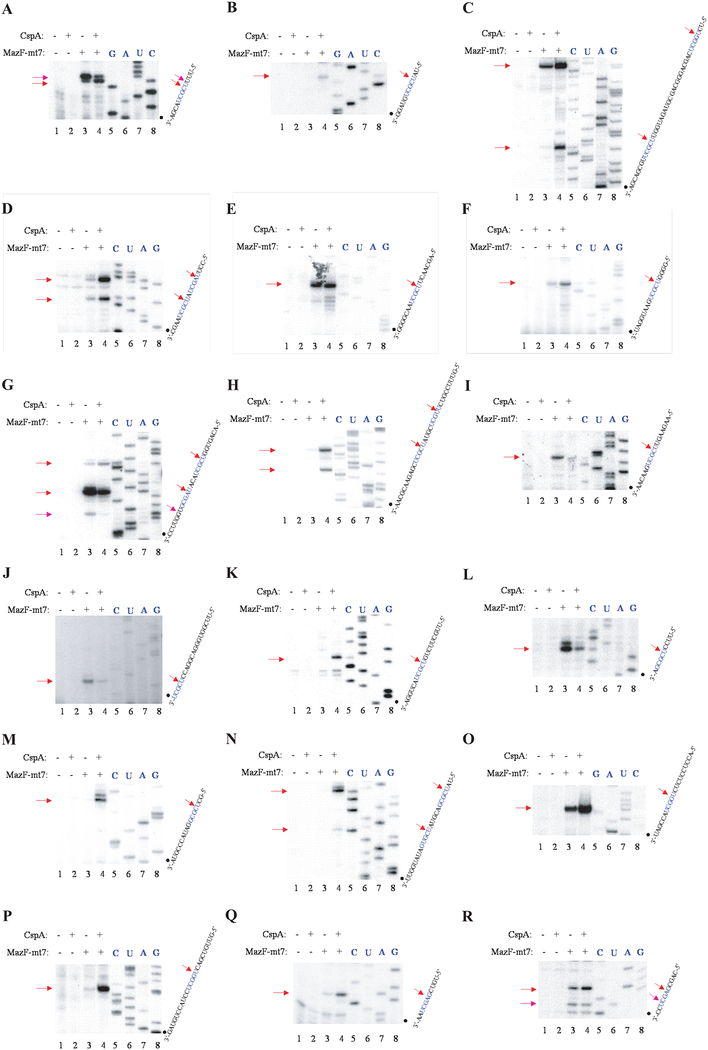
To validate the general applicability of the method, we used MS2 RNA-CspA system to identify the specific cleavage sites of MazF-mt7. Using purified N-terminal His-tagged MazF-mt7, primer extension experiments were carried out and the results are shown in Figure 4A to R and summarized in Table 4. Most of the cleavage sites contain the sequence UCGCU (Figure 4A–K), where MazF-mt7 cleaves between first U and second C. Some cleavage sites have a one-base mismatch from the consensus UCGCU (Figure4 C, D, H and L-P), and a few cleavage sites have a two-base mismatch (Figure4 G, N, Q, R). All these cleavage sites, however, share the central G residue and most of them also have a C residue following the central G residue. MazF-mt7 thus is an mRNA interferase that recognizes a five-base UˇCGCU sequence; however it appears to be less stringent than MazF-mt3.
Figure 4. Cleavage of MS2 RNA by MazF-mt7.
A-O, In vitro cleavage of the MS2 RNA with (His)6MazF-mt7. Lane 1, control reaction in which no enzymes were added; lane 2, control reaction in which only CspA protein was added; lane 3, MS2 RNA incubated only with (His)6MazF-mt3; lane 4, MS2 RNA incubated with (His)6MazF-mt3 and CspA protein. Cleavage sites are indicated by red arrows (strong cleavage sites) or pink arrows (weak cleavage sites) on the RNA sequence and were determined using the RNA ladder shown on the right.
Table 4.
Cleavage sites of MazF-mt7 in MS2 RNA
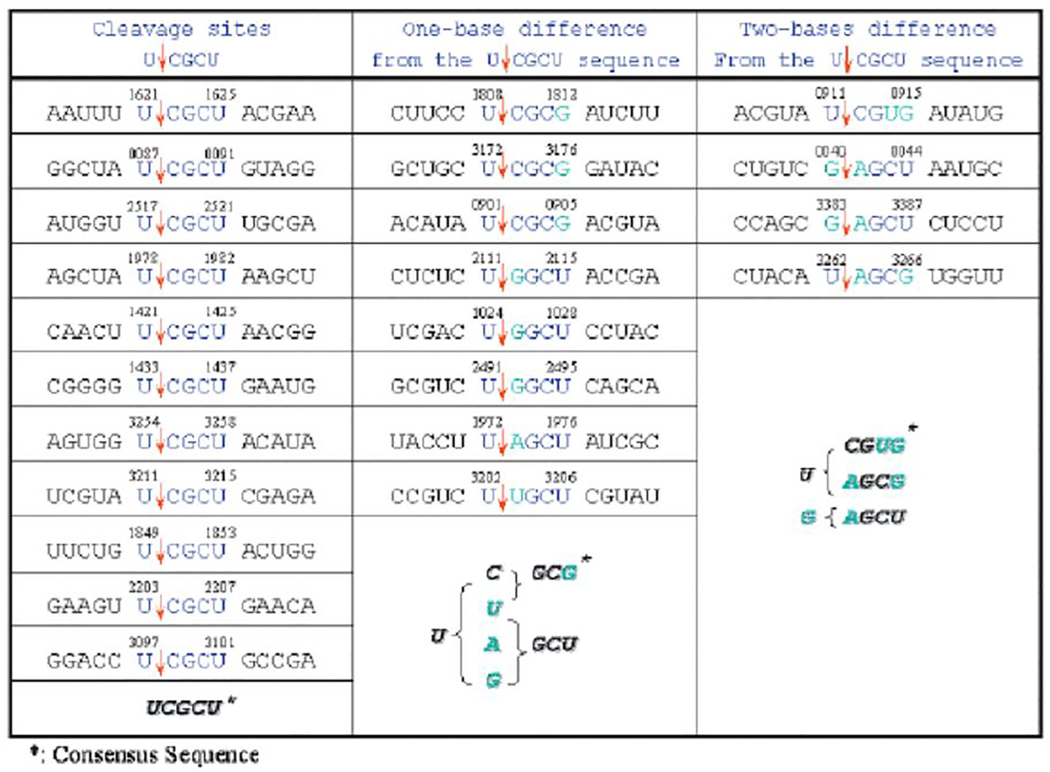
|
The target pentad sequence for MazF-mt7 is underrepresented in PE and PPE genes in M. tuberculosis–
Similar bioinformatics analysis to that described above was carried out to search for M. tuberculosis mRNAs that are resistant or hypersusceptible to cleavage by MazF-mt7 (see Supplementary Data 2). In the top ten MazF-mt7 resistant genes (Table 5), there are four genes belonging to the PE or PPE family. Three out of ten are PPE_MPTR and one out of ten is a PE_PGRS. Most significantly, both PPE55 and PPE56 are resistant not only to MazF-mt7 but also to MazF-mt3. PPE 55 is known to be highly immunogenic, and is expressed during incipient, subclinical M. tuberculosis infection. PPE 56 is highly homologous (67%) to PPE55 (Singh et al., 2005). These data suggest that the expression of different PE and PPE family proteins may be regulated at certain stages of infection by mRNA interferases and raises the possibility that regulation of protein expression by TA systems may play a role in M. tuberculosis pathogenesis.
Table 5.
The top 10 MazF-mt7-resistant genes in the M. tuberculosis genome
| CDS position | length | expected motif counts | actual motif counts | probability | locus | gene |
|---|---|---|---|---|---|---|
| complement(3755952..3767102) | 11151 | 12.73 | 4 | 0.00452 | Rv3350c | PPE56 |
| complement(366150..372764) | 6615 | 6.53 | 1 | 0.01893 | Rv8384c | PPE5 |
| complement(3743711..3753184) | 9474 | 10.93 | 4 | 0.01579 | Rv3347c | PPE55 |
| 1287328..1291026 | 3699 | 3.65 | 0 | 0.02603 | Rv1161 | narG |
| complement(2801254..2806236) | 4983 | 2.71 | 0 | 0.06642 | Rv2490c | PE_PGRS43 |
| complement(3007236..3009209) | 1974 | 2.69 | 0 | 0.06794 | Rv2690c | |
| complement(1663215..1666046) | 2832 | 2.46 | 0 | 0.08551 | Rv1475c | acn |
| complement(1490117..1492312) | 2196 | 2.41 | 0 | 0.08926 | Rv1326c | glgB |
| 895828..898084 | 2265 | 2.35 | 0 | 0.09519 | Rv0803 | purL |
| complement(3774871..3777501) | 2631 | 2.33 | 0 | 0.09676 | Rv3365c |
Discussion
In the present report, we first developed a general method for the determination of cleavage specificities of mRNA interferases using the 3.5-kb MS2 RNA and CspA, an RNA chaperone. In our earlier work with MazF (Zhang et al., 2003), ChpBK (Zhang et al., 2005), PemK (Zhang et al., 2004) and MazF-mt1 and MazF-mt6 (Zhu et al., 2006), in vivo primer extension was sufficient to determine their cleavage site specificities, since all these mRNA interferases recognize only three base sequences in mRNAs. On the other hand, we were unable to determine the cleavage site specificity for MazF-mt3 by in vivo primer extension experiments, since very few cleavage sites were found. We reasoned that this might be due to a longer recognition sequence for MazF-mt3 cleavage. Using the 3.5-kb MS2 RNA, we indeed identified several MazF-mt3 cleavage sites. The further identification of the sequences recognized by MazF-mt7 supports the general applicability of the method for the determination of the cleavage site specificities for mRNA interferases that recognize four- or five-base sequences. It should be emphasized that the use of CspA is very important to remove the secondary structures in the RNA, to allow the mRNA interferase access to the entire 3.5 kb RNA. Using this technology, we were able to identify two specific five-base sequences, CUˇCCU and UUˇCCU, for MazF-mt3, and also UˇCGCU for MazF-mt7. As previously reported (Zhu et. al. 2006) the MazF-mt3 has a putative 11-residue S1-S2 loop similar to E. coli MazF, whereas the MazF-mt7 has a much shorter (only 4 residues) S1-S2 loop. Since the S1-S2 loop has been implicated in stabilizing the interaction of MazF with its mRNA substrates, the S1-S2 loop structures of MazF-mt3 and –mt7 are consistent with our observation. Although both MazF-mt3 and MazF-mt7 target pentad sequences, the MazF-mt3 has a higher specificity for sequence reorganization in comparison to MazF-mt7.
Discovery of mRNA interferases having higher cleavage specificities raises intriguing questions about their function. These enzymes have been proposed to be activated in response to stresses, causing a global shutdown of translation by degradation of mRNA’s (Inouye, 2006). This global response, may lead to a transient state of metabolic “dormancy” or eventual cell death as an adaptive response at the cell or population level. In addition, mRNA interferases with greater specificity such as MazF-mt3 may regulate specific mRNAs in the cell either negatively (by cleaving them) or positively (by not being able to cleave them). This differential susceptibility to cleavage of individual mRNAs by more specific mRNA interferases, may allow a focused response to specific stresses or environmental conditions, rather than the generalized response attributed to TA systems encoding mRNA interferases with three base target sequences that have been characterized to date.
The pathogen M. tuberculosis contains a remarkably large number of TA systems (as many as 60) including at least seven MazF homologues (Golby et al., 2007; Pandey and Gerdes, 2005) in contrast to the non-pathogenic mycobacterium, M. smegmatis. Despite a much larger genome (7 vs. 4.4 megabases) M. smegmatis has only two TA systems including one MazF homologue. These facts suggest an intriguing hypothesis regarding the role of TA systems in M. tuberculosis: Individual TA system may be differentially activated in response to different environmental conditions in the host during the course of infection. Furthermore, individual mRNA interferases may uniquely target specific groups of mRNAs as part of an adaptive response, and thus contribute to the pathogenesis of this organism.
Though this hypothesis remains to be tested, our finding that a subset of the PE and PPE family genes encode mRNAs that may be relatively protected from degradation by MazF-mt3 and MazF-mt7 is consistent with this prediction. Though the function of most genes in this family is obscure at present, a number of these proteins have been shown to localize to the mycobacterial cell wall/cell surface, and several have been shown to provoke an immune response in vivo or play a role in host-pathogen interactions (Basu et al., 2007; Brennan and Delogu, 2002; Denny and Smith, 2004; Mishra et al., 2007), In addition, it has been shown that members of these families are differentially expressed in different tissues or at distinct times during growth in vitro (Delogu et al., 2006; Dheenadhayalan et al., 2006). Thus, to the extent that MazF-mt3 and MazF-mt7 can affect the production of specific PE or PPE family proteins in vivo, these mRNA interferases could play a role in the pathogenesis of tuberculosis.
It is very interesting that the target pentad sequences of MazF-mt3 and MazF-mt7 are underrepresented in the PPE_MPTR subfamily. Members of this group are present in multiple copies only in M. tuberculosis and its close relatives, and are totally absent in the fast-growing mycobacteria, including M. smegmatis. This finding suggests that these genes have been positively selected during the evolution of these slow-growing pathogens, raising an interesting possibility that the target specificity of these mRNA interferases might have played a role in the evolutionary process to expand the PE/PPE family in the slow-growing pathogenic mycobacteria.
As many PE family proteins (notably the PE_PGRS group) are rich in glycine and alanine, the low frequency of MazF-mt3 and MazF-mt7 sequences could be secondary to codon usage driven by processes independent of MazF-mt3 and MazF-mt7 targeting. The fact that only a small fraction of the more than 168 PE and PPE family genes have significantly fewer cleavage sites than predicted, however, suggests that MazF-mt3 and MazF-mt7 interferase activities may degrade these mRNAs to a lesser extent than those encoding other PE or PPE family proteins.
The present work demonstrates a novel and generalizable approach to the identification of mRNA interferases target sequences, including those with sequence requirements longer than the three base targets previously recognized. The identification of the pentad target sequences required by MazF-mt3 and MazF-mt7 suggests the possibility of differential mRNA degradation as a novel regulatory mechanism for adaptation to environmental conditions. This work thus opens up a new avenue for determining of the sequence specificity of the full complement of mRNA interferases in M. tuberculosis and other organisms, and for investigating whether sequence-specific mRNA degradation plays a regulatory role in microbial adaptation, including adaptation that contributes to virulence in pathogenic microorganisms such as M. tuberculosis.
Experimental Procedures
Strains and Plasmids —
E. coli BL21 (DE3) strain was used for recombinant protein expression. Plasmid pET-28a-MazF-mt3 and -mt7 were constructed from pET-28a (Novagen) to express (His)6MazF-mt3, and –mt7, respectively.
Purification of (His)6 tagged MazF-mt3 and MazF-mt7 in E. coli —
MazF-mt3 and MazF-mt7 (His)6 tagged at the N-terminal end were purified from strain BL21(DE3) carrying pET-28a –MazF-mt3 and MazF-mt7 using Ni-NTA resin (Qiagen) as described previously (Zhang et al., 2003).
Purification of CspA protein from E. coli —
CspA was purified as described previously (Chatterjee et al., 1993).
Primer Extension Analysis in vitro —
For primer extension analysis of mRNA cleavage sites in vitro, the full-length MS2 mRNAs were partially digested with or without purified toxin protein (MazF-mt3 or MazF-mt7), and with or without purified CspA protein at 37°C for 15 min. The digestion reaction mixture (10 μl) consisted of 0.8 μg of MS2 RNA substrate, 0.0625 μg of MazF-mt3(His)6 or MazF-mt7(His)6, 32μg CspA and 0.5 μl of RNase inhibitor (Roche) in 10 mM Tris-HCl (pH 7.8). Primer extension was carried out at 47°C for 1 h in 20 μl of the reaction mixture as described previously (Zhang et al., 2005). The reactions were stopped by adding 12 μl of sequence loading buffer (95% formamide, 20 mM EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol EF). The samples were incubated at 90°C for 5 min prior to electrophoresis on a 6% polyacrylamide and 36% urea gel. The primers used for primer extension analysis of the MS2 mRNA are listed in Table 1. The primers were 5’-labeled with [γ−32P]ATP using T4 polynucleotide kinase.
Table 1.
The primers used for primer extension analysis of the MS2 RNA.
| Primer Name | Sequence | Primer Name | Sequence |
|---|---|---|---|
| B3 | 5′-AGCACACCCACCCCGTTTAC-3′ | E | 5′-GAAGATCAATACATAAAGAG-3′ |
| B 4 | 5′-GGTCCGTCCCACCGAAGAAC-3′ | E1 | 5′-TGCATTGCCTTAACAATAAG-3′ |
| B5 | 5′-AGAACTTGCGTTCTCGAGCG-3′ | E2 | 5′-TACAGGTTACTTTGTAAGCC-3′ |
| B7 | 5′-TATAACGCGCACGCCGGCGG-3′ | G | 5′-GAGCCGTTGCCTGATTAATG-3′ |
| B8 | 5′-CGGAGTCTTGGTGTATACCG-3′ | H | 5′-AGCGTCAACGCTTATGATGG-3′ |
| C2 | 5′-GCGGATACGATCGAGATATG-3′ | I | 5′-AGCATCCCACGGGGGCCG-3′ |
| C3 | 5′-GCGCACATTGGTCTCGGACC-3′ | J | 5′-GGTTCAAGATACCTAGAGAC-3′ |
| D | 5′-CCGCTCTCAGAGCGCGGGGG-3′ | J1 | 5′-GACGGCCATCTAACTTGATG-3’ |
| D1 | 5′-GCAATTGATTGGTAAATTTC-3′ | K1 | 5′-CTTCGGTCGACGCCCGGTTC-3′ |
| D2 | 5′-TCTCTATTTATCTGACCGCG-3′ | L1 | 5′-TAGCCATGGTAGCGTCTCGC-3′ |
| D5 | 5′-AATTCGTCCCTTAAGTAAGC-3′ | L2 | 5′-CCTATCAAGGGTACTAAAAG-3′ |
Bioinformatic Analysis of the Frequencies of MazF-mt3 or -mt7 Motifs in Mycobacterium tuberculosis Coding Sequences —
We retrieved the genomic sequence of M. tuberculosis H37Rv from NCBI RefSeq (Accession NC_000962), and extracted all coding sequences (CDS) from the record. We first calculate the nucleotide composition of each CDS. The probability, p, of the two motifs CUCCU or UUCCU appearing anywhere in the CDS is:
Let L be the length of the CDS. Then the expected number, E, of the motifs in the CDS is:
Let K be the actual number of the motifs in the CDS. Then the probability, P, of having K or fewer motifs in the CDS is:
| Equation (1). |
A CDS with a very small P value suggests that it may have evolved to eliminate the two motifs from its sequence.
Bioinformatic Analysis of the over-representation of the PE family among the ORFs where the numbers of MazF-mt3 cleavage sites are less than expected —
The NCBI RefSeq record shows that Mycobacterium tuberculosis has 3,989 ORFs. The PE and PPE families together contain around 168 genes (http://genolist.pasteur.fr/TubercuList/). From the proceeding analysis, we identify a set of genes where the numbers of cleavage sites are significantly less than expected. Let n be the number of such significant genes, and let m be the number of PE genes among these n genes. Then the significance of the over-representation of the PE family follows the hypergeometric distribution:
| Equation (2). |
Supplementary Material
Acknowledgments
This work was partially supported by a research fund from Takara-Bio. Inc. Japan to IM and by a grant from the National Institute of Allergy and Infectious Disease to RNH.
References
- Basu S, Pathak SK, Banerjee A, Pathak S, Bhattacharyya A, Yang Z, Talarico S, Kundu M, and Basu J (2007) Execution of macrophage apoptosis by PE_PGRS33 of Mycobacterium tuberculosis is mediated by Toll-like receptor 2-dependent release of tumor necrosis factor-alpha. J Biol Chem 282: 1039–1050. [DOI] [PubMed] [Google Scholar]
- Brennan MJ, and Delogu G (2002) The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol 10: 246–249. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Jiang W, Emerson SD, and Inouye M (1993) The backbone structure of the major cold-shock protein CS7.4 of Escherichia coli in solution includes extensive beta-sheet structure. J Biochem (Tokyo) 114: 663–669. [DOI] [PubMed] [Google Scholar]
- Chon H, Tsunaka Y, Haruki M, Morikawa M, and Kanaya S (2002) Site-specific cleavage of MS2 RNA by a thermostable DNA-linked RNase H. Protein Eng 15: 683–688. [DOI] [PubMed] [Google Scholar]
- Delogu G, Sanguinetti M, Pusceddu C, Bua A, Brennan MJ, Zanetti S, and Fadda G (2006) PE_PGRS proteins are differentially expressed by Mycobacterium tuberculosis in host tissues. Microbes Infect 8: 2061–2067. [DOI] [PubMed] [Google Scholar]
- Denny PW, and Smith DF (2004) Rafts and sphingolipid biosynthesis in the kinetoplastid parasitic protozoa. Mol Microbiol 53: 725–733. [DOI] [PubMed] [Google Scholar]
- Dheenadhayalan V, Delogu G, Sanguinetti M, Fadda G, and Brennan MJ (2006) Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J Bacteriol 188: 3721–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Donegan NP, Memmi G, and Cheung AL (2007) Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J Bacteriol 189: 8871–8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, and Warren RM (2006) Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol 6: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golby P, Hatch KA, Bacon J, Cooney R, Riley P, Allnutt J, Hinds J, Nunez J, Marsh PD, Hewinson RG, and Gordon SV (2007) Comparative transcriptomics reveals key gene expression differences between the human and bovine pathogens of the Mycobacterium tuberculosis complex. Microbiology 153: 3323–3336. [DOI] [PubMed] [Google Scholar]
- Inouye M (2006) The discovery of mRNA interferases: Implication in bacterial physiology and application to biotechnology. J Cell Physiol 209: 670–676. [DOI] [PubMed] [Google Scholar]
- Jiang W, Hou Y, and Inouye M (1997) CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem 272: 196–202. [DOI] [PubMed] [Google Scholar]
- Mishra KC, de Chastellier C, Narayana Y, Bifani P, Brown AK, Besra GS, Katoch VM, Joshi B, Balaji KN, and Kremer L (2007) The Mycobacterium tuberculosis triacylglycerol hydrolase LipY: functional role of the PE domain and immunogenicity. Infect Immun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey DP, and Gerdes K (2005) Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33: 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Dong Y, Patibandla SA, McMurray DN, Arora VK, and Laal S (2005) Immunogenicity of the Mycobacterium tuberculosis PPE55 (Rv3347c) protein during incipient and clinical tuberculosis. Infect Immun 73: 5004–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Zhang J, Liu M, Woychik NA, and Inouye M (2005) Single protein production in living cells facilitated by an mRNA interferase. Mol Cell 18: 253–261. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Y, and Inouye M (2003) Characterization of the interactions within the mazEF addiction module of Escherichia coli. J Biol Chem 278: 32300–32306. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Y, Zhu L, Suzuki M, and Inouye M (2004) Interference of mRNA function by sequence-specific endoribonuclease PemK. J Biol Chem 279: 20678–20684. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, and Inouye M (2003) MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell 12: 913–923. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hara H, Kato I, and Inouye M (2005) Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem 280: 3143–3150. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhu L, Zhang J, and Inouye M (2005) Characterization of ChpBK, an mRNA interferase from Escherichia coli. J Biol Chem 280: 26080–26088. [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, Rubin H, and Inouye M (2006) Characterization of mRNA interferases from Mycobacterium tuberculosis. J Biol Chem. 2006 Apr 12; 281:18638–18643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.