Simulations associate urbanization with earlier peaks, higher peak prevalence, and shifting bimodality of pandemics in Australia.
Abstract
We examine salient trends of influenza pandemics in Australia, a rapidly urbanizing nation. To do so, we implement state-of-the-art influenza transmission and progression models within a large-scale stochastic computer simulation, generated using comprehensive Australian census datasets from 2006, 2011, and 2016. Our results offer a simulation-based investigation of a population’s sensitivity to pandemics across multiple historical time points and highlight three notable trends in pandemic patterns over the years: increased peak prevalence, faster spreading rates, and decreasing spatiotemporal bimodality. We attribute these pandemic trends to increases in two key quantities indicative of urbanization: the population fraction residing in major cities and international air traffic. In addition, we identify features of the pandemic’s geographic spread that we attribute to changes in the commuter mobility network. The generic nature of our model and the ubiquity of urbanization trends around the world make it likely for our results to be applicable in other rapidly urbanizing nations.
INTRODUCTION
The global population is both highly urbanized and rapidly urbanizing, with people around the world flocking to cities more quickly every year since 1950 (1). This critical trend has inspired a vigorous research effort toward understanding the economic, environmental, and social implications of global urbanization (2–4). To further this effort, we present findings on the relationship between urbanization trends and pandemic influenza sensitivity.
In general, urbanization is known to worsen epidemics through a variety of mechanisms. In developing nations, this is well documented and is associated with high-density living conditions combined with concentrated poverty and poor sanitation (5). In the developed world, where urban living has been the norm for the last century, the direct effects of further population concentration into cities on potential pandemic dynamics are less obvious. The concentration of health care facilities, combined with increased economic opportunities, tends to increase the general quality of public health (6). However, with respect to communicable disease, the concentration of the workforce in central business districts, combined with suburban sprawl, can produce large hubs in the commuter interaction network that could potentially lead to faster proliferation of infectious disease between work and home (7, 8). In addition, city growth is coupled to increased air traffic and connectivity, which could be expected to increase both the probability of intercity disease spread and the potential for the disease to arrive from overseas because of international traffic (9).
Our study focuses on Australia, a highly urbanized nation for which comprehensive census data spanning the last decade have been made publicly available. We used these data to calibrate a nation-level model of pandemic influenza spread and to investigate the surrogate population’s vulnerability to the contagion over a period of rapid urbanization.
Australia has a total urban population fraction of about 90%, with more than half of the country’s population located within only a few urban centers. Over time, this trend has led to substantial infrastructural stress, including increased health care economic burden. In particular, the cost of influenza in Australia is estimated to be between AU$828 and AU$884 million annually (10). Rapidly escalating epidemics directly affect other aspects of life. Individual behavior changes, and the resilience of critical infrastructure is called into question (11, 12). Impacts are quickly seen also in labor supply and productivity, in the movement of goods and services across regions, and in the sustainability of consumer and investor confidence. Arguably the most acute aspect of an epidemic or pandemic crisis is the strain on medical infrastructure. In the last decade, Australia’s hospital beds have been maintained at approximately 2.5 per 1000 individuals, leading major hospitals to regularly operate from 90 to 100% or higher capacity. The Australian Medical Association has assessed the condition as unacceptable, but hospital capacity continues to lag behind demand (13). Because of the constant (low) per-capita bed numbers and the country’s aging population, any relative increases in disease prevalence are substantial from the perspective of hospital overcrowding and the related adverse health outcomes for patients and staff, including increased patient mortality (14).
Despite its relative isolation, Australia was not spared from the H1N1 influenza (“swine flu”) pandemic in 2009 (15). Since then, the prevalence of seasonal influenza in Australia has been increasing on average. The country experienced a particularly severe season in 2017, with levels approaching those of 2009 (16). To make matters worse, a recent analysis has assessed Australia’s pandemic preparedness as sorely lacking (17). On the basis of these general trends, we can expect future pandemics to be more damaging. Our motivation for the present study is the prospect of linking long-term structural and social trends to Australia’s apparently increasing susceptibility to contagion (in this case, the influenza virus).
There are many existing methods and tools for modeling epidemics and assisting in crisis response and preparedness planning (7, 18). However, none has been used to provide a high-resolution comparative analysis of pandemic trends across multiple historical time points. Here, we accomplish this by simulating the spread of influenza through a stochastically generated population mimicking the Australian Bureau of Statistics (ABS) censuses of 2006, 2011, and 2016. We conduct our simulations at the community and national level in a way that rigorously accounts for salient features of the contagion, such as prevalence dynamics and spatiotemporal structure. This approach allows us to investigate the role of the slowly evolving population distribution and social interaction network over which the virus spreads, independently of the viral characteristics. Such a separation of contagion and population properties is impossible in field studies because both are intrinsically dynamic (19).
Our results highlight three notable trends in epidemic patterns over the years, which are independent of the specific simulation parameters defining the influenza virus: (i) increased peak prevalence, (ii) faster spreading rates (earlier epidemic peak), and (iii) decreasing spatiotemporal bimodality. These results have important implications: The first point predicts increased intensity of health crises in infrastructural terms, with more people simultaneously expressing flu symptoms during the peak of the epidemic; the second point indicates shorter periods during which detection and response strategies would have to be implemented for effective mitigation; and the third point pertains to the bimodal character of the epidemic, which occurs in two waves—the first in cities near international airports (where the pandemic is introduced) and the second in rural areas and cities without international airports. In this case, decreasing bimodality occurs as the first mode subsumes the second, contributing to the severity of the epidemic at its peak.
Bimodal H1N1 outbreaks were observed around the world during the 2009 global H1N1 pandemic (20–24). However, despite its ubiquity, the mechanisms behind bimodality remain elusive. Although community network topology has been suggested as an important potential cause (25, 26), the confluence of environmental factors and sporadic interventions makes it impossible to isolate the effects of community structure. In addition, bimodal epidemics have not been observed in previous simulation studies based on real-world mobility networks and disease characteristics [see, for example, (27, 28)].
SUMMARY OF METHODS
In general terms, our simulation strategy is similar to others in the literature (28–30) and is based primarily on a model that is the subject of our previous publication, calibrated to the Australian demographics and mobility (31). For the present work, we made several minor improvements to the process by which the sample population is generated (these are summarized in Methods).
We initialize our model by creating stochastically generated sample populations for each year based on Australian census data from 2006, 2011, and 2016. For each year, we then simulate scenarios in which the pandemic reaches Australia from overseas, continuously seeding the epidemic within 50 km of international airports with probability proportional to the average number of incoming passengers at the corresponding airport. The simulation continues for 180 “days,” each with daytime and nighttime components during which individuals interact at workplaces and within residential communities, respectively. Here, continuous seeding means that we infect people at random in the seeding regions at the beginning of each day of the simulation, providing a small but continuous stream of new infections from abroad.
The disease then spreads in households, neighborhoods, schools, and workplaces based on the disease progression and transmission models validated in our previous work (31). These models are similar to those developed for other countries (29, 32), with transmission probability dependent on context (location and demographics) and infection status (viral titers and symptom expression). Whenever possible, relative transmission and contact probabilities were derived from field studies (27, 28).
For the results reported here, we set the transmissibility of the disease to achieve a basic reproductive ratio Ro = 2 (calibrated for 2006) and held all parameters constant while varying the demographic inputs (33). All results were averaged over five different sample populations for each year, with 30 pandemic instances each, for a total of 150 pandemic instances for each year. Results for other Ro values, along with standard deviations (SDs) of incidence, prevalence, and attack rate, are shown in figs. S1 to S3. (Note that because of the stochastic nature of our seeding protocol, there is a low probability that the contagion will not spread in the population, giving prevalence values near zero. In our entire set of simulations, this occurred a total of three times: once for the 2006 population with Ro = 2, once for the 2006 population with Ro = 1.25, and once for the 2011 population with Ro = 1. For the results presented, these three instances were omitted in the analysis).
RESULTS AND DISCUSSION
Our results indicate a concerning progression in the population’s response to pandemic influenza since 2006. The character of this progression is shown by several metrics in Fig. 1. The incidence (number of newly symptomatic individuals) on each day of our simulated epidemics is shown in Fig. 1A and indicates a steadily increasing peak infection rate. The total prevalence (number of symptomatic individuals) in the population as a function of time is shown in Fig. 1B and illustrates a corresponding trend in peak prevalence. The accumulated attack rate (total number of people affected) in Fig. 1C shows only minor changes in the total proportion of people infected, despite the shifts to larger, earlier epidemic peaks. The progressions of peak day and peak prevalence are given in Fig. 1D.
Fig. 1. The ensemble average of incidence of new infection, prevalence of infected agents, and cumulative infection temporal dynamics for simulated influenza in 2006, 2011, and 2016.
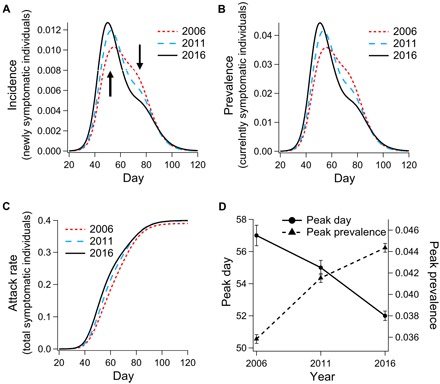
Comparison of simulation results for (A) incidence, (B) prevalence, (C) accumulated incidence (attack rate), and (D) trend in dynamics of peak day and prevalence; error bars (± SEM) designate standard error of the mean.
At first glance, the stability in overall attack rate (Fig. 1C) looks reassuring. This implies that the social network of Australia is not changing in a way that leads to a larger percentage of people becoming exposed to potential infection. A pessimistic but realistic explanation for this result is that almost everyone in the simulation is eventually exposed to potential infection in all years. Regardless of the explanation for consistent attack rates, the change in shape of the incidence and prevalence curves leads to some concerning implications. Two of these are illustrated in Fig. 1D, which traces a correlation between earlier peak days (delay between epidemic onset and peak prevalence) and greater peak prevalence. These results indicate that the virus is spreading more quickly in the population, leading to greater stress on the medical infrastructure and shorter periods of time to respond after the disease is detected.
For Australia, no empirical estimates exist for the probability of hospitalization, given pandemic influenza infection, because the 2009 H1N1 clinical attack rate is unknown. However, on the basis of the documented absolute number of 4855 hospitalized cases in 2009, we can estimate the hospitalization probability, given infection, for a reasonable range of attack rate values. Bounding the estimated 2009 attack rate between 10 and 40% translates to hospitalization rates of 0.22 and 0.056% in the infected population, respectively. Applying these rates to our incidence curves in 2006 and 2016, we estimate the 10-year increase in the number of additional hospitalizations during the peak 3 weeks of the pandemic to be in the range between 664 and 2658. Both of these numbers are large enough to contribute substantial stress to emergency departments across the country and would represent a notable increase in hospitalizations due to influenza. In reality, real hospitalization rates depend enormously on the symptom severity associated with the strain and on the levels of pre-existing respiratory health conditions in the population, neither of which is treated explicitly in our model.
In all years, the model output shows strong bimodality in the histogram of local peak prevalence dates, which corresponds to a transition from an initial wave in urban regions with seeding locations around international airports to a second wave affecting areas not directly connected to these seed regions (Fig. 2). This bimodality is apparent on the level of Statistical Area level 2 (SA2) regions (see the maps in Fig. 2). In 2006, the initial wave is confined almost exclusively to major cities and the surrounding suburbs. In subsequent years, the trend is an increasing number of rural regions peaking in the first wave. [Note that SA2 is analogous to the Statistical Local Area partitions used in 2006 and earlier years. The hierarchy of partitions used for the 2006, 2011, and 2016 censuses can all be viewed on the ABS website (34)].
Fig. 2. Maps and histograms demonstrating geographic and temporal bimodality in epidemic spread.
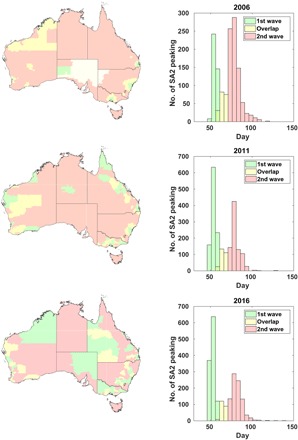
The histograms represent the number of statistical areas (SA2) experiencing peak disease prevalence on a given day. The colors correspond to heuristic classification; green bars indicate the first wave, yellow bars are undetermined, and red bars indicate the second wave. The colors on the map correspond to those in the histogram and demonstrate the geographic distribution of each pandemic wave.
Bimodality is also detectable on the state level. For example, comparing the 2006 total incidence in New South Wales (which is home to the largest international airport, located in Sydney) to that of South Australia (which receives lower levels of international traffic) indicates, unsurprisingly, that states with large international airports tend to contribute to the initial wave, while the states with lower international traffic are affected later and contribute to the second wave (Fig. 3A). The bimodal dynamics are acutely visible in time series of regional prevalence (please refer to movies S1 to S3 for dynamic visualizations of disease spread).
Fig. 3. Analysis of bimodality in disease prevalence.
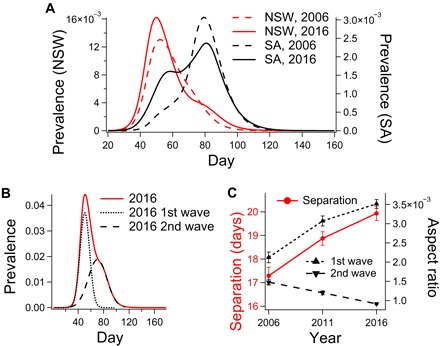
(A) The state-level prevalence for New South Wales (NSW) and South Australia (SA), comparing prevalence curves for 2006 and 2016. (B) National prevalence for 2016 and the two Gaussian curves used to fit the data. (C) Increasing interpeak separation, increasing aspect ratio of the first mode, and decreasing aspect ratio of the second mode (error bars: ±SEM).
To quantify the progression of the bimodal character that is qualitatively observed in Fig. 2, we fit the prevalence data for each year to pairs of Gaussian curves corresponding to the first and second epidemic waves (χ2 = [6.63, 6.62, 26.7] × 10−6 for 2006, 2011, and 2016, respectively). This fitting procedure is illustrated for 2016 in Fig. 3B, while the trends in interpeak separation and peak aspect ratio (the ratio of peak height and SD) are shown in Fig. 3C. These indicate progressive sharpening of the first wave and broadening of the second wave. This trend corresponds to an increased separation of the two peaks, which also manifests as an overall decrease in geographic synchrony: [0.071, 0.057, 0.052] d−1 for 2006, 2011, and 2016, respectively (quantified here as the reciprocal SDs of the histograms of local peak days shown in Fig. 2).
In our assessment, there are two primary mechanisms responsible for these trends. The first is that international air traffic (number of arrivals) increases with time (see Table 1), which tends to promote the initial wave by introducing a more potent seed infection into the network near urban centers. The second is that the population has become more concentrated in urban regions, close to the international airports where the pandemic is introduced. This trend is clearly observable in plots of population growth comparing urban and rural regions (Fig. 4) (35). The confluence of increased disease influx coupled with a population located closer to the influx point provides a simple but reasonable explanation for the progression in simulated pandemic trends observed across years.
Table 1. Average daily incoming international air traffic.
| Airport | State | Year | ||
| 2006 | 2011 | 2016 | ||
| Sydney | New South Wales | 13,214 | 15,995 | 19,991 |
| Melbourne | Victoria | 5,923 | 8,557 | 12,802 |
| Brisbane | Queensland | 5,053 | 5,946 | 7,299 |
| Perth | Western Australia | 2,766 | 4,512 | 5,906 |
| Gold Coast | Queensland | 285 | 1,044 | 1,435 |
| Adelaide | South Australia | 492 | 766 | 1,170 |
| Cairns | Queensland | 1,186 | 707 | 824 |
| Darwin | Northern Territory | 160 | 356 | 355 |
| Townsville | Queensland | 0 | 11 | 39 |
Fig. 4. Variation in population growth rates across years for different levels of region remoteness.

(A) Population growth by remoteness. The urban population has been increasing steadily, while the rural population declines in both relative (all nonurban areas) and absolute (remote regions) terms. (B to D) Time series representations of urban (B), nonurban (C), and remote (D) populations since 2008. (E and F) Time series of relative urban population fractions computed against nonurban (E) and remote (F) populations.
The role of seeding is demonstrated in Fig. 5, which shows the results of control studies in which the 2006 and 2011 populations were seeded with 2016 international passenger traffic (Fig. 5, A and B). The residuals between these and the 2016 prevalence levels are shown in Fig. 5C and show peaks corresponding approximately to the peaks of the second epidemic waves in both 2006 and 2011 (Fig. 5C, black and orange dashed lines, respectively).
Fig. 5. The role of seeding.
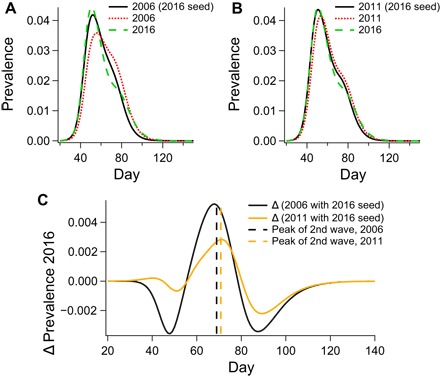
(A and B) Raw prevalence curves demonstrating the effect of applying the seeding conditions of 2016 on the topologies of 2006 (A) and 2011 (B). (C) The residual between prevalence in 2016 and prevalence in 2006 (black curve) and 2011 (orange curve) when seeded with 2016 airport traffic. The dashed vertical lines in (C) indicate the maxima of Gaussian fits for the second epidemic wave.
The coincidence of these residual peaks and the second epidemic waves indicates that the seeding conditions have a larger impact on the first wave than on the second. The control for 2011 has an almost identical prevalence profile to that of 2016 during the peak of the epidemic. On the other hand, seeding does not account for the decrease in the intensity of the second pandemic wave from year to year, a trend that we ascribe to increased urbanization.
In our model, the epidemic can only spread outside of the seeding regions through the work and school commuter networks, which are known to be important to influenza propagation (36–38). Therefore, the inclusion of rural regions outside of seeding zones in the first wave for 2011 and 2016 pandemics (clearly observable in the maps; Fig. 2) indicates that the travel-to-work (TTW) network is partially responsible for the increase in the first pandemic wave and the decrease of the second. However, the concentration of the urban population within the seeding regions makes this contribution relatively minor because direct seeding and local transmission dominates the dynamics. Although it is beyond the scope of the present work, the coupled effect of urbanization trends and mobility network evolution on epidemic spread will be the subject of future investigations. Contact network properties are known to be essential factors in contagion spread (39). It is feasible that contact network evolution could be decoupled from the urbanization process to stall or reverse the observed trends in pandemic dynamics. Through this line of inquiry, we hope to design network-based pandemic intervention and urban engineering strategies that will make rapidly urbanizing societies more resilient to pandemics (40).
CONCLUSION
By applying a high-fidelity epidemic simulation of the spread of influenza through the Australian population of 2006, 2011, and 2016, respectively, we have observed an increase in pandemic spreading rate and peak prevalence, combined with decreasing urban-rural bimodality. These trends are associated with increased international air travel and a more urbanized population distribution. The net effect of these two coacting mechanisms is a shift from a highly bimodal epidemic (both temporally and geographically) to an increasingly monomodal dynamics where the initial wave dominates. The mobility network extends this phenomenon outside the urban seeding regions. Incidentally, although we observe a net decrease in geographic synchrony over the years investigated here, the observed trends appear to predict a future increase in national epidemic synchrony, as the second wave becomes negligible and the initial wave becomes more prominent and peaked. The trend in peak prevalence is particularly worrying, as it indicates a nonlinear increase in strain on medical infrastructure that current public health policy does not take into account with fixed per-capita hospital capacity. Mechanistically, increased air traffic accounts for the trend of earlier peak dates and increased magnitude of the first pandemic wave, while the commuter network and shifting population distribution are responsible for the systematic decrease in the intensity of the second pandemic wave. All three of these causal factors are associated with increased urbanization. To our knowledge, this work provides the first example of (i) pandemic simulations across years comparing historical time points and (ii) epidemic bimodality mechanistically associated with an empirical interaction network representing a real social system.
METHODS
Data preprocessing
Census data used to produce the sample populations were preprocessed to remove nongeographical census regions and ensure consistency across different spatial scales. Australian census data are publicly available through the ABS’ online platform Census TableBuilder. To generate our sample populations, we used a subset of demographic data relating to the following characteristics of interest: dwelling location, employment status/work location, household composition, sex, and age. To gather this information, we accessed the following datasets: (i) statistical area level 1 (SA1) [usual residence (UR)] by AGEP (population count by age) and SEXP (population count by sex), (ii) SA1 (UR) by CDCF (count of dependent children in family) and NPRD (number of persons usually resident in dwelling), and (iii) SA1 (UR) by employment destination zone (DZN) [place of work (POW)].
All exported data are subject to perturbation to preserve the anonymity of individuals as per the Australia Census and Statistics Act 1905. The relative effect of perturbation becomes more prominent with the increased parsing of data. This effect is most severe in the TTW networks, as these break the population into the smallest groups.
The net effect of these perturbations can lead to inconsistencies between data amalgamated for different spatial partitions. For example, accumulating commuter data on the level of (small) SA1 partitions cannot reproduce the ABS-provided statistics over (larger) SA2 partitions and can lead to the creation of nonexistent edges in the SA2 (UR) to SA2 (POW) TTW network.
To avoid artificial links in the disease transmission network, we removed those SA1 to DZN entries that could not be accounted for on the level of SA2 [i.e., that produced nonexistent edges when amalgamated to the scale of SA2 (UR) to SA2 (POW)]. Because of a change in the ABS procedure for introducing perturbations into the 2016 data, some additional processing was required to ensure consistency between 2011 and 2016. This procedure involved the recovery of TTW network edges by sampling over several additional ABS datasets. We will release this modified dataset and outline our sampling procedure in detail in our forthcoming publication (41).
Initialization
We began by generating sample populations based on census data from the three years investigated. The datasets that inform our sampling procedure describe local area populations on the order of several hundred people (SA1 in both 2016 and 2011, and Census District in 2006). These datasets provide population (e.g., age, sex, and employment status) and housing (e.g., household size and composition) statistics. These were used as (dependent) probability density functions in the stochastic generation of households and agents, respectively. Additional details of the population generation procedure can be found in the Supplementary Materials. We positioned schools pseudodeterministically based on their postal code as reported by the Australian Curriculum, Assessment and Reporting Authority (ACARA), a noncensus dataset that contains the most complete information available on school enrollment numbers and locations since 2008 (note that we used 2008 school locations and enrollments in place of 2006 data that were not available) (42). We then assigned students to schools based on the proximity rules described in our previous work (31).
Seeding the pandemic
To realistically model the domestic spread of the disease, we assumed that the Australian population is exposed to the strain once it is a global pandemic. Following the approach of Germann et al. (28), we modeled this influx of disease by introducing the pandemic to local areas within 50 km of international airports every day. This dynamic seeding procedure infects the population within the seeding zone proportionally to the average daily incoming number of passengers reported by the Bureau of Infrastructure, Transport and Regional Economics (43). Full details of the dynamic seeding procedure can be found in section 3.4 of (31). Although the seeding procedure between years is methodologically identical, differences in international airport traffic between years (see Table 1) mean that the dynamic seeding procedures are crucially different.
Disease transmission
The pandemic can spread within households, neighborhoods, schools, and workplaces according to the transmission probabilities set out in our previous work (31). During each 24-hour period, a daytime phase occurs during which infected individuals can spread the infection to others in their working groups comprising approximately 10 others working in the same “destination zone” [these groups include classes, grades, and schools for teachers and students, which have more than 10 people; see (31)]. This is followed by a nighttime phase during which the infection can spread within households, household clusters, and neighborhoods. The pandemic can only spread between statistical areas during the daytime phase, as this is the largest geographical area of interaction in the nighttime phase. Further details of the transmission model are given in the Supplementary Materials.
Supplementary Material
Acknowledgments
We are grateful to T. Germann, J. T. Lizier, P. Pattison, E. Y. Erten, M. Gambhir, and S. Leeder for helpful discussions on agent-based simulation of pandemic influenza. Funding: This work was supported by the Australian Research Council Discovery Project DP160102742. Author contributions: M.Pr. and M.Pi. conceived the project; C.Z., K.M.F., N.H., and M.Pr. wrote the manuscript; C.Z. and K.M.F. performed the simulations and analyzed the data; N.H. and O.M.C. assisted with modeling and data preprocessing. All authors contributed to discussions regarding the interpretation of results. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/12/eaau5294/DC1
Movie S1. The color mapped, temporal evolution of the average simulated prevalence for 2006, using the SA2 partition.
Movie S2. The color mapped, temporal evolution of the average simulated prevalence for 2011, using the SA2 partition.
Movie S3. The color mapped, temporal evolution of the average simulated prevalence for 2016, using the SA2 partition.
Fig. S1. Incidence proportion for various Ro values.
Fig. S2. Prevalence proportion for various Ro values.
Fig. S3. Attack rate for various Ro values.
Model description
REFERENCES AND NOTES
- 1.United Nations Department of Economic and Social Affairs, Population Division, World Urbanization Prospects: The 2014 Revision, Highlights (United Nations, 2014).
- 2.Wigginton N. S., Fahrenkamp-Uppenbrink J., Wible B., Malakoff D., Cities are the future. Science 352, 904–905 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Seto K. C., Ramankutty N., Hidden linkages between urbanization and food systems. Science 352, 943–945 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Bloom D. E., Canning D., Fink G., Urbanization and the wealth of nations. Science 319, 772–775 (2008). [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization, Our cities, our health, our future: Acting on social determinants for health equity in urban settings. Report to the WHO Commission on Social Determinants of Health from the Knowledge Network on Urban Settings (World Health Organization, 2008).
- 6.Dye C., Health and urban living. Science 319, 766–769 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Eubank S., Guclu H., Kumar V. S. A., Marathe M. V., Srinivasan A., Toroczkai Z., Wang N., Modelling disease outbreaks in realistic urban social networks. Nature 429, 180–184 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Yashima K., Sasaki A., Epidemic process over the commute network in a metropolitan area. PLOS ONE 9, e98518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brueckner J. K., Airline traffic and urban economic development. Urban Stud. 40, 1455–1469 (2003). [Google Scholar]
- 10.A. T. Newall, P. A. Scuffham, B. Hodgkinson, Economic Report into the Cost of Influenza to the Australian Health System (National Institute of Clinical Studies, 2007). [Google Scholar]
- 11.O’Rourke T. D., Critical infrastructure, interdependencies, and resilience. Bridge 37, 22–29 (2007). [Google Scholar]
- 12.Boin A., McConnell A., Preparing for critical infrastructure breakdowns: The limits of crisis management and the need for resilience. J. Contingen. Crisis Manage. 15, 50–59 (2007). [Google Scholar]
- 13.Australian Medical Association, Public Hospital Report Card 2010. An AMA Analysis of Australia’s Public Hospital System (Australian Medical Association, 2010).
- 14.Sprivulis P. C., Da Silva J.-A., Jacobs I. G., Jelinek G. A., Frazer A. R. L., The association between hospital overcrowding and mortality among patients admitted via Western Australian emergency departments. Med. J. Aust. 184, 208–212 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Bishop J. F., Murnane M. P., Owen R., Australia’s winter with the 2009 pandemic influenza A (H1N1) virus. N. Engl. J. Med. 361, 2591–2594 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Australian Government Department of Health, Australian Influenza Surveillance Report and Activity Updates (2017); www.health.gov.au/flureport.
- 17.Itzwerth R., Moa A., MacIntyre C. R., Australia’s influenza pandemic preparedness plans: An analysis. J. Public Health Policy 39, 111–124 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Nsoesie E. O., Brownstein J. S., Ramakrishnan N., Marathe M. V., A systematic review of studies on forecasting the dynamics of influenza outbreaks. Influenza Other Respi. Viruses 8, 309–316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrova V. N., Russell C. A., The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 16, 47–60 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Chowell G., Viboud C., Munayco C. V., Gómez J., Simonsen L., Miller M. A., Tamerius J., Fiestas V., Halsey E. S., Laguna-Torres V. A., Spatial and temporal characteristics of the 2009 A/H1N1 influenza pandemic in Peru. PLOS ONE 6, e21287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhry A., Singh S., Khare S., Rai A., Rawat D. S., Aggarwal R. K., Chauhan L. S., Emergence of pandemic 2009 influenza A H1N1, India. Indian J. Med. Res. 135, 534–537 (2012). [PMC free article] [PubMed] [Google Scholar]
- 22.Echevarría-Zuno S., Mejía-Aranguré J. M., Mar-Obeso A. J., Grajales-Muñíz C., Robles-Pérez E., González-León M., Ortega-Alvárez M. C., González-Bonilla C. R., Rascón-Pacheco R. A., Borja-Aburto V. H., Infection and death from influenza A H1N1 virus in Mexico: A retrospective analysis. Lancet 374, 2072–2079 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Donaldson L. J., Rutter P. D., Ellis B. M., Greaves F. E. C., Mytton O. T., Pebody R. G., Yardley I. E., Mortality from pandemic A/H1N1 2009 influenza in England: Public health surveillance study. Br. Med. J. 339, b5213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayward A. C., Fragaszy E. B., Bermingham A., Wang L.-L., Copas A. J., Edmunds W. J., Ferguson N. M., Goonetilleke N., Harvey G., Kovar J., Lim M. S. C., McMichael A. J., Millett E. R. C., Nguyen-Van-Tam J. S., Nazareth I., Pebody R., Tabassum F., Watson J. M., Wurie F. B., Johnson A. M., Zambon M. C., Comparative community burden and severity of seasonal and pandemic influenza: Results of the Flu Watch cohort study. Lancet Respir. Med. 2, 445–454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrera-Valdez M. A., Cruz-Aponte M., Castillo-Chavez C., Multiple outbreaks for the same pandemic: Local transportation and social distancing explain the different “waves” of A-H1N1pdm cases observed in México during 2009. Math. Biosci. Eng. 8, 21–48 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Hoen A. G., Hladish T. J., Eggo R. M., Lenczner M., Brownstein J. S., Meyers L. A., Epidemic wave dynamics attributable to urban community structure: A theoretical characterization of disease transmission in a large network. J. Med. Internet Res. 17, e169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cauchemez S., Bhattarai A., Marchbanks T. L., Fagan R. P., Ostroff S., Ferguson N. M., Swerdlow D.; Pennsylvania H1N1 Working Group , Role of social networks in shaping disease transmission during a community outbreak of 2009 H1N1 pandemic influenza. Proc. Natl. Acad. Sci. U.S.A. 108, 2825–2830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Germann T. C., Kadau K., Longini I. M. Jr., Macken C. A., Mitigation strategies for pandemic influenza in the United States. Proc. Natl. Acad. Sci. U.S.A. 103, 5935–5940 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longini I. M. Jr., Nizam A., Xu S., Ungchusak K., Hanshaoworakul W., Cummings D. A. T., Halloran M. E., Containing pandemic influenza at the source. Science 309, 1083–1087 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Halloran M. E., Ferguson N. M., Eubank S., Longini I. M. Jr., Cummings D. A. T., Lewis B., Xu S., Fraser C., Vullikanti A., Germann T. C., Wagener D., Beckman R., Kadau K., Barrett C., Macken C. A., Burke D. S., Cooley P., Modeling targeted layered containment of an influenza pandemic in the United States. Proc. Natl. Acad. Sci. U.S.A. 105, 4639–4644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cliff O. M., Harding N., Piraveenan M., Erten E. Y., Gambhir M., Prokopenko M., Investigating spatiotemporal dynamics and synchrony of influenza epidemics in Australia: An agent-based modelling approach. Simul. Model. Pract. Theory 87, 412–431 (2018). [Google Scholar]
- 32.Chao D. L., Halloran M. E., Obenchain V. J., Longini I. M. Jr., FluTE, a publicly available stochastic influenza epidemic simulation model. PLOS Comput. Biol. 6, e1000656 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diekmann O., Heesterbeek J. A. P., Metz J. A. J., On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous populations. J. Math. Biol. 28, 365–382 (1990). [DOI] [PubMed] [Google Scholar]
- 34.Australian Bureau of Statistics, ABS Maps (2017); http://stat.abs.gov.au/itt/r.jsp?ABSMaps.
- 35.Australian Bureau of Statistics, Regional Population Growth, Australia 2016-17, Table 01. Estimated Resident Population by Remoteness Area (2018).
- 36.Yang Y., Sugimoto J. D., Halloran M. E., Basta N. E., Chao D. L., Matrajt L., Potter G., Kenah E., Longini I. M. Jr., The transmissibility and control of pandemic influenza A (H1N1) virus. Science 326, 729–733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brockmann D., Helbing D., The hidden geometry of complex, network-driven contagion phenomena. Science 342, 1337–1342 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Viboud C., Bjørnstad O. N., Smith D. L., Simonsen L., Miller M. A., Grenfell B. T., Synchrony, waves, and spatial hierarchies in the spread of influenza. Science 312, 447–451 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Pastor-Satorras R., Castellano C., Van Mieghem P., Vespignani A., Epidemic processes in complex networks. Rev. Mod. Phys. 87, 925 (2015). [Google Scholar]
- 40.Piraveenan M., Prokopenko M., Hossain L., Percolation centrality: Quantifying graph-theoretic impact of nodes during percolation in networks. PLOS ONE 8, e53095 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.K. M. Fair, C. Zachreson, M. Prokopenko, Creating a surrogate commuter network from Australian Bureau of Statistics census data; https://arxiv.org/abs/1808.09267 (2018). [DOI] [PMC free article] [PubMed]
- 42.Australian Curriculum, Assessment and Reporting Authority, ACARA Data Access Program (2017); www.acara.edu.au/contact-us/acara-data-access.
- 43.Australian Government Department of Infrastructure, Regional Development and Cities, Airport traffic data (2018); https://bitre.gov.au/publications/ongoing/airport_traffic_data.aspx.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/12/eaau5294/DC1
Movie S1. The color mapped, temporal evolution of the average simulated prevalence for 2006, using the SA2 partition.
Movie S2. The color mapped, temporal evolution of the average simulated prevalence for 2011, using the SA2 partition.
Movie S3. The color mapped, temporal evolution of the average simulated prevalence for 2016, using the SA2 partition.
Fig. S1. Incidence proportion for various Ro values.
Fig. S2. Prevalence proportion for various Ro values.
Fig. S3. Attack rate for various Ro values.
Model description


