To improve the precision of targeted and conventional therapy, the molecular profiles of gastroesophageal tumors were assessed, with the aim of comparing the molecular characteristics of esophageal adenocarcinoma, esophageal squamous cell carcinoma, and gastric adenocarcinoma. Results are reported in this article.
Keywords: Gastroesophageal cancers, Squamous cell carcinoma, Adenocarcinomas, Next‐generation sequencing
Abstract
Background.
Gastroesophageal cancers are often grouped together even though cancers that originate in the esophagus often exhibit different histological features, geographical distribution, risk factors, and clinical characteristics than those originating in the stomach. Herein, we aimed to compare the molecular characteristics of three different gastroesophageal cancer types: esophageal squamous cell carcinoma (ESCC), esophageal adenocarcinoma (EAC), and gastric adenocarcinoma (GAC).
Subjects, Materials, and Methods.
In total, 3,342 gastroesophageal cancers were examined. Next‐generation sequencing was performed on genomic DNA isolated from formalin‐fixed paraffin‐embedded tumor samples using the NextSeq platform. Tumor mutational burden was measured by counting all nonsynonymous missense mutations, and microsatellite instability was examined at over 7,000 target microsatellite loci. Immunohistochemistry and in situ hybridization techniques were also performed.
Results.
When compared with EAC and GAC, ESCC showed significantly lower mutational rates within APC, ARID1A, CDH1, KRAS, PTEN, and SMAD4, whereas more frequent mutations were observed in BAP1, CDKN2A, FOXO3, KMT2D, MSH6, NOTCH1, RB1, and SETD2. Human epidermal growth receptor 2 (HER2) overexpression was observed in 13% of EAC compared with 6% of GAC and 1% of ESCC (p < .0001). Compared with EAC and GAC, ESCC exhibited higher expression of programmed death‐ligand 1 (PD‐L1) (27.7% vs. 7.5% vs. 7.7%, p < .0001). We observed that FGF3, FGF4, FGF19, CCND1 (co‐localized on 11q13), and FGFR1 were significantly more amplified in ESCC compared with EAC and GAC (p < .0001).
Conclusion.
Molecular comparisons between ESCC, EAC, and GAC revealed distinct differences between squamous cell carcinomas and adenocarcinomas in each platform tested. Different prevalence of HER2/neu overexpression and amplification, and immune‐related biomarkers between ESCC, EAC, and GAC, suggests different sensitivity to HER2‐targeted therapy and immune checkpoint inhibition. These findings bring into question the validity of grouping patients with EAC and ESCC together in clinical trials and provide insight into molecular features that may represent novel therapeutic targets.
Implications for Practice.
This study highlights the genomic heterogeneity of gastroesophageal cancers, showing striking molecular differences between tumors originating from different locations. Moreover, this study showed that esophageal squamous cell carcinomas exhibit a unique molecular profile, whereas gastric adenocarcinomas and esophageal adenocarcinomas have some similarities, supporting the fact that adenocarcinomas and squamous cell carcinomas are completely different diseases, irrespective of the tumor location. This raises the question of whether treatment of gastroesophageal tumors should be determined according to histological subtype and molecular targets rather than anatomical site. These findings provide insights that could enable physicians to better select patients and inform therapeutic choices in order to improve clinical outcome.
摘要
背景。 虽然起源于食管的肿瘤与起源于胃部的肿瘤在组织学特征、区域分布、危险因素和临床特征等往往表现不同,但胃食管肿瘤往往被归为一类。在此,我们比较了三种不同类型的胃食管肿瘤的分子特征:食管鳞状细胞癌 (ESCC)、食管腺癌 (EAC) 和胃腺癌 (GAC)。
受试者、材料及方法。 共3 342例胃食管肿瘤患者接受了检查。利用 NextSeq 平台,从福尔马林固定石蜡包埋肿瘤标本中分离出基因组 DNA,进行了下一代测序。通过计数所有非同义错义突变来测量肿瘤突变负担,并在7 000多个靶微卫星位点上检测微卫星不稳定性。同时也使用了免疫组化和原位杂交技术。
结果。 与EAC 和 GAC相比, ESCC在 APC、ARID1A、CDH1、KRAS、PTEN和 SMAD4中的突变率明显降低,而突变更频繁地出现在BAP1、CDKN2A、FOXO3、KMT2D、MSH6、NOTCH1、RB1和 SETD2。观察到 EAC 的人表皮生长因子受体2(HER2) 超表达为13%,而 GAC 为6%,ESCC 为1%(p<0.0001)。与 EAC 和 GAC 相比,ESCC 表现为程序性死亡配体1(PD‐L1)(27.7% vs. 7.5% vs. 7.7%,p<0.000 1)高表达。与 EAC 和 GAC 相比,我们观察到 FGF3、FGF4、FGF19、CCND1 (共定位于11q13)及FGFR1 在 ESCC 中明显扩增 (p<0.0001)。
结论。 通过 ESCC、EAC 和 GAC 的分子比较显示,在每一个平台的测试中,鳞状细胞癌和腺癌之间有明显的差异。ESCC、EAC 和 GAC 之间的 HER2/neu 过表达和扩增以及与免疫相关的生物标记物的不同发生率表明,对 HER2 靶向治疗和免疫检查点抑制剂具有不同的敏感性。这些发现引起了人们对临床试验中将 EAC 和 ESCC 患者归为一组的有效性的质疑,并对可能代表新治疗靶点的分子特征提出了深刻见解。
实践意义: 这项研究突出了胃食管肿瘤的基因组异质性,显示源自不同部位的肿瘤之间有明显的分子差异。此外,本研究还发现食管鳞状细胞癌具有独特的分子特征,而胃腺癌和食管腺癌有一些相似之处,在不考虑肿瘤部位的情况下,这支持了腺癌和鳞状细胞癌是完全不同的疾病这一事实。这就提出了一个问题:是否应该根据组织学亚型和分子靶点而不是解剖部位来确定胃食管肿瘤的治疗方案。这些发现提出的深刻见解使医生能够更好地甄别患者,并告知治疗选项,以改善临床效果。
Introduction
In 2017 in the U.S., there are expected to be about 28,000 and 16,940 individuals newly diagnosed with gastric and esophageal cancer, respectively, along with 10,960 and 15,690 deaths from these diseases [1]. Gastroesophageal cancers are often grouped together, although cancers that originate from the esophagus and stomach exhibit different histological features, geographical distribution, risk factors, molecular profiles, and clinical characteristics.
Esophageal cancers comprise two main histological types: squamous cell carcinoma (ESCC) and adenocarcinomas (EAC). The former is more prevalent in the upper and mid‐esophagus, whereas the latter predominates in the lower esophagus. The incidence of ESCC has been decreasing in the U.S. over the last few decades, in part due to reductions of well‐established, behavioral risk factors such as tobacco and alcohol consumption. However, an increase in the incidence of EAC has been observed, most likely due to the increasing rates in Western countries of obesity and gastroesophageal reflux disease (GERD), the main cause of Barrett's esophagus, which is considered the precancerous lesion of EAC [2].
The majority of gastric cancers are adenocarcinomas (GAC). GAC can be further divided into intestinal and diffuse types according to Lauren's classification [3]. These subtypes have distinct characteristics in terms of cells morphology, pathogenesis, and molecular and epidemiological features [4]. Gastric cancer incidence rates have declined in most parts of the world, although tumors in the cardia have displayed the opposite trend, mostly due to the increasing rates of obesity and GERD, which are the main risk factors associated with this type of cancer [5].
An unresolved issue is an appropriate demarcation between gastric and esophageal adenocarcinomas; thus, tumors arising in this anatomic area are currently grouped together and termed gastroesophageal junction (GEJ) cancers. In fact, in the revised American Joint Committee on Cancer staging system, cancers involving GEJ that have their epicenter within the proximal 2 cm of the cardia (Siewert types I/II) are classified as esophageal cancers, whereas cancers with an epicenter more than 2 cm distal from the GEJ are staged using the stomach cancer TNM and stage groups [6]. However, this anatomical approach to classification does not take genomic differences in tumor types into account, and, therefore, it remains controversial.
In order to better classify gastric and esophageal cancers, The Cancer Genome Atlas (TCGA) consortium extensively characterized the genetic landscape of these diseases [7], [8]. Gastric cancers were subdivided into four groups according to the presence or absence of Epstein‐Barr virus (EBV) infection, and three genetic determinants: microsatellite instability (MSI), genome stability, and chromosomal instability, which may serve as a guide for novel targeted therapy [7]. In addition, it has been shown that the histological subtypes of EAC and ESCC are distinct in their molecular characteristics across all platforms tested [8]. Comprehensive molecular studies in the research setting, using multiple test platforms by TCGA, have suggested that molecular alterations in tumors of the upper gastrointestinal tract lead to significant heterogeneity that is not reflected by histologic features. For instance, ESCC shares molecular similarities with head and neck squamous cell cancer (HNSCC), whereas EAC exhibits features of the chromosomal instability subtype of gastric cancer that is prevalent in tumors arising in the GEJ/cardia [8]. These findings raise the issue of whether it is still advisable to combine EAC and ESCC in clinical trials.
In an attempt to improve the precision of targeted and conventional therapy, we have assessed the molecular profiles of these tumors, aiming to compare the molecular characteristics of EAC, ESCC, and GAC. We explored potentially targetable biomarkers in each of these cancer groups, using a commercially available test platform, which could be translated into different treatment strategies. In addition, we aimed to compare molecular differences between intestinal and diffuse gastric cancer, with the goal of providing insights into molecular features that may represent an opportunity for the development of novel therapeutic agents.
Subjects, Materials, and Methods
Patients
Patients with gastroesophageal tumors profiled by Caris Life Sciences between 2009 and September 2017 were deidentified, and their specimens were retrospectively analyzed for molecular alterations. Tumor histology and diagnoses were taken from submitted pathology reports and confirmed by board‐certified pathologists.
Next‐Generation Sequencing
Next‐generation sequencing (NGS) was performed on genomic DNA isolated from formalin‐fixed paraffin‐embedded (FFPE) tumor samples using NextSeq platform (Illumina, Inc., San Diego, CA). We did not have available matched normal tissue for sequencing. A custom‐designed SureSelect XT assay was used to enrich 592 whole‐gene targets (Agilent Technologies, Santa Clara, CA). All variants were detected with >99% confidence based on allele frequency and amplicon coverage with an average sequencing depth of coverage of >500 and with an analytic sensitivity of 5%. Tumor enrichment was achieved by harvesting targeted tissue by manual microdissection performed on all cases prior to molecular testing.
Tumor Mutational Burden and Microsatellite Instability by NGS
Tumor mutational burden (TMB) was measured (592 genes and 1.4 megabases sequenced per tumor) by counting all nonsynonymous missense mutations found per tumor that had not been previously described as germline alterations. The threshold to define TMB‐high was greater than or equal to 17 mutations/megabase and was established by comparing TMB with MSI by Fragment analysis (FA) in colorectal cancer (CRC) cases, based on reports of TMB having high concordance with MSI in CRC. MSI was examined at over 7,000 target microsatellite loci and compared with the reference genome hg19 from the University of California, Santa Cruz Genome Browser database. Copy number variation (CNV) was tested by NGS and was determined by comparing the depth of sequencing of genomic loci to a diploid control as well as the known performance of these genomic loci. Calculated gains ≥6 copies were considered amplified.
Immunohistochemistry Analysis
Immunohistochemistry (IHC) was performed on full FFPE sections of glass slides. Slides were stained using automated staining techniques, per the manufacturer's instructions, and were optimized and validated per Clinical Laboratory Improvement Amendments/CAO and International Organization for Standarization (ISO) requirements. Staining was scored for intensity (0 = no staining; 1+ = weak staining; 2+ = moderate staining; 3+ = strong staining) and staining percentage (0%–100%). Results were categorized as positive or negative by defined thresholds specific to each marker based on published clinical literature that associates biomarker status with patient responses to therapeutic agents. A board‐certified pathologist evaluated all IHC results independently. The primary antibodies used were ERCC1 (8F1), RRM1 (polyclonal), human epidermal growth receptor 2 (HER2; 4B5), epidermal growth factor receptor (EGFR; 31G7), PTEN (6H2.1), TLE3 (polyclonal), TOPO1 (1D6), TUBB3 (polyclonal), and P‐glycoprotein (PGP; C494). Chromogenic in situ hybridization (CISH) was used for HER2/neu amplification (INFORM HER2 Dual ISH DNA Probe Cocktail), and amplification was defined as HER2/chr17 ratio ≥2.0. The primary antibody for programmed death‐ligand 1 (PD‐L1) was SP142, and the staining was regarded as positive if its intensity on the membrane of the tumor cells was ≥2+ and the percentage of positively stained cells was >5%.
Results
Patient Characteristics
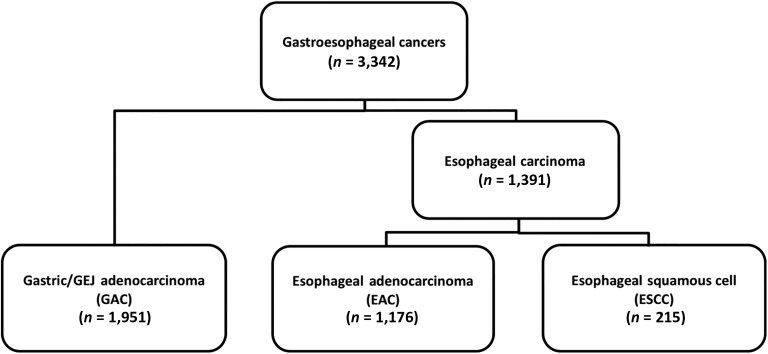
A total of 3,342 tumors were investigated: 1,391 esophageal tumors and 1,951 gastric tumors. The tumors originating in the esophagus were further classified based on histological subtype: 215 tumors were classified as ESCC and 1,176 tumors as EAC (Fig. 1). All gastric/GEJ tumors were GACs.
Figure 1.
Consolidated Standards of Reporting Trials diagram.
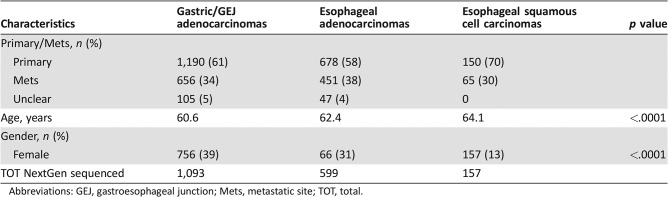
All three cohorts of tumors were more common in males than females, with a significantly higher prevalence in males of esophageal tumors (p < .0001) compared with gastric adenocarcinomas (Table 1).
Table 1. Patient characteristics.
Abbreviations: GEJ, gastroesophageal junction; Mets, metastatic site; TOT, total.
Mutational Profile and Copy Number Variations via Next‐Generation Sequencing
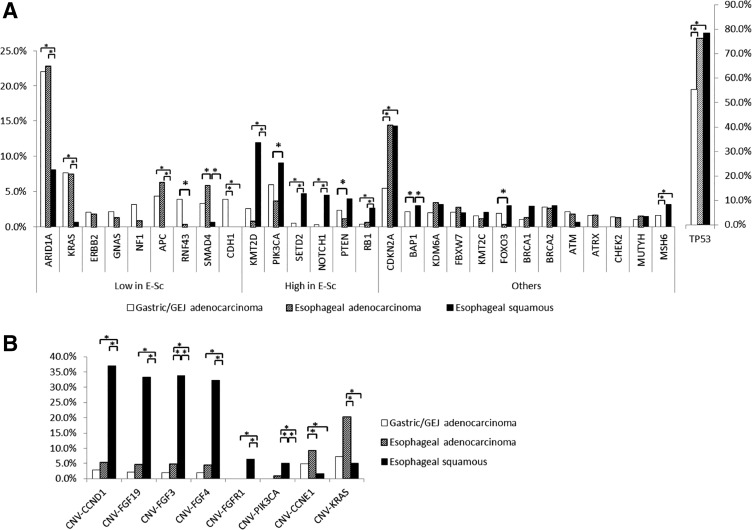
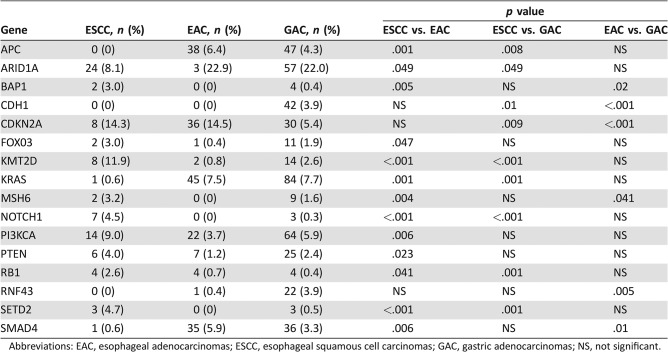
NGS was performed on 157 ESCC, 599 EAC, and 1,093 GAC tumors. As shown in Figure 2A and in Table 2, ESCC exhibits a different molecular profile compared with EAC and GAC. Indeed, ESCC showed lower mutational rates in ARID1A, KRAS, APC, PTEN, SMAD4, and CDH1, whereas more frequent mutations were observed in KMT2D, SETD2, NOTCH1, RB1, CDKN2A, BAP1, FOXO3, and MSH6 compared with EAC and GAC. On the other hand, when we compared EAC and GAC, only a few genes showed different mutational rates (BAP1, CDH1, CDKN2A, MSH6, RNF43, and SMAD4).
Figure 2.
Next‐generation sequencing comparison between esophageal squamous carcinomas, esophageal adenocarcinomas, and gastric/GEJ adenocarcinomas. (A): Mutational frequency. (B): Copy number variations analysis.
Abbreviations: *, significantly different mutation rate; GEJ, gastroesophageal junction.
Table 2. Molecular differences between ESCC, EAC, and GAC via next‐generation sequencing.
Abbreviations: EAC, esophageal adenocarcinomas; ESCC, esophageal squamous cell carcinomas; GAC, gastric adenocarcinomas; NS, not significant.
We further analyzed the CNV differences between these three cohorts (Fig. 2B). We observed that FGF3, FGF4, FGF19, and CCND1 co‐localized on 11q13 and were significantly more amplified in ESCC compared with EAC and GAC (respectively: FGF3, 33.9% vs. 4.8% vs. 2.1%, p < .0001; FGF4, 32.3 vs. 4.4% vs. 2.1%, p < .0001; FGF19, 33.3% vs. 4.5% vs. 2.2%, p < .0001; CCND1, 37.1% vs. 5.2% vs. 2.8%, p < .0001). ESCC also showed more amplified FGFR1 (6.5% vs. 0.0% vs. 0.0%, p < .0001) and PIK3CA (5.0% vs. 0.8% vs. 0.0%, p = .002 and p < .0001) than EAC and GAC, respectively. Finally, EAC showed more amplified CCNE1 compared with GAC and ESCC (9.2% vs. 4.9% vs. 1.6%, p = .019 and p = .044, respectively).
Comparisons of Immune‐Related Biomarkers (MSI‐H, TMB, and PD‐L1)
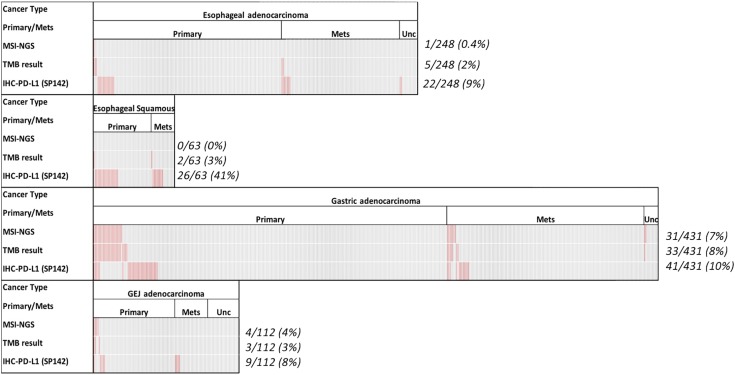
Figure 3 depicts the OncoPrint showing the prevalence of immune‐related biomarkers in the three different cohorts. We have recently examined the molecular landscape of TMB, MSI‐high (MSI‐H), and PD‐L1 among gastrointestinal tumors. We observed that GEJ exhibit an immune profile that is different from gastric and esophageal adenocarcinomas [9]; therefore, in the current analysis, we compared GAC, GEJ, EAC, and ESCC. MSI‐H and TMB‐high (TMB‐H) are more prevalent in GAC compared with ESCC and EAC. Specifically, MSI‐H status was shown in 7% of GAC, 4% in GEJ adenocarcinomas, 0.4% in EAC, and 0% in ESCC. TMB‐H was observed in 8% of GAC, 3% of GEJ adenocarcinomas, 2% of EAC, and 3% of ESCC. On the other hand, PD‐L1 overexpression was more prevalent in ESCC (41%) compared with GAC (9%) and EAC (2%). However, none of these differences were significantly different. Overall, immune‐related biomarkers are more prevalent in primary tumors than metastatic tumors. Specifically, MSI‐H prevalence was 5.7% in primary/local (31/541) and 2.4% (7/294) in metastatic sites (p = .027).
Figure 3.
OncoPrint showing the landscape of immune checkpoint inhibitor‐associated markers in the subtypes of gastroesophageal cancers. Every column indicates a tumor: Red indicates MSI‐NGS high, TMB high, or PD‐L1 overexpression; gray indicates un‐notable data.
Abbreviations: GEJ, gastroesophageal junction; IHC, immunohistochemistry; Mets, metastatic site; MSI, microsatellite instability; NGS, next‐generation sequencing; PD‐L1, programmed death‐ligand 1; TMB, tumor mutational burden; Unc, unclear.
Protein Expression and Gene Amplification
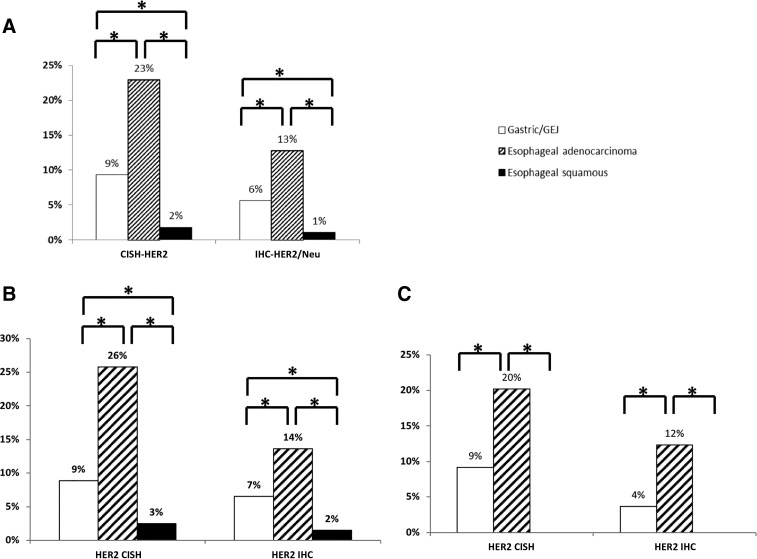
We further analyzed HER2 overexpression via IHC and amplification via CISH. As shown in Figure 4, HER2 overexpression and amplification are significantly higher in EAC than in GAC or ESCC (the lowest). Specifically, HER2 overexpression was observed in 13% of EAC compared with 6% of GAC and 1% of ESCC (p < .0001), whereas HER2 amplification was seen in 23% of EAC compared with 9% of GAC and 2% of ESCC (p < .0001; Fig. 4A). Of note, when we excluded MSI‐H tumors and tumors with diffuse histology, these results remained significant (supplemental online Tables 1, 2). Thus, we analyzed HER2 overexpression and amplification in primary and metastatic tumors. Interestingly, we observed the same prevalence and differences, regardless of tumor location (Fig. 4B, 4C).
Figure 4.
HER2 amplification and overexpression in the three cancer types. (A): All tumors. (B): Primary tumors. (C): Metastatic tumors.
Abbreviations: *, significantly different mutation rate; CISH, chromogenic in situ hybridization; HER2, human epidermal growth receptor 2; IHC, immunohistochemistry.
We then analyzed differences in protein expression between the three cohorts (supplemental online Fig. 1). Compared with EAC and GAC, ESCC displayed higher expression of ERCC1 (ESCC vs. EAC vs. GAC: 64.5% vs. 37.0% vs. 32.5%; p < .0001), EGFR (88.5% vs. 68.5% vs. 69.2%; p = .004), PD‐L1 (27.7% vs. 7.5% vs. 7.7%, p < .0001), PTEN (68.7% vs. 52.0% vs. 52.2%; p = .0003), RRM1 (56.0% vs. 37.9% vs. 32.9%; p = .002 and p < .0001, respectively), TLE3 (69.8% vs. 33.9% vs. 31.1%; p < .0001), and TOPO1 (76.1% vs. 67.0% vs. 62.7%; p = .015 and p = .0003, respectively). On the other hand, PGP showed lower expression in ESCC compared with EAC and GAC (9.3% vs. 50.9% vs. 46.9%; p < .0001).
Comparison Between Intestinal and Diffuse Gastric Cancers
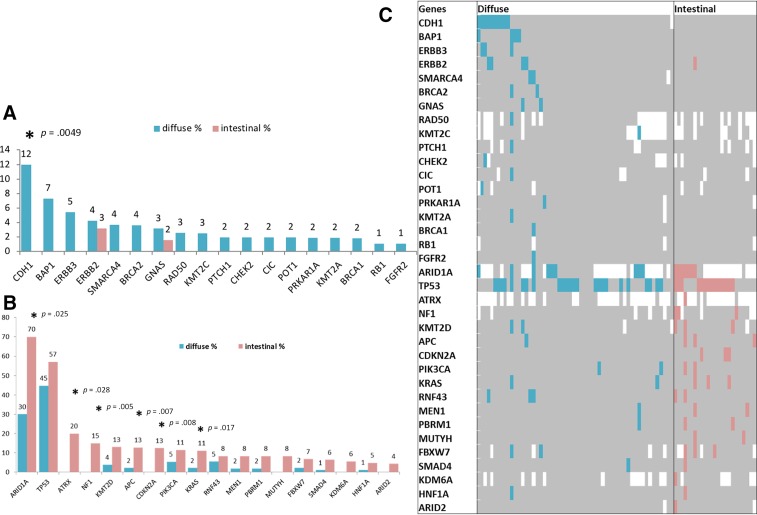
Among our gastric cohort, 296 gastric adenocarcinomas with annotated histology (diffuse subtype, n = 181; intestinal subtype, n = 115) were analyzed. Patients with diffuse type cancers were younger than those with intestinal subtype cancers (mean age, 58 vs. 67.5 years, p < .0001). The majority of patients with a diffuse subtype were female compared with those with an intestinal subtype (51% vs. 35%, p = .0051; supplemental online Table 3). The most frequently mutated genes in intestinal subtype cancers were ARID1A (70%), TP53 (57%), ATRX (20%), and NF1 (15%), whereas the most frequent mutations in diffuse cancers were TP53 (45%), ARID1A (30%), CDH1 (12%), BAP1 (7%), and RNF43 (5%). The intestinal subtype had a significantly higher mutation rate than the diffuse subtype in ARID1A (70.0% vs. 30.0%, p = .025), ATRX (20.0% vs. 0.0%, p = .028), NF1 (15.0% vs. 0.0%; p = .005), APC (13% vs. 2%, p = .007), CDKN2A (13.0% vs. 0.0%, p = .008), and KRAS (11.0% vs. 2.0%, p = .017), whereas the diffuse subtype had a higher rate of CDH1 (12.0% vs. 0.0%, p = .0049; Fig. 5). There was no significant difference in PD‐L1 tumor expression (diffuse, 3%; intestinal, 9%). Accordingly, MSI‐H and TMB‐high was seen in 5% and 4% of diffuse cancers and 13% and 8% of intestinal tumors, respectively, although no significant differences were observed (supplemental online Fig. 1). Finally, when compared with diffuse subtype, intestinal subtype exhibited greater HER2 amplification (29% vs. 3%, p < .0001; supplemental online Fig. 3). Of note, the lack of EBV evaluation may have impacted the results.
Figure 5.
Next‐generation sequencing reveals significant difference between mutational landscapes of diffuse (DS) and intestinal (IS) subtypes. (A): Gene mutations seen more frequently in DS than in IS. (B): Gene mutations seen more frequently in IS than in DS. For A and B: Y axis indicates the mutation frequency (%); data are from both TruSEQ and NextSEQ; significant differences are indicated by an asterisk with the p value calculated by chi‐square test. (C): Mutation pattern seen in DS and IS tumors tested with NextSEQ panel. Blue and red indicate the pathogenic and presumed pathogenic mutations; gray indicates wild type or nonpathogenic variants, whereas blank indicates unavailable data points.
Discussion
In the era of precision medicine, optimizing therapeutics and drug combinations for a small subset of patients based on their tumor molecular characteristics is of potential importance in order to improve outcomes and minimize exposure to unnecessary toxicities. For this reason, several studies [7], [8], [10] have attempted to characterize the molecular profile of these extremely heterogeneous diseases, although none of these have been incorporated into clinical practice to guide treatment choice. In fact, to date, the only available biomarkers to guide therapeutic decisions include HER2 amplification for trastuzumab [11], MSI‐H, or mismatch repair deficient (dMMR), and PD‐L1 expression (>1%) for pembrolizumab [12]. Ramucirumab [13], an anti‐vascular endothelial growth factor antibody, and nivolumab (approved in Asia) [14] do not require biomarker selection. Moreover, all these targeted agents are available only for GAC and GEJ metastatic adenocarcinomas; thus, ESCC patients are not eligible for these treatments, underlining the urgent need for more therapeutic options for these patients.
Our study highlights the genomic heterogeneity of gastroesophageal cancers, showing differences between tumors originating from different locations in all the platforms tested. These data suggest the presence of lineage‐specific alterations that drive progression in EAC, ESCC, and GAC. Moreover, we showed that ESCC exhibit a unique molecular profile, whereas GAC and EAC harbor some similarities, supporting the fact that adenocarcinomas and squamous cell carcinoma are completely different diseases, irrespective of the tumor location.
In the esophageal tumor cohort investigated in this study, distinct proteomic and genomic differences were seen between patients with ESCC and EAC.
ESCC exhibited a unique molecular profile compared with the other cohorts analyzed. For instance, genes on the MAPK pathway (KRAS, ERBB2, GNAS, NF1) and WNT pathway activation (APC, RNF43) are less frequently mutated in ESCC compared with the other cohorts, whereas mutations in chromatin modifier genes (KMT2D, SETD2), PI3K pathway (PIK3CA, PTEN), NOTCH1, and RB1 are more frequent (Fig. 2A; Table 2). Our findings are consistent with previous reports [15], [16], [17] and shed light on molecular alterations that may contribute to ESCC tumorigenesis, and they might represent the basis for the development of novel drugs. These differences suggest that ESCC and EAC should not be considered as one disease and that they should not be grouped together in clinical trials.
Herein, we observed that 6% of ESCC had amplification of FGFR1, as presented in prior studies [18], [19] (Fig. 2B). Given the established roles of aberrant FGFR signaling in oncogenesis, FGFR‐targeted agents may be used to dwarf tumor growth, target angiogenesis, and reverse acquired resistance to anticancer agents [20]. Furthermore, in our study, ESCC exhibited a significantly higher amplification of CCND1 in comparison with its adenocarcinoma counterparts (Fig. 2B). CCND1 encodes for the cyclin D1 protein involved in G1/S cell cycle transition and may be a potential biomarker of sensitivity to cyclin‐dependent kinase (CDK) inhibitors (e.g., palbociclib) [21]. Although monotherapy with CDK inhibitors has shown only modest activity in gastroesophageal cancers in a small phase II trial [22], the combination with other therapeutic strategies may be of potential benefit and should be tested in future clinical studies [23]. Interestingly, protein expression of PGP is more than fivefold higher in adenocarcinomas than in squamous cell tumors, suggesting that adenocarcinomas carry a significantly higher multidrug resistance phenotype [24]. Our findings support previous reports in which PGP expression was associated with chemo resistance and higher recurrence rate in gastric cancer [25].
Before pembrolizumab was approved for the treatment of gastric and GEJ adenocarcinomas, using PD‐L1 expression as a predictive biomarker, the only validated predictive biomarker available was the HER2/neu amplification for trastuzumab [11]. Herein, we showed that HER2 protein expression via IHC and gene amplification via CISH is observed almost exclusively in adenocarcinomas (Fig. 4). Despite different results reported previously [26], we show a near absence of HER2 expression/amplification in ESCC. These results are in accordance with the TCGA report of a 3% HER2 amplification in ESCC and 32% in EAC [8]. The lower positivity rate shown in our study reflects the difference in platforms used: The American Society of Clinical Oncology/College of American Pathologists‐required thresholds used here (IHC: 3+, 10% and CISH: HER2/CEP17 ratio of 2.0) provide a more precise evaluation of patients who could benefit from treatment. Additionally, we also observed that among adenocarcinomas, HER2‐positive rate is higher in EAC than in GAC (p < .0001; Fig. 4). Interestingly, the same pattern of HER2/neu overexpression and amplification was observed in both primary and metastatic sites (Fig. 4B, 4C).
When we analyzed the landscape of immune‐related biomarkers (Fig. 3), higher expression of PD‐L1 was observed in ESCC (41%) compared with EAC (9%) and GAC (10%). Of note, pembrolizumab has the indication only for PD–L1‐positive (combined positive score ≥1) gastric and GEJ adenocarcinomas. However, the efficacy of immune checkpoint inhibitors in squamous cell tumors (e.g., HNSCC [27], [28], squamous non‐small cell lung cancer [29], and anal canal [30]) has been established, and nivolumab showed promising activity with a manageable safety profile for patients with treatment‐refractory advanced ESCC [31], suggesting that these patients may potentially benefit from immune checkpoint inhibitor treatment. MSI‐H status is a positive predictive biomarker of benefit from immunotherapy and should be tested in all gastrointestinal cancers patients, as this group of patients, although very small, may experience durable responses, as reported in several clinical trials that have led to the U.S. Food and Drug Administration approval of pembrolizumab for the treatment of patients with unresectable or metastatic, MSI‐H, or dMMR solid tumors. TMB‐high has been associated with improved sensitivity to immune checkpoint inhibitors [32], [33]; therefore, it may be a useful biomarker, together with PD‐L1 and MSI‐H/dMMR status, to better select those patients who will benefit from immunotherapy. It is noteworthy that it has recently been shown that enhanced T cell response and prolonged survival are associated with unique epitope “quality,” instead of the “quantity” of mutation load [34], opening new horizons in this field. Additionally, several other biomarkers have been shown to be potential predictive factors of response to immune checkpoint inhibitors, such as EBV and human papillomavirus status: Indeed, EBV‐positive low‐mutation‐burden gastric cancers are a subset of microsatellite‐stable gastric cancers that may respond to immune checkpoint therapy [35].
Finally, we analyzed the molecular differences between intestinal and diffuse gastric cancers (Fig. 5). Patients with diffuse subtype tumors were significantly more likely to be female and younger compared with those with an intestinal subtype, reflecting the well‐known distributions of Lauren's classification [36]. A different molecular profile observed with NGS analysis, as well as a different protein expression with IHC analysis, indicate different carcinogenic pathways and biology, which may be the underlying cause of potential differences in response to therapy. Of interest, intestinal subtype tumors exhibited a higher prevalence of immune‐related biomarkers, such as MSI‐H, TMB‐high, and PD‐L1 overexpression, compared with the diffuse subtype, as well as a greater HER2 amplification compared with the diffuse subtype, highlighting a potential higher sensitivity to immune checkpoint inhibitors and to anti‐HER2 targeted therapy.
Gastroesophageal cancers are widely heterogeneous diseases, not only between different histologies and tumor locations, as we reported in this study, but also within the same patient, between primary tumor and metastatic lesions [37]. This is the case even in patients who have not received prior systemic therapy, suggesting that high gastroesophageal cancer heterogeneity is likely a natural feature of this malignancy, rather than a consequence of clonal selection arising from therapeutic pressure [38].
We certainly acknowledge that our study has some limitations, such as the retrospective nature of the analysis and the lack of clinical and outcome data for these patients that did not allow us to correlate biomarkers with outcomes. Furthermore, the evaluation of PD‐L1 in intestinal and diffuse tumors should ideally be stratified according to MSI status; however, the numbers were too small to allow for such analysis.
Conclusion
Molecular comparisons between ESCC, EAC, and GAC revealed distinct separation between squamous cell carcinomas and adenocarcinomas in each platform tested. Here, we showed that ESCC exhibits a unique molecular profile that differs from EAC and GAC, while exhibiting molecular similarity with HNSCC [39], whereas EAC showed stronger similarity with GAC. This raises the question of whether treatment of gastroesophageal tumors should be determined according to histological subtype rather than anatomical site. These findings also provide insights into the different prevalence of HER2/neu overexpression and amplification and of immune‐related biomarkers between ESCC, EAC, and GAC, suggesting different sensitivity to HER2‐targeted therapy and immune checkpoint inhibitors. Moreover, among the gastric cancer cohort, the intestinal subtype exhibited a higher prevalence of immune‐related biomarkers as well as a greater HER2 amplification compared with the diffuse subtype. In the present study, we examined the molecular profiles of these tumors in a large patient cohort with gastroesophageal cancers and attempted to portray the differences between these three tumor types, trying to offer a more comprehensive understanding of the underlying biology, which could enable us to better select patients and inform therapeutic choices in order to improve clinical outcome. Further studies are warranted to deeply investigate potential novel targets for therapeutic development and to shed light on the heterogeneity of these tumors, even within the same patient.
See http://www.TheOncologist.com for supplemental material available online.
Author Contributions
Conception/design: Mohamed E. Salem, Alberto Puccini
Provision of study material or patients: Mohamed E. Salem, Alberto Puccini, Joanne Xiu, Derek Raghavan, Heinz‐Josef Lenz, W. Michael Korn, Anthony F. Shields, Philip A. Philip, John L. Marshall, Richard M. Goldberg
Collection and/or assembly of data: Mohamed E. Salem, Alberto Puccini, Joanne Xiu
Data analysis and interpretation: Mohamed E. Salem, Alberto Puccini, Joanne Xiu, Derek Raghavan, Heinz‐Josef Lenz, W. Michael Korn, Anthony F. Shields, Philip A. Philip, John L. Marshall, Richard M. Goldberg
Manuscript writing: Mohamed E. Salem, Alberto Puccini, Joanne Xiu, Derek Raghavan, Heinz‐Josef Lenz, W. Michael Korn, Anthony F. Shields, Philip A. Philip, John L. Marshall, Richard M. Goldberg
Final approval of manuscript: Mohamed E. Salem, Alberto Puccini, Joanne Xiu, Derek Raghavan, Heinz‐Josef Lenz, W. Michael Korn, Anthony F. Shields, Philip A. Philip, John L. Marshall, Richard M. Goldberg
Disclosures
Joanne Xiu: Caris Life Sciences (E, OI); Derek Raghavan: Caris Life Sciences (SAB), Gerson Lehrman, Pfizer (C/A); W. Michael Korn: Caris Life Sciences (E, OI), Merck, Eli Lilly and Company (SAB); Anthony F. Shields: Caris Life Sciences (RF, travel expenses); John L. Marshall: Caris Life Sciences (SAB, C/A, H). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Domper Arnal MJ, Ferrandez Arenas A, Lanas Arbeloa A. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol 2015;21:7933–7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lauren P. The two histological main types of gastric carcinoma: Diffuse and so‐called intestinal‐type carcinoma. An attempt at a histo‐clinical classification. Acta Pathol Microbiol Scand 1965;64:31–49. [DOI] [PubMed] [Google Scholar]
- 4. Shah MA, Khanin R, Tang L et al. Molecular classification of gastric cancer: A new paradigm. Clin Cancer Res 2011;17:2693–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karimi P, Islami F, Anandasabapathy S et al. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rice TW, Ishwaran H, Ferguson MK et al. Cancer of the esophagus and esophagogastric junction: An eighth edition staging primer. J Thorac Oncol 2017;12:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network . Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network . Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salem ME, Puccini A, Grothey A et al. Landscape of tumor mutation load, mismatch repair deficiency, and PD‐L1 expression in a large patient cohort of gastrointestinal cancers. Mol Cancer Res 2018;16(suppl 5):805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cristescu R, Lee J, Nebozhyn M et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–456. [DOI] [PubMed] [Google Scholar]
- 11. Bang YJ, Van Cutsem E, Feyereislova A et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): A phase 3, open‐label, randomised controlled trial. Lancet 2010;376:687–697. [DOI] [PubMed] [Google Scholar]
- 12. Fuchs CS, Doi T, Jang RW et al. KEYNOTE‐059 cohort 1: Efficacy and safety of pembrolizumab (pembro) monotherapy in patients with previously treated advanced gastric cancer. J Clin Oncol 2017;35(suppl 15):4003a. 29040031 [Google Scholar]
- 13. Fuchs CS, Tomasek J, Yong CJ et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro‐oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo‐controlled, phase 3 trial. Lancet 2014;383:31–39. [DOI] [PubMed] [Google Scholar]
- 14. Kang YK, Boku N, Satoh T et al. Nivolumab in patients with advanced gastric or gastro‐oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO‐4538‐12, ATTRACTION‐2): A randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet 2017;390:2461–2471. [DOI] [PubMed] [Google Scholar]
- 15. Cheng C, Zhou Y, Li H et al. Whole‐genome sequencing reveals diverse models of structural variations in esophageal squamous cell carcinoma. Am J Hum Genet 2016;98:256–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao YB, Chen ZL, Li JG et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097–1102. [DOI] [PubMed] [Google Scholar]
- 17. Lin DC, Hao JJ, Nagata Y et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 2014;46:467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. von Loga K, Kohlhaussen J, Marx AH et al. Abstract 3024: FGFR1 amplification is linked to the squamous cell carcinoma subtype in esophageal carcinoma. Cancer Res 2013;73:3024–3024. [Google Scholar]
- 19. Dienstmann R, Rodon J, Prat A et al. Genomic aberrations in the FGFR pathway: Opportunities for targeted therapies in solid tumors. Ann Oncol 2014;25:552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Touat M, Ileana E, Postel‐Vinay S et al. Targeting FGFR signaling in cancer. Clin Cancer Res 2015;21:2684–2694. [DOI] [PubMed] [Google Scholar]
- 21. O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol 2016;13:417–430. [DOI] [PubMed] [Google Scholar]
- 22. Karasic TB, O'Hara MH, Teitelbaum UR et al. Phase II trial of palbociclib in patients with advanced esophageal or gastric cancer. J Clin Oncol 2018;36(suppl 4):68a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou J, Wu Z, Wong G et al. CDK4/6 or MAPK blockade enhances efficacy of EGFR inhibition in oesophageal squamous cell carcinoma. Nat Commun 2017;8:13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Higgins CF. Multiple molecular mechanisms for multidrug resistance transporters. Nature 2007;446:749–757. [DOI] [PubMed] [Google Scholar]
- 25. Shi WJ, Gao JB. Molecular mechanisms of chemoresistance in gastric cancer. World J Gastrointest Oncol 2016;8:673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhan N, Dong WG, Tang YF et al. Analysis of HER2 gene amplification and protein expression in esophageal squamous cell carcinoma. Med Oncol 2012;29:933–940. [DOI] [PubMed] [Google Scholar]
- 27. Ferris RL, Blumenschein G, Jr, Fayette J et al. Nivolumab for recurrent squamous‐cell carcinoma of the head and neck. N Engl J Med 2016;375:1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seiwert TY, Burtness B, Mehra R et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE‐012): An open‐label, multicentre, phase 1b trial. Lancet Oncol 2016;17:956–965. [DOI] [PubMed] [Google Scholar]
- 29. Brahmer J, Reckamp KL, Baas P et al. Nivolumab versus docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N Engl J Med 2015;373:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morris VK, Salem ME, Nimeiri H et al. Nivolumab for previously treated unresectable metastatic anal cancer (NCI9673): A multicentre, single‐arm, phase 2 study. Lancet Oncol 2017;18:446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kudo T, Hamamoto Y, Kato K et al. Nivolumab treatment for oesophageal squamous‐cell carcinoma: An open‐label, multicentre, phase 2 trial. Lancet Oncol 2017;18:631–639. [DOI] [PubMed] [Google Scholar]
- 32. Rizvi NA, Hellmann MD, Snyder A et al. Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science 2015;348:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD‐1 inhibition. N Engl J Med 2017;377:2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Balachandran VP, Luksza M, Zhao JN et al. Identification of unique neoantigen qualities in long‐term survivors of pancreatic cancer. Nature 2017;551:512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Panda A, Mehnert JM, Hirshfield KM et al. Immune activation and benefit from avelumab in EBV‐positive gastric cancer. J Natl Cancer Inst 2017;110:316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu MZ, Cai MY, Zhang DS et al. Clinicopathological characteristics and prognostic analysis of Lauren classification in gastric adenocarcinoma in China. J Transl Med 2013;11:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pectasides E, Stachler MD, Derks S et al. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov 2018;8:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sundar R, Tan P. Genomic analyses and precision oncology in gastroesophageal cancer: Forwards or backwards? Cancer Discov 2018;8:14–16. [DOI] [PubMed] [Google Scholar]
- 39. Dubot C, Bernard V, Sablin MP et al. Comprehensive genomic profiling of head and neck squamous cell carcinoma reveals FGFR1 amplifications and tumour genomic alterations burden as prognostic biomarkers of survival. Eur J Cancer 2018;91:47–55. [DOI] [PubMed] [Google Scholar]