Abstract
Zinc is an essential cofactor for many proteins. A key mechanism of zinc homeostasis during deficiency is “zinc sparing” in which specific zinc-binding proteins are repressed to reduce the cellular requirement. In this report, we evaluated zinc sparing across the zinc proteome of Saccharomyces cerevisiae. The yeast zinc proteome of 582 known or potential zinc-binding proteins was identified using a bioinformatics analysis that combined global domain searches with local motif searches. Protein abundance was determined by mass spectrometry. In zinc-replete cells, we detected over 2500 proteins among which 229 were zinc proteins. Based on copy number estimates and binding stoichiometries, a replete cell contains ~9 million zinc-binding sites on proteins. During zinc deficiency, many zinc proteins decreased in abundance and the zinc-binding requirement decreased to ~5 million zinc atoms per cell. Many of these effects were due at least in part to changes in mRNA levels rather than simply protein degradation. Measurements of cellular zinc content showed that the level of zinc atoms per cell dropped from over 20 million in replete cells to only 1.7 million in deficient cells. These results confirmed the ability of replete cells to store excess zinc and suggested that the majority of zinc-binding sites on proteins in deficient cells are either unmetalated or mismetalated. Our analysis of two abundant zinc proteins, Fba1 aldolase and Met6 methionine synthetase, supported that hypothesis. Thus, we have discovered widespread zinc sparing mechanisms and obtained evidence of a high accumulation of zinc proteins that lack their cofactor during deficiency.
Graphical Abstract

We cataloged the zinc proteome of Saccharomyces cerevisiae and analyzed how it responds to changing zinc ststus.
INTRODUCTION
Zinc is an essential catalytic and/or structural cofactor for many proteins. Approximately 9% of genes in eukaryotic organisms and ~5% of prokaryotic genes encode proteins that bind zinc to become functional (1). The abundance and importance of zinc-dependent proteins is reflected in the concept of the “zinc quota”. The zinc quota is defined as the amount of zinc in a cell grown under a given condition (2). The “minimum zinc quota” is the lowest amount of zinc per cell that allows for optimal growth. The minimum zinc quota varies widely for different organisms and has been experimentally determined to be ~105 atoms of zinc per cell in E. coli, ~107 atoms in yeast, and ~108 atoms in mammalian cells (2–5). Many studies have indicated that the level of labile or exchangeable zinc in cells is very low and the great majority is tightly bound by the proteins that require this metal for function (6–8). Therefore, the minimum zinc quota is likely dictated by the number of zinc-binding sites on proteins that require the metal for optimal cellular physiology.
Organisms have evolved with many mechanisms of zinc homeostasis. During times of excess zinc exposure, these mechanisms limit uptake and promote efflux to maintain the intracellular metal content at tolerable levels (9, 10). They also control the generation of intracellular zinc stores in the form of either organellar or buffered cytosolic (e.g. metallothionein) pools that are available for later use (3, 11). During zinc deficiency, homeostatic mechanisms work to maintain zinc levels at the minimum zinc quota (12). These mechanisms increase zinc uptake, decrease zinc efflux, and mobilize the release of zinc from intracellular storage sites. An additional mechanism of zinc homeostasis during deficiency has been called “zinc sparing”, i.e. reducing the levels of specific zinc-binding proteins to decrease the cellular zinc requirement (13). In some cases, reduced production of a zinc-dependent protein is compensated by increased synthesis of a zinc-independent paralog. In many bacterial species, for example, several zinc-dependent ribosomal subunits are repressed during deficiency and corresponding zinc-independent subunits are upregulated to take their place (14–16). In this way, cells can reduce their total requirement for zinc and prioritize the distribution of the limited supply of this nutrient to more critical sites and functions.
In Saccharomyces cerevisiae, the Zap1 transcription factor is the central regulator of zinc homeostasis (12, 17). Zap1 is a transcriptional activator protein whose activity is low in zinc-replete cells and high in deficient cells. Zap1 increases the expression of many genes including those that encode zinc uptake transporters in the plasma membrane. In addition, Zap1 increases expression of organellar transporters that control the levels of zinc in intracellular compartments such as the vacuole and the endoplasmic reticulum. In addition to maintaining zinc homeostasis, Zap1 also regulates genes involved in adapting cellular processes to the challenges of zinc deficiency. For example, the CKI1 and EKI1 genes are induced by Zap1 to maintain phospholipid synthesis (18, 19). The CTT1 catalase gene is also induced by Zap1 and this response is likely to eliminate the oxidative stress that arises during zinc deficiency (20, 21).
Among the adaptive responses to zinc deficiency, Zap1 induces expression of the TSA1 gene. Tsa1 is a dual function protein that acts as a peroxidase and as a “holdase”-type protein chaperone (22). Tsa1 function is essential for growth of zinc-deficient cells. Our initial studies indicated a role of the Tsa1 peroxidase activity in protecting zinc-deficient cells against an elevated level of reactive oxygen species (21). Subsequently, we found that the Tsa1 protein chaperone activity was more important than the peroxidase function for zinc-deficient growth (23). Zinc-deficient cells lacking Tsa1 chaperone activity accumulated elevated levels of stress-responsive protein chaperones, suggesting elevated unfolded protein accumulation and a corresponding heat shock response. Also consistent with this hypothesis, zinc-deficient tsa1Δ cells accumulated distinct cytoplasmic foci marked by the Hsp104 disaggregase chaperone. These foci resembled the “IPOD” compartment that accumulates in cells accumulating abundant unfolded proteins (24). Our findings for Tsa1 suggested that zinc-deficient cells accumulate unfolded zinc-dependent proteins because they lack their metal cofactor needed for folding and stability. Under these conditions, the holdase function of the Tsa1 chaperone may stabilize zinc apoproteins and shield them from misfolding and aggregation until zinc supplies increase.
Zap1 also controls an important zinc sparing mechanism involving the abundant zinc-binding alcohol dehydrogenases Adh1 and Adh3 (25). Under zinc-replete conditions, the ADH1 gene is expressed and its protein product accumulates to high levels. Under deficient conditions, the ADH1 promoter is repressed by an intergenic noncoding regulatory RNA under the control of Zap1. This mechanism allows for a transcriptional activator to repress expression of a target gene. The less abundant Adh3 alcohol dehydrogenase is regulated in a similar manner. At the same time, Zap1 induces expression of the ADH4 gene. ADH4 encodes an alternative alcohol dehydrogenase that accumulates to lower levels than Adh1 and whose activity is dependent on zinc or possibly iron (26, 27). While Adh1 and Adh3 require two zinc atoms per monomer for function, Adh4 requires only one metal ion and is not as highly expressed as Adh1, thereby providing for a net reduction in the zinc requirement of the cell during deficiency regardless of which metal it uses.
Zap1-independent mechanisms of zinc sparing in S. cerevisiae have also been discovered. For example, RNA polymerase I (RNAPI) is targeted for degradation in zinc-deficient cells (28). In replete cells, RNAPI large subunit Rpa190 is ubiquitinated and retained in the nucleus and nucleolus where it transcribes ribosomal RNA genes. Under zinc-deficient conditions, Rpa190 becomes deubiquitinated, RNAPI is exported from the nucleus and taken up into the vacuole where it is degraded by vacuolar proteases. Similarly, the zinc-binding vacuolar alkaline phosphatase Pho8 is also targeted for degradation in zinc-deficient cells through a mechanism that is independent of Zap1 (29). The zinc released into the vacuole by these mechanisms is likely transported back to the cytosol where it is used by other proteins for their function.
In this study, we performed an analysis of the yeast proteome in replete cells and cells transitioning to conditions of severe zinc deficiency. A major goal of this study was to identify additional examples of zinc sparing in the yeast zinc proteome. A second goal was to test the hypothesis raised by our studies of Tsa1 that zinc-deficient cells accumulate high levels of zinc apoproteins that require protein chaperones for their stability in the absence of their metal cofactor. We describe an extensive catalog of predicted zinc proteins in the yeast proteome and their abundance and distribution in zinc-replete conditions. During the transition to deficiency, we found that decreased accumulation of zinc-binding proteins is widespread and many effects are mediated at least in part by changes in mRNA levels rather than simply degradation of apoproteins. In addition, we present evidence that the accumulation of apoproteins is high during deficiency and that the majority of zinc sites in a cell are not occupied by zinc under these conditions. Thus, we provide unique insights into the economy of zinc in a eukaryotic cell.
RESULTS
Cataloging the zinc proteome of S. cerevisiae.
To determine the effects of zinc status on the abundance of zinc-binding proteins, we first cataloged the proteins that coordinate that ion. Proteins encoded by the yeast genome that are predicted to bind zinc were identified by a bioinformatics analysis that combined global domain searches with local motif searches. Based on this analysis, we identified 571 known or likely zinc-binding proteins in yeast (Table S1). To this group, we added several transporter proteins (Zrt1, Zrt2, Fet4, Pho84, Zrt3, Zrc1, Cot1, Zrg17, Msc2) that have been implicated to transport zinc either exclusively or among known substrates. Also included was Zps1, an accessory protein for zinc uptake that is the S. cerevisiae ortholog of C. albicans “zincophore” Pra1 (30). The yeast genome also encodes two metallothioneins, Cup1 and Crs5, that confer copper resistance. While both also bind zinc in vitro, only Crs5 contributes to zinc resistance when metal levels are high (31, 32). Therefore, Crs5 was included in the zinc proteome and Cup1 was not. These additions raised the total number of predicted zinc-binding proteins in yeast to 582. We refer to this catalog of known or likely zinc-binding proteins as the “zinc proteome”. Their number represents ~10% of the total yeast proteome and this is similar to the prevalence of zinc-binding proteins encoded by other eukaryotic genomes (1). In addition, 45 other proteins were identified that may bind zinc based on structural data but these proteins lacked published references to support that zinc is the native metal ion. Therefore, we were unable to assess the functional relevance of zinc binding by members of this group. While not included in the analysis of the zinc proteome described in this study, this list of additional potential zinc-binding proteins is included in Table S2.
The catalog of yeast zinc proteins is a useful tool to study the role of zinc in a eukaryotic cell. We examined the zinc proteome of yeast from a number of perspectives to characterize the diverse functions, types of zinc binding, and subcellular distributions of these proteins. Figure 1A–D summarizes classifications of the predicted zinc proteome based on the number of genes in each category that encode those proteins. When classified by the general function of the zinc cofactor, the most abundant group of these genes encode proteins with structural zinc sites (70%) followed by proteins that bind zinc as a catalytic cofactor (18%) (Fig. 1A). Some genes, specifically members of the alcohol dehydrogenase family, encode proteins that bind two zincs with one serving a structural role and the other acting in catalysis; these comprise 2% of the total. The remainder encode zinc transport proteins (2%) and genes whose protein products met our search criteria but the function of their metal cofactor has not yet been determined (8%).
Figure 1.
The zinc proteome of S. cerevisiae. The zinc proteome was classified based on general cofactor function (A, E), enzyme classification (B, F), structural zinc-binding site (C, G), and subcellular localization (D, H). In panels A-D, proportions were determined based on the number of genes encoding proteins in each group. In panels E-H, proportions were determined based on protein copy number in each group in replete cells.
Focusing on the 116 genes encoding proteins for which zinc plays a catalytic role (including those having both catalytic and structural zinc sites), enzymes in all six general enzyme classes were found (Fig. 1B). The majority of these genes encode hydrolases with smaller numbers of the other classes represented. Similar distributions of genes in these enzyme classes has been found for the predicted zinc proteomes of other organisms (33). For proteins in which zinc plays structural roles, several commonly shared motifs were observed (Fig. 1C). These included the C2H2-like zinc fingers and Zn2Cys6 zinc fingers most often found in DNA-binding transcription factors (34). Additional structural zinc-binding motifs observed included zinc ribbons, treble clef motifs, and zinc necklace domains (35). Some proteins bind multiple zinc atoms and have more than one type of site.
While it has been long recognized that zinc-binding proteins are present in many different compartments, the distribution of specific proteins has not been determined for an entire zinc proteome. Because previous studies have determined the subcellular distribution of the majority of yeast proteins (36, 37), we could assign subcellular locations to all but 8% of the predicted zinc proteome. The majority of zinc proteome genes (74%) encode nuclear and/or cytosolic proteins (Fig. 1D). Less common were genes encoding proteins found in the secretory pathway (ER, Golgi, endosome), mitochondria, vacuole, peroxisomes, plasma membrane, and cell wall.
Mass spectrometry analysis of the predicted zinc proteome in replete cells.
To estimate the absolute abundance of zinc-binding proteins in replete cells, quantitative label-free mass spectrometry was performed on total protein samples from cells grown to exponential phase under zinc-replete conditions (i.e. LZM supplemented with 100 μM ZnCl2) (38, 39). The resulting peak intensities were converted to protein copy number per cell using the “Proteomic Ruler” method (40). Of the ~6000 total genes in yeast, we obtained copy number estimates of 2582 gene products in zinc-replete cells. The median coefficient of variation was 12% for each set of replicates demonstrating the high quality of the mass spectrometry data (Fig. S1). From these data, we estimated the total number of proteins per cell to be ~9.2 × 107, which is similar to estimates made using other methods (41, 42). Of the 582 proteins in the predicted zinc proteome, we estimated the copy number of 229 (39%). These proteins added up to 7.6 × 106 zinc-binding proteins per cell or ~8% of the total protein number. Those zinc proteins not detected in this analysis likely represent proteins of low abundance and therefore would not contribute greatly to this estimate of total zinc protein number. Abundance data for the full proteome are provided in Table S3.
In zinc-replete cells, the abundance of zinc proteins delineated by general functional classifications (Fig. 1E) was roughly similar to the distribution based on gene number. The majority of zinc protein molecules in a cell had structural sites followed by enzymes that use zinc for catalysis. The proteins with both catalytic and structural sites made up a much larger fraction of the total zinc protein abundance (17%) than reflected by their gene number (2%) due to the high level of alcohol dehydrogenases (Adh1, Adh3, etc.). An impact of the high abundance of alcohol dehydrogenases was also observed when grouping by enzyme class (Fig. 1F) with oxidoreductases (including the alcohol dehydrogenases) making up a much larger proportion of protein copy number (41%) than was observed based on gene number (11%). Similarly, lyase protein copy number was more abundant (38%) than their gene number (9%) largely due to the high abundance of the glycolytic enzyme aldolase (Fba1). While hydrolases are very numerous at the gene level, they were consistently of low abundance and accounted for only 9% of detectable proteins that use zinc as a catalytic cofactor.
Considering structural motifs, zinc ribbons were a large fraction primarily due to the high number of ribosomal subunits with this motif (Fig. 1G). The high abundance of alcohol dehydrogenases also had a large impact on the fraction of proteins with zinc necklace motifs. Also indicated by the data is the low relative abundance at the protein level of many other classes of zinc motifs. For example, many of the DNA-binding C2H2 and Zn2Cys6 zinc finger proteins were not detected by mass spectrometry suggesting their abundance is very low. Finally, when considering the subcellular distribution of zinc-binding proteins, the vast majority are found in the cytosol (Fig. 1H). While cytosolic zinc proteins represent only 17% of genes, 89% of zinc protein abundance is cytosolic (not including the 2% of proteins with both nuclear and cytosolic distributions). The nucleus, mitochondria, and other compartments contain far lower levels of zinc-binding proteins.
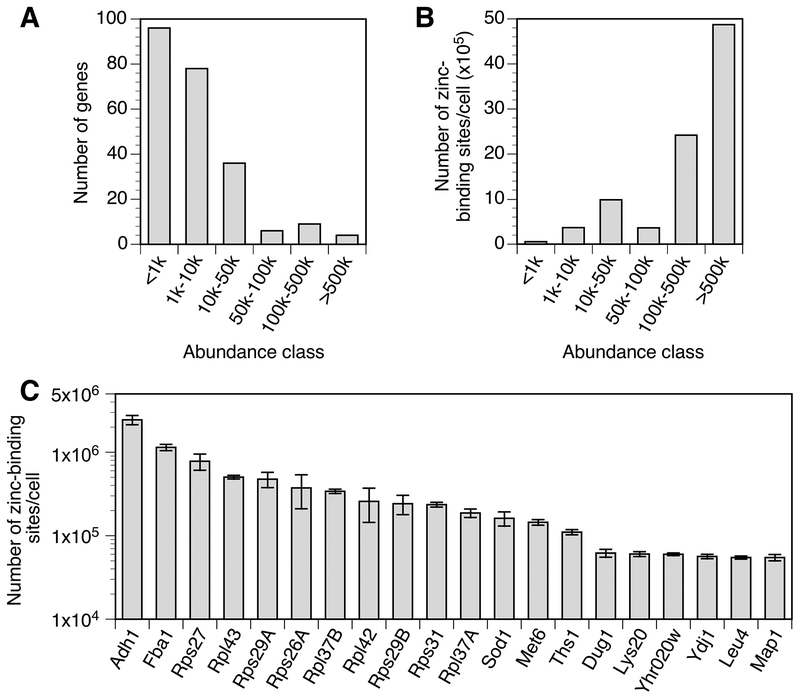
The detectable zinc-binding proteins ranged from fewer than 10 to over 106 copies per cell. Most of these proteins were of relatively low abundance (<10,000 copies per cell) and only a few were of very high abundance (>100,000 copies per cell) (Fig. 2A). When these protein copy numbers were translated into number of zinc-binding sites based on their predicted stoichiometries of metal binding, it was clear that a small number of highly abundant proteins dominate the total zinc requirement of a replete cell (Fig. 2B). In fact, the twenty most abundant zinc proteins accounted for almost 90% of the total zinc requirement of the cell (Fig. 2C). These included Adh1 alcohol dehydrogenase, Fba1 aldolase, several zinc-binding ribosomal subunits, Sod1 superoxide dismutase, and Met6 methionine synthetase. Based on our analysis, the total number of zinc-binding sites in a replete cell was calculated to be 9 × 106. Notably, this number is very close to the experimentally determined minimum zinc quota of a yeast cell (3). Note that transporter proteins were not included in this estimate of zinc-binding sites because they only transiently interact with zinc. Unsurprisingly, neither Crs5 nor Cup1 metallothioneins were detected; these proteins are induced by copper treatment and are expressed at very low levels in untreated cells (43).
Figure 2.
Abundance classes of the zinc proteome in replete cells. A) The number of genes encoding proteins in each abundance class is plotted. B) The number of predicted zinc atoms bound by proteins in each abundance class is plotted. C) The 20 proteins most abundant proteins in the zinc proteome are plotted with their predicted number of bound zinc atoms. Some ribosomal subunits are encoded by gene pairs, e.g. RPS27A and RPS27B, whose proteins are not distinguishable by mass spectrometry; these are labeled without specific reference to the paralogous genes. The error bars represent ± 1 S.D. calculated from 4 biological replicates
The response of the yeast proteome to zinc deficiency.
To determine the effects of zinc deficiency on the total proteome and specifically the zinc proteome of yeast, label-free quantitative proteomics was performed on total protein samples from cells transitioning from growth in zinc-replete to deficient conditions. Cells grown to exponential phase in a zinc-replete medium (LZM + 100 μM ZnCl2, i.e. the same cells sampled above) were transferred to a zinc-deficient medium (LZM + 1 μM ZnCl2) and cells were harvested after 4, 8, 12, and 16 hours after that transition. These times correspond to approximately 2, 3, 4 and 5 generations of growth. LC-MS peak intensities were converted to protein copy number using the Proteomics Ruler method.
The time course data are also provided in Table S3 and the effect of zinc deficiency on the total proteome is plotted in Figure 3. Cluster analysis was performed to identify cohorts of similarly affected proteins and four clusters were identified. Proteins not included in these four clusters did not share significantly enriched gene ontology (GO) terms. Clusters 1 and 2 consisted of proteins that decreased in abundance during zinc deficiency with cluster 1 showing a larger fold decrease. Cluster 3 increased during zinc deficiency while cluster 4 showed a small increase or no change. Figure 3 displays some of the significant GO terms that were found for these clusters. A complete list of GO terms is available in Table S4. In cluster 1, down-regulated proteins were related to ribosomal proteins, ribosome biogenesis, and cytoplasmic translation. Cluster 2 included many terms found in cluster 1 including protein biosynthesis, ribosome biogenesis, translation, and also unfolded protein binding and chaperone proteins. These results suggest that zinc deficiency causes a systemic decrease in protein synthesis and protein chaperone capacity. Cluster 3 contains up-regulated proteins related to oxidation-reduction processes, mitochondrial function, and vesicle-mediated transport. Up-regulated proteins in cluster 4 are related to proteasome activity, actin binding, and glycolysis. Many of these changes may help mitigate the deficit of zinc.
Figure 3.
The response of the total proteome to the transition from zinc-replete to deficient conditions. Zinc-replete cells were transferred to a zinc-deficient medium and harvested for proteomics analysis after 4, 8, 12, and 16 h. The resulting changes in protein abundance relative to replete conditions for 2,119 proteins are plotted as a heat map. Cluster analysis was performed and four clusters (1–4) were identified. Gene ontology (GO) analysis of each cluster was then performed and significant terms observed are plotted with the −log of their p-values in each histogram color-coded for their corresponding cluster. Full GO results are provided Table S4.
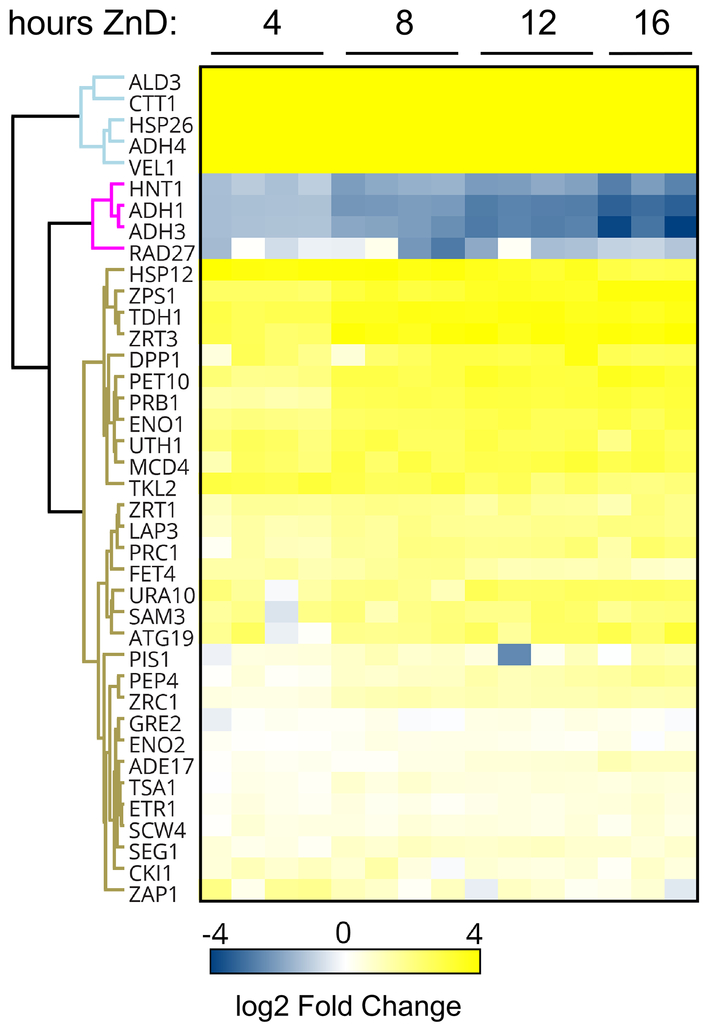
Many of the effects of zinc deficiency on the total proteome are likely due to indirect consequences of metal status. To focus on responses that are directly controlled by zinc status, we examined the abundance of proteins known or hypothesized to be regulated by the Zap1 transcription factor. Based on several published studies, Zap1 is thought to regulate transcription of 87 genes in response to zinc deficiency (20, 44–49). For the majority of those genes, including the zinc transporters ZRT1, ZRT3, and FET4, Zap1 activation increases gene expression and protein abundance during zinc deficiency. For a small number of other target genes, Zap1 represses gene expression and thereby reduces protein accumulation during deficiency. Of the 87 known or predicted Zap1-regulated genes, protein abundance was measurable for 39 of their encoded proteins and the effect of zinc deficiency on their levels is depicted in Figure 4. For most Zap1 target genes, the abundance of their protein products increased in zinc deficiency, consistent with the action of Zap1 as a transcriptional activator. For some proteins, e.g. Ald3, Ctt1, Hsp26, and Adh4, the response to deficiency was immediate and strong. For other proteins, the response was much slower and/or of less dynamic range (e.g. Pep4 and Zrc1). In contrast, four proteins (Adh1, Adh3, Rad27, Hnt1) showed decreased protein abundance during the transition to deficiency. Adh1 and Adh3 were previously shown to be controlled by Zap1-regulated non-coding RNAs that repress promoter function (25). The mechanisms controlling Rad27 and Hnt1 are currently being studied. These varied results demonstrate the diversity of responses of the proteins encoded by genes regulated by Zap1.
Figure 4.
The response of proteins encoded by the Zap1 regulon to the transition from zinc-replete to deficient conditions. During the transition to zinc-deficient conditions, the resulting changes in protein abundance relative to replete conditions for 39 proteins are plotted as a heat map. Proteins affected similarly were identified by cluster analysis.
The response of the predicted zinc proteome to zinc deficiency.
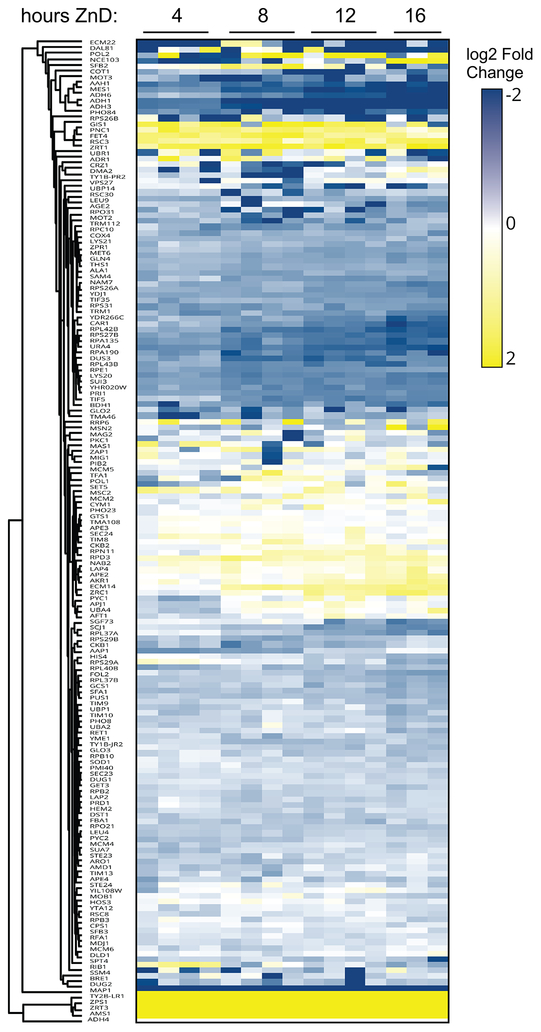
Of the 229 zinc-binding proteins that were measured in replete cells, 199 of those had sufficient data (measurements at T0 and at least two subsequent time points) to assess the effects of zinc deficiency on their abundance (Table S3). In addition, there were 13 zinc-binding proteins that were not detected at T0 but were detected at two or more subsequent timepoints. Similarly, there were 30 zinc-binding proteins detectable at T0 that were not detected at later time points suggesting a marked loss of protein abundance. As shown in Figure 5, of the proteins in the predicted zinc proteome that were detected by mass spectrometry, many more proteins decreased in abundance during the transition from zinc-replete to deficient conditions than increased. Using a fold change of 1.5 or greater and a false discovery rate of 0.05, we found 31 proteins that increased in abundance and more than twice that number of proteins (68) that decreased in response to zinc deficiency. Among those proteins that increased in abundance were several zinc transport proteins (Zrt1, Fet4, Zrt3, Zrc1, Zps1) that are regulated by Zap1. The level of the Msc2 zinc transporter of the endoplasmic reticulum, while not Zap1 regulated (50), also increased in zinc-deficient cells. Also among the increasing proteins was the Adh4 alcohol dehydrogenase, which is activated by Zap1 to likely replace the lost activity of the repressed Adh1 and Adh3 proteins. In addition, many proteins involved in chromatin modification (e.g. Rpd3, Hda1, Hst2, Set3, Gis1, Rsc3), components of the basal transcription machinery (e.g. Sua7, Brf1, Tfa1, Rpb9), and numerous proteases (e.g. Ape2, Ape3, Dpp3, Rpn11) increased in abundance in response to zinc deficiency. Furthermore, the gag-pol fusion proteins of transposable elements Ty1 and Ty2 increased. Increased levels of these Ty-encoded proteins have been observed under other stress conditions (51–53).
Figure 5.
The response of proteins in the zinc proteome to the transition from zinc-replete to deficient conditions. During the transition to zinc-deficient conditions, the resulting changes in protein abundance relative to replete conditions for 159 proteins are plotted as a heat map. Proteins affected similarly were identified by cluster analysis.
Many more zinc-binding proteins that we detected decreased in abundance during deficiency. These included zinc-binding ribosomal subunits and several ribosome biogenesis proteins (e.g. Bud20, Nmd3, Reh1), general translation factors (e.g. Tif5, Tif35, Sui3), tRNA synthetases and tRNA modifying proteins (e.g. Ths1, Mes1, Dus3), several RNA polymerase subunits and general transcription factors (e.g. Rpa12, Rpa135, Rpa190, Pri1, Taf1) and six alcohol dehydrogenases (Adh1, Adh3, Adh5, Adh6, Bdh1, Sfa1). The effect of zinc deficiency on the abundance of several specific examples is shown in Figure 6. To determine if any of the observed changes in protein abundance were the result, at least in part, of altered transcription, we performed quantitative RT-PCR analysis of mRNA levels of 28 of the affected proteins in replete cells and in cells after 8 and 16 h of zinc deficiency. Those data are shown for specific examples in Figure 6 and the full results are reported in Table 1. For 16 of the tested proteins, their decreased abundance during deficiency had some element of transcriptional control (p < 0.05). For example, the effect of Zap1 on ADH1 and ADH3 expression in zinc deficiency were observed in the RT-PCR results (Fig. 6). Our analysis also suggested that the decreases of Adh5, Adh6, and Bdh1 alcohol dehydrogenases also occur at least in part at the transcriptional level.
Figure 6.
The effect of zinc deficiency on the abundance of example zinc proteins and their mRNA. For each, the effect on the transition from replete to deficient conditions is plotted relative to protein copy number (left panels, gray columns) and mRNA abundance (right panels, filled columns, A.U. = arbitrary units). Protein levels are from 3–4 replicates per timepoint and the mRNA levels were determined by quantitative RT-PCR (n = 3). The error bars indicate ± 1 S.D.
Table 1.
Protein copy number and RNA abundance for example zinc proteome members under zinc-replete (0 h) and deficient (8 and 16 h) conditions. Protein copy numbers are from mass spectrometry analysis and mRNA abundance was determined by quantitative RT-PCR (n = 3). The ratios of levels at 16 h and 0 h are reported and the p-values were calculated using Student’s t-test.
| Protein Copy Number (per cell) | RNA Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein/Gene | 0 h | 8 h | 16 h | Ratio of T16/T0 | p-value | 0 h | 8 h | 16 h | Ratio of T16/T0 | p-value |
| Zrt1 | 29467 | 104388 | 99222 | 3.36 | 0.001 | 1.444 | 7.064 | 11.028 | 7.64 | 0.00005 |
| Zrt3 | 627 | 7351 | 8247 | 13.20 | 0.001 | 0.520 | 5.261 | 5.422 | 10.43 | 0.000009 |
| Adh1 | 1222338 | 283955 | 132595 | 0.11 | 0.000006 | 30.58 | 3.11 | 3.81 | 0.12 | 0.04 |
| Adh3 | 21531 | 5370 | 1905 | 0.09 | 0.00009 | 0.31 | 0.16 | 0.21 | 0.66 | 0.002 |
| Adh5 | 2659 | 0 | 0 | N/A | N/A | 0.53 | 0.24 | 0.36 | 0.67 | 0.01 |
| Adh6 | 14959 | 6939 | 2649 | 0.18 | 0.00005 | 0.80 | 0.17 | 0.17 | 0.21 | 0.0003 |
| Bdh1 | 13002 | 6165 | 6047 | 0.47 | 0.01 | 0.23 | 0.04 | 0.09 | 0.40 | 0.0005 |
| Aah1 | 3344 | 1206 | 801 | 0.24 | 0.01 | 0.29 | 0.29 | 0.23 | 0.77 | 0.09 |
| Bud20 | 2552 | 0 | 0 | N/A | N/A | 0.06 | 0.10 | 0.08 | 1.40 | 0.05 |
| Car1 | 12831 | 8155 | 4461 | 0.35 | 0.006 | 0.05 | 0.03 | 0.03 | 0.65 | 0.03 |
| Dus3 | 3684 | 1471 | 1200 | 0.33 | 0.02 | 0.04 | 0.04 | 0.03 | 0.78 | 0.3 |
| Fcy1 | 15308 | 0 | 0 | N/A | N/A | 0.83 | 0.38 | 0.52 | 0.63 | 0.02 |
| Map1 | 27378 | 1034 | 2405 | 0.09 | 0.00007 | 0.76 | 0.78 | 0.84 | 1.11 | 0.6 |
| Mes1 | 28274 | 8847 | 6347 | 0.22 | 0.001 | 0.59 | 0.12 | 0.14 | 0.23 | 0.002 |
| Met6 | 144826 | 101595 | 86610 | 0.60 | 0.003 | 1.18 | 0.55 | 0.88 | 0.75 | 0.1 |
| Nmd3 | 3559 | 1398 | 1720 | 0.48 | 0.01 | 0.51 | 0.25 | 0.26 | 0.50 | 0.003 |
| Reh1 | 356 | 141 | 200 | 0.56 | N/A | 0.23 | 0.11 | 0.23 | 1.03 | 0.6 |
| Rpa12 | 3567 | 0 | 0 | N/A | N/A | 0.72 | 0.28 | 0.36 | 0.51 | 0.001 |
| Rpa135 | 5595 | 2543 | 1844 | 0.33 | 0.003 | 0.24 | 0.22 | 0.18 | 0.76 | 0.2 |
| Rpa190 | 8784 | 3709 | 2580 | 0.29 | 0.0003 | 0.35 | 0.29 | 0.31 | 0.89 | 0.2 |
| Rpe1 | 13710 | 6477 | 6076 | 0.44 | 0.01 | 0.14 | 0.12 | 0.12 | 0.81 | 0.06 |
| Sui3 | 27585 | 12387 | 11510 | 0.42 | 0.009 | 1.26 | 0.38 | 0.51 | 0.40 | 0.0002 |
| Ths1 | 55203 | 36381 | 31764 | 0.58 | 0.005 | 1.40 | 0.49 | 0.82 | 0.59 | 0.0001 |
| Tif35 | 24715 | 14869 | 12549 | 0.51 | 0.006 | 3.65 | 1.34 | 1.76 | 0.48 | 0.0003 |
| Tif5 | 35778 | 18271 | 17712 | 0.50 | 0.005 | 0.43 | 0.23 | 0.26 | 0.61 | 0.03 |
| Ura4 | 23647 | 11292 | 8225 | 0.35 | 0.006 | 0.83 | 0.13 | 0.22 | 0.26 | 0.0007 |
| Ycr087c-A | 2431 | 0 | 0 | N/A | N/A | 0.30 | 0.15 | 0.12 | 0.40 | 0.000008 |
| Ydj1 | 28211 | 19306 | 13749 | 0.49 | 0.002 | 0.46 | 0.34 | 0.42 | 0.91 | 0.3 |
| Yhr020w | 59964 | 29010 | 26517 | 0.44 | 0.002 | 0.36 | 0.22 | 0.29 | 0.80 | 0.2 |
| Zpr1 | 5310 | 4095 | 3285 | 0.62 | 0.02 | 0.20 | 0.21 | 0.16 | 0.77 | 0.1 |
In contrast, the mRNA encoding 12 of the proteins that decrease in zinc-deficient cells did not decrease during this transition. For example, RPA190 mRNA was not reduced despite a large decrease in protein (Fig. 6). This result is consistent with previous studies showing that Rpa190 is targeted for degradation specifically under zinc-deficient conditions (28). Other examples of post-transcriptional effects newly identified in this analysis are Map1, Bud20, and Ydj1. These data indicated that there is a transcriptional component to the decreased accumulation of many, but not all, zinc proteins during the transition to zinc deficiency.
Estimates of in vivo zinc-binding site number and zinc content suggest significant deficits in zinc metalation during deficiency.
One purpose of the widespread decrease in zinc-binding proteins during deficiency may be zinc sparing, i.e. the regulated decrease in the zinc requirement of the cell. Figure 7A shows the relative zinc-sparing effects for zinc-binding proteins grouped by their general function. Decreases in ribosome subunits and ribosomal biogenesis factors and decreases in alcohol dehydrogenase abundance reduce the zinc requirement to the greatest degree while other functional groups contribute to a lesser extent. The hypothesis of targeted zinc sparing suggested that the decrease in zinc proteins is significantly greater than the effects observed for the total proteome. During the transition to zinc deficiency, the total proteome decreased 24% from 9.2 × 107 copies per cell to 7.0 × 107 copies (Fig. 7B). Similarly, the copy number of non-zinc proteins also decreased 24% from 8.4 × 107 copies per cell to 6.5 × 107. These decreases may reflect the decreased translation capacity of zinc-deficient cells (Fig. 3) and/or the results of autophagic degradation, which was previously shown to be induced by zinc deficiency (54–56). In contrast, the effect of zinc deficiency on the zinc proteome was even more striking with protein copy number dropping from 7.5 × 106 to 4.5 × 106 copies per cell (39%). While regulation of specific zinc proteins during the transition to deficiency may occur for many reasons, these data suggest that widespread zinc-sparing mechanisms are at work to specifically decrease the zinc demand of a cell during deficiency.
Figure 7.
Zinc sparing is widespread in the zinc proteome. A) 120 members of the zinc proteome that decrease during zinc deficiency were grouped by general function and plotted as the number of zinc-binding sites in each group under replete (filled columns) and deficient (i.e. 16 h) (gray columns) conditions. B) The effect of zinc deficiency on the copy number per cell of the total proteome (circles), the total proteome not including the zinc proteome (triangles), and the zinc proteome (squares) is plotted. C) The estimated number of zinc atoms bound by the zinc proteome (squares) is plotted relative to the number of zinc atoms per cell as determined by ICP-AES (circles). The error bars indicate ± 1 S.D calculated from 3–4 biological replicates.
Factoring in their predicted stoichiometries of zinc binding, the abundance of zinc proteins in replete cells translates into 9 × 106 zinc-binding sites on proteins per cell (Fig. 7C). By 16 h after the transition to zinc deficiency, the number of zinc sites decreased to 4.7 × 106 sites. Thus, the zinc requirement was found to drop after 16 h of zinc deficiency by over 4 × 106 zinc sites per cell or ~45% of the sites found in replete cells. To determine the amount of zinc available in deficient cells to meet this demand, zinc abundance was determined using ICP-AES. Total zinc in replete cells grown under these conditions was measured to be 2.3 × 107 atoms per cell (Fig. 7C). This value is approximately twice the estimated number of zinc-binding sites on proteins. By 16 h of zinc deficiency, zinc levels dropped to only 1.7 × 106 atoms per cell, i.e. ~30% of the estimated number of zinc-binding sites on proteins. These results suggested that despite the large decrease in zinc protein number described above, a zinc-deficient cell grown under these conditions accumulates a very high level of apoproteins or, alternatively, zinc proteins that are mismetalated with some other cation.
Analyses of zinc-binding proteins indicate reduced zinc metalation during deficiency.
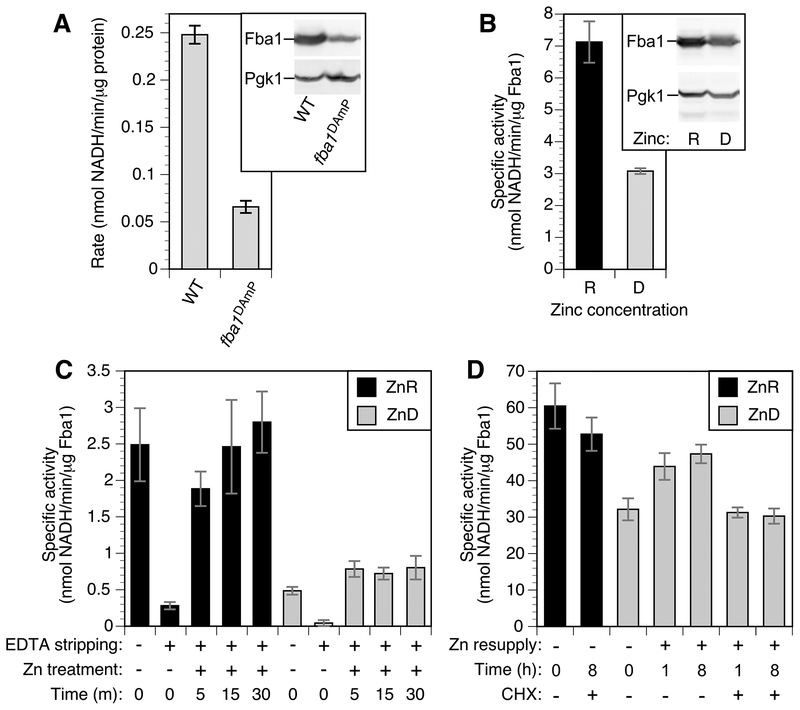
To assess to what degree zinc metalation is disrupted during deficiency, we focused our analysis on the Fba1 aldolase protein. This glycolytic enzyme is the second most abundant protein in the zinc proteome of replete cells (Fig. 2) and the most abundant zinc protein in deficient cells. Fba1 levels were similar in replete and deficient cells with approximately 1.1 × 106 copies per cell. Thus, the zinc requirement of this enzyme alone is over half of the entire zinc content of a deficient cell. Given these factors, we hypothesized that most Fba1 molecules are either unmetalated or mismetalated during zinc deficiency. To test this hypothesis, we used an enzymatic assay of aldolase activity in permeabilized cells. We confirmed the specificity of the assay for Fba1 aldolase activity using zinc-replete wild-type cells and fba1DAmP mutant cells that have lower FBA1 expression (57). Immunoblots indicated that the fba1DAmP strain accumulated ~25% of the wild-type level of protein and the measured enzyme activity showed a similar decrease (Fig. 8A). FBA1 is essential for viability so a null allele could not be used for this experiment.
Figure 8.
Effects of zinc status on Fba1 aldolase activity. A) Aldolase activity in wild-type (BY4741) and fba1DAmP cells normalized to total protein. The inset shows an immunoblot of Fba1 protein level detected with anti-Fba1 and anti-Pgk1 in 20 μg of total protein. B) Aldolase specific activity in zinc-replete (R) and deficient (D) BY4741 cells normalized to Fba1 protein levels. The inset shows an immunoblot of Fba1 protein level detected as described for panel A. C) Aldolase specific activity in permeabilized replete (ZnR) and deficient (ZnD) cells before and after in vitro EDTA stripping and zinc resupply for the times indicated. D) Aldolase specific activity in zinc-replete and deficient cells following zinc resupply in vivo with and without cycloheximide treatment (100 μg/ml). The error bars indicate ± 1 S.D (n = 3).
To determine the effect of zinc status on Fba1 function, aldolase specific activity normalized to Fba1 protein levels as determined by immunoblotting, was measured in cells after growth in zinc-replete and deficient conditions (Fig. 8B). Aldolase specific activity was high in zinc-replete cells and greatly reduced in deficient cells. These results were consistent with our hypothesis of reduced zinc metalation during deficiency. To further test this hypothesis, we determined whether zinc added back in vitro restored aldolase activity. EDTA treatment of permeabilized zinc-replete cells reduced aldolase activity and zinc subsequently added back restored activity to the full activity of untreated cells (Fig. 8C). In contrast, zinc addition to EDTA-stripped deficient cells for 5, 15, or 30 minutes did not restore activity to replete levels. These data suggest that zinc-deficient cells contain a pool of inactive aldolase that cannot be metalated efficiently with zinc. This conclusion was further supported by analysis of Fba1 activity following zinc addition in vivo. When zinc-deficient cells were supplemented with zinc in vivo prior to permeabilization, much of the missing Fba1-normalized activity was recovered after 1 h of zinc treatment and ~80% of full activity was observed after 8 h (Fig. 8D). However, this restoration of activity was completely blocked when the translation inhibitor cycloheximide was added prior to zinc addition. These results suggested that the inactive Fba1 aldolase in zinc-deficient cells cannot be activated with zinc supplementation and new protein synthesis is required to restore full aldolase activity.
To assess the metalation state of aldolase in vivo, we adapted a method previously used to probe zinc binding by proteins in vitro (58–60). N-ethylmaleimide (NEM) is a cell-permeable reagent that alkylates free cysteine thiol groups in proteins. Reactivity of thiols with NEM is greatly reduced by metal binding to those residues or, for non-ligand cysteines, when the thiol group is inaccessible to solvent. Thus, reactivity to NEM can be used to assess both zinc binding to cysteine ligands and zinc-dependent conformation changes that affect the accessibility of non-ligand cysteines. In our modified procedure, cells growing in culture are treated with NEM to modify reactive cysteine thiols in vivo, lysed, proteins denatured, and then treated with PEG-maleimide, which modifies unreacted thiols that had been protected from in vivo NEM modification. Thiols alkylated by NEM are no longer reactive to PEG-maleimide whereas sites that were not alkylated become modified with the PEG reagent. While NEM modification only slightly alters protein molecular mass and is not detectable by immunoblotting, PEG-maleimide modification increases the apparent molecular mass on SDS-PAGE by ~15 kDa per moiety and is therefore readily detected by immunoblotting. Thiols that are oxidized in vivo are also not modified by NEM; these can be identified as being protected from NEM modification in vivo, reactive to PEG-maleimide in vitro following DTT treatment, but not reactive with PEG-maleimide without pre-treatment with reductant.
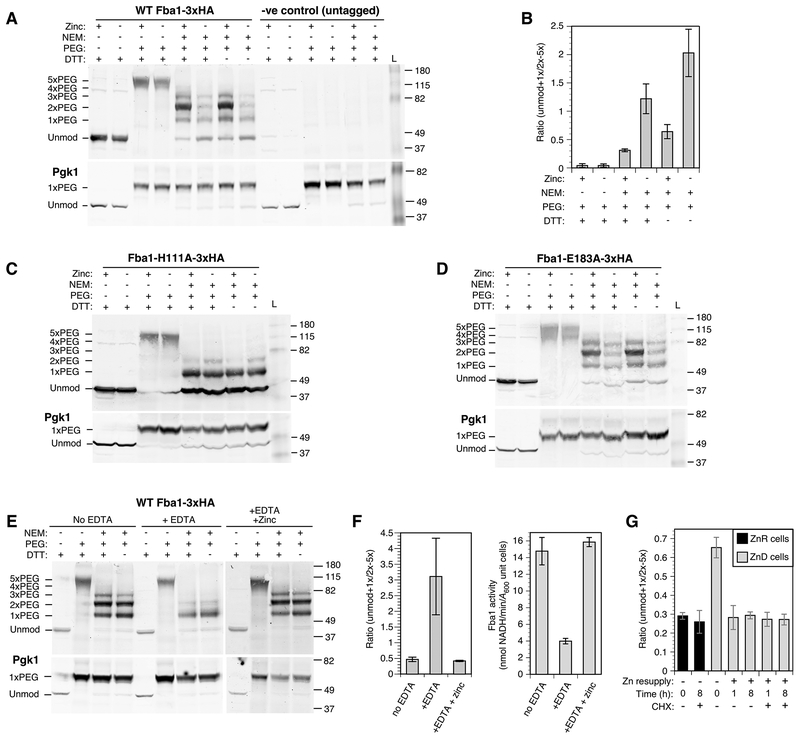
Fba1 contains five cysteine residues. While none of these are directly involved in zinc binding, we found that their accessibility to modification by NEM is altered by zinc status. Without modification, Fba1 tagged with the hemagglutinin antigen epitope (Fba1-3×HA) migrates at ~40 kDa on SDS-PAGE (Fig. 9A). When treated with PEG-maleimide alone following denaturation, the Fba1-HA protein shifts in mobility to ~115 kDa, corresponding to the addition of five PEG moieties. When zinc-replete cells were treated with NEM in vivo prior to PEG-maleimide treatment in vitro, 1–4 cysteines were protected from NEM reaction to varying degrees with most copies having two NEM-resistant cysteines. In zinc-deficient cells, NEM sensitivity increased as indicated by the decreased abundance of 2–4 × PEG-modified forms and increased abundance of proteins with 0–1 × PEG moieties added. This result indicates a zinc-dependent conformational change in vivo that changes the sensitivity of one or more cysteines to NEM. The effect on NEM sensitivity was observed both with and without DTT treatment confirming that protection in vivo was not due to thiol oxidation. To quantify these effects, band intensities were measured and the ratio of 0–1 × PEG-modified forms vs. 2–4 × PEG-modified forms was determined (Fig. 9B). This analysis demonstrated higher levels of PEG modification (i.e. NEM resistance) in replete cells and lower levels (i.e. NEM sensitivity) in deficient cells. In contrast, no effect of zinc was observed on the NEM sensitivity of the single cysteine in Pgk1; this cysteine is protected from NEM modification in vivo due to solvent inaccessibility but is modified with PEG-maleimide following protein denaturation (Fig. 9A). Modification of Pgk1 with PEG-maleimide is highly efficient and provides a positive control for this reaction.
Figure 9.
In vivo analysis of zinc binding by Fba1. A) NEM/PEG-maleimide analysis of wild-type (BY4742) cells expressing Fba1-3×HA or an untagged Fba1 allele. Zinc-replete (+) or deficient (–) cells were treated with and without NEM, proteins harvested, and then treated with or without DTT prior to PEG-maleimide treatment. The positions of molecular mass markers (kDa) and of unmodified and PEG-modified forms of Fba1-3×HA are indicated. Pgk1, which has a single cysteine that is insensitive to zinc supply, was used as a control for the efficiency of PEG labeling. B) Quantitation of the results in panel A. The ratios of unmodified + 1× PEG-modified to 2×-5× PEG-modified forms are shown and the error bars indicate ± 1 S.D (n = 3). C,D) NEM/PEG-maleimide analysis of wild-type (BY4742) cells expressing Fba1-H111A-3×HA or Fba1-E183A-3×HA as described for panel A. E) NEM/PEG-maleimide analysis of permeabilized wild-type (BY4742) cells expressing Fba1-3×HA following stripping with EDTA and reloading with zinc. F) Quantitation of the results in panel E. The ratios of unmodified + 1× PEG-modified to 2×-5× PEG-modified forms are shown (left panel) is compared with Fba1 activity determined from the same samples (right panel). The error bars indicate ± 1 S.D (n = 3). G) NEM/PEG analysis of wild-type (BY4743) cells expressing Fba1-3×HA grown in zinc-replete (ZnR) or deficient (ZnD) and following resupply with zinc (100 μM in LZM) in vivo for 1 and 8 h, with and without cycloheximide treatment (100 μg/ml). The ratios of unmodified + 1× PEG-modified to 2×-5× PEG-modified forms are shown and the error bars indicate ± 1 S.D (n = 3).
To assess whether the observed conformational change observed for the wild-type Fba1-3×HA protein was due to zinc binding in vivo, we mutated the histidine zinc ligand at position 111 to alanine (H111A). No zinc-responsive changes in NEM reactivity were then observed (Fig. 9C). A similar analysis of a mutation in glutamate 183 (E183A) supported this conclusion. The E183A mutation disrupts enzyme activity without affecting metal binding. Despite its lack of activity, the same zinc-dependent conformational change observed for the wild type protein was seen with this mutant Fig. 9D). To further confirm that the observed conformational changes were due to the presence or absence of zinc binding, we performed the experiment in permeabilized replete cells using stripped and re-metalated Fba1-3×HA. Without EDTA stripping, PEG-maleimide reactivity was high indicating NEM resistance (Fig. 9E). When Fba1-3×HA was stripped of metal with EDTA treatment, NEM resistance was reduced and re-addition of zinc restored protection from NEM modification. These effects are quantitated in Figure 9F and the reciprocal effect of EDTA and zinc on Fba1 enzyme activity is shown. Again, no effect of EDTA or zinc treatment was observed on the PEG sensitivity of Pgk1. These results support the conclusion that the change in NEM sensitivity is directly due to zinc binding in the Fba1 active site.
To test whether the in vivo dependence of Fba1 reactivation on protein synthesis (Fig. 8) was due to slow re-metalation with zinc, we used the NEM/PEG-maleimide method. Cells were grown in zinc-replete and deficient conditions. The deficient cells were then resupplied with zinc for 1 or 8 h in the presence or absence of cycloheximide. In vivo NEM treatment was carried out prior to zinc addition and at subsequent timepoints and the cells were then processed for PEG-maleimide modification. In the absence of cycloheximide, full re-metalation was apparent after 1 h of zinc treatment (Fig. 9G). Surprisingly, apparent re-metalation was also detected in cycloheximide-treated cells after 1 h and 8 h despite the absence of increased activity. These results suggest that while a substantial fraction of Fba1 protein is not metalated in zinc-deficient cells, it is re-metalated rapidly in vivo and some other factor limits its activity. We hypothesize that the protein may be damaged or modified in some way.
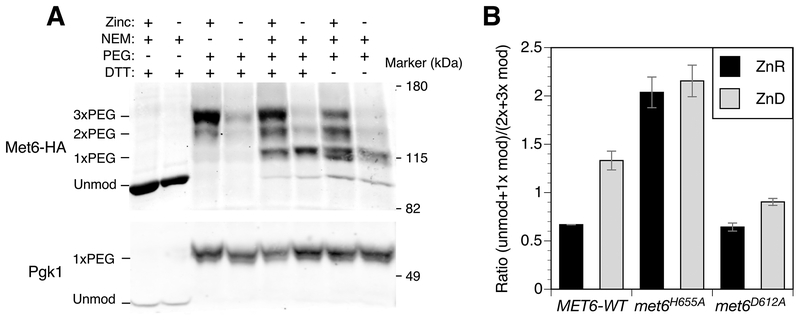
Our analysis of Fba1 identified a zinc-responsive conformational change that affects the accessibility of non-ligand cysteines to NEM modification. To test for effects of zinc status specifically on zinc-binding cysteine ligands in a protein, we tested the in vivo NEM sensitivity of Met6 methionine synthetase. Met6 accumulates to ~145,000 molecules per cell in zinc-replete cells and ~87,000 in deficient cells. Fungal methionine synthetases contain three cysteine residues, two of which (C657 and C737) are homologous to the highly conserved residues Cys659 and Cys739 shown to bind zinc in the C. albicans Met6 protein (61). NEM treatment of zinc-replete cells followed by in vitro treatment with PEG-maleimide demonstrated protection of 1, 2, or 3 cysteines, with most Met6 proteins having three protected cysteines (Fig. 10A). Under zinc-deficient conditions, most proteins had only one protected cysteine. This result is consistent with zinc binding protecting the two cysteine ligands while the third thiol is constitutively protected in vivo. Omitting the in vitro DTT reduction step had little effect on the pattern of bands detected indicating that the protective effect of zinc was not conferred by Met6 cysteine oxidation.
Figure 10.
In vivo analysis of zinc binding by Met6. A) NEM/PEG-maleimide analysis of wild-type (BY4742) cells expressing Met6-HA. Zinc-replete (+) or deficient (–) cells were treated with and without NEM, proteins harvested, and then treated with and without DTT prior to PEG-maleimide treatment. The positions of molecular mass markers (kDa) and unmodified and PEG-modified forms of Met6-HA are shown. Pgk1 was detected as a control for PEG labeling efficiency. B) Quantification of NEM/PEG-maleimide analysis of wild-type (BY4742) cells expressing Met6-HA, Met6H655A-HA or Met6D612A-HA as described for panel A. The ratios of unmodified + 1 × PEG-modified to 2–3 × PEG-modified forms are shown and the error bars indicate ± 1 S.D (n = 3).
This conclusion was supported by the analysis of a mutation that disrupts one of the non-cysteine zinc ligands in the metal site. S. cerevisiae His 655 is homologous to H657 of the C. albicans and S. pombe Met6 proteins, which was identified as zinc binding by structural analysis (62, 63). An H655A mutation resulted in constitutive NEM sensitivity (Fig. 10B). In contrast, a mutation in Asp 612 (D612A, homologous to Asp614 in S. pombe), which inactivates Met6 by disrupting homocysteine binding (63) had no effect on cysteine sensitivity to NEM in replete conditions, indicating unaltered zinc binding. In deficient conditions, the D612A mutant showed increased NEM protection when compared with the wild-type, suggesting that substrate binding affects zinc lability. Overall however, these observations indicate that the effect of the zinc ligand mutation on NEM sensitivity was not due simply to loss of enzyme activity but reflected a loss of zinc from the active site. Our results are consistent with Met6 accumulating in a zinc-metalated form in replete cells and is only partially metalated in deficient cells.
DISCUSSION
In this report, we cataloged the zinc proteome of yeast, and determined the abundance of many of its proteins, their response to zinc deficiency, and estimated the metalation state of those proteins in zinc-replete and deficient cells. This analysis has provided unprecedented insights into the cellular economy of zinc under replete and deficient conditions. The majority of zinc proteins in yeast bind zinc as a structural cofactor and includes proteins with C2H2 zinc fingers, Zn2C6 zinc fingers, zinc ribbon domains, and other motifs. These proteins play key roles in gene regulation, transcription, translation, protein degradation, and many other functions. The zinc proteome also plays many catalytic roles and includes enzymes in all six general classes of enzymes. The catalog of the yeast zinc proteome we generated illustrates the diversity of function of this essential cofactor within a eukaryotic cell. We recognize, however, that this is likely not a perfect list of all zinc proteins in yeast. Zinc may play regulatory roles as a second messenger to control different aspects of cell physiology (64, 65). Given that such regulatory sites are probably of lower affinity and greater lability, they may not have been identified in our analysis of stable motifs and domains. Second, some enzymes (i.e. “cambialistic” proteins) are functional with different metal cofactors and the specific metals that metalate those enzymes likely depend on the cellular conditions of metal homeostasis and other factors (66). Thus, to refer to such proteins as “zinc proteins” may not accurately reflect their cofactor requirements. Despite these caveats, however, the catalog of predicted zinc-binding proteins in yeast provides an essential tool for future studies of zinc function, trafficking, and homeostasis.
Using mass spectrometry we measured the abundance of almost half of the entire yeast proteome. Focusing on the zinc proteome, this analysis indicated the existence of 9 × 106 total zinc sites in replete cells. This proteomics-based estimate is remarkably similar to our experimentally determined measurement of the vacuolar and non-vacuolar pools of zinc under these same growth conditions (3). Cells grown in LZM supplemented with 100 μM ZnCl2 contain 2.3 × 107 zinc atoms per cell. When the Zrc1 and Cot1 vacuolar zinc transporters were mutated so that zinc could no longer be stored in the vacuole, the accumulation of zinc in the cell dropped to ~1 × 107 zinc atoms per cell. This result indicated that ~60% of the total zinc in a replete cell grown under these conditions is stored in the vacuole. Because labile zinc levels in cells are known to be very low, it is likely that the non-vacuolar zinc atoms are bound by proteins and this conclusion is very consistent with the number of zinc sites we found in these cells using proteomics. Our previous studies also established that the minimum zinc quota of a yeast cell was ~1 × 107 zinc atoms per cell. This comparison suggests that the zinc proteome abundance of a replete cell is tuned for optimal growth and even slight deficits that decrease full metalation inhibits cell growth. One caveat to this analysis is that we were only able to measure the abundance of about half of the proteins that comprise the zinc proteome. The proteins missing from our analysis are likely to be of very low abundance and therefore may contribute little to our calculations of the cellular zinc requirement. A result very different from ours was obtained from a similar study of the Gram-negative bacterium Cupriavidus metallodurans where it was found that replete cells contain a large excess of zinc-binding sites relative to the minimum zinc quota (67). A high number of zinc sites may add to zinc resistance of this bacterium by serving as a sink for excess metal. Organisms such as S. cerevisiae are not adapted to high zinc levels and we suspect it is more indicative of what occurs in most other organisms. A viable alternative hypothesis is that C. metallodurans has an excess of zinc sites to serve as a storage pool of the metal in the absence of intracellular organelles.
The total proteome and the zinc proteome of replete cells changes markedly during the transition to deficient conditions. Among the zinc proteins that increased in abundance were several transporter proteins induced by Zap1 to maintain zinc homeostasis. We also observed increased levels of several proteins involved in transcription and chromatin modification. These changes may represent compensatory regulatory responses to loss of these activities due to reduced metalation. The increased abundance of these zinc proteins was more than offset by the decreased abundance of many other zinc proteins such that the net effect was a decrease in the total zinc requirement of the cell by ~45%. We had anticipated that proteasome-mediated degradation of apoproteins may be a common effect during the transition to zinc deficiency and there are some potential cases of this in our results. For example, Map1 aminopeptidase levels drop dramatically in zinc-deficient cells with no detectable change in mRNA abundance. In addition, macroautophagy is induced during zinc deficiency and this process may decrease protein abundance more generally (54–56). However, it is clear that changes in mRNA levels play at least some role in the down-regulation of numerous zinc proteins. While these regulatory effects may occur for many reasons, we hypothesize that a common purpose is to reduce the zinc requirement of the cell, i.e. zinc sparing.
One specific group of proteins that had a great impact on the cellular zinc requirement are the alcohol dehydrogenases. There are 13 different zinc-dependent alcohol dehydrogenases in yeast and we could measure the abundance of seven, Adh1, Adh3, Adh4, Adh5, Adh6, Bdh1, and Sfa1. All but Adh4 decrease in zinc-deficient cells for a net decrease in the zinc requirement of ~9 × 105 zinc atoms per cell. As mentioned above, Adh4 is induced and Adh1 and Adh3 are repressed by Zap1 (25). How are the other four Adh enzymes shut off in response to zinc deficiency? A clue for how Adh6 is regulated comes from our recent analysis of transcription start sites in the yeast genome. Loss of ADH6 gene expression coincides with the induction of an antisense transcript that initiates downstream of the ADH6 open reading frame (50). While we have not mapped the 3’ end of this antisense transcript, it may pass through the open reading frame and ADH6 promoter and thereby interfere with ADH6 transcription. This antisense transcript is likely to be Zap1-dependent because there is a consensus Zap1 binding site, 5’-ACCTTAAAGGT-3’, located ~130 bp upstream of where the antisense RNA initiates transcription. Notably, a similar mechanism of antisense regulation was discovered controlling the ADH1 gene of S. pombe in response to zinc (68). While ADH5 and BDH1 also show decreased mRNA abundance, we have no clues as yet of how that occurs. While we have not directly examined SFA1 mRNA levels, it was not detected as zinc regulated in previous transcriptome studies (20, 44). These observations suggest that Sfa1 protein levels decrease due to a post-transcriptional mechanism.
The broad effect of zinc deficiency on the abundance of proteins that comprise the translational machinery of the cell is also striking. Several zinc-dependent ribosomal subunits are decreased in expression. In addition, zinc-binding proteins of the ribosome biogenesis (RiBi) regulon, including ribosome biogenesis factors, translation initiation factors, and tRNA synthetases and modifying enzymes, also decrease (69). Summing all of these proteins, the estimated number of zinc-binding sites drops in deficiency by ~2.2 × 106 zinc atoms per cell. This collectively represents about half of the zinc sparing that we observed across the entire zinc proteome. This effect extends far beyond the zinc-dependent proteins and includes a large number of zinc-independent translation machinery proteins as well (Fig. 3). It has been previously shown that expression of ribosomal proteins and the RiBi regulon is controlled in response to growth rate such that slower growing cells have reduced levels of these proteins than do faster growing cells (69, 70). Given that zinc deficiency results in slower growth, the underlying mechanisms linking growth rate to gene expression likely plays some role in their decreased abundance we observed in our studies.
An additional mechanism regulating the translational machinery in response to zinc deficiency is suggested by the studies from Chanfreau and colleagues who showed that RNAPI is specifically targeted for degradation during zinc deficiency (28). Decreased RNAPI levels decrease 5.8S, 18S, and 25S rRNA synthesis, which would then trigger decreased ribosomal protein and RiBi regulon expression. It was previously suggested that targeted degradation of RNAPI was a zinc sparing response because of the zinc no longer required for this specific RNA polymerase complex. Based on our results, we estimate that the number of zinc atoms that are directly spared by degrading the five zinc-binding RNAPI subunits would be ~30,000 total. While not an insignificant amount, it represents only about 1% of the total zinc sparing that we observed. Mutant cells defective for RNAPI degradation are hypersensitive to EDTA, suggesting a greater disruption in zinc homeostasis than is expected from the level of zinc sparing provided by degrading this protein complex alone. Therefore, we suggest that the effect of RNAPI degradation on zinc sparing may be much more extensive because of the resulting effects on the expression of ribosomal subunits and the RiBi regulon. In other words, zinc-responsive RNAPI degradation may be the master switch that controls the bulk of the zinc sparing response in zinc-deficient cells.
Despite the massive decrease in the number of zinc proteins that occurs during deficiency, we found that the number of zinc atoms that cells accumulated under these conditions was not sufficient to metalate even the reduced zinc requirement. In fact, we estimated that 70% of the zinc sites on proteins in a zinc-deficient cell were either not metalated (i.e. apoproteins) or mismetalated with a different cation. There is some question about how many potential zinc-binding ribosomal subunits in eukaryotes actually bind zinc in vivo (71). Nonetheless, if those proteins are removed from our calculations, the level of zinc atoms per cell are still insufficient for full metalation. Our previous studies of Tsa1 suggested that this protein’s chaperone function was critical to tolerate this stress and that zinc apoproteins are the likely clients of Tsa1 (23). We tested the hypothesis of accumulated apoproteins for two abundant zinc proteins, Fba1 and Met6. Despite little change in protein levels, Fba1 enzyme activity was greatly reduced in zinc-deficient cells compared to replete cells. In addition, using a novel application of thiol-reactive agents to study in vivo metalation, we found that Fba1 in cells underwent a conformational change that was dependent on zinc availability and zinc ligands. These results suggested that Fba1 is not fully metalated in zinc-deficient cells. It was therefore surprising that reintroduction of zinc appeared to rapidly metalate the protein but did not restore enzyme activity to replete levels and new protein synthesis was required to restore full activity. It is conceivable that the Fba1 protein is somehow damaged or modified (e.g. by phosphorylation) in zinc-deficient cells such that new Fba1 protein is required to restore activity. One possible mechanism of damage is glycation which has been previously observed for yeast Fba1 (72). Our studies of Met6 also indicated the accumulation of apoproteins during zinc deficiency. For Met6, we could probe reactivity of the cysteine ligands involved in direct zinc binding. These studies suggested that the many Met6 molecules in a cell are also unmetalated. It is conceivable that Fba1 and/or Met6 are mismetalated but if so, binding of that other cation fails to produce the same conformational change as zinc does for Fba1 and fails to protect the bound ligand from reaction with NEM in Met6. It was also surprising that many chaperones, co-chaperones, and chaperonin proteins (e.g. Ssa2, Ydj1, Scj1, Cct2) decreased in abundance during zinc deficiency (Fig. 3, cluster 2) when our other results indicated an increase in apoprotein accumulation and unfolded protein stress. We note that many of the proteins that decreased in abundance during deficiency are “foldase”-type chaperones while Tsa1 is a “holdase”-type protein. These results make biological sense because apoproteins cannot be fully folded without resupply of their metal cofactor and foldase chaperones would not suffice.
CONCLUSIONS
In this report, we have cataloged the zinc proteome of yeast and determined the effects of zinc deficiency on the abundance of these proteins. This analysis highlighted the diverse functions of zinc proteins and mapped in detail the subcellular requirement of this essential metal nutrient. Our studies demonstrated that the majority of zinc proteins detected decrease in abundance during deficiency and this apparent zinc-sparing response is due at least in part to transcriptional regulation of many of their respective genes. Our results also indicated that a surprisingly high number of zinc sites are not metalated with zinc during deficiency. Future studies will address the role of Tsa1 as a chaperone of these abundant zinc apoproteins. Future studies will also address the prevalence of zinc apoproteins more broadly and determine whether all proteins are similarly affected by zinc deficiency or whether some proteins can compete for zinc better than others. It will be exciting to learn whether prioritization of zinc distribution occurs in zinc trafficking among apoproteins.
EXPERIMENTAL
Strains and growth conditions-
Yeast were grown in rich (YPD), synthetic defined (SD), or low zinc medium (LZM), as previously described (73). LZM contains 20 mM citrate and 1 mM EDTA to buffer pH and zinc availability. Glucose (2%) was the carbon source for all experiments. LZM + 1 μM ZnCl2 was routinely used as the zinc-deficient condition, and LZM + 100 μM ZnCl2 as the replete condition. The yeast strains used in this work were BY4743 (MATa/MATα his3/his3 leu2/leu2 ura3/ura3 lys2/LYS2 met15/MET15), BY4742 (MATa his3 leu2 ura3 lys2), BY4741 (MATα his3 leu2 ura3 met15), and BY4741 fba1DAmP (Thermo Fisher Scientific) (57).
Cataloging the zinc proteome of yeast-
Using the approach described in Valasatava et al. (74), we created two libraries of Hidden Markov Model profiles (75): a library of zinc-binding Pfam domains (76), and a library of zinc-binding structural motifs. The Pfam domain library was created by merging two lists: first, a list of Pfam domains with known 3D structure that contain a zinc-binding site extracted from MetalPDB (77) in which each of these domains could be associated with the residues responsible for zinc binding and with their positions within the domain sequence and second, a list of Pfam domains without a known 3D structure but annotated as zinc-binding obtained by text mining of the annotations in the Pfam database. The procedure resulted in a set of 573 Pfam domains: 541 with an associated zinc-containing 3D structure, and an additional 32 annotated as zinc-binding domains. The library of zinc-binding structural motifs was created by splitting into fragments the zinc-binding sites stored in MetalPDB as of June 2017, as described in Rosato et al. (35). Only one representative was kept for zinc-binding sites that, though found in different PDB structures, fall in the same position of the same protein domain. Sites that are not physiologically relevant based on literature inspection (e.g., zinc-substituted structures, spurious sites) were manually removed from the dataset. This procedure resulted in a library of 6450 zinc-binding motifs derived from 2651 zinc-binding sites. An additional library of 339 zinc-binding motifs was compiled separately because the native metal ion of their corresponding proteins is still under investigation.
The zinc proteome of yeast was obtained by using the hmmscan tool (75) to search each yeast sequence for the profiles contained in the two libraries. A yeast sequence was identified as a potential zinc-binding site if at least one of the following conditions was verified: A) The profiles of all the fragments of a given site matched the sequence with an e-value lower than 10−3 and the corresponding ligands are conserved in the sequence. B) The profile of a domain with associated ligands matched the sequence with an e-value lower than 10−5 and the ligands are conserved in the sequence. C) The profile of a domain with no associated ligands matched the sequence with an e-value lower than 10−7. These predictions were integrated by adding the proteins annotated as zinc-binding in the UniProt database (78). In total, 571 yeast proteins were identified as zinc binding using the 6450 motifs (Table S1). An additional set of 45 potential zinc-binding proteins were identified with the 339 input sites as able to bind zinc but for which the identity of the native metal ion is still unknown (Table S2). For each of these proteins, the subcellular location and enzyme EC number were retrieved from UniProt. For each site contained in these proteins, the information on zinc function, number of zinc ions per monomer, and structural zinc-site classification (34) was imported from the site in the library that yielded the best match in the search, and then manually checked.
Mass spectrometry analysis and protein copy number estimations-
Mass spectrometry for proteomics analysis was performed on four biological replicates each for times 0, 4, 8, and 12 h of zinc deficiency and three biological replicates for 16 h of deficiency. Frozen cell pellets of various cell counts were lysed in 300 μl cold methanol. Precipitated proteins were separated by centrifugation for 10 minutes at 13,400 × g and 4°C. The protein pellet was resuspended in 200 μl lysis buffer (8 M urea, 20 mM TCEP, 80 mM chloroacetamide, 100 mM Tris pH 8) and diluted with 1 ml 100 mM Tris pH 8. Protein digestion was performed overnight with trypsin (4 μg) before centrifuging for 5 minutes at 10,000 × g. The resulting supernatant was de-salted with Strata C18 solid phase extraction cartridges and quantified using Pierce Quantitative Colorimetric Peptide Assay (Thermo Fisher Scientific). Peptides were then dried in a vacuum centrifuge before resuspension in 0.2% formic acid.
Samples were analyzed using a LC/MS instrument comprising an Orbitrap Fusion Lumos tribrid mass spectrometer (Thermo Fisher Scientific). Mobile phase A consisted of 0.2% formic acid in water and mobile phase B consisted of 0.2% formic acid in acetonitrile. A 75-minute gradient ranging from 0% to 50% B was used spanning a total runtime of 90 minutes. Analytes were injected onto a 1.7-micron C18 column packed in-house to a length of 35 cm and heated to 60°C. Survey scans of peptide precursors were collected from 350–1350 Th with an AGC target of 1,000,000 and a resolution of 60,000 in the orbitrap followed by HCD MS/MS turbo scans taken in the ion trap.
The resulting LC-MS proteomic data were processed using Maxquant software version 1.5.2.8 and searched against a Saccharomyces cerevisiae database downloaded from Uniprot on 8/10/16. The digestion enzyme was set to trypsin with up to two missed cleavages, carbamidomethylation of cysteines as a fixed modification, and oxidation of methionines and protein N-terminal acetylation as variable modifications. The match between runs feature was utilized to decrease missing data values within the data set. Precursor mass tolerance was 20 ppm and product ions were searched at 4.5 ppm tolerances. Peptides were filtered to a 1% FDR and combined to protein groups based on the rules of parsimony. Protein copy number calculations were performed in Perseus using the Proteomic Ruler plugin (40). This method uses the peak intensities of histone proteins, which are proportional to DNA content, to estimate protein abundance on a per cell basis. Statistically significant effects were defined as those proteins with 1.5-fold or greater changes and a p-value of <0.05 after Benjamini-Hochberg correction. The proteomics data was deposited in CHORUS (https://chorusproject.org) under Project ID number 1530. Hierarchical cluster was performed using Euclidean distance with Perseus (79). Gene Ontology analysis of four clusters produced from hierarchical clustering was performed using DAVID (80).
Quantitative RT-PCR analysis-
Quantitative analysis of gene expression by RT-PCR was performed as previously described (81). Assays were performed on three biological replicates. The sequences of the primer pairs used are provided in Table S5. Gene expression was calculated relative to the average Ct values for three control genes (TAF10, ACT1 and CMD1) selected from multiple candidate genes tested for their highly stable expression under the conditions used in our experiments (data not shown).
ICP-AES analysis-
Culture aliquots were washed three times in cold water, pelleted, and frozen in liquid nitrogen. Total zinc was determined using inductively-coupled plasma atomic emission spectrometry (ICP-AES) on six biological replicates for each timepoint. Cell pellets were desiccated by incubation at 60°C for 12–18 h and subsequent dry weights recorded. The dried yeast pellets were acid digested in 250 μl OmniTrace 70% HNO3 (EMD Chemicals) at 60°C for 12–18 h with 150–200 rpm orbital shaking. The acid lysates were then diluted to 5% HNO3 with OmniTrace water (EMD Chemicals) and analyzed by ICP-AES (5100 SVDV, Agilent Technologies). The ICP-AES instrument was calibrated using National Institute of Standards and Technology (NIST)-traceable elemental standards and validated using NIST-traceable 1577b bovine liver reference material. The detection range for zinc was 0.005–5 parts per million with inter-assay precision <10%. Cesium (50 ppm) was used for ionization suppression and yttrium (5 ppm) was used as an internal standard for all samples. All reagents and plasticware were certified or routinely tested for trace metal work. Zinc content was determined using native software (ICPExpert) and normalized to the cell number in each sample.
Protein extraction and immunoblotting-
Yeast protein extracts were prepared for immunoblotting with a TCA extraction protocol as previously described (82). SDS-PAGE and immunoblotting was conducted using a Li-Cor Odyssey infrared dye detection system as previously described (82). Antibodies used were anti-HA (12CA5, Roche product 11583816001), anti-Fba1 (a gift from Dr. Magdalena Boguta) (83), and anti-Pgk1 (Abcam product 22C5D8, lot # GR166098). IR 680 dye-labeled secondary anti-mouse antibody (product 680LT, lot # C30605–02) was obtained from Li-Cor.
Assay of Fba1 aldolase activity-
Fba1 enzyme activity was determined using either permeabilized yeast cells or cell lysates. An assay developed by Freire was modified for use with 96-well plates (84). After harvest, cells were washed once with equal volume of ice-cold deionized water containing 1 mM EDTA, twice with ice-cold deionized water, and resuspended in 0.1 M MES-NaOH pH 6.5 buffer. Cell densities of each sample were normalized to A595 = 1.5, and 0.01% digitonin (w/v) was added to permeabilize the cells. After incubating at 30°C in a shaking water bath for 10 minutes, cells were placed on ice and then washed twice, resuspended with ice-cold 25 mM KH2PO4/K2HPO4 pH 7.4 buffer to a density of A595 = 0.5. A standard assay of aldolase activity contained 10 units of triose-phosphate isomerase (TPI), 4 units of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in a 25 mM KH2PO4/K2HPO4 pH 7.4 buffer containing 5 mM sodium arsenate, and 5 mM of NAD+ and 20 μl of permeabilized cells. The assay was started by adding 5 mM of the substrate, fructose 1,6-bisphosphate (FBP), and incubated at 30°C in a temperature-controlled plate reader. The absorbance at 340 mm of NADH generated by the assay was recorded at 1-minute intervals for up to 20 minutes. The recorded A340 values were graphed and the linear portion of the graph was used to calculate the rates of Fba1 aldolase activity using an NADH standard curve following normalization to the cell density (A600) in each reaction. Alternatively, some assays were conducted using 1–10 μg of protein lysate per reaction and activity was normalized to total protein level. Cell pellets were resuspended in 0.5 ml of 25 mM KH2PO4/K2HPO4 pH 7.4 buffer containing 1 mM PMSF and 1 × EDTA-free protease inhibitor mix (Roche) and transferred to 1.5 ml tubes. A 0.2 ml volume of glass beads was added and the cells disrupted by vortexing for 5 minutes at 4°C. The homogenate was centrifuged at 16,000 × g for 10 minutes at 4°C and the supernatant stored on ice. The protein concentration was determined using a Bio-Rad DC kit against a BSA standard. To determine specific activity of Fba1 (in nmol NADH/minute/μg Fba1 protein), A340 absorbance values were converted to NADH concentration using a standard curve and resulting activity values (nmol NADH/minute/μg total protein) were normalized to Fba1-HA protein level as determined by immunoblotting, taking into account the value for Fba1 abundance in zinc replete cells that we determined by mass spectrometry (i.e. 1% of total protein). This normalization was not performed in cases where Fba1 abundance was known to be stable during the experiment (e.g. Figure 9F). For some experiments, zinc was stripped from Fba1 by treating permeabilized cells with 50 mM EDTA and 5 mM FBP in a 30°C shaking water bath for 30 minutes. The cells were then washed five times with ice-cold 25 mM KH2PO4/K2HPO4 buffer containing 1 μM EDTA to prevent any zinc remetalation. Zinc was added back to aliquots of stripped cells by treating with 20 μM ZnCl2 in 30°C shaking water bath [for 5, 15, and 30 minutes] this detail should go in the figure legend?. The cells were then washed three times with ice-cold 25 mM KH2PO4/K2HPO4 buffer containing 1 μM EDTA and resuspended at A595 = 0.5 for the assay.
Plasmid constructions-
All plasmids were constructed by homologous recombination in yeast. Plasmids expressing Fba1 with a triple repeat of the hemagglutinin antigen (HA) epitope fused to the C-terminus were constructed by PCR-amplification of wild-type or mutant FBA1 promoter and coding sequence and fused to the HA tags and terminator of the low copy episomal plasmid YCp-ZRC1-HA digested with BamHI and EcoRI. To generate a wild-type tagged plasmid, a fragment was amplified with the oligonucleotides Fbacds-ha (5’-TAGCCCGCATAGTCAGGAACATCGTATGGGTATAAAGTGTTAGTGGTACGGAAAGT-3’) and Fba1pr5’ (5’-CACGACGTTGTAAAACGACGGCCAGTGAATTCTGACGCAAGCCCTAAGAA-3’) and used to gap repair YCp-ZRC1-HA. To introduce the H265A mutation, a 5’-most fragment was generated using the oligonucleotides H-Arev (5’-TGGAATTCTTGGACAGTAGAACCGGAACCACCGGCGAAGACCAAGA-3’) and Fba1pr5’, and a 3’ -most fragment with H-Acomp (5’- GGTGGTTCCGGTTCTACTGT-3’) and Fbacds-ha. The two fragments were combined with gapped YCp-ZRC1-HA for yeast transformation. To construct the H111A and E183A mutants, a PCR fragment was amplified from the corresponding mutant versions of pFL44L-FBA1 using the oligonucleotides Fba1pr5’ and Fbacds-ha prior to gap repair. All plasmids were fully sequenced to verify the mutations. Functionality of the wild-type epitope-tagged plasmid was verified by complementation of aldolase enzyme activity when expressed in an fba1DAmP mutant strain (data not shown). Fba1-3×HA accumulated to a similar level as untagged Fba1 when both forms were coexpressed and detected with anti-Fba1, and the tagged protein had similar specific activity to untagged (data not shown), indicating that the epitope tags had no major effect on stability or function. The Met6-HA construct and inactivated mutant versions were constructed similarly to Fba1-HA using gap repair. The wild-type Met6 plasmid (YCpMet6-HA) was constructed by amplifying Met6 from BY4742 genomic DNA using met6cds3’ (5’-CACGACGTTGTAAAACGACGGCCAGTGAATTCCACAGCCATTCAACTCAG-3’) and Met6cds3’ (5-TAGCCCGCATAGTCAGGAACATCGTATGGGTAATTCTTGTATTGTTCACGGA-3’) primers, followed by gap repair of YCp-ZRC1-HA digested with EcoRI and BamHI. To construct the H655A mutant, a fragment containing the mutation was generated by PCR amplification of YCpMet6-HA using the oligonucleotides met65’−1 (5’-GGCTGACAAGGATTCTCT-3’) and met6H-A3’ (5’-CATCAGCATCCAAAGCCTTGATATGGTTTGGATCCAAGTCAGAGTAACAGAAAGCAGAGTGTATTTGAGTCTTGTT3’). This fragment was used to gap repair YCpMet6-HA digested with SacI and BamHI. To construct the D612A mutant, a fragment was amplified from YCpMet6-HA with the met65’−1 and met6DA3’ (CTTCTCTTAAAGCTGGTTCAgCAACTTGGATAACCTTGATACCGGCAGCT) oligonucleotides, and a second fragment was amplified with the met6da5’ (TATCCAAGTTGcTGAACCAGCTTTAAGAGAAGGTTTACCATTGAGAGAAGGTA) and met6cds3’ oligonucleotides. The two fragments were combined and used to gap repair YCpMet6-HA digested with SacI and BamHI. Wild-type and mutant plasmids were fully sequenced, and the effect of the mutations on activity was verified by complementation of the methionine auxotrophy of a met6 deletion mutant.
NEM/PEG maleimide analysis-
To identify cysteine residues showing zinc-dependent reactivity with N-ethylmaleimide (NEM) in vivo, 5 ml cultures of yeast were grown in zinc-replete (LZM + 100 μM ZnCl2) or deficient (LZM + 1 μM ZnCl2) medium to log phase (A595 0.3–0.4) and harvested by centrifugation. Cells were washed twice with ice-cold 1 × PBS + 1 mM EDTA and resuspended in 5 ml PBS + 1 mM EDTA. A solution of 1 M NEM in 100% ethanol was added to 5 ml of cells to give a final concentration of 5 mM NEM. For a negative control, the same volume of 100% ethanol was added to an identical aliquot of cells. After 30 min incubation at 30° with shaking, the cultures were harvested by centrifugation and washed twice with 1 × PBS. Protein was extracted using the TCA method (23) and redissolved in buffer A (200 mM Tris base, 1% SDS, 1 mM EDTA). After measurement of protein concentration (DC protein assay, Bio-Rad), aliquots of protein were processed to modify cysteines with mPEG-5kDa (Sigma) as previously described (23). Briefly, aliquots of 500 μg protein were treated with 20 mM DTT for 10 min at 65°C to reduce disulfide bonds, then reprecipitated by adding 1/10 volume 100% TCA. Precipitated samples were centrifuged and washed twice with acetone to remove TCA, then redissolved in buffer B (100 mM Tris-Cl pH 7.4, 2% SDS, 1 mM EDTA) + 5 mM PEG-maleimide (mPEG). After overnight incubation at 30°C, 10–30 μg of each protein sample was analyzed by SDS-PAGE and immunoblotting to determine the degree of mPEG modification of cysteine residues. To determine the degree to which cysteines were normally oxidized in vivo (and thus unavailable for reaction with NEM), some control samples were not treated with DTT prior to mPEG treatment.
Supplementary Material
Figure S1. Evaluation of proteomics reproducibility.
Table S1. The zinc proteome of S. cerevisiae.
Table S2. Other potential zinc-binding proteins of S. cerevisiae.
Table S3. Full results of the proteomics analysis.
Table S4. Gene ontology analysis of the total proteome clusters.
Table S5. Primers used for quantitative RT-PCR analysis.
SIGNIFICANCE TO METALLOMICS.
Zinc is a common protein cofactor and zinc deficiency is often faced by many organisms. How do cells adapt to conditions of zinc deficiency to maintain function of their many zinc-dependent proteins (i.e. their “zinc proteome”)? To answer this question, we cataloged the zinc proteome of Saccharomyces cerevisiae, determined the effect of deficiency on zinc protein abundance, and assessed the metalation state of the zinc proteome during deficiency. This analysis revealed many new mechanisms cells use to reduce their requirement for this essential nutrient.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grant R01-GM56285 (DJE) and grants P41-GM10853 and R35GM118110 (JJC). The authors also acknowledge the support and the use of resources of Instruct-ERIC, a Landmark ESFRI project, and specifically the CERM/CIRMMP Italy Centre.
Footnotes
CONFLICTS OF INTEREST
There are no conflicts of interest to declare.
REFERENCES
- 1.Andreini C, Banci L, Bertini I, Rosato A. Zinc through the three domains of life. J Proteome Res. 2006;5(11):3173–8. [DOI] [PubMed] [Google Scholar]
- 2.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292(5526):2488–92. [DOI] [PubMed] [Google Scholar]
- 3.MacDiarmid CW, Gaither LA, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000;19(12):2845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmiter RD, Findley SD. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 1995;14(4):639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suhy DA, Simon KD, Linzer DI, O’Halloran TV. Metallothionein is part of a zinc-scavenging mechanism for cell survival under conditions of extreme zinc deprivation. J Biol Chem. 1999;274(14):9183–92. [DOI] [PubMed] [Google Scholar]
- 6.Dittmer PJ, Miranda JG, Gorski JA, Palmer AE. Genetically encoded sensors to elucidate spatial distribution of cellular zinc. J Biol Chem. 2009;284(24):16289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fierke CA, Thompson RB. Fluorescence-based biosensing of zinc using carbonic anhydrase. Biometals. 2001;14(3–4):205–22. [DOI] [PubMed] [Google Scholar]
- 8.Vinkenborg JL, Koay MS, Merkx M. Fluorescent imaging of transition metal homeostasis using genetically encoded sensors. Curr Opin Chem Biol. 2010;14(2):231–7. [DOI] [PubMed] [Google Scholar]
- 9.Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol. 2017;15(6):338–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kambe T, Tsuji T, Hashimoto A, Itsumura N. the physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol Rev. 2015;95(3):749–84. [DOI] [PubMed] [Google Scholar]
- 11.Krezel A, Maret W. The functions of metamorphic metallothioneins in zinc and copper metabolism. Int J Mol Sci. 2017;18(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eide DJ. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem. 2009;284(28):18565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merchant SS, Helmann JD. Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv Microbial Physiol. 2012;60:91–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panina EM, Mironov AA, Gelfand MS. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci USA. 2003;100(17):9912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nanamiya H, Akanuma G, Natori Y, Murayama R, Kosono S, Kudo T, et al. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol Microbiol. 2004;52(1):273–83. [DOI] [PubMed] [Google Scholar]
- 16.Gabriel SE, Helmann JD. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J Bact. 2009;191(19):6116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi S, Bird AJ. Zinc’ing sensibly: controlling zinc homeostasis at the transcriptional level. Metallomics. 2014;6(7):1198–215. [DOI] [PubMed] [Google Scholar]
- 18.Kersting MC, Carman GM. Regulation of the Saccharomyces cerevisiae eki1-encoded ethanolamine kinase by zinc depletion. J Biol Chem. 2006;281(19):13110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soto A, Carman GM. Regulation of the Saccharomyces cerevisiae CKI1-encoded choline kinase by zinc depletion. J Biol Chem. 2008;283(15):10079–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CY, Bird AJ, Chung LM, Newton MA, Winge DR, Eide DJ. Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae. BMC Genomics. 2008;9:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu CY, Bird AJ, Winge DR, Eide DJ. Regulation of the yeast TSA1 peroxiredoxin by ZAP1 is an adaptive response to the oxidative stress of zinc deficiency. J Biol Chem. 2007;282(4):2184–95. [DOI] [PubMed] [Google Scholar]
- 22.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117(5):625–35. [DOI] [PubMed] [Google Scholar]
- 23.Macdiarmid CW, Taggart J, Kerdsomboon K, Kubisiak M, Panascharoen S, Schelble K, et al. Peroxiredoxin chaperone activity is critical for protein homeostasis in zinc-deficient yeast. J Biol Chem. 2013;188:31313–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454(7208):1088–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bird AJ, Gordon M, Eide DJ, Winge DR. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 2006;25(24):5726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williamson VM, Paquin CE. Homology of Saccharomyces cerevisiae ADH4 to an iron-activated alcohol dehydrogenase from Zymomonas mobilis. Mol Gen Genet. 1987;209(2):374–81. [DOI] [PubMed] [Google Scholar]
- 27.Drewke C, Ciriacy M. Overexpression, purification and properties of alcohol dehydrogenase IV from Saccharomyces cerevisiae. Biochim Biophys Acta. 1988;950(1):54–60. [DOI] [PubMed] [Google Scholar]
- 28.Lee YJ, Lee CY, Grzechnik A, Gonzales-Zubiate F, Vashisht AA, Lee A, et al. RNA polymerase I stability couples cellular growth to metal availability. Mol Cell. 2013;51(1):105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao W, Ellis C, Steffen J, Wu CY, Eide DJ. Zinc status and vacuolar zinc transporters control alkaline phosphatase accumulation and activity in Saccharomyces cerevisiae. Mol Microbiol. 2009;72(2):320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Citiulo F, Jacobsen ID, Miramon P, Schild L, Brunke S, Zipfel P, et al. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog. 2012;8(6):e1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagani A, Villarreal L, Capdevila M, Atrian S. The Saccharomyces cerevisiae Crs5 Metallothionein metal-binding abilities and its role in the response to zinc overload. Mol Microbiol. 2007;63(1):256–69. [DOI] [PubMed] [Google Scholar]
- 32.Palacios O, Atrian S, Capdevila M. Zn− and Cu−thioneins: a functional classification for metallothioneins? J Biol Inorg Chem. 2011;16(7):991–1009. [DOI] [PubMed] [Google Scholar]
- 33.Andreini C, Bertini I. A bioinformatics view of zinc enzymes. J Inorg Biochem. 2012;111:150–6. [DOI] [PubMed] [Google Scholar]
- 34.Andreini C, Bertini I, Cavallaro G. Minimal functional sites allow a classification of zinc sites in proteins. PloS One. 2011;6(10):e26325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosato A, Valasatava Y, Andreini C. Minimal functional sites in metalloproteins and their usage in structural bioinformatics. Int J Mol Sci. 2016;17(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–91. [DOI] [PubMed] [Google Scholar]
- 37.Koh JL, Chong YT, Friesen H, Moses A, Boone C, Andrews BJ, et al. CYCLoPs: A comprehensive database constructed from automated analysis of protein abundance and subcellular localization patterns in Saccharomyces cerevisiae. G3. 2015;5(6):1223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richards AL, Hebert AS, Ulbrich A, Bailey DJ, Coughlin EE, Westphall MS, et al. One-hour proteome analysis in yeast. Nat Prot. 2015;10(5):701–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stefely JA, Kwiecien NW, Freiberger EC, Richards AL, Jochem A, Rush MJP, et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat Biotechnol. 2016;34(11):1191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wisniewski JR, Hein MY, Cox J, Mann M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol Cell Proteom. 2014;13(12):3497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–41. [DOI] [PubMed] [Google Scholar]
- 42.Milo R What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays. 2013;35(12):1050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen LT, Howard WR, Strain JJ, Winge DR, Culotta VC. Enhanced effectiveness of copper ion buffering by CUP1 compared with CRS5 metallothionein in Saccharomyces cerevisiae. J Biol Chem. 1996;271:18514–9. [DOI] [PubMed] [Google Scholar]
- 44.Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci USA. 2000;97(14):7957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han SH, Han GS, Iwanyshyn WM, Carman GM. Regulation of the PIS1-encoded phosphatidylinositol synthase in Saccharomyces cerevisiae by zinc. J Biol Chem. 2005;280(32):29017–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Nicola R, Hazelwood LA, De Hulster EA, Walsh MC, Knijnenburg TA, Reinders MJ, et al. Physiological and transcriptional responses of Saccharomyces cerevisiae to zinc limitation in chemostat cultures. Appl Environ Microbiol. 2007;73(23):7680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu CY, Roje S, Sandoval FJ, Bird AJ, Winge DR, Eide DJ. Repression of sulfate assimilation is an adaptive response of yeast to the oxidative stress of zinc deficiency. J Biol Chem. 2009;284(40):27544–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh N, Yadav KK, Rajasekharan R. ZAP1-mediated modulation of triacylglycerol levels in yeast by transcriptional control of mitochondrial fatty acid biosynthesis. Mol Microbiol. 2016;100(1):55–75. [DOI] [PubMed] [Google Scholar]
- 49.Soto-Cardalda A, Fakas S, Pascual F, Choi HS, Carman GM. Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast. J Biol Chem. 2012;287(2):968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu YH, Taggart J, Song PX, MacDiarmid C, Eide DJ. An MSC2 promoter-lacz fusion gene reveals zinc-responsive changes in sites of transcription initiation that occur across the yeast genome. PloS One. 2016;11(9):e0163256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanley D, Fraser S, Stanley GA, Chambers PJ. Retrotransposon expression in ethanol-stressed Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2010;87(4):1447–54. [DOI] [PubMed] [Google Scholar]
- 52.Todeschini AL, Morillon A, Springer M, Lesage P. Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25(17):7459–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bradshaw VA, McEntee K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol Gen Genet. 1989;218(3):465–74. [DOI] [PubMed] [Google Scholar]
- 54.Kawamata T, Horie T, Matsunami M, Sasaki M, Ohsumi Y. Zinc starvation induces autophagy in yeast. J Biol Chem. 2017;292(20):8520–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bucci MD, Weisenhorn E, Haws S, Yao Z, Zimmerman G, Gannon M, et al. An autophagy-independent role for atg41 in sulfur metabolism during zinc deficiency. Genetics. 2018;208(3):1115–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwanyshyn WM, Han GS, Carman GM. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc. J Biol Chem. 2004;279(21):21976–83. [DOI] [PubMed] [Google Scholar]
- 57.Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Meth. 2008;5(8):711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Apuy JL, Busenlehner LS, Russell DH, Giedroc DP. Ratiometric pulsed alkylation mass spectrometry as a probe of thiolate reactivity in different metalloderivatives of Staphylococcus aureus pI258 CadC. Biochemistry. 2004;43(13):3824–34. [DOI] [PubMed] [Google Scholar]
- 59.Apuy JL, Chen X, Russell DH, Baldwin TO, Giedroc DP. Ratiometric pulsed alkylation/mass spectrometry of the cysteine pairs in individual zinc fingers of MRE-binding transcription factor-1 (MTF-1) as a probe of zinc chelate stability. Biochemistry. 2001;40(50):15164–75. [DOI] [PubMed] [Google Scholar]
- 60.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom Rev. 2009;28(5):785–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ubhi D, Kago G, Monzingo AF, Robertus JD. Structural analysis of a fungal methionine synthase with substrates and inhibitors. J Mol Biol. 2014;426(8):1839–47. [DOI] [PubMed] [Google Scholar]
- 62.Ubhi D, Kavanagh KL, Monzingo AF, Robertus JD. Structure of Candida albicans methionine synthase determined by employing surface residue mutagenesis. Arch Biochem Biophys. 2011;513(1):19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prasannan P, Suliman HS, Robertus JD. Kinetic analysis of site-directed mutants of methionine synthase from Candida albicans. Biochem Biophys Res Comm. 2009;382(4):730–4. [DOI] [PubMed] [Google Scholar]
- 64.Hara T, Takeda TA, Takagishi T, Fukue K, Kambe T, Fukada T. Physiological roles of zinc transporters: molecular and genetic importance in zinc homeostasis. J Phys Sci. 2017;67(2):283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Que EL, Bleher R, Duncan FE, Kong BY, Gleber SC, Vogt S, et al. Quantitative mapping of zinc fluxes in the mammalian egg reveals the origin of fertilization-induced zinc sparks. Nat Chem. 2015;7(2):130–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barnett JP, Millard A, Ksibe AZ, Scanlan DJ, Schmid R, Blindauer CA. Mining genomes of marine cyanobacteria for elements of zinc homeostasis. Front Microbiol. 2012;3:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herzberg M, Dobritzsch D, Helm S, Baginsky S, Nies DH. The zinc repository of Cupriavidus metallidurans. Metallomics. 2014;6(11):2157–65. [DOI] [PubMed] [Google Scholar]
- 68.Ehrensberger KM, Mason C, Corkins ME, Anderson C, Dutrow N, Cairns BR, et al. Zinc-dependent regulation of the Adh1 antisense transcript in fission yeast. J Biol Chem. 2013;288(2):759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de la Cruz J, Gomez-Herreros F, Rodriguez-Galan O, Begley V, de la Cruz Munoz-Centeno M, Chavez S. Feedback regulation of ribosome assembly. Curr Gen. 2018;64(2):393–404. [DOI] [PubMed] [Google Scholar]
- 70.Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21(6):855–63. [DOI] [PubMed] [Google Scholar]
- 71.Brandes N, Reichmann D, Tienson H, Leichert LI, Jakob U. Using quantitative redox proteomics to dissect the yeast redoxome. J Biol Chem. 2011;286(48):41893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gomes RA, Vicente Miranda H, Silva MS, Graca G, Coelho AV, Ferreira AE, et al. Yeast protein glycation in vivo by methylglyoxal. Molecular modification of glycolytic enzymes and heat shock proteins. FEBS J. 2006;273(23):5273–87. [DOI] [PubMed] [Google Scholar]
- 73.Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci U S A. 1996;93(6):2454–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valasatava Y, Rosato A, Banci L, Andreini C. MetalPredator: a web server to predict iron-sulfur cluster binding proteomes. Bioinformatics. 2016;32(18):2850–2. [DOI] [PubMed] [Google Scholar]
- 75.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–63. [DOI] [PubMed] [Google Scholar]
- 76.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Putignano V, Rosato A, Banci L, Andreini C. MetalPDB in 2018: a database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2018;46(D1):D459–D64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46(5):2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Meth. 2016;13(9):731–40. [DOI] [PubMed] [Google Scholar]
- 80.Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 81.MacDiarmid CW, Taggart J, Jeong J, Kerdsomboon K, Eide DJ. Activation of the yeast UBI4 polyubiquitin gene by Zap1 via an intragenic promoter is critical for zinc-deficient growth. J Biol Chem. 2016;291:18880–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.MacDiarmid CW, Taggart J, Kerdsomboon K, Kubisiak M, Panascharoen S, Schelble K, et al. Peroxiredoxin chaperone activity is critical for protein homeostasis in zinc-deficient yeast. J Biol Chem. 2013;288(43):31313–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciesla M, Mierzejewska J, Adamczyk M, Farrants AK, Boguta M. Fructose bisphosphate aldolase is involved in the control of RNA polymerase III-directed transcription. Biochim Biophys Acta. 2014;1843(6):1103–10. [DOI] [PubMed] [Google Scholar]
- 84.Freire AP, Martins AM, Cordeiro C. An experiment illustrating metabolic regulation in situ using digitonin permeabilized yeast cells. Biochem Educ. 1998;26:161–3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Evaluation of proteomics reproducibility.
Table S1. The zinc proteome of S. cerevisiae.
Table S2. Other potential zinc-binding proteins of S. cerevisiae.
Table S3. Full results of the proteomics analysis.
Table S4. Gene ontology analysis of the total proteome clusters.
Table S5. Primers used for quantitative RT-PCR analysis.