Abstract
Two decades ago, the recognition of protein misfolding and aggregate accumulation as defining features of neurodegenerative disease set the stage for a thorough examination of how protein quality control is maintained in neurons and in other nonneuronal cells in the central nervous system (CNS). Autophagy, a pathway of cellular self-digestion, has emerged as especially important for CNS proteostasis, and autophagy dysregulation has been documented as a defining feature of neurodegeneration in Alzheimer’s disease (AD), Parkinson’s disease (PD), and Huntington’s disease (HD). Transcription factor EB (TFEB) is one of the main transcriptional regulators of autophagy, as it promotes the expression of genes required for autophagosome formation, lysosome biogenesis, and lysosome function, and it is highly expressed in CNS. Over the last 7 years, TFEB has received considerable attention and TFEB dysfunction has been implicated in the pathogenesis of numerous neurodegenerative disorders. In this review, we delineate the current understanding of how TFEB dysregulation is involved in neurodegeneration, highlighting work done on AD, PD, HD, X-linked spinal & bulbar muscular atrophy, and amyotrophic lateral sclerosis. Because TFEB is a central node in defining autophagy activation status, efforts at understanding the basis for TFEB dysfunction are yielding insights into how TFEB might be targeted for therapeutic application, which may represent an exciting opportunity for the development of a treatment modality with broad application to neurodegeneration.
Keywords: neurodegeneration, autophagy, transcription factor EB, Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, polyglutamine, spinal & bulbar muscular atrophy, amyotrophic lateral sclerosis, lysosome, synucleinopathy
Neuron survival depends upon maintaining efficient protein quality control. Autophagy is an evolutionarily conserved lysosomal degradation pathway, which is highly active in neurons and functions to eliminate aggregate-prone proteins and dysfunctional mitochondria, both of which are hallmarks of many neurodegenerative disorders, including Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease (Figure 1). Indeed, recent evidence strongly suggests that neuronal autophagy is both a target of neurodegeneration, based upon strong evidence for autophagy dysfunction early on in disease pathogenesis, and an effector during neurodegeneration, as demonstrated by a significant propensity for neurodegenerative disease-associated mutations to occur within genes that encode component proteins and regulators of the autophagy pathway.
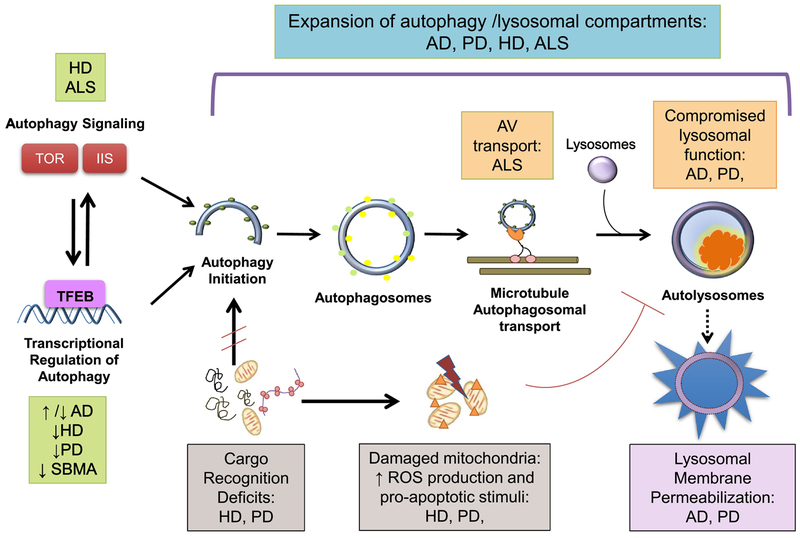
Figure 1. Autophagy Dysfunction in Neurodegenerative Disease.
Increased numbers of autophagic vesicles (AVs) are a common finding for many neurodegenerative disorders, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), X-linked spinal & bulbar muscular atrophy (SBMA), and amyotrophic lateral sclerosis (ALS). However, different molecular mechanisms account for the observed autophagy pathway dysfunction in these neurodegenerative disorders. 1) Autophagy signaling pathways are impaired in HD, ALS, and in the aging brain, with the mechanistic target of rapamycin (TOR) and insulin/IGF1 signaling (IIS) being significantly impacted. 2) Autophagy transcriptional network activity decreases in HD, PD, and in the aging brain. Contradictory to these findings, some evidence suggests up-regulation of autophagy genes in AD. Alterations in the master autophagy regulator transcription factor E-B (TFEB) signaling have been reported for PD, HD, and SBMA. 3) Deficits in cargo recognition, particularly of defective mitochondria, occur in HD and PD. The accumulation of damaged mitochondria generates an increased burden of reactive-oxygen species (ROS), which compromises lysosomal function and culminates in lysosomal permeabilization and leakage of lysosomal contents into the cytoplasm. Accumulation of ‘empty’ autophagosomes in HD also compromises neuronal homeostasis. 4) Despite normal autophagy induction and autophagosome formation, impaired maturation and microtubule-transport of autophagic vacuoles (AVs) towards lysosomes is a feature of ALS. 5) Lysosomal dysfunction contributes to autophagy dysregulation in AD and HD, especially as proteostasis clearance mechanisms decline with age. Accumulation of undigested cargo in the lysosomal lumen (e.g. organelles, liposfuscin) further impairs the lysosome’s degradative ability, enhancing lysosomal membrane destabilization and leakage.
Transcription factor EB (TFEB) has arisen as a master regulator of lysosome function and autophagy. As a key transcription factor modulating fundamental cellular homeostasis pathways, a rather complex, but still incompletely defined regulatory network governs TFEB activity and function. Although considerable work has sought to understand the role of TFEB in cellular metabolism (Martina and Puertollano, 2013; Medina et al., 2015; Settembre et al., 2013; Settembre et al., 2012), only recently have we begun to explore TFEB involvement in central nervous system (CNS) homeostasis. TFEB is widely expressed in the CNS (Decressac et al., 2013; Reddy et al., 2016; Wang et al., 2016b), as it is active in both neurons and astrocytes (Decressac et al., 2013; Xiao et al., 2014), and TFEB localization, activity, and function are altered in neurodegenerative disease (Cortes et al., 2014b; Decressac et al., 2013; Tsunemi et al., 2012; Wang et al., 2016b). In this review, we delineate the current understanding of TFEB’s role in neurodegenerative disease, and we examine if TFEB could represent a potential therapeutic target for treating these devastating diseases.
How is TFEB activity regulated?
TFEB is a member of the basic helix-loop-helix leucine-zipper family of transcription factors, and normally localizes to the cytosol, but translocates into the nucleus under starvation conditions or when lysosomal function is compromised (Sardiello et al., 2009; Settembre et al., 2011). TFEB overexpression induces increased numbers of autophagosomes, promotes the generation of new lysosomes, and increases autophagic flux. Through the CLEAR (Coordinated Lysosomal Enhancement And Regulation) signaling network, TFEB regulates expression of autophagy-lysosomal genes and promotes cellular clearance and metabolism (Sardiello et al., 2009; Settembre et al., 2011). Indeed, in addition to autophagy and lysosome related genes, deeper examination of the CLEAR gene network highlights pathways involved in cellular catabolism, mitochondrial function, and cellular stress responses (Palmieri et al., 2011). In agreement with this, TFEB was recently shown to directly regulate lipid catabolism and degradation in the liver (Settembre et al., 2013), and to promote glucose homeostasis and mitochondrial biogenesis in skeletal muscle (Mansueto et al., 2017). Cellular context may thus play an important role in determining the specific targets of TFEB-mediated transcription, highlighting the need for in-depth analysis of CNS-specific TFEB targets to decipher the role of TFEB in neurodegeneration.
Despite its key role as a master transcription factor modulating fundamental cellular homeostatic pathways, the regulatory network that governs TFEB activity is complex and not yet fully defined. TFEB is subject to post-translational modifications, which directly regulate its protein interactions and subcellular localization. Our current understanding of TFEB biology posits phosphorylation as the major driver of TFEB activity. In particular, phosphorylation of Serine 211 plays crucial role in determining the status of TFEB activation (Martina et al., 2012; Roczniak-Ferguson et al., 2012), facilitating interaction with cytosolic chaperone 14–3–3 and retaining TFEB in the cytoplasm. Phosphorylation of Serine 142 may also be required for TFEB cytosolic retention (Settembre et al., 2011; Settembre et al., 2012). Elegant in vitro studies have identified nutrient-sensing cellular kinases that directly phosphorylate TFEB at these residues, including the all-important mechanistic target of rapamycin complex 1 (mTORC1) and possibly extracellular signal-regulated kinase 2 (also known as MAPK1) (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2013; Settembre et al., 2011). Very recently, MAP4K3 was found to be a key upstream regulator of TFEB, linking amino acid supply to TFEB activation status (Hsu et al., 2018). MAP4K3 phosphorylates TFEB on Serine 4, and this phosphorylation event is required for mTORC1 to phosphorylate TFEB on its Serine 211 residue to insure its complete inactivation via cytosolic sequestration with 14–3–3 (Hsu et al., 2018).
Upon starvation or lysosomal stress, mTORC1 is inactivated and lysosomal calcium release activates the TFEB-targeting phosphatase calcineurin. This signaling cascade culminates in TFEB dephosphorylation, promoting its nuclear translocation and subsequent gene transcription of TFEB target genes in the CLEAR network (Medina et al., 2015). TFEB has arisen as a central hub of bioenergetics signaling, coordinating transcription of the CLEAR network to manage cellular metabolic demand (Palmieri et al., 2011; Settembre et al., 2012). More recently, acetylation of TFEB at Lysine 116 was discovered to be a novel post-translational modifier of TFEB function in microglia (Bao et al., 2016). Analysis by a variety of predictive databases has identified other potential phosphorylation-permissive residues in the amino acid sequence of TFEB, most of which remain currently unstudied. Examination of the contribution (if any) of post-translational modifications of these novel residues to TFEB homeostasis, and their role in misfolded protein clearance and metabolism in neurons and other CNS cell types will be of paramount importance.
What is the function of TFEB in the CNS?
Although considerable work has been performed to understand the role of TFEB in cellular metabolism (Martina and Puertollano, 2013; Medina et al., 2015; Settembre et al., 2013; Settembre et al., 2012), we have only recently begun to explore its potential involvement in neuronal homeostasis. TFEB is highly expressed in the CNS (Decressac et al., 2013; Reddy et al., 2016; Wang et al., 2016b), and is known to be active in both neurons and astrocytes (Decressac et al., 2013; Xiao et al., 2014). Importantly, TFEB activity and localization are altered in neurodegenerative disease (Cortes et al., 2014b; Decressac et al., 2013; Tsunemi et al., 2012; Wang et al., 2016b), implicating impaired TFEB function in the defective cellular clearance phenotypes linked to disease pathogenesis. Modest overexpression of TFEB can be achieved without significant neurotoxicity (Pi et al., 2017), permitting various studies of TFEB activity modulation in the murine CNS in models of neurodegenerative disease (Decressac et al., 2013; Pi et al., 2017; Polito et al., 2014; Xiao et al., 2014). To date, mutations in TFEB have only been associated with renal cell carcinoma in human patients (Kauffman et al., 2014), although a recent GWAS linked an intronic region variant of TFEB to late-onset Alzheimer’s disease risk in African-American populations (Mez et al., 2017). Thus far no one has examined the effect of this particular variant on TFEB function, but the detection of this potential modifier leaves open the prospect of additional disease-associated TFEB genetic alterations.
ALZHEIMER’S DISEASE
Alzheimer’s Disease (AD) is a very common cause of dementia, and is characterized by age-related brain degeneration resulting in inexorably progressive cognitive and behavioral impairment. A hallmark of AD brain pathology is intraneuronal fibrillary tangles, composed of hyperphosphorylated microtubule-associated-protein Tau (p-Tau) and neuritic plaques, which are extracellular deposits of amyloid Beta-42 (Aβ42) peptide. Inherited mutations in the genes encoding amyloid-precursor protein (APP), presenilin 1 and presenilin 2 account for familial cases of AD, with affected brain regions displaying profound transcriptional dysregulation of autophagy networks and wide-spread accumulation of autophagic vacuoles, often carrying undigested cargo (Lipinski et al., 2010; Wolfe et al., 2013). The current working model for autophagy dysfunction in AD posits that compromised lysosomal function (caused by genetic AD-linked mutations and/or age-related or environmental-induced alterations in proteostasis) creates a bottle-neck in autophagic flux, directly leading to the pathological accumulation of immature autophagy vesicles and endo-lysosomal intermediates (Bordi et al., 2016; Cataldo et al., 1996; Cataldo et al., 2000; Wolfe et al., 2013).
The role of TFEB in AD pathogenesis is currently the subject of intense study. Although important alterations of TFEB function and localization have been reported in models of AD, available evidence for direct TFEB involvement in AD remains highly correlative and sometimes contradictory. Analysis of AD patient lymphocytes and monocytes (immune cells that migrate to damaged CNS regions and likely regulate AD progression) has revealed markedly decreased TFEB expression, suggesting that TFEB dysfunction may underlie the lysosomal deficits observed in AD (Tiribuzi et al., 2014). In agreement with this conclusion, subcellular fractionation of AD patient brain samples documented a selective loss of nuclear TFEB (Wang et al., 2016b), with a strong inverse correlation between the extent of pathology and levels of hippocampal nuclear TFEB (Figure 2). Similar nuclear exclusion of TFEB was recently reported in an in vitro model of double presenilin knock-out cells (Reddy et al., 2016). Furthermore, CLEAR network activity is significantly reduced in AD patient fibroblasts and iPSC-derived human AD neurons (Figure 3), indicating that TFEB cytosolic retention may contribute to AD pathogenesis (Reddy et al., 2016). Computational modeling and DNA pull-down binding assays suggest that APOE ɛ4, the most well-known major risk factor allele for late-onset AD, may compete with TFEB for binding to CLEAR elements in the promoters of the SQSTM, MAP1LC3B, and LAMP2 genes (Parcon et al., 2017), all established targets of TFEB transactivation and all key players in autophagy function (Figure 2). This finding is quite significant, as APOE ε4/ε4 carriers have earlier disease onset and markedly increased protein aggregation in comparison to AD patients carrying other APOE alleles, which strongly suggests inhibition of the CLEAR network in APOE ε4/ε4 AD patients. In agreement with this hypothesis, neuron-specific TFEB excision in mice yields Aβ and paired helical filament (PHF) p-Tau accumulation in the brain, which is highly reminiscent of neuropathological findings in human AD patients (Reddy et al., 2016). However, to date there are no studies directly addressing the contribution of TFEB dysfunction to AD. The effect of modulation of TFEB on AD neurodegeneration and neuropathology, either by cell-type specific knock-out or overexpression in the CNS, may yield important insights into the contribution of TFEB dyshomeostasis to AD pathogenesis.
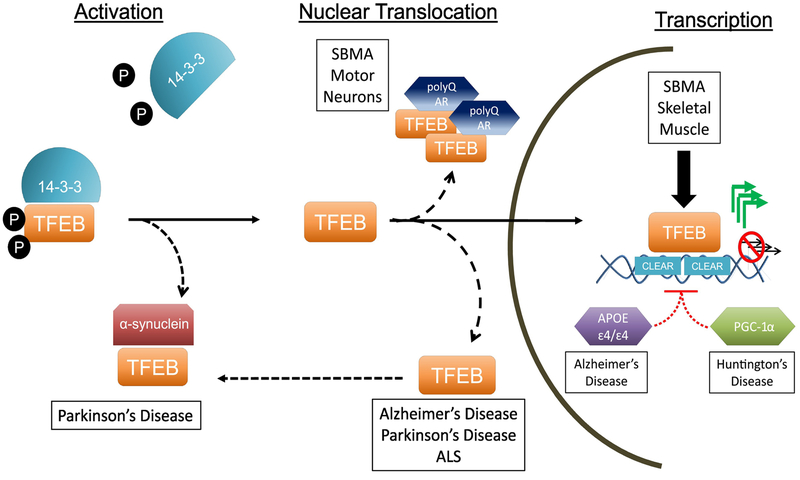
Figure 2. TFEB dysregulation in Neurodegenerative Disease.
TFEB function is tightly regulated at three distinct steps: its own activation, nuclear translocation, and transcription activity at its target genes. In its inactive form, phosphorylated TFEB interacts with 14–3–3 proteins, remaining sequestered in the cytosol. Upon activation (under conditions of lysosomal stress or mTOR inhibition), TFEB is dephosphorylated and dissociates from the 14–3–3 complex, unmasking its nuclear localization signal. TFEB can now translocate into the nucleus, driving transcription of the CLEAR network of target genes. Several of these steps have been reported to be dysfunctional in neurodegenerative disease, including TFEB sequestration (in PD), nuclear exclusion (in SBMA, AD, PD, and ALS) and transcription incompetence (in SBMA, AD, and HD). These observations indicate that TFEB dysregulation is a defining feature of neurodegenerative proteinopathies.
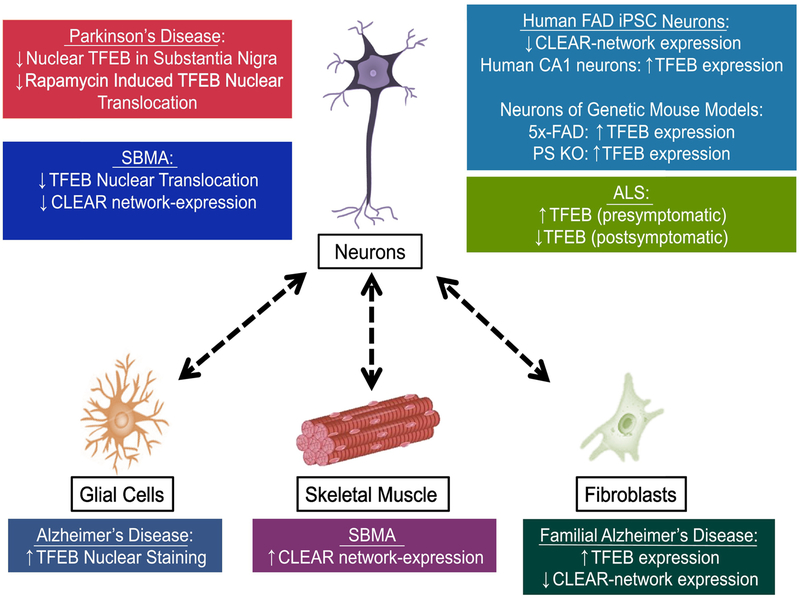
Figure 3. Non cell-autonomous dysregulation of TFEB in neurodegeneration.
Non cell-autonomous neurotoxicity, wherein non-neuronal cells contribute to neuron dysfunction in neurodegeneration, has emerged as a major feature of disease pathogenesis in most disorders. Many studies have documented cell-type specific dysregulation of TFEB in neurodegenerative disease, indicating that cellular context plays an important role in TFEB biology. This is especially relevant for Alzheimer’s disease and SBMA, where different disease-relevant tissues display diametrically opposite patterns of TFEB dysregulation. Understanding the cell-type specific regulation of TFEB function, and the signaling pathways that communicate TFEB status between different tissues, may yield important targets for neurodegenerative disease therapy development. FAD: Familial Alzheimer’s Disease; iPSC: induced pluripotent stem cell
In contrast to observations of reduced TFEB activity, AD patient-derived fibroblasts carrying the familial presenilin-1 A246E mutation displayed increased TFEB expression (Coffey et al., 2014). Another study of presenilin-1 conditional knock-out mice further revealed significant upregulation of a subset of CLEAR network genes, although no changes in TFEB levels were detected (Zhang et al., 2012). Similar transcriptome upregulation of TFEB targets was reported in 5x-FAD mice, which carry five familial AD mutations (Landel et al., 2014). Recent transcriptome analysis of human CA1 pyramidal neurons in the hippocampus of AD subjects by laser capture microdissection showed increased expression of TFEB and TFE3, a closely related member of the MitF transcription factor family, as well as increased expression of autophagy gene targets (Bordi et al., 2016). These findings strongly support TFEB gain-of-function in AD, particularly in the affected brain regions (Figure 3).
TFEB dysfunction in AD: Correlation or Causation?
While contradictory, these two sets of opposing results may not be mutually exclusive, as familial AD presenilin mutations may impair lysosomal function by producing chronic lysosome alkalinization (Coffey et al., 2014; Lee et al., 2010; Zhang et al., 2012), which can trigger TFEB engagement and activation as a compensatory mechanism (Sardiello et al., 2009; Settembre et al., 2011). Whether secondary (or parallel) age-associated TFEB nuclear exclusion and concomitant loss-of-function also occur in sporadic AD is currently unknown, and should be investigated. Furthermore, TFEB activation status is highly cell-type specific in neurodegeneration (Cortes et al., 2014b), indicating that TFEB function may differ in adjacent cell types, requiring isolation of cellular CNS populations to permit a thorough dissection of TFEB dysfunction in AD. Indeed, it is not unprecedented for TFEB function to be totally opposite in communicating non-neuronal cells and in neurons, as this has been documented in X-linked spinal & bulbar muscular atrophy between motor neurons and skeletal muscle (Cortes et al., 2014b). Finally, analysis of post-mortem AD patient brain tissue represents a very late stage of disease, associated with catastrophic neuronal collapse. TFEB overactivation may thus reflect a final cellular response that does not drive disease pathogenesis perse.
Glycogen synthase kinase 3 (GSK3), a serine-threonine kinase, has been proposed to play a role in AD pathogenesis by directly phosphorylating APP and tau to promote Aβ42 production (Blalock et al., 2004; Parr et al., 2012). GSK was identified as a putative regulator of TFEB activity in 2011 (Settembre et al., 2011), and GSK inhibitor VIII can increase nuclear TFEB accumulation and reduce APP levels in vitro (Parr et al., 2012). Although these findings implicate TFEB dysfunction mediated by altered GSK activity in AD, further studies are necessary to determine if TFEB is a direct target of GSK-mediated phosphorylation, if APP reduction by GSK is TFEB dependent, and most importantly, if this pathway is truly relevant to in vivo AD pathogenesis.
Therapeutic Targeting of TFEB in AD
Both the Aβ42 peptide and p-Tau are substrates for autophagy-mediated degradation (Berger et al., 2006; Caccamo et al., 2010), suggesting autophagy modulation may be beneficial in AD. For example, lentiviral delivery of Beclin-1 (an autophagy initiation factor also known as ATG6) to the brain of APP mice significantly decreased intracellular Aβ42 and amyloid plaque accumulation, whereas reduction of Beclin-1 worsened AD and autophagy phenotypes in APP/Beclin-1 heterozygous null mice (Pickford et al., 2008). Treatment with Rapamycin, a well-known mTOR inhibitor and therefore autophagy inducer, decreased hippocampal Aβ and Tau pathology, activated autophagy in the CNS, and improved learning and memory deficits in 3xTg-AD mice (Caccamo et al., 2010). Rapamycin also decreased tau toxicity in a Drosophila model of tauopathy, improving survival and degenerative eye phenotypes in flies, while enhancing mutant tau protein clearance through autophagy (Berger et al., 2006). Although the specific role of TFEB in these studies was not examined, mTOR inhibition is a key determinant of TFEB nuclear translocation and activation (Settembre et al., 2011). Furthermore, these studies highlight the responsiveness of AD brains to autophagy induction in mice, suggesting that autophagy modulation deserves further consideration for AD therapy development.
Despite the results of autophagy modulation by altering Beclin-1 gene dosage and inhibiting mTOR, it is important to realize that autophagy transcription networks are profoundly dysregulated in AD (Lipinski et al., 2010), with hyperactivation of the key autophagy inhibitor mTOR (Caccamo et al., 2010; Lafay-Chebassier et al., 2005), suggesting that the autophagy pathway itself is affected in AD pathogenesis. Extensive biochemical and ultrastructural analysis has revealed that autophagy loss-of-function based upon impaired degradation of autophagic cargo in lysosomes (Boland et al., 2008; Lee et al., 2010; Nixon et al., 2005; Yang et al., 2014). Thus, neuroprotective induction of autophagy in AD brains may necessitate correction of these underlying cargo clearance deficits, but only if the entire autophagy pathway, from initial vesicle formation to lysosome fusion and lysosomal degradative function, is boosted. As many currently available autophagy treatments only target autophagy activation, an inability to increase overall lysosomal clearance is likely required for therapeutic efficacy. TFEB thus should be an ideal therapeutic target, as TFEB upregulates autophagy induction and promotes increased lysosomal numbers, function and clearance (Sardiello et al., 2009). In agreement with this, TFEB has shown promise in models of AD and related tauopathies. CNS delivery of TFEB effectively reduced neurofibrillary tangle pathology and rescued behavioral phenotypes, synaptic deficits, and neurodegeneration in the rTg4510 mouse model of tauopathy (Polito et al., 2014). TFEB overexpression appeared to selectively target toxic p-Tau species, but had no deleterious effects on healthy neurons (Polito et al., 2014). Similar specificity for TFEB-mediated degradation of toxic Tau species in vivo was reported in a genetic neuron-specific TFEB over-expression model (Wang et al., 2016a). In this system, neuron-targeted TFEB overexpression in the P301S tauopathy mouse model resulted in significant reductions of toxic p-Tau and lipofuscin levels in the cortex and hippocampus, together with significantly improved performance in memory and learning tests (Wang et al., 2016a).
As noted above, TFEB status can differ between non-neuronal cells and neurons, so what have studies of TFEB modulation in non-neuronal cells shown? Interestingly, nonneuronal cells in AD may also benefit from TFEB induction. Astrocytes play a key role in neuron survival, regulating synaptic clearance and neural function, and their function is altered during AD. Astrocyte-specific delivery of TFEB to the hippocampus of APP/PS1 transgenic mice reduced interstitial Aβ levels and hippocampal plaque load by promoting Aβ uptake and clearance in TFEB-overexpressing astrocytes (Xiao et al., 2014), suggesting that TFEB can facilitate astrocytic Aβ clearance in vivo (Figure 3). Microglia, the resident macrophages of the CNS, play a key role in surveying the brain and are recruited to sites of amyloid plaque deposition in AD (Meyer-Luehmann et al., 2008). Deacetylation of TFEB by SIRT1 enhances lysosomal biogenesis and microglial degradation of fibrillary Aβ and elicits reduction of amyloid plaque in ex vivo brain slices of APP/PS1 mice (Bao et al., 2016). But recent work indicates that excessive autophagy activation in microglia, due to altered metabolic regulation and impaired mTORC1 function, could be deleterious in AD (Ulland et al., 2017), underscoring the complexity of autophagy dysregulation in different cell types in neurodegeneration. TFEB nuclear staining in AD hippocampus appears to be more prominent in glia (Bordi et al., 2016), perhaps to support scavenging of neuronal debris and protein aggregation, though overactivation of TFEB due to aberrant mTORC1 signaling also deserves consideration. In any event, the contribution of astroglial cell types to the pathology of AD is under intense investigation, but it is tempting to speculate that non-neuronal cells may be more potent entities in regulating Aβ-clearance than neurons. Astrocytes even appear to play a role in early stage AD, clearing soluble Aβ from the synaptic cleft, while microglia can clear fibrillar Aβ42 at later stages of AD pathology. These findings support a potential therapeutic approach of coordinated TFEB upregulation, where directed TFEB induction in different cell types could be pursued at different disease stages.
The discovery of novel small molecule inducers of TFEB has generated great interest in the AD field. Activation of TFEB through chemical inhibition of GSK3 promotes lysosomal clearance of APP and toxic C-terminal APP fragments (Parr et al., 2012), suggesting that TFEB induction of lysosomal clearance can effectively clear AD-related proteotoxic insults. In agreement with this, hydroxypropyl-β-cyclodextrin, a recently identified chemical activator of TFEB (Song et al., 2014), improved the clearance of toxic proteins and clinical symptoms in AD mice expressing two APP-disease associated mutations (Yao et al., 2012). Similar neuroprotective effects mediated by TFEB activation in an APP/PS1 transgenic mouse model were recently reported for gypenoside XVII, a compound found in ginseng (Meng et al., 2016). Furthermore, in addition to the lysosomal clearance benefits resulting from TFEB induction, other TFEB actions may be highly relevant for AD therapy benefit. Increased expression of lysosomal proton pumps, known targets of TFEB-mediated transcription (Sardiello et al., 2009), may potentially rectify the impaired lysosomal acidification observed in familial AD (Lee et al., 2010). TFEB also induces lysosomal calcium release (Medina et al., 2015), a process known to be disrupted in AD. Interestingly, TFEB may generate an auto-regulatory loop through up-regulation of the PTEN kinase and concomitant inhibition of Akt and mTOR, culminating in even further TFEB activation (Polito et al., 2014). Since PTEN is essential for neural differentiation and synaptic plasticity, TFEB-mediated PTEN up-regulation of PTEN could further benefit AD neurons.
PARKINSON’S DISEASE AND RELATED SYNUCLEINOPATHIES
Parkinson’s disease (PD) is a neurodegenerative disorder primarily affecting dopaminergic neurons in the substantia nigra. A major hallmark of PD is the accumulation of proteinaceous cytoplasmic inclusions, known as Lewy bodies, in nigral dopaminergic neurons, mainly composed of misfolded and aggregated α-synuclein (Kalia and Lang, 2016). Autophagy-lysosome deficiencies are a feature of PD (Figure1), with reports of accumulation of autophagic vesicles and impaired lysosomal function (Dehay et al., 2010; Winslow et al., 2010). A number of molecular players in PD pathology have also been reported to regulate the autophagy pathway itself, including α-synuclein, parkin, DJ-1, and LRRK2 (Kalia and Lang, 2016).
Similar to AD, TFEB appears to be selectively excluded from the nucleus in postmortem substantia nigra of PD patients and is closely linked to a progressive decline in markers of lysosomal function, suggesting TFEB mislocalization occurs in PD. Alpha-synuclein shares both structural and functional homology with 14–3–3 proteins (Ostrerova et al., 1999), a family of chaperones known to associate with TFEB and promote its retention in the cytosol (Martina et al., 2012). Interestingly, α-synuclein and TFEB coimmunoprecipitated in vivo in a rat model of PD-like neurodegeneration (Decressac et al., 2013), providing a mechanistic link between α-synuclein toxicity and impaired TFEB function, as α-synuclein may physically interact with TFEB, sequestering it away from its canonical 14–3–3 partners and preventing its transactivation and nuclear import (Figure 2). In agreement with this hypothesis, rapamycin treatment of Parkin Q311X mutant mice, which display α-synuclein pathology, revealed impaired TFEB nuclear translocation and function in the substantia nigra (Siddiqui et al., 2015), suggesting that an aberrant α-synuclein – TFEB interaction may be a common feature in various forms of α-synuclein toxicity. Similarly reduced TFEB nuclear translocation responses were also reported in a cellular model of X-linked spinal & bulbar muscular atrophy (Cortes et al., 2014b) (discussed in detail below), highlighting TFEB sequestration from the nucleus as a potentially shared mechanism contributing to different neurodegenerative disorders. In support of this model, one study found that TFEB function and subcellular localization were unchanged between control and PD ventral tegmental areas (Decressac et al., 2013), brain regions commonly resistant to PD neurodegeneration. Further experimentation is necessary, and could evaluate the TFEB interactome in PD brain regions alternatively sensitive or resistant to α-synuclein toxicity.
Alpha-synuclein is a known substrate for autophagy-mediated degradation, and lysosomal function is impaired in various models of PD. For example, in MPTP-induced dopaminergic neurotoxicity in mice, accumulation of autophagic vacuoles and domapinergic cell death were preceded by a marked decrease in lysosomal numbers, ultimately culminating in lysosomal permeabilization and defective cellular clearance (Dehay et al., 2010). Induction of lysosomal biogenesis in nigral dopaminergic neurons by viral TFEB overexpression rescued lysosomal breakdown and autophagic vacuole accumulation, achieving significant neuroprotection and reduction of α–synuclein aggregates in dopaminergic neurons (Dehay et al., 2010). Rapamycin treatment also attenuated PD-related dopaminergic neurodegeneration in this model and restored lysosomal levels and function (Dehay et al., 2010). 2-Hydroxypropyl-β-cyclodextrin, an activator of TFEB (Song et al., 2014), was recently shown to promote autophagic clearance of α-synuclein in vitro (Kilpatrick et al., 2015), and treatment with CCI-79, an FDA-approved mTOR inhibitor, blocked progression after disease onset in a rat model of dopaminergic α–synuclein toxicity, concomitant with restoration of TFEB nuclear localization (Decressac et al., 2013). These findings are encouraging in terms of potential therapeutic application, as an effective intervention treatment after disease onset would be a valuable advance.
Loss-of-function mutations in the mitochondrial kinase PINK1 and the cytosolic E3 ligase Parkin cause early onset familial PD, and are associated with the accumulation of damaged mitochondria (Kalia and Lang, 2016). The PINK1/Parkin pathway functions to selectively target and eliminate damaged mitochondria via mitochondrial autophagy (mitophagy), although the downstream steps between PINK1/Parkin activation and mitophagy remain poorly understood. Induction of mitophagy with oligomycin/antimycin treatment, a classical method for mitochondrial depolarization, induces TFEB nuclear translocation in a PINK1/Parkin dependent manner (Nezich et al., 2015). Intriguingly, this pathway also requires intact Atg5 function to operate and appears to be independent of mTORC1 signaling (Nezich et al., 2015). It is currently unclear how this novel mitophagy-TFEB signaling axis might be involved in PD pathogenesis.
Impaired mitophagy is observed in Parkin Q311X mice, and is associated with reduced TFEB expression (Siddiqui et al., 2015). Interestingly, analysis of the CLEAR network revealed that mitochondrial proteins, such as TFAM and Nrf1, are targets of TFEB-mediated transactivation (Palmieri et al., 2011). Expression of these mitochondrial factors is significantly reduced in Parkin Q311X mice, and can be rescued with rapamycin treatment (Siddiqui et al., 2015). Indeed, analysis of Parkin Q311X substantia nigra also exhibited reduced activity of the PGC-1α/TFEB pathway (Siddiqui et al., 2015), which was attributed to dysregulation of the transcription repressor PARIS. These results indicate that TFEB is both a target of regulation and an effector of regulation in terms of mitochondrial biology, as PGC-1 α can promote TFEB expression and TFEB can transactivate genes that encode mitochondrial proteins (Settembre et al., 2013; Tsunemi et al., 2012). Similar deficits of the PGC-1α/TFEB axis have been reported in Huntington’s disease (Tsunemi et al., 2012), further highlighting the central role of TFEB signaling in neurodegeneration.
The exact role of TFEB dysfunction in PD and its potential as a therapeutic target are still under investigation. However, two early-onset, genetic versions of PD known as X-linked parkinsonism with spasticity (XPDS) and Kufor-Rakeb syndrome are caused by mutations in subunits of the vacuolar ATPase, showing a direct link between lysosomal acidification and the development of PD-like neuropathology (Kalia and Lang, 2016). This suggests that TFEB, with its transcriptional control of cellular clearance through regulation of autophagy and its regulation of lysosomal acidification by controlling the expression of genes that encode ion pumps in the lysosome membrane, is an especially appealing target for therapy development in PD.
POLYGLUTAMINE REPEAT EXPANSION DISEASES
Polyglutamine repeat diseases are adult-onset, progressive neurodegenerative disorders caused by the expansion of a CAG repeat tract residing in the coding region of the involved gene. The respective resulting proteins thus contain expanded polyglutamine (polyQ) tracts, and elicit a gain-of-function toxic effect due to misfolding and accumulation of the mutant disease protein. As the disease repeat length increases, age of onset inversely correlates with the size of the mutant CAG expansion tract. There are nine recognized CAG-polyQ disorders, including Huntington’s disease (HD), X-linked spinal and bulbar muscular atrophy (SBMA), dentatorubralpalludoluysian atrophy (DRPLA), and six spinocerebellar ataxias (SCA1, 2, 3, 6, 7 & 17). The causative mutant proteins display broad patterns of expression and thus can be readily detected in a variety of cell types, both within the CNS and outside it. However, despite widespread expression, all polyQ disorders exhibit selective patterns of neurotoxicity, targeting specific neuron populations in each disorder. Autophagy dysfunction is observed in polyglutamine diseases, suggesting common mechanisms of pathogenesis leading to neuronal demise (Figure 1).
Huntington’s Disease
Huntington’s disease (HD), the most common polyQ disease, is an autosomal dominant neurodegenerative disorder characterized by involuntary motor movement, cognitive decline, and psychiatric illness. HD is caused by a CAG trinucleotide repeat expansion (> 36 CAG repeats) in the amino-terminal region of the huntingtin (Htt) protein. Various lines of evidence strongly implicate dysfunctional autophagy as part of HD pathology and suggest that autophagy dysregulation may contribute to polyQ-Htt neurotoxicity (Cortes and La Spada, 2014). One study proposed a defect in autophagy cargo recognition, most likely mediated through an aberrant interaction between autophagy cargo-receptors and polyQ-Htt protein (Martinez-Vicente et al., 2010). Interestingly, the Htt protein appears to be an autophagy pathway protein itself, mas Htt has been proposed to act as an autophagy adaptor protein, serving as a scaffold for the ULK1 (Ochaba et al., 2014) and Atg1 autophagy initiation complexes (Rui et al., 2015).
The first evidence of TFEB dysregulation in HD was reported in 2011. TFEB expression and function was shown to be impaired in the striatum of HD N171–82Q transgenic mice (Tsunemi et al., 2012), caused by inhibition of PGC1α-mediated transcription. TFEB induction was sufficient to drastically reduce polyQ-Htt aggregation in vitro, and PGC-1α overexpression enhanced polyQ-Htt turnover and eliminated protein aggregation in the striatum of HD mice by promoting TFEB transcription and activation (Tsunemi and La Spada, 2012) (Figure 2). The relationship between TFEB and PGC-1α is turning out to be rather complicated, as TFEB exerts transcriptional control of lipid catabolism by directly activating PGC-1α transcription (Settembre et al., 2013), resulting in a dual feedback loop of bidirectional activation between TFEB and PGC1α. Interestingly, one of the key clinical features of HD patients is bioenergetics dysregulation, including significant weight loss and muscle atrophy, raising the possibility that polyQ-Htt – PGC-1α transcription interference of TFEB activity may contribute to these observed metabolic manifestations of HD, but this phenomenon requires further study.
Full length and amino-terminal truncated polyQ-Htt are efficiently degraded by the autophagy pathway. Indeed, autophagy modulation has been a long-standing target for HD therapy development, and important benefits, including amelioration of behavioral motor abnormalities and neuropathology, have been achieved in various HD animal models. Promising compounds include CCI-779, a known mTOR inhibitor (Ravikumar et al., 2004), as well as the mTOR-independent autophagy activators trehalose (Tanaka et al., 2004) and rilmenidine (Rose et al., 2010). Both CCI-779 and trehalose are weak activators of TFEB, but the role of TFEB on the observed neuroprotective effects was not addressed. TFEB overexpression can promote efficient clearance of polyQ-Htt in vitro (Sardiello et al., 2009; Tsunemi et al., 2012), and more recently in the striatum of zQ175 HD mice (Vodicka et al., 2016).
X-linked Spinal and Bulbar Muscular Atrophy
X-linked Spinal and Bulbar Muscular Atrophy (SBMA), also known as Kennedy’s disease, is a neuromuscular disorder caused by expansion of a CAG triplet in the first exon of the Androgen Receptor (AR) gene. SBMA is characterized by adult-onset proximal muscle weakness due to lower motor neuron degeneration in the spinal cord and brain stem.
Exhaustive analysis of stable cell lines, transgenic mice, and patient-derived neuronal progenitor cells (NPCs) recently uncovered a profound transcriptional inhibition of TFEB signaling in SBMA (Cortes et al., 2014b). Interestingly, while SBMA cells and motor neurons appear to be competent for autophagy initiation and autophagosome formation, they fail to successfully complete autophagic degradation, indicating compromised lysosomal function. We identified a novel interaction between TFEB and AR, suggesting that TFEB dysregulation by polyQ-AR might account for autophagic flux impairments present in SBMA (Figure 2). Importantly, we restored autophagy flux by over-expressing TFEB in patient-derived NPCs (Cortes et al., 2014b), highlighting the potential for TFEB modulation as an important target for therapy development in SBMA and other disorders characterized by inhibition of autophagic flux. Indeed, independent treatment of SBMA transgenic mice expressing AR97Q with paneoflorin, a plant extract, partly exerted therapeutic effects on behavioral and pathological neuromuscular phenotypes by strongly upregulating TFEB expression (Tohnai et al., 2014). Importantly, we also found evidence of an interaction between normal Q-length AR and TFEB, and we detected enhanced TFEB signaling and increased autophagy pathway activity when normal AR is overexpressed. (Cortes et al., 2014b). Our data suggest that AR can normally interact with TFEB to promote its activity, functionally and spatially regulating TFEB in response to testosterone. As AR normally interacts with many transcription co-regulators and studies of a SBMA fly model indicate that polyQ-AR may promote neurotoxicity by reducing the function of the co-regulators with which it interacts (Nedelsky et al., 2010), reduced availability of a co-activator protein shared by AR and TFEB may result in decreased TFEB transactivation function in SBMA. A reciprocal feedback loop wherein AR can also promote TFEB transcription in prostate cancer was described recently, suggesting a complex, androgen-dependent transcription regulation of autophagy/lysosomal genes (Blessing et al., 2017). However, the role of this novel signaling pathway to SBMA disease pathogenesis remains to be explored.
Skeletal muscle plays a primary role in SBMA pathogenesis, superseding motor neurons as the key site of polyQ-AR toxicity (Cortes et al., 2014a; Lieberman et al., 2014). Interestingly, while TFEB activity in SBMA motor neurons and patient-derived NPCs was significantly reduced, analysis of quadriceps muscle samples from symptomatic SBMA YAC AR100 transgenic mice yielded an opposite and dramatic up-regulation of TFEB target genes (Cortes et al., 2014b), consistent with studies in SBMA knock-in AR113Q mice (Chua et al., 2014). This suggests a muscle-specific process of supraphysiological induction of TFEB in diseased SBMA muscle cells (Figure 3). Since uncontrolled autophagy is thought to underlie muscle wasting in models of muscular dystrophy (Sandri et al., 2013), excessive activation of autophagy could be contributing to SBMA skeletal muscle phenotypes. In agreement with this hypothesis, global reduction of autophagic activity by Beclin-1 haploinsufficiency in SBMA knock-in AR113Q mice increased skeletal muscle fiber size and significantly extended lifespan in this model (Yu et al., 2011). Feeding SBMA AR113Q knock-in mice a high-fat diet reduced excessive muscle TFEB activity, rescuing several of the metabolic alterations in SBMA skeletal muscle (Rocchi et al., 2016). This is the first evidence of dietary manipulation as a therapeutic approach to modify TFEB function, and provides a proof-of-concept for therapy development in SBMA and other diseases associated with TFEB dysfunction.
The mechanisms responsible for the different aspects of TFEB dysregulation by polyQAR in different tissue types remain unknown, but raise concern that systemic delivery of autophagy therapies could have deleterious effects in SBMA. Understanding the crosstalk between SBMA skeletal muscle and motor neurons, and identifying key players that regulate TFEB activity in a tissue-specific manner will be essential for rationale autophagy therapy development to proceed. Importantly, however, the non-cell autonomous nature of motor neuron toxicity in SBMA and the accessibility of skeletal muscle will facilitate delivery of drugs to treat degenerating motor neurons.
AMYOTROPHIC LATERAL SCLEROSIS
Amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease, is the most common motor neuron disorder in adults, and is characterized by selective loss of motor neurons in the motor cortex, brainstem, and spinal cord. ALS leads to weakness and atrophy of limb and respiratory muscles, ultimately causing death. Although most ALS cases are “sporadic” have no known etiology, up to 10% of cases are familial. These familial cases are due to mutations in a number of genes, including especially superoxide dismutase 1 (SOD1), TAR DNA binding protein (TDP-43), fused-insarcoma/translated-in-sarcoma (FUS/TLS), and chromosome 9 open-reading frame 72 (C9orf72) (Ramesh and Pandey, 2017). Interestingly, mutations in several autophagy genes have been linked to ALS, including the autophagy adaptors p62 (Sequesterome 1, SQSTM1) and optineurin (OPTN), the proteostasis and stress-granule regulator valosin-containing protein (VCP), autophagy receptor ubiquilin-2 (UBQLN2), and the autophagy regulatory TANK-binding kinase 1 (TBK1) (Peters et al., 2015).
In ALS, multiple studies of post-mortem patient tissue have uncovered evidence for autophagy dysregulation in motor neurons (Ramesh and Pandey, 2017). Autophagosome accumulations are frequently seen in degenerating motor neurons of sporadic and familial ALS patients, and are often found adjacent to p62-positive inclusions (Li et al., 2008; Morimoto et al., 2007; Sasaki, 2011). Accumulation of damaged mitochondria is also observed in ALS, although unlike HD or AD, the damaged mitochondria are usually found within autophagic vacuoles (Liu et al., 2004; Vande Velde et al., 2008). This suggests that autophagy dysfunction in ALS involves a distinct mechanism, as cargo recognition processes are intact. Instead, defects in axonal transport or autophagosome-lysosome fusion may account for the accumulation of autophagic vacuoles (Ligon et al., 2005; Williamson and Cleveland, 1999) (Figure 1). However, evidence of decreased mTOR signaling has been documented in ALS mice (Hetz et al., 2009; Li et al., 2008; Morimoto et al., 2007), consistent with supraphysiological activation of autophagy. Reflecting the complex mechanisms underlying autophagy dysfunction in ALS disease pathogenesis, autophagy targeting therapies have shown mixed results in animal models, with some reports showing improvement and others showing no benefit or a significant worsening of neuromuscular disease (Ramesh and Pandey, 2017).
A key neuropathological hallmark of familial ALS1 that contributes to the progressive loss of motor neurons is abnormal accumulation of mutant SOD1 protein aggregates. Recently, analysis of pre- and post-symptomatic spinal cord lysates from ALS SOD1-G39A transgenic mice revealed stage-dependent alterations in TFEB expression (Chen et al., 2015). TFEB was up-regulated in the early stage of disease, but then down-regulated at the middle and end stages of disease in what appeared to be a neuron-specific manner (Chen et al., 2015). Overexpression of TFEB in vitro increased cell survival and proliferation by increasing Beclin-1 expression, although the effect of autophagy on SOD1-aggregation clearance in this system was not examined. Nonetheless, dysregulation of TFEB localization has been reported in subcellular fractionation analysis of ALS patient brain samples, where nuclear TFEB levels were reduced as much as 60% (Wang et al., 2016b). These findings need to be reproduced in a larger series of sporadic and familial ALS cases, which may represent two different pathogenic entities; however, nuclear exclusion of TFEB may be a prominent feature of ALS disease pathogenesis.
Although C9orf72 has been the subject of intensive study since discovery of the disease-causing hexanucleotide repeat expansion in 2011, a complete understanding of the cellular functions of C9orf72 protein remains elusive. Recent evidence suggests that C9orf72 may negatively regulate autophagy (Amick et al., 2016; Sellier et al., 2016; Sullivan et al., 2016; Ugolino et al., 2016). Two independent C9orf72 knock-out mouse models displayed increased autophagy in the liver, spleen, and brain (Sullivan et al., 2016; Ugolino et al., 2016), supporting a role for C9orf72 protein in autophagy regulation. In one knock-out model, this autophagy increase was associated with elevations in TFEB protein expression in the brain, although this was attributed to by inhibition of mTOR activity (Amick et al., 2016; Ugolino et al., 2016), rather than a direct effect of the C9orf72 protein on TFEB. C9orf72 was reported to localize to lysosomes in response to amino acid depletion (Amick et al., 2016), suggesting a potential novel role for C9orf72 on energy sensing and cellular metabolism. This is consistent with recent data suggesting TDP-43, another ALS-associated gene, acts as a negative regulator of autophagy and TFEB activity (Xia et al., 2016). However, the validity of this discovery and its relevance to ALS disease pathogenesis are still unknown, but remain a topic of keen interest. On the other hand, the C9orf72 protein may act as a positive regulator of autophagy by physically interacting with the Ulk1 autophagy initiation complex (Webster et al., 2016), suggesting a complex, perhaps dual relationship between C9orf72 protein and autophagy. Indeed, these studies collectively identified a novel C9orf72 interactor known as SMCR8 as necessary for the observed autophagy effects (Amick et al., 2016; Sullivan et al., 2016; Yang et al., 2016), although the relevance of this interaction and its effect on autophagy in motor neuron disease pathogenesis remain unclear. There is an obvious need for further studies to clarify the nature of autophagy dysfunction in C9orf72 FALS as well as the normal function of the C9orf72 gene product, and then determine whether and how TFEB fits into the ALS pathogenic cascade for this form of FALS.
Despite uncertainty as to the status of the autophagy pathway in C9orf72 ALS, recent studies in other forms of genetic ALS have found that autophagy-mediated clearance of disease-associated protein aggregates can be beneficial, but these effects can be achieved independent of TFEB function. For example, induction of the heat shock protein B8 mediates the clearance of TDP-43 without changing the expression or localization of TFEB in vitro (Crippa et al., 2016). MASK, a novel autophagy regulator, can mitigate eye degeneration in a Drosophila model of FUS neurodegeneration, boosting expression of v-ATPase subunits in a TFEB-independent manner (Zhu et al., 2017). Furthermore, dietary restriction corrected autophagy defects and partially restored overall neurological function in a dynactin G59S ALS mouse model, likely through TFEB activation, although the role of TFEB in the neuroprotection was not examined (Wiesner et al., 2015). TBK1 is required for efficient recruitment of autophagy adaptors and proper degradation of mitochondria through autophagy (mitophagy). ALS-associated mutations in TBK1 disrupt its interaction with optinuerin, suggesting that impaired mitophagy may underlie TBK1-ALS disease pathogenesis (Oakes et al., 2017). To date, there is no evidence of altered TFEB activity in the context of TBK1-ALS, however. Thus, while the study of TFEB dysregulation in motor neuron disease is a topic of great interest, the exact role of autophagy in ALS disease pathogenesis is complicated, as exemplified by the contradicting results of preclinical studies in ALS animal models. Future studies to define specific mechanisms of autophagy dysfunction in ALS are needed to clarify these inconsistent and often discouraging results to determine if TFEB or even autophagy itself will be viable targets for therapeutic consideration.
CONCLUDING REMARKS
Autophagy dysfunction is a defining hallmark of almost all neurodegenerative diseases characterized by the accumulation of toxic protein aggregates. TFEB dysfunction also appears to be a shared mechanism in the neurodegenerative proteinopathies, including especially AD, PD, and HD, although the contribution of TFEB dysregulation to disease pathogenesis needs to be better delineated in most neurodegenerative diseases. TFEB activity can be modulated in the brain via viral or genetic overexpression, achieving significant amelioration of pathological and behavioral phenotypes in numerous and varied animal models of neurodegenerative disease (Bao et al., 2016; Decressac et al., 2013; Wang et al., 2016a; Xiao et al., 2014). Yet there is much about the basic biology of TFEB that remains unknown. For example, although it has long been known that phosphorylation is a major regulator of TFEB localization and function (Martina et al., 2012; Sardiello et al., 2009; Settembre et al., 2012), acetylation may also act as a powerful modulator of TFEB activity. TFEB is deacetylated by Sirtuin 1 in the nucleus, enhancing lysosome biogenesis in microglia exposed to fibrillar Aβ (Bao et al., 2016). Furthermore, studies of TFEB’s post-translational modifications in health and disease may shed light on TFEB function, and could yield appealing strategies for therapy development. Finally, numerous reports suggest the existence of distinct, cell-type specific TFEB regulatory networks, most likely due to differential interactomes in each cellular milieu. These neuronal and astroglial TFEB interactome networks are under investigation, and should inform our understanding of TFEB’s physiological functions in the CNS. The identification of novel chemical activators targeting TFEB function suggests that TFEB is a ‘druggable’ target (Meng et al., 2016; Song et al., 2014), and with the recent discovery of a TFEB-activating compound with blood-brain-barrier penetration and bioactivity (Song et al., 2016), the field is set to test TFEB modulation as a tractable neurotherapeutic intervention. Indeed, C1, a curcumin-derived compound, achieves robust TFEB activation in the mammalian CNS independent of mTOR, thus bypassing many of the undesirable side effects of long-term mTOR inhibition. Importantly, C1 was found to be active via oral delivery, further facilitating its use in human patients and making it an attractive candidate for therapy development. Of course, much of the regulatory biology of TFEB involves interactions and modifications occurring at the lysosome, underscoring the importance of better understanding how this increasingly important organelle is wired into the TFEB regulatory cascade. Toward that end, recent work on the Transient Receptor Potential Mucolipin 1 (TRPML1) channel, which regulates calcium ion efflux from the lysosome and was found to be a potent activator of TFEB (Di Paola et al., 2017), underscores that more definitive insight into TFEB regulatory biology could yield even more appealing targets for physiologically relevant TFEB and autophagy modulation. Hence, we predict that the next decade will witness an even greater acceleration of efforts to delineate TFEB’s role in the autophagy dysregulation occurring in neurodegeneration with an eye toward opportunities for leveraging gained knowledge into successful neurotherapeutic agents.
ACKNOWLEDGMENTS
Our work on TFEB and autophagy is supported by funding from the National Institutes of Health (R01 AG033082 to A.R.L., R01 NS100023 to A.R.L., and RF1 AG057264 to C.J.C. & A.R.L.) and the Muscular Dystrophy Association (A.R.L.).
REFERENCES
- Amick J, Roczniak-Ferguson A, Ferguson SM, 2016. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell. 27, 3040–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Zheng L, Zhang Q, Li X, Zhang X, Li Z, Bai X, Zhang Z, Huo W, Zhao X, et al. , 2016. Deacetylation of TFEB promotes fibrillar Abeta degradation by upregulating lysosomal biogenesis in microglia. Protein Cell. 7, 417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, Rubinsztein DC, 2006. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 15, 433–42. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW, 2004. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 101, 2173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blessing AM, Rajapakshe K, Reddy Bollu L, Shi Y, White MA, Pham AH, Lin C, Jonsson P, Cortes CJ, Cheung E, et al. , 2017. Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression. Autophagy. 13, 506–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA, 2008. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 28, 6926–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi M, Berg MJ, Mohan PS, Peterhoff CM, Alldred MJ, Che S, Ginsberg SD, Nixon RA, 2016. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy. 12, 2467–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A, Majumder S, Richardson A, Strong R, Oddo S, 2010. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: effects on cognitive impairments. J Biol Chem. 285, 13107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Hamilton DJ, Barnett JL, Paskevich PA, Nixon RA, 1996. Abnormalities of the endosomal-lysosomal system in Alzheimer’s disease: relationship to disease pathogenesis. Adv Exp Med Biol. 389, 271–80. [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA, 2000. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 157, 277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu H, Guan Y, Wang Q, Zhou F, Jie L, Ju J, Pu L, Du H, Wang X, 2015. The altered autophagy mediated by TFEB in animal and cell models of amyotrophic lateral sclerosis. Am J Transl Res. 7, 1574–87. [PMC free article] [PubMed] [Google Scholar]
- Chua JP, Reddy SL, Merry DE, Adachi H, Katsuno M, Sobue G, Robins DM, Lieberman AP, 2014. Transcriptional activation of TFEB/ZKSCAN3 target genes underlies enhanced autophagy in spinobulbar muscular atrophy. Human molecular genetics. 23, 1376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey EE, Beckel JM, Laties AM, Mitchell CH, 2014. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience. 263, 111–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes CJ, La Spada AR, 2014. The many faces of autophagy dysfunction in Huntington’s disease: from mechanism to therapy. Drug discovery today. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes CJ, Ling SC, Guo LT, Hung G, Tsunemi T, Ly L, Tokunaga S, Lopez E, Sopher BL, Bennett CF, et al. , 2014a. Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron. 82, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes CJ, Miranda HC, Frankowski H, Batlevi Y, Young JE, Le A, Ivanov N, Sopher BL, Carromeu C, Muotri AR, et al. , 2014b. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nature Neuroscience. 17, 1180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa V, D’Agostino VG, Cristofani R, Rusmini P, Cicardi ME, Messi E, Loffredo R, Pancher M, Piccolella M, Galbiati M, et al. , 2016. Transcriptional induction of the heat shock protein B8 mediates the clearance of misfolded proteins responsible for motor neuron diseases. Sci Rep. 6, 22827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A, 2013. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A. 110, E1817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Bove J, Rodriguez-Muela N, Perier C, Recasens A, Boya P, Vila M, 2010. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 30, 12535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paola S, Scotto-Rosato A, Medina DL, 2017. TRPML1: The Ca((2+))retaker of the lysosome. Cell Calcium. [DOI] [PubMed] [Google Scholar]
- Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, Martinez G, Cuervo AM, Brown RH, Glimcher LH, 2009. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 23, 2294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CL, Lee EX, Gordon KL, Paz EA, Shen WC, Ohnishi K, Meisenhelder J, Hunter T, La Spada AR, 2018. MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat Commun. 9, 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia LV, Lang AE, 2016. Parkinson disease in 2015: Evolving basic, pathological and clinical concepts in PD. Nat Rev Neurol. 12, 65–6. [DOI] [PubMed] [Google Scholar]
- Kauffman EC, Ricketts CJ, Rais-Bahrami S, Yang YF, Merino MJ, Bottaro DP, Srinivasan R, Linehan WM, 2014. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nature Reviews Urology. 11, 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick K, Zeng Y, Hancock T, Segatori L, 2015. Genetic and chemical activation of TFEB mediates clearance of aggregated alpha-synuclein. PLoS One. 10, e0120819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafay-Chebassier C, Paccalin M, Page G, Barc-Pain S, Perault-Pochat MC, Gil R, Pradier L, Hugon J, 2005. mTOR/p70S6k signalling alteration by Abeta exposure as well as in APP-PS1 transgenic models and in patients with Alzheimer’s disease. Journal of neurochemistry. 94, 215–25. [DOI] [PubMed] [Google Scholar]
- Landel V, Baranger K, Virard I, Loriod B, Khrestchatisky M, Rivera S, Benech P, Feron F, 2014. Temporal gene profiling of the 5XFAD transgenic mouse model highlights the importance of microglial activation in Alzheimer’s disease. Mol Neurodegener. 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, et al. , 2010. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 141, 1146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang X, Le W, 2008. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 4, 290–3. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Yu Z, Murray S, Peralta R, Low A, Guo S, Yu XX, Cortes CJ, Bennett CF, Monia BP, et al. , 2014. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell reports. 7, 774–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon LA, LaMonte BH, Wallace KE, Weber N, Kalb RG, Holzbaur EL, 2005. Mutant superoxide dismutase disrupts cytoplasmic dynein in motor neurons. Neuroreport. 16, 533–6. [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J, 2010. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 107, 14164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lillo C, Jonsson PA, Vande Velde C, Ward CM, Miller TM, Subramaniam JR, Rothstein JD, Marklund S, Andersen PM, et al. , 2004. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 43, 5–17. [DOI] [PubMed] [Google Scholar]
- Mansueto G, Armani A, Viscomi C, D’Orsi L, De Cegli R, Polishchuk EV, Lamperti C, Di Meo I, Romanello V, Marchet S, et al. , 2017. Transcription Factor EB Controls Metabolic Flexibility during Exercise. Cell Metab. 25, 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R, 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Puertollano R, 2013. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 200, 475–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM, 2010. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 13, 567–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. , 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 17, 288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Luo Y, Liang T, Wang M, Zhao J, Sun G, Sun X, 2016. Gypenoside XVII Enhances Lysosome Biogenesis and Autophagy Flux and Accelerates Autophagic Clearance of Amyloid-beta through TFEB Activation. J Alzheimers Dis. 52, 1135–50. [DOI] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, Koenigsknecht-Talboo J, Holtzman DM, Bacskai BJ, Hyman BT, 2008. Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer’s disease. Nature. 451, 720–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mez J, Chung J, Jun G, Kriegel J, Bourlas AP, Sherva R, Logue MW, Barnes LL, Bennett DA, Buxbaum JD, et al. , 2017. Two novel loci, COBL and SLC10A2, for Alzheimer’s disease in African Americans. Alzheimers & Dementia. 13, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto N, Nagai M, Ohta Y, Miyazaki K, Kurata T, Morimoto M, Murakami T, Takehisa Y, Ikeda Y, Kamiya T, Abe K, 2007. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 1167, 112–7. [DOI] [PubMed] [Google Scholar]
- Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, Taylor JP, 2010. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 67, 936–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich CL, Wang C, Fogel AI, Youle RJ, 2015. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J Cell Biol. 210, 435–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM, 2005. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 64, 113–22. [DOI] [PubMed] [Google Scholar]
- Oakes JA, Davies MC, Collins MO, 2017. TBK1: a new player in ALS linking autophagy and neuroinflammation. Mol Brain. 10, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochaba J, Lukacsovich T, Csikos G, Zheng S, Margulis J, Salazar L, Mao K, Lau AL, Yeung SY, Humbert S, et al. , 2014. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci U S A. 111, 16889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrerova N, Petrucelli L, Farrer M, Mehta N, Choi P, Hardy J, Wolozin B, 1999. alpha-Synuclein shares physical and functional homology with 14–3–3 proteins. J Neurosci. 19, 5782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A, 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 20, 3852–66. [DOI] [PubMed] [Google Scholar]
- Parcon PA, Balasubramaniam M, Ayyadevara S, Jones RA, Liu L, Shmookler Reis RJ, Barger SW, Mrak RE, Griffin WST, 2017. Apolipoprotein E4 inhibits autophagy gene products through direct, specific binding to CLEAR motifs. Alzheimers Dement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C, Carzaniga R, Gentleman SM, Van Leuven F, Walter J, Sastre M, 2012. Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-beta precursor protein. Molecular and cellular biology. 32, 4410–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters OM, Ghasemi M, Brown RH Jr., 2015. Emerging mechanisms of molecular pathology in ALS. J Clin Invest. 125, 1767–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi HF, Li M, Tian L, Yang ZQ, Yu ZP, Zhou Z, 2017. Enhancing lysosomal biogenesis and autophagic flux by activating the transcription factor EB protects against cadmium-induced neurotoxicity. Scientific Reports. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T, 2008. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 118, 2190–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito VA, Li H, Martini-Stoica H, Wang B, Yang L, Xu Y, Swartzlander DB, Palmieri M, di Ronza A, Lee VM, et al. , 2014. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO molecular medicine. 6, 1142–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh N, Pandey UB, 2017. Autophagy Dysregulation in ALS: When Protein Aggregates Get Out of Hand. Front Mol Neurosci. 10, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC, 2004. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 36, 585–95. [DOI] [PubMed] [Google Scholar]
- Reddy K, Cusack CL, Nnah IC, Khayati K, Saqcena C, Huynh TB, Noggle SA, Ballabio A, Dobrowolski R, 2016. Dysregulation of Nutrient Sensing and CLEARance in Presenilin Deficiency. Cell Rep. 14, 2166–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi A, Milioto C, Parodi S, Armirotti A, Borgia D, Pellegrini M, Urciuolo A, Molon S, Morbidoni V, Marabita M, et al. , 2016. Glycolytic-to-oxidative fiber-type switch and mTOR signaling activation are early-onset features of SBMA muscle modified by high-fat diet. Acta Neuropathol. 132, 127–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM, 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 5, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C, Menzies FM, Renna M, Acevedo-Arozena A, Corrochano S, Sadiq O, Brown SD, Rubinsztein DC, 2010. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington’s disease. Hum Mol Genet. 19, 2144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, et al. , 2015. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 17, 262–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Coletto L, Grumati P, Bonaldo P, 2013. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. Journal of cell science. 126, 5325–33. [DOI] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. , 2009. A gene network regulating lysosomal biogenesis and function. Science. 325, 473–7. [DOI] [PubMed] [Google Scholar]
- Sasaki S, 2011. Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 70, 349–59. [DOI] [PubMed] [Google Scholar]
- Sellier C, Campanari ML, Julie Corbier C, Gaucherot A, Kolb-Cheynel I, Oulad-Abdelghani M, Ruffenach F, Page A, Ciura S, Kabashi E, Charlet-Berguerand N, 2016. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 35, 1276–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. , 2013. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nature cell biology. 15, 647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. , 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332, 1429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, et al. , 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J. 31, 1095–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A, Bhaumik D, Chinta SJ, Rane A, Rajagopalan S, Lieu CA, Lithgow GJ, Andersen JK, 2015. Mitochondrial Quality Control via the PGC1alpha-TFEB Signaling Pathway Is Compromised by Parkin Q311X Mutation But Independently Restored by Rapamycin. J Neurosci. 35, 12833–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JX, Sun YR, Peluso I, Zeng Y, Yu X, Lu JH, Xu Z, Wang MZ, Liu LF, Huang YY, et al. , 2016. A novel curcumin analog binds to and activates TFEB in vitro and in vivo independent of MTOR inhibition. Autophagy. 12, 1372–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Wang F, Lotfi P, Sardiello M, Segatori L, 2014. 2-Hydroxypropyl-beta-cyclodextrin promotes transcription factor EB-mediated activation of autophagy: implications for therapy. J Biol Chem. 289, 10211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, Hu F, 2016. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 4, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, Kurosawa M, Nekooki M, Nukina N, 2004. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 10, 148–54. [DOI] [PubMed] [Google Scholar]
- Tiribuzi R, Crispoltoni L, Porcellati S, Di Lullo M, Florenzano F, Pirro M, Bagaglia F, Kawarai T, Zampolini M, Orlacchio A, 2014. miR128 up-regulation correlates with impaired amyloid beta(1–42) degradation in monocytes from patients with sporadic Alzheimer’s disease. Neurobiology of aging. 35, 345–56. [DOI] [PubMed] [Google Scholar]
- Tohnai G, Adachi H, Katsuno M, Doi H, Matsumoto S, Kondo N, Miyazaki Y, Iida M, Nakatsuji H, Qiang Q, et al. , 2014. Paeoniflorin eliminates a mutant AR via NF-YA-dependent proteolysis in spinal and bulbar muscular atrophy. Hum Mol Genet. 23, 3552–65. [DOI] [PubMed] [Google Scholar]
- Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, Lazarowski ER, Damian VA, Masliah E, La Spada AR, 2012. PGC-1alpha Rescues Huntington’s Disease Proteotoxicity by Preventing Oxidative Stress and Promoting TFEB Function. Sci Transl Med. 4, 142ra97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, La Spada AR, 2012. PGC-1alpha at the intersection of bioenergetics regulation and neuron function: from Huntington’s disease to Parkinson’s disease and beyond. Progress in neurobiology. 97, 142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, Brady NR, Hamacher-Brady A, Wang J, 2016. Loss of C9orf72 Enhances Autophagic Activity via Deregulated mTOR and TFEB Signaling. PLoS Genet. 12, e1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland TK, Song WM, Huang SC, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, et al. , 2017. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell. 170, 649–663 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Velde C, Miller TM, Cashman NR, Cleveland DW, 2008. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proc Natl Acad Sci U S A. 105, 4022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodicka P, Chase K, Iuliano M, Tousley A, Valentine DT, Sapp E, Kegel-Gleason KB, Sena-Esteves M, Aronin N, DiFiglia M, 2016. Autophagy Activation by Transcription Factor EB (TFEB) in Striatum of HDQ175/Q7 Mice. J Huntingtons Dis. 5, 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang R, Carrera I, Xu S, Lakshmana MK, 2016a. TFEB Overexpression in the P301S Model of Tauopathy Mitigates Increased PHF1 Levels and Lipofuscin Puncta and Rescues Memory Deficits. eNeuro. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang R, Xu S, Lakshmana MK, 2016b. Transcription Factor EB Is Selectively Reduced in the Nuclear Fractions of Alzheimer’s and Amyotrophic Lateral Sclerosis Brains. Neurosci J. 2016, 4732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, Myszczynska MA, Higginbottom A, Walsh MJ, Whitworth AJ, et al. , 2016. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 35, 1656–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner D, Sinniger J, Henriques A, Dieterle S, Muller HP, Rasche V, Ferger B, Dirrig-Grosch S, Soylu-Kucharz R, Petersen A, et al. , 2015. Low dietary protein content alleviates motor symptoms in mice with mutant dynactin/dynein-mediated neurodegeneration. Hum Mol Genet. 24, 2228–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson TL, Cleveland DW, 1999. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat Neurosci. 2, 50–6. [DOI] [PubMed] [Google Scholar]
- Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, et al. , 2010. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 190, 1023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe DM, Lee JH, Kumar A, Lee S, Orenstein SJ, Nixon RA, 2013. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. The European journal of neuroscience. 37, 1949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q, Wang H, Hao Z, Fu C, Hu Q, Gao F, Ren H, Chen D, Han J, Ying Z, Wang G, 2016. TDP-43 loss of function increases TFEB activity and blocks autophagosomelysosome fusion. EMBO J. 35, 121–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Burchett JM, Schuler DR, Cirrito JR, et al. , 2014. Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J Neurosci. 34, 9607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DS, Stavrides P, Saito M, Kumar A, Rodriguez-Navarro JA, Pawlik M, Huo C, Walkley SU, Cuervo AM, Nixon RA, 2014. Defective macroautophagic turnover of brain lipids in the TgCRND8 Alzheimer mouse model: prevention by correcting lysosomal proteolytic deficits. Brain : a journal of neurology. 137, 3300–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, Chen JF, 2016. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv. 2, e1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ho D, Calingasan NY, Pipalia NH, Lin MT, Beal MF, 2012. Neuroprotection by cyclodextrin in cell and mouse models of Alzheimer disease. J Exp Med. 209, 2501–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Wang AM, Adachi H, Katsuno M, Sobue G, Yue Z, Robins DM, Lieberman AP, 2011. Macroautophagy is regulated by the UPR-mediator CHOP and accentuates the phenotype of SBMA mice. PLoS genetics. 7, e1002321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Garbett K, Veeraraghavalu K, Wilburn B, Gilmore R, Mirnics K, Sisodia SS, 2012. A role for presenilins in autophagy revisited: normal acidification of lysosomes in cells lacking PSEN1 and PSEN2. J Neurosci. 32, 8633–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Zhang S, Tian X, Wu C, 2017. Mask mitigates MAPT- and FUS-induced degeneration by enhancing autophagy through lysosomal acidification. Autophagy. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]