Abstract
Phenotypic diversity helps populations persist in changing and often unpredictable environments. One diversity-generating strategy is for individuals to switch randomly between phenotypic states such that one subpopulation has high fitness in the present environment, and another subpopulation has high fitness in an environment that might be encountered in the future. This sort of biological bet hedging can be found in all domains of life. Here we discuss a recently described example from the bacterium Escherichia coli. When exposed to both oxygen and trimethylamine oxide (TMAO), E. coli hedges its bets on the possibility of oxygen loss by generating high cell-to-cell variability in the expression of the TMAO respiratory system. If oxygen is rapidly depleted from the environment, only those cells that had been expressing the TMAO respiratory system at high levels can continue to grow. This particular bet-hedging scheme possesses some unusual characteristics, most notably the decoupling of gene expression noise from the mean expression level. This decoupling allows bacteria to sense oxygen and regulate the amount of variability in TMAO reductase expression (that is, to turn bet hedging on or off) without having to adjust the mean TMAO reductase expression level. In this review, we discuss the features of the TMAO signaling pathway that permit the decoupling of gene expression noise from mean and the regulation of bet hedging. We also highlight some open questions regarding the TMAO respiratory system and its regulatory architecture that may be relevant to many signaling systems.
Keywords: anaerobic respiration, bet hedging, noise, oxygen, trimethylamine oxide, two-component signaling
Introduction
Survival in an uncertain world requires adaptability and anticipation, the complementary abilities to respond to and prepare for change. Bacteria, like all organisms, possess these capabilities. Bacterial adaptation has long been appreciated, and our understanding of how it occurs has greatly increased over the last several decades with the help of single-cell studies. The analysis of properties such as gene expression status, physiological state, and protein localization in individual cells in a population has revealed numerous examples of phenotypic diversification—the manifestation of more than one phenotype across a population of genetically identical cells. In many cases such cell-to-cell variability appears to function as a kind of bacterial “anticipatory behavior” in which a diverse population is more likely to contain individuals that can survive a sudden change in the environment than a homogeneous population (Freddolino and Tavazoie 2012; Ackermann 2015). We recently showed that Escherichia coli cells anticipate a rapid decrease in oxygen availability by randomly pre-inducing expression of the trimethylamine oxide (TMAO) respiratory system (Carey et al. 2018). The mechanism through which cell-to-cell variability in TMAO reductase expression is generated allows the variance in expression to be regulated independently from the mean and involves the propagation of molecular noise through a signal transduction pathway.
Playing the Odds
Anticipatory phenotypic diversification is commonly known as bet hedging. A population that hedges its bets protects itself against unpredictable future events by harboring individuals that are optimally adapted for life in an environment to which the population may be exposed in the future rather than the environment to which it is exposed at present (de Jong et al. 2011; Norman et al. 2015; Martins and Locke 2015). The maladapted individuals suffer reduced fitness as long as the phenotype/environment mismatch persists. However, should the population experience a rapid environmental shift, those individuals that had been preadapted to the new environment can thrive even though the rest of the population suffers. This strategy allows the population to survive chance events that, because of their rapidity or severity, would be difficult to contend with by post hoc adaptation. There are specific quantitative requirements that must be met for a behavior to conform to the formal mathematical definition of bet hedging, but very few experimental studies in microbiology have included sufficient analysis to fulfill this definition (Philippi and Seger 1989; de Jong et al. 2011; Simons 2011; Viney and Reece 2013; Grimbergen et al. 2015). Informally, then, the term “bet hedging” is often used to describe behavior that appears to be qualitatively consistent with the formal definition even when a complete quantitative analysis has not been performed.
Bet-hedging behaviors in bacteria can be roughly placed into two categories, bimodal or unimodal, by the pattern of phenotypic diversification exhibited by a population. In the bimodal pattern the population bifurcates into subpopulations with distinct phenotypes, whereas in the unimodal pattern the population exhibits a broad distribution of phenotypes that does not resolve into clearly demarcated subpopulations (Garcia-Bernardo and Dunlop 2016). Examples of the bimodal pattern include sporulation (Veening et al. 2008) and competence (Maamar and Dubnau 2005; Smits et al. 2005) in Bacillus subtilis, persister cell formation in E. coli (Balaban et al. 2004; Hõrak and Tamman 2017), and stringent response activation in Mycobacterium smegmatus (Sureka et al. 2008). Fewer examples of the unimodal pattern have been described, but these include transient antibiotic resistance (Meouche et al. 2016) and antibiotic-induced acid resistance (Mitosch et al. 2017) in E. coli. It is worth noting that the distinction between these patterns is not always clear, as categorization depends to some extent on what variables are used to assess phenotype. In addition, random switching between phenotypes can produce either a bimodal or unimodal distribution, depending on how long an individual and its offspring occupy each phenotype before switching to the other; low-frequency switching leads to a bimodal distribution and high-frequency switching leads to a unimodal distribution (Thomas et al. 2014). Random switching between phenotypes is a very common feature in bacterial bet-hedging strategies but is not required. Asymmetric cell division, for instance, can generate subpopulations that are differently fit in different environments (Ratcliff and Denison 2010).
Several excellent reviews have been published containing much more in-depth treatments of bet hedging in general (see, for instance, Grimbergen et al. 2015, Simons 2011, de Jong et al. 2011, Philippi and Seger 1989, and Childs et al. 2010). Here we focus on a specific system in E. coli that we have shown can allow a population of aerobically growing cells to hedge its bets on a rapid transition to anoxic conditions (Carey et al. 2018). This system enables the use of trimethylamine oxide (TMAO) as a terminal electron acceptor for respiration and is encoded by the torCAD operon (Méjean et al. 1994; McCrindle et al. 2005). TMAO respiration has a much lower energetic yield than aerobic respiration, so the observation that torCAD is expressed in the presence of oxygen—and at roughly the same mean level as in the absence of oxygen—was surprising (Ansaldi et al. 2007), especially considering that no other alternative respiratory system in E. coli is known to be significantly expressed during aerobic growth. Curiously, although oxygen does not affect mean torCAD expression, it does affect the variance around the mean (Roggiani and Goulian 2015). In the absence of oxygen individual cells all express torCAD at approximately the same level, but in the presence of oxygen torCAD is expressed with exceptionally high cell-to-cell variability. This variability features a high switching frequency and follows a broad, unimodal distribution as described above. When an aerobically growing population is shifted to an anaerobic environment, only those cells with high torCAD expression at the time of the shift continue to grow anaerobically—a behavior consistent with bet hedging (Carey et al. 2018).
Sensing and Signaling
TMAO activates transcription of the torCAD operon through a signaling system consisting of three proteins: TorT senses the presence of TMAO in the periplasm, TorS transmits this information across the cell membrane and phosphorylates TorR, and phosphorylated TorR activates transcription from the torCAD promoter (Figure 1) (Simon et al. 1994; Baraquet et al. 2006). The TorT/TorS/TorR system belongs to a class of regulatory systems called two-component systems. As with TorT/TorS/TorR, many of these systems have more than two components, but the name stems from the shared core architecture of a histidine kinase (in this case, TorS) that phosphorylates a response regulator (TorR), which then goes on to effect some physiological change, usually by regulating gene expression. In the TorT/TorS/TorR system, signal sensing and transduction across the membrane are relegated to different proteins (TorT and TorS, respectively) that function as a complex. A cell’s responsiveness to TMAO depends on the abundance of TorT and TorS such that a cell with an excess of TorT relative to TorS fully activates torCAD transcription, while a cell with an excess of TorS relative to TorT responds weakly, if at all, to the presence of TMAO (Roggiani and Goulian 2015). The sensitivity of the system output (torCAD transcription) to the relative amounts of TorT and TorS is significantly enhanced by the bifunctionality of TorS: in the absence of TorT-TMAO stimulation, TorS is not simply inert. Instead, it dephosphorylates TorR, thereby shutting off torCAD transcription (Figure 1) (Ansaldi et al. 2001). Many histidine kinases can receive signals from multiple sources, and there is at least one protein other than TorT that feeds information to TorS and regulates its activity. This secondary signal comes from TorC, the cytochrome component of TMAO reductase. TorC requires heme cofactors (Méjean et al. 1994; Sanders et al. 2010), and if lacking these cofactors it interacts with TorS and prevents it from phosphorylating TorR, thereby blocking further expression of torCAD and negatively regulating its own expression (Figure 1) (Ansaldi et al. 1999; Gon et al. 2001).
Fig. 1.
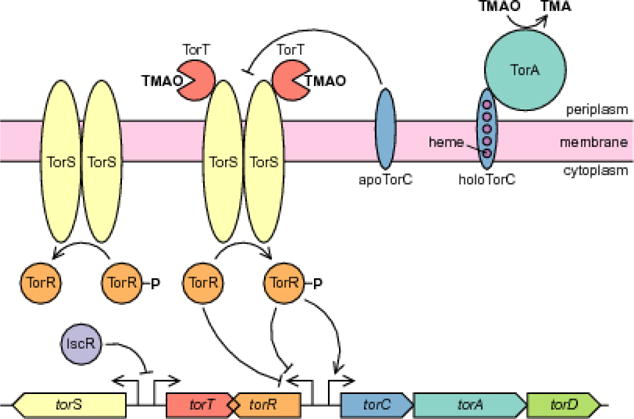
Regulation of the trimethylamine oxide (TMAO) respiratory system in E. coli. TorT detects TMAO in the periplasm and causes TorS to phosphorylate TorR. Phosphorylation of TorR requires a three-step intermolecular phosphorelay (not pictured) between the subunits of the TorS dimer before the final phosphotransfer to TorR (Jourlin et al. 1997; Ansaldi et al. 2001). Phosphorylated TorR activates transcription of the torCAD operon, which encodes the TMAO reductase complex TorCA. TorD is a chaperone for TorA maturation and export to the periplasm (Pommier et al. 1998; Ilbert et al. 2003; Jack et al. 2004). TorCA catalyzes the terminal reduction step of TMAO respiration, converting TMAO into trimethylamine (TMA). TorS dephosphorylates TorR-P when not interacting with TorT-TMAO, thereby deactivating torCAD transcription. Expression of torR is negatively autoregulated by both phosphorylated and unphosphorylated TorR (Ansaldi et al. 2000). TorC also participates in negative autoregulation, with apoTorC (the immature form lacking its heme cofactors) inhibiting TorS and preventing TorR phosphorylation. Transcription of torS and torT is repressed by the transcription factor IscR, which is more abundant in aerobic conditions
Bifunctionality of histidine kinases is extremely common in two-component systems, with the histidine kinase dephosphorylating its cognate response regulator in the absence of an inciting signal (Gao and Stock 2009). Two opposing reactions, the phosphorylation and dephosphorylation of the response regulator, are thus carried out by a single enzyme, with the direction of the reaction being dictated by information received (or not received) by the sensor domain(s) of the enzyme. Because the TorT/TorS/TorR system assigns sensing to one protein subunit (TorT) and signaling to another (TorS), the ratio of TorT to TorS molecules in a cell sets the net direction and rate of TorR phosphorylation in a TMAO-replete environment. While most multiprotein complexes in bacteria are encoded in operons, which provide transcriptional coordination of the various subunits, TorT and TorS are encoded by genes independently transcribed from separate promoters (Figure 1). This decoupled transcription enables transcriptional noise to influence the ratio of TorT to TorS and is critical for the generation of highly variable expression from the torCAD promoter.
Harnessing Noise
Biological noise can be loosely defined as random variability in a biological process originating in the intrinsic random behavior of molecules (Balázsi et al. 2011; Bidnenko and Bidnenko 2018). Our model of the TorT/TorS/TorR system posits that cells harness transcriptional and partitioning noise in TorT and TorS to generate and regulate the high cell-to-cell variability in torCAD expression that permits bet hedging (Carey et al. 2018). To generate variability during aerobic growth, cells express TorT and TorS at exceptionally low levels so that noise in the ratio of TorT to TorS leads to considerable fluctuations in the extent of TorR phosphorylation and results in noisy torCAD expression. To generate uniform torCAD expression when oxygen is absent, cells need only increase the expression of TorT and TorS to levels where gene expression noise and random partitioning have a negligible effect on the TorT-to-TorS ratio. Published reports indicate that the average number of TorT and TorS proteins per cell is indeed very low, with only a few copies of each present during aerobic growth (Taniguchi et al. 2010; Li et al. 2014). We have shown that transcription of torT and torS increases during anaerobic growth (Carey et al. 2018), which implies a correlative increase in the number of TorT and TorS proteins. The system appears to be organized such that increased anaerobic expression sufficiently increases mean TorT and TorS levels to where transcriptional and/or partitioning noise no longer contribute much to the output, leading to uniform expression of torCAD.
That noise in mRNA and protein levels decreases as the mean increases is a phenomenon well supported by theory and experiment (Ozbudak et al. 2002; Elowitz et al. 2002; Paulsson 2004; Bar-Even et al. 2006; Dar et al. 2016). Noise can therefore readily be modulated by changing mean expression. However, regulating noise independently from the mean requires more effort, and E. coli appears to have achieved this for torCAD by placing the important source of noise upstream from torCAD in the signaling pathway. Cells can regulate noise in TorT and TorS levels by regulating mean torT and torS transcription, and our model is that this noise reaches the torCAD promoter via the TorT-to-TorS ratio without direct regulation of mean torCAD expression being necessary. We were able to show that mean torT and torS expression is regulated by oxygen through the transcription factor IscR, which binds to a shared regulatory site between the genes and represses their transcription (Figure 1) (Carey et al. 2018). IscR levels are oxygen-sensitive, with the concentration of IscR higher in aerobic conditions than in anaerobic conditions (Giel et al. 2006; Giel et al. 2013; Mettert and Kiley 2014). When IscR is more abundant, torT and torS are more repressed, which leads to increased noise in the relative levels of TorT and TorS.
Perspectives
The mechanism we have described can account for a decoupling of the control of variance from control of the mean, but it does not explain why mean torCAD expression does not significantly change between aerobic and anaerobic conditions. This phenomenon could emerge spontaneously from the properties of the known regulators, or there could be additional layers of regulation that are actively involved in holding the mean steady. This question has been outstanding since our first description of oxygen-dependent variability in torCAD transcription (Roggiani and Goulian 2015), but it has been made all the more intriguing by our observation that the threshold for growth upon a transition to anaerobiosis aligns very closely with the population mean (Carey et al. 2018). The threshold for growth also closely matches the mean level of anaerobic torCAD expression, implying that in anaerobic conditions cells synthesize the minimal amount of TorCAD that they need to carry out respiratory growth. If this is true, why do aerobic cells transiently express torCAD at levels much higher than the mean? Perhaps this extremely “bursty” expression pattern reduces the fitness cost of aerobic torCAD expression in some way we have not yet been able to detect.
We presume that there must be a fitness cost associated with aerobic torCAD expression or else its expression would be unregulated. However, none of our efforts have revealed such a cost. Fitness is a measure of reproductive success, and in laboratory studies of bacteria growth rate is a convenient measure of fitness. Neither direct measurements of growth rate nor co-culture competition experiments revealed convincing evidence that growth rate is negatively affected by aerobic torCAD expression—even if we force the expression of torCAD in the absence of TMAO. However, there are two major limitations of laboratory fitness experiments. First, the sensitivity to small fitness differences is poor. Small differences that are very important over long, evolutionary time scales may be undetectable over the short duration of laboratory experiments. Executing long-term fitness experiments in the lab also presents its own set of challenges, both practically in terms of the time required and, more significantly, as bacteria readily evolve to adapt to growth in the lab setting (Wiser et al. 2013), easily obscuring the phenotype that was originally under study. Second, we can never know what environmental pressures shaped the evolution of the regulatory system that generates variable torCAD expression, and we can only guess at what laboratory conditions would most closely approximate the conditions in which such behavior occurs in nature. In short, lack of laboratory evidence of a fitness cost is not evidence against the existence of a fitness cost.
Furthermore, bet hedging and its associated fitness costs may only be part of the story. Phenotypic diversity in microbial populations is associated with a kaleidoscope of social behaviors (West et al. 2006). It may be the case, for instance, that aerobic TMAO reduction is altruistic, meaning that it exerts a toll on the individual cell performing the reaction but benefits the population as a whole. It has been reported that aerobic TMAO reduction can protect a growing population against acidification of the growth medium (Bordi et al. 2003; Ansaldi et al. 2007). This phenomenon is consistent with altruistic behavior, wherein some cells in the population transiently take on the burden of producing trimethylamine (the product of TMAO respiration) and, by doing so, help the entire population by counteracting acidification.
As illustrated by these comments, there are still many specific aspects of TMAO respiration and its regulation that are open for exploration. Taking a wider view, the mechanism in this system that allows cells to regulate gene expression noise independently from the mean may well be a general scheme found in signaling network architectures. Only a few features seem to be required for producing this behavior: the system output should depend on the relative, not absolute, abundance of two proteins, and the mean expression levels of those two proteins should be regulated such that the low end of the range is in the high-noise domain. This architecture as it occurs in the TorT/TorS/TorR system appears to enable a bet-hedging strategy, but it could be useful in many other systems in which regulating phenotypic diversity is desirable. With genetic tools becoming increasingly sophisticated and single-cell studies becoming increasingly common, we expect we will soon be learning of other fascinating contexts in which biology has employed this mechanism.
Footnotes
ORCID for Jeffrey N. Carey: 0000-0002-5105-0265
ORCID for Mark Goulian: 0000-0003-0076-023X
References
- Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol. 2015;13:497–508. doi: 10.1038/nrmicro3491. [DOI] [PubMed] [Google Scholar]
- Ansaldi M, Bordi C, Lepelletier M, Méjean V. TorC apocytochrome negatively autoregulates the trimethylamine N-oxide (TMAO) reductase operon in Escherichia coli. Mol Microbiol. 1999;33:284–295. doi: 10.1046/j.1365-2958.1999.01468.x. [DOI] [PubMed] [Google Scholar]
- Ansaldi M, Jourlin-Castelli C, Lepelletier M, et al. Rapid dephosphorylation of the TorR response regulator by the TorS unorthodox sensor in Escherichia coli. J Bacteriol. 2001;183:2691–2695. doi: 10.1128/JB.183.8.2691-2695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansaldi M, Simon G, Lepelletier M, Méjean V. The TorR high-affinity binding site plays a key role in both torR autoregulation and torCAD operon expression in Escherichia coli. J Bacteriol. 2000;182:961–966. doi: 10.1128/jb.182.4.961-966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansaldi M, Théraulaz L, Baraquet C, et al. Aerobic TMAO respiration in Escherichia coli. Mol Microbiol. 2007;66:484–494. doi: 10.1111/j.1365-2958.2007.05936.x. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, et al. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Balázsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- Baraquet C, Théraulaz L, Guiral M, et al. TorT, a member of a new periplasmic binding protein family, triggers induction of the Tor respiratory system upon trimethylamine N-oxide electron-acceptor binding in Escherichia coli. J Biol Chem. 2006;281:38189–38199. doi: 10.1074/jbc.M604321200. [DOI] [PubMed] [Google Scholar]
- Bidnenko E, Bidnenko V. Transcription termination factor Rho and microbial phenotypic heterogeneity. Curr Genet. 2018;64:541–546. doi: 10.1007/s00294-017-0775-7. [DOI] [PubMed] [Google Scholar]
- Bordi C, Théraulaz L, Méjean V, Jourlin-Castelli C. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol Microbiol. 2003;48:211–223. doi: 10.1046/j.1365-2958.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- Carey JN, Mettert EL, Roggiani M, et al. Regulated stochasticity in a bacterial signaling network permits tolerance to a rapid environmental change. Cell. 2018;173:196–207.e14. doi: 10.1016/j.cell.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs DZ, Metcalf CJE, Rees M. Evolutionary bet-hedging in the real world: empirical evidence and challenges revealed by plants. Proc Biol Sci. 2010;277:3055–3064. doi: 10.1098/rspb.2010.0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar RD, Shaffer SM, Singh A, et al. Transcriptional Bursting Explains the Noise-Versus-Mean Relationship in mRNA and Protein Levels. PLoS ONE. 2016;11:e0158298. doi: 10.1371/journal.pone.0158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong IG, Haccou P, Kuipers OP. Bet hedging or not? A guide to proper classification of microbial survival strategies. Bioessays. 2011;33:215–223. doi: 10.1002/bies.201000127. [DOI] [PubMed] [Google Scholar]
- El Meouche I, Siu Y, Dunlop MJ. Stochastic expression of a multiple antibiotic resistance activator confers transient resistance in single cells. Sci Rep. 2016;6:19538. doi: 10.1038/srep19538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Freddolino PL, Tavazoie S. Beyond homeostasis: a predictive-dynamic framework for understanding cellular behavior. Annual Review of Cell and Developmental Biology. 2012;28:363–384. doi: 10.1146/annurev-cellbio-092910-154129. [DOI] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bernardo J, Dunlop MJ. Phenotypic Diversity Using Bimodal and Unimodal Expression of Stress Response Proteins. Biophys J. 2016;110:2278–2287. doi: 10.1016/j.bpj.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giel JL, Giel JL, Rodionov D, et al. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- Giel JL, Nesbit AD, Mettert EL, et al. Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe-2S]-IscR in Escherichia coli. Mol Microbiol. 2013;87:478–492. doi: 10.1111/mmi.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gon S, Jourlin-Castelli C, Théraulaz L, Méjean V. An unsuspected autoregulatory pathway involving apocytochrome TorC and sensor TorS in Escherichia coli. Proceedings of the National Academy of Sciences. 2001;98:11615–11620. doi: 10.1073/pnas.211330598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimbergen AJ, Siebring J, Solopova A, Kuipers OP. Microbial bet-hedging: the power of being different. Curr Opin Microbiol. 2015;25:67–72. doi: 10.1016/j.mib.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Hõrak R, Tamman H. Desperate times call for desperate measures: benefits and costs of toxin-antitoxin systems. Curr Genet. 2017;63:69–74. doi: 10.1007/s00294-016-0622-2. [DOI] [PubMed] [Google Scholar]
- Ilbert M, Méjean V, Giudici-Orticoni M-T, et al. Involvement of a mate chaperone (TorD) in the maturation pathway of molybdoenzyme TorA. J Biol Chem. 2003;278:28787–28792. doi: 10.1074/jbc.M302730200. [DOI] [PubMed] [Google Scholar]
- Jack RL, Buchanan G, Dubini A, et al. Coordinating assembly and export of complex bacterial proteins. EMBO J. 2004;23:3962–3972. doi: 10.1038/sj.emboj.7600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourlin C, Ansaldi M, Méjean V. Transphosphorylation of the TorR response regulator requires the three phosphorylation sites of the TorS unorthodox sensor in Escherichia coli. J Mol Biol. 1997;267:770–777. doi: 10.1006/jmbi.1997.0919. [DOI] [PubMed] [Google Scholar]
- Li G-W, Burkhardt D, Gross C, Weissman JS. Quantifying Absolute Protein Synthesis Rates Reveals Principles Underlying Allocation of Cellular Resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins BMC, Locke JCW. Microbial individuality: how single-cell heterogeneity enables population level strategies. Curr Opin Microbiol. 2015;24:104–112. doi: 10.1016/j.mib.2015.01.003. [DOI] [PubMed] [Google Scholar]
- McCrindle SL, Kappler U, McEwan AG. Microbial dimethylsulfoxide and trimethylamine-N-oxide respiration. Adv Microb Physiol. 2005;50:147. doi: 10.1016/S0065-2911(05)50004-3. [DOI] [PubMed] [Google Scholar]
- Mettert EL, Kiley PJ. Coordinate regulation of the Suf and Isc Fe-S cluster biogenesis pathways by IscR is essential for viability of Escherichia coli. J Bacteriol. 2014;196:4315–4323. doi: 10.1128/JB.01975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méjean V, Iobbi-Nivol C, Lepelletier M, et al. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- Mitosch K, Rieckh G, Bollenbach T. Noisy Response to Antibiotic Stress Predicts Subsequent Single-Cell Survival in an Acidic Environment. Cell Syst. 2017;4:393–403.e5. doi: 10.1016/j.cels.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Norman TM, Lord ND, Paulsson J, Losick R. Stochastic Switching of Cell Fate in Microbes. Annu Rev Microbiol. 2015;69:381–403. doi: 10.1146/annurev-micro-091213-112852. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, et al. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- Philippi T, Seger J. Hedging one's evolutionary bets, revisited. Trends Ecol Evol (Amst) 1989;4:41–44. doi: 10.1016/0169-5347(89)90138-9. [DOI] [PubMed] [Google Scholar]
- Pommier J, Méjean V, Giordano G, Iobbi-Nivol C. TorD, a cytoplasmic chaperone that interacts with the unfolded trimethylamine N-oxide reductase enzyme (TorA) in Escherichia coli. J Biol Chem. 1998;273:16615–16620. doi: 10.1074/jbc.273.26.16615. [DOI] [PubMed] [Google Scholar]
- Ratcliff WC, Denison RF. Individual-Level Bet Hedging in the Bacterium Sinorhizobium meliloti. Curr Biol. 2010;20:1740–1744. doi: 10.1016/j.cub.2010.08.036. [DOI] [PubMed] [Google Scholar]
- Roggiani M, Goulian M. Oxygen-Dependent Cell-to-Cell Variability in the Output of the Escherichia coli Tor Phosphorelay. J Bacteriol. 2015;197:1976–1987. doi: 10.1128/JB.00074-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C, Turkarslan S, Lee D-W, Daldal F. Cytochrome c biogenesis: the Ccm system. Trends in Microbiology. 2010;18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G, Méjean V, Jourlin C, et al. The torR gene of Escherichia coli encodes a response regulator protein involved in the expression of the trimethylamine N-oxide reductase genes. J Bacteriol. 1994;176:5601–5606. doi: 10.1128/jb.176.18.5601-5606.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons AM. Modes of response to environmental change and the elusive empirical evidence for bet hedging. Proc Biol Sci. 2011;278:1601–1609. doi: 10.1098/rspb.2011.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Eschevins CC, Susanna KA, et al. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol. 2005;56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- Sureka K, Ghosh B, Dasgupta A, et al. Positive Feedback and Noise Activate the Stringent Response Regulator Rel in Mycobacteria. PLoS ONE. 2008;3:e1771. doi: 10.1371/journal.pone.0001771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Taniguchi Y, Choi PJ, et al. Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Popović N, Grima R. Phenotypic switching in gene regulatory networks. Proc Natl Acad Sci USA. 2014;111:6994–6999. doi: 10.1073/pnas.1400049111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Veening J-W, Stewart EJ, et al. Bet-hedging and epigenetic inheritance in bacterial cell development. Proceedings of the National Academy of Sciences. 2008;105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney M, Reece SE. Adaptive noise. Proc Biol Sci. 2013;280:20131104–20131104. doi: 10.1098/rspb.2013.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- Wiser MJ, Ribeck N, Lenski RE. Long-Term Dynamics of Adaptation in Asexual Populations. Science. 2013;342:1364–1367. doi: 10.1126/science.1243357. [DOI] [PubMed] [Google Scholar]


