Abstract
The massive and irrational use of antibiotics in livestock productions has fostered the occurrence and spread of resistance to “old class antimicrobials.” To cope with that phenomenon, some regulations have been already enforced in the member states of the European Union. However, a role of livestock animals in the relatively recent alerts on the rapid worldwide increase of resistance to last-choice antimicrobials as carbapenems is very unlikely. Conversely, these antimicrobials are increasingly administered in veterinary hospitals whose role in spreading bacteria or mobile genetic elements has not adequately been addressed so far. A cross-sectional study was carried out on 105 hospitalized and 100 non-hospitalized pets with the aim of measuring the prevalence of carbapenem-resistant Gram-negative bacteria (GNB) colonizing dogs and cats, either hospitalized or not hospitalized and estimating the relative odds. Stool samples were inoculated on MacConkey agar plates containing 1 mg/L imipenem which were then incubated aerobically at 37°C ± 1 for 48 h. Isolated bacteria were identified first by Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and were confirmed by 16S rRNA sequencing. The genetic basis of resistance was investigated using PCR methods, gene or whole genome sequencing (WGS). The prevalence of pets harboring carbapenem-resistant bacteria was 11.4 and 1.0% in hospitalized and not-hospitalized animals, respectively, with an odds ratio of 12.8 (p < 0.01). One pet carried two diverse isolates. Overall, 14 gram-negative non-fermenting bacteria, specifically, one Acinetobacter radioresistens, five Acinetobacter baumannii, six Pseudomonas aeruginosa and two Stenotrophomonas maltophilia were isolated. The Acinetobacter species carried acquired carbapenemases genes encoded by blaNDM-1 and blaOXA-23. In contrast, Pseudomonas phenotypic resistance was associated with the presence of mutations in the oprD gene. Notably, inherent carbapenem-resistant isolates of S. maltophilia were also resistant to the first-line recommended chemotherapeutic trimethoprim/sulfamethoxazole. This study estimates the risk of colonization by carbapenem-resistant non-fermenting GNB in pets hospitalized in veterinary tertiary care centers and highlights their potential role in spreading resistance genes among the animal and human community. Public health authorities should consider extending surveillance systems and putting the release of critical antibiotics under more strict control in order to manage the infection/colonization of pets in veterinary settings.
Keywords: one health, carbapenem resistance, Acinetobacter radioresistens, dogs and cats, hospitalization, veterinary tertiary care facilities, blaNDM-1blaOXA-23
Introduction
Among the large quantities of antimicrobials given to animals each year, only a fractional amount is reserved to pets (Guardabassi et al., 2004; European Medicines Agency, 2015). The latter estimate is expected to be undervalued since injectable molecules and human specialties are not adequately taken into account (European Medicines Agency, 2015). Even more interesting, it is the qualitative point of view; indeed, whereas in husbandry almost exclusively “old class antibiotics” are used; in veterinary tertiary care facilities (VTCF) for pets, also antimicrobials critically important for human medicine as carbapenems are administered in certain critical cases. In humans, Enterobacteriaceae and non-fermenting gram-negative bacteria (GNB) Pseudomonas aeruginosa, Acinetobacter spp. and Stenotrophomonas maltophilia account for the majority of the pandemic carbapenem resistance threat. The occurrence of carbapenem-resistant bacteria in veterinary medicine has already been brought to the attention of the scientific community due to the severe impact that this phenomenon may cause (Endimiani et al., 2011; Shaheen et al., 2013; Stolle et al., 2013). Resistant organisms are transmitted among pets, owners and veterinary staff and, in this way, spread to the community. (Boerlin et al., 2001; Guardabassi et al., 2004; Leite-Martins et al., 2014; Yao et al., 2016).
To date, the assessment of public health risk posed by VTCFs has not adequately been addressed and only scarce but relevant information is available (Boerlin et al., 2001; Pulss et al., 2018). Certainly, veterinary hospitals share some risk factors with human hospitals, such as the use of antibiotics, complex treatments, prolonged hospitalization of critically ill or immunocompromised patients, and the presence of intensive care units. These factors favor nosocomial infections, and the exchange of antimicrobial-resistant bacteria and genetic determinants of resistance (Poirel et al., 2008). In human hospitals, a particular concern has recently arisen, based on the evidence that the spread of resistant bacteria is not restricted to clonal outbreaks but also due to asymptomatic carriage (Cerqueira et al., 2017).
With the aim to refine the focus of antibiotic resistance in pets hospitalized in VTCFs, a cross-sectional study was thus carried out to investigate the prevalence of carbapenem-resistant GNB in pets admitted to VTCFs. To this aim, dogs and cats admitted to two different VTCFs in the Bologna area (Northern Italy) were included. In addition, privately owned pets living in the same area and not hospitalized were also investigated and compared in order to assess the relative odds of being colonized after hospitalization.
Materials and Methods
Experimental Design and Animals
This cross-sectional study was carried out within the Bologna area from November 2014 to January 2015. Feces from 105 hospitalized pets (61 dogs and 44 cats) and 100 non-hospitalized pets (73 dogs and 27 cats) were collected. Hospitalized pets from two different facilities, both of which were characterized by the presence of intensive care units and referral practices, were included consecutively after at least 48 h of hospitalization. The non-hospitalized pets were from 100 distinct owners, had never been hospitalized and had not been given antibiotics for at least 2 months; they will hereafter be defined as “general population.” All the private owners of the animals considered in this study signed an informed consent form to authorize the rectal swab sampling.
Isolation and Identification of Enteric Bacteria
Rectal swabs were used for collecting the fecal material from the pets and were immediately placed in the transport medium Cary-Blair Medium (Oxoid). Within 24 h, the swabs were soaked in 1 mL of sterile saline, vortexed, and 0.1 mL was inoculated on plates of MacConkey Agar (Oxoid) containing 1 mg/L imipenem (Sigma-Aldrich). The plates were incubated aerobically at 37°C ± 1 for 48 h.
All the different morphotypes of colonies grown in the same plate were subcultivated to obtain pure culture. Isolates were identified firstly by Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (bioMérieux) and successively confirmed by partial sequencing of the 16S rRNA gene (Brosius et al., 1978).
Antimicrobial Susceptibility
For all isolates, the minimum inhibitory concentrations (MICs) of imipenem and meropenem was determined using E-test strips (bioMérieux) following manufacturer instructions. P. aeruginosa, S. maltophilia, and Acinetobacter spp. were considered resistant to imipenem and meropenem at values ≥ 8 mg/L as suggested by the EUCAST (2016). The MIC of colistin were determined by broth microdilution using a breakpoint > 2 mg/L (EUCAST, 22 march 2016). Disk diffusion was used for testing all other antibiotics (Supplementary Material) and interpreted according to Clinical and Laboratory Standards Institute standard (CLSI 2014 M100-S24).
Molecular Characterization of Antimicrobial Resistance
The isolates were screened for the presence of the following genes coding for carbapenem-hydrolyzing enzymes: blaV IM, blaNDM-1, blaIMP, blaKPC, and blaOXA-48-like using a multiplex polymerase chain reaction (PCR) assay as previously described (Doyle et al., 2012). In addition, a real-time PCR was used for detecting blaOXA-23 (Supplementary Material) followed by direct Sanger sequencing of the amplicons as a confirmatory assay. The presence of AmpC was carried out using a previously described multiplex PCR (Pérez-Pérez and Hanson, 2002) while extended-spectrum β-lactamases (ESBL) genes were detected using a multiplex PCR with subsequent melt curve analysis suitable to identify blaSHV, blaTEM, and blaCTX-M gene targets (Supplementary Material).
Two PCR reactions were used to confirm the presence of chromosomally encoded metallo β-lactamases (MBL) in S. maltophilia (Supplementary Material); in particular, two PCR assays were carried out targeting an internal sequence of the L1 gene and a sequence located within the region bridging the transcription factor encoding gene ampR and the inducible β-lactamase L2 gene (Gould et al., 2006; Okazaki and Avison, 2008). Furthermore, trimethoprim/sulfamethoxazole (SXT) resistance was investigated using primers pairs for sul1, sul2, and dfrA genes as well as class 1, 2, and 3 integrons (Hu et al., 2011; Supplementary Material).
To investigate whether blaNDM-1 was located in transposon Tn125, a PCR was carried out at 5′ terminus with primers fwd_ISAba125 and NDM-reverse and at the 3′ terminus of the transposon with the fwd primer annealing on F_Δpac _and rev_ISAba125 (Supplementary Material). Finally, the oprD gene of P. aeruginosa was amplified using outer primers, amplifying the almost complete sequence of the gene followed by sequencing of the amplicon by internal primers (Supplementary Material).
Whole Genome Sequencing
Since, to the best of our knowledge, Acinetobacter radioresistens carrying chromosomally encoded blaNDM-1 has not previously been reported, the whole genome of the isolate was sequenced to better characterize the resistance pattern. Also the five isolates of A. baumannii were fully sequenced to characterize the blaOXA-23 genetic environment, the complete resistome profile and to assess the Multi Locus Sequence Type as well as the clonal relationship. For Whole Genome Sequencing (WGS), genomic DNA was extracted from overnight broth culture in Brain Heart Infusion (Oxoid, Basingstoke, United Kingdom) using the MagAttract HMW DNA kit (Qiagen, Milan, Italy). The DNA samples were quantified with a fluorescent nucleic acid dye (Picogreen; Invitrogen, Paisley, United Kingdom), and libraries were prepared using the Nextera XT DNA Sample Preparation Kit (Illumina, Inc., San Diego, CA, United States). Sequencing was carried out on an Illumina MiSeq platform with 2 × 250 bp paired-end runs. The sequence quality of the reads was evaluated using FastQC (Andrews, 2016). The reads were assembled with SPAdes 3.7.1. The ResFinder web server1 using a threshold of 100% identity for the genes encoding β-lactamases and 98% identity for all other genes and CARD2 (Jia et al., 2017) using the Resistance Gene Identifier v 4.0.2 were used to search for antimicrobial resistance genes in the Acinetobacter spp. draft genomes.
MLST
The Sequence Type of A. baumannii isolates was inferred from assembled contigs using the Multi Locus Sequence Typing application 1.8 (Assembled Genomes/Contigs) (Larsen et al., 2012).
In addition, to further analyze the genetic relatedness of the apparently clonal A. baumannii isolates, an alternative approach to the MLST, the wgMLST was carried out by genome-wide gene-by-gene comparisons of a dataset of 5633 genes retrieved from the whole genome assemblies using the Bionumerics 7.6 package software (Applied Maths) using either an assembly free and an assembly based algorithm in Bionumerics engine calculation. The isolate 92A IMI was assumed as index isolate for comparison. The results were graphed as dendrogram with percentage of homology (Supplementary Figure S1).
PFGE Typing
For clonality assessment, A. baumannii isolates were typed using a rapid PFGE protocol with ApaI (40 U/plug) (Promega) as a restriction enzyme according to Durmaz (Durmaz et al., 2009).
Nucleotide Accession Number
The raw sequence data of Acinetobacter radioresistens and of five Acinetobacter baumannii isolates were deposited in the GenBank database under BioProject accession PRJNA344732 and PRJNA437120, respectively.
Results
Thirteen out of 205 pets carried carbapenem-resistant bacteria with a prevalence estimate of 6.3% [95% confidence interval (CI): 3.4–10.6%]. Of these, 12 (8 dogs and 4 cats) were hospitalized pets [prevalence estimate 11.4% (95% CI: 6.0–19.1%)] and 1 dog was from the general population [prevalence estimate 1.0% (95% CI: 0.0–5.4%)]. The odds ratio of the hospitalized pets was 12.8 (95% CI: 1.6–100.2; p < 0.01). In one hospitalized cat, two different carbapenem-resistant bacteria were isolated; therefore, 14 isolates were found in total. They included one Acinetobacter radioresistens, five A. baumannii, six P. aeruginosa, and two S. maltophilia (Supplementary Table S1). The MICs of imipenem for the Acinetobacter species ranged from 16 mg/L to more than 32 mg/L, for the P. aeruginosa they were 16 mg/L and for the S. maltophilia they were > 32 mg/L (Table 1). All bacteria showed multiple resistance, however, in all cases, the bacteria were susceptible to colistin (MIC < 2 mg/L). Carbapenemase resistance genes were detected in all isolates except P. aeruginosa.
Table 1.
Characteristics of the isolates resistant to carbapenems obtained from feces of dogs and cats.
| Isolate | Organism | MIC of imipenem (mg/L) | MIC of meropenem (mg/L) | Resistance determinant | Dog/Cat (source) | Hospitalized | |
|---|---|---|---|---|---|---|---|
| Carpapem-resistance | Other β-lactamases | ||||||
| 1A IMI | Acinetobacter radioresistens | >32 | >32 | blaNDM-1 | blaOXA-23 | Dog | Yes |
| 87A IMI | Acinetobacter baumannii (ST2) | >32 | >32 | blaOXA-23 | blaOXA-66,blaTEM, AmpC | Cat | Yes |
| 92A IMI | Acinetobacter baumannii (ST2) | 16 | >32 | blaOXA-23 | blaOXA-66,blaTEM, AmpC | Dog | Yes |
| 108A IMI | Acinetobacter baumannii (ST2) | 16 | >32 | blaOXA-23 | blaOXA-66,blaTEM, AmpC | Cat | Yes |
| 115A IMI | Acinetobacter baumannii (ST2) | >32 | >32 | blaOXA-23 | blaOXA-66,blaTEM, AmpC | Dog | Yes |
| 213A IMI | Acinetobacter baumannii (ST2) | 16 | >32 | blaOXA-23 | blaOXA-66,blaTEM, AmpC | Cat∗ | Yes |
| 3 A IMI | Pseudomonas aeruginosa | 16 | 8 | oprD | blaSHV AmpC | Dog | Yes |
| 110A IMI | Pseudomonas aeruginosa | 16 | 4 | oprD | blaSHV CTX-M | Dog | Yes |
| 111A IMI | Pseudomonas aeruginosa | 16 | 8 | ND | blaSHV | Dog | Yes |
| 117A IMI | Pseudomonas aeruginosa | 16 | 2 | oprD | blaTEM | Dog | Yes |
| 121A IMI | Pseudomonas aeruginosa | 16 | 4 | oprD | AmpC | Dog | Yes |
| 131A IMI | Pseudomonas aeruginosa | 16 | 8 | ND | blaSHV AmpC | Cat | Yes |
| 207A IMI | Stenotrophomonas maltophilia | >32 | >32 | L1 | Dog | No | |
| 213B IMI | Stenotrophomonas maltophilia | >32 | >32 | L1 | Cat∗ | Yes | |
MIC, Minimum Inhibitory Concentration; ND, not determined; ST, Sequence type (Pasteur scheme). ∗Indicates the same animal.
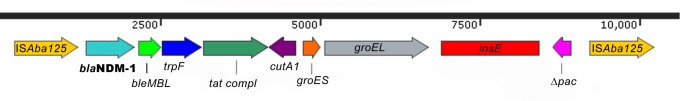
A. radioresistens carried both blaNDM-1 and blaOXA-23. The blaNDM-1 gene was chromosomally located and organized in a composite transposon encompassing blaNDM-1, bleMBL, trpF, tat, cutA1, groES, groEL, insE, and Δpac genes bracketed between a pair of ISAba125. The presence of a mobile element with IS elements was further confirmed by a specific PCR assay designed to investigate the genetic environment of blaNDM-1 gene (Figure 1).
FIGURE 1.
Acinetobacter radioresistens blaNDM-1 genetic environment. The schematic presentation of the A. radioresistens blaNDM-1 transposon Tn125 encompassing blaNDM-1, bleMBL, trpF, tat, cutA1, groES, groEL, insE, and Δpac genes bracketed between a pair of ISAba125 is drawn above. Red rectangles indicate the position of the amplicons.
The blaOXA-23 lacked ISAba1, ISAba4 (Turton et al., 2006; Corvec et al., 2007; Poirel et al., 2008; Karthikeyan et al., 2010; Smet et al., 2012) or ISAcsp2 (Poirel et al., 2012) upstream insertion sequences according to ISfinder3. These IS sequences are known to provide the effective promoter to the hydrolyzing enzyme encoding gene blaOXA-23 in A. baumannii for an elevated level of expression (Turton et al., 2006). Other genetic determinants of resistance included aminoglycoside resistance encoding genes strA [aph(3″)-ib] and strB [aph(6)-1d], sulphonamide resistance gene sul2, bleomycin resistant protein BRP and tetracycline resistance gene tet.
The A. radioresistens whole genome was sequenced, and the reads showed a coverage of 293X and a guanine-cytosine (GC) content of 42%. The assembled genome encountered 81 contigs with N50 of 113445 and mean contig size of 38028.383 bp. The genome length was 3.080 Mbp.
All the A. baumannii strains carried both intrinsic blaOXA-66 and acquired blaOXA-23 Class D Oxacillinases. However, while blaOXA-23 was embedded in a transposon bracketed within two ISAba1 insertion sequence which provide a strong promoter leading to a sustained expression of oxacillinase hydrolyzing enzyme, the blaOXA-66 lacks an upstream IS sequence (Figure 2).
FIGURE 2.
Acinetobacter baumannii blaOXA-23 genetic environment. The schematic presentation of the A baumannii blaOXA-23 transposon Tn2006 encompassing blaOXA-23, and few ORFs bracketed between ISAba1 colored in yellow. Flanking ORFs are colored in purple.
Additionally, besides oxacillinases, other β-lactamases were present in all isolates of A. baumannii. Acinetobacter derived cephalosporinase blaADC-73 (AmpC allele 20) with an upstream ISAba1 in the promoter region and a likely sustained expression was present. Furthermore, A. baumannii isolates carried blaTEM-1D embedded in a single copy IS26- blaTEM-1D-Tn3-IS26, likely responsible for the sulbactam resistance phenotype (Krizova et al., 2013; Yang et al., 2018).
Overall, the antimicrobial resistance profile of the five A. baumannii isolates showed an extensively drug-resistant (XDR) phenotype, i.e., non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (Supplementary Table S2; Magiorakos et al., 2012). The comprehensive resistance determinants are enlisted in Supplementary Table S3.
The two S. maltophilia isolates carried inducible chromosomal encoded L1 metallo-β-lactamase with carbapenem hydrolyzing activity and its regulatory element ampR (Gould et al., 2006; Okazaki and Avison, 2008). Notably, both S. maltophilia isolates were resistant to SXT due to the presence of both sul1 and sul2 genes associated with class 1 integron mobile element.
Finally, carbapenem-resistant P. aeruginosa were negative to carbapenemase genes but carried either blaSHV, and/or blaTEM, and/or blaCTX-M β-lactamase associated with an oprD gene with truncating mutations in 4 out of 6 of these isolates (Tables 1, 2).
Table 2.
oprD mutations in Pseudomonas aeruginosa isolates resistant to carbapenems.
| Isolate | oprD mutation | Resulting change in the protein |
|---|---|---|
| 3A IMI | c.604ins (IS256 family transposase ISPa1328) | Protein truncated |
| 110A IMI | c.840_854delGGGCGCGTATACCCTinsCGGC | Frameshift and premature stop codon; Predicted protein of 313 aa |
| 111A IMI | c.875 T > C | p.292P > L |
| 117A IMI | c.970delA | Frameshift and premature stop codon; Predicted protein of 344 aa |
| 121A IMI | c.1244G > A | Nonsense mutation and premature stop codon; Predicted protein of 414 aa |
| 131A IMI | c.154G > A; c.397A > C; | p.52D > N p.132K > Q |
Strain used for comparison: P. aeruginosa ATCC 27853 (GenBank Accession Number KF649209.1). Missense mutations with respect to the reference strain are indicated only in those isolates without truncating mutations.
All A. baumannii strains showed the same macrorestriction profile by PFGE (Supplementary Figure S2). All isolates were assigned to ST2 (Pasteur’s scheme) or ST451 (Oxford’s scheme). Using the complete panel of the coded alleles a UPGMA tree was calculated in Bionumerics 7.6 software (Supplementary Figure S1). The wgMLST showed that all isolates except one were closely related with homology > 99.8%. (<6 diverse allele). The only isolate 108A IMI seemed more dissimilar with an homology of 98.8%. The most closely related isolates were sampled on the same day (87A IMI and 92A IMI) or 2 months apart (115A IMI and 213A IMI) (Supplementary Figure S1).
Discussion
This study findings focuses on Acinetobacter spp. as the main emerging threat to public health associated with the hospitalization in VTCFs. Notably, we identified for the first time to the best of our knowledge, a carbapenem resistant A. radioresistens isolate carrying a blaNDM-1 gene within a Tn125 mobile element. The same composite Tn125 has already been reported in other Acinetobacter species (Pfeifer et al., 2011; Bonnin et al., 2012; Dortet et al., 2012; Fu et al., 2012; Chatterjee et al., 2016) but not yet in A. radioresistens. The presence of a Class B MBL in a mobile element is a concern since it could be readily transferred to other bacteria residing in the intestine, such as Enterobacteriaceae. Its potential to serve as a reservoir for worldwide dissemination of blaNDM should be given outstanding attention and should not be undervalued in view of the fact that was isolated in a veterinary and not human tertiary care settings.
Conversely, A. radioresistens was found to carry blaOXA-23 without ISAb1 sequences and, as a consequence, the hydrolyzing oxacillinase was not expressed at high level. Overall, A. radioresistens was multi-drug resistant being not susceptible to quinolones, tetracycline and trimethoprim/sulfamethoxazole though it remained susceptible to amikacin and gentamicin, which are not affected by Aminoglycoside O-phosphotransferases strA and strB, as well as to ampicillin/sulbactam and colistin.
Noteworthy, the resistome of the five A. baumannii isolates showed an XDR phenotype (Supplementary Table S2) including non-susceptibility to quinolones, gentamicin, amikacin and sulbactam with only colistin as putative effective treatment. It is worth mentioning that also 16S rRNA methylase gene armA recently reported in nosocomial outbreaks in Italy and responsible for resistance to aminoglycosides including gentamicin and amikacin, was found (Brigante et al., 2012). For these features, the A. baumannii isolates represent an actual hazard retaining high epidemic potential.
A. baumannii is reported as a frequent cause of nosocomial infections with increased mortality worldwide (Higgins et al., 2010). Very recently, single cases and small case series of clinical isolates of carbapenem resistant A. baumannii of different sequence types and carrying blaOXA-23 were reported (Pomba et al., 2014; Ewers et al., 2016, 2017; Lupo et al., 2017). Those authors focused the attention on companion animals as potential public health hazard. Also a remarkably high rate of asymptomatic pet carriers of carbapenem-resistant A. baumannii ST25 (2.7%) has been reported in France and pointed out pets as possible reservoir of community acquired infections (Hérivaux et al., 2016).
All the A. baumannii isolates in VTCFs in our study belong to ST2. In Italy, besides, the endemic so called “Italian strain” ST78, the ST2 strains belonging to International Clonal lineage II has progressively become dominant as a cause of epidemic outbreaks in hospitals and is even replacing the ST78 strains. Both strains share great ability to adhere to cells, to invade and survive within pneumocytes as well as to form biofilm on abiotic surface (Ambrosi et al., 2017) hence, replacing of the ST78 may be associated with the acquisition of ST2 strains of highly expressed blaOXA-23 oxacillinase (D’Arezzo et al., 2011; Brigante et al., 2012; Mezzatesta et al., 2012). Indeed, in last years, nosocomial outbreaks in Italy are almost all ascribed to ST2 blaOXA-23 producing strains (Carretto et al., 2011; Agodi et al., 2014; Principe et al., 2014; Perilli et al., 2015; Piana et al., 2015). BlaOXA-23 producing strains were not reported in a survey, which had investigated nosocomial A. baumannii isolated in 2007 in the Bologna Area, while they were reported in another survey carried out later in 2011 (Carretto et al., 2011; Principe et al., 2014).
Within-hospital selective pressure clearly plays a role in favoring the emergence of MDR but there is also inter-hospital interconnection modalities (i.e., patients transfer) which explains the simultaneous outbreaks of related resistant A. baumannii strains (Karkada et al., 2011). How the VTCFs/ICUs fit in this model is far from being understood but the evidence that the strains isolated in veterinary and human medical settings in the same geographical area worth to be examined as well as the role of the community.
When investigating the clonal relatedness, all A. baumannii isolates showed the same PFGE pattern. Also wgMLST confirmed the high relatedness of at least of four of five of the isolates whereas one isolate showed less similarity, however, the complexity in interpreting the genomic variability is challenging, since no threshold guidelines could be applied and a case-by-case interpretation is still used in this matter (Halachev et al., 2014; Higgins et al., 2017; Shaheen et al., 2018); Since this study did not deal with an epidemic outbreak, the A. baumannii isolates found during the survey may represent the genetic evolution over time of originally the same clone which has colonized the medical environment for its ability to grow on abiotic surface, has accumulated genetic variations making it to diverge from the original clone. Eventually, the knowledge of the frequency rate of mutation accumulation in A. baumannii would lead to infer the precise time of colonization.
Unlike Acinetobacter isolates, P. aeruginosa-associated carbapenem resistance is linked to the loss of oprD (Lee and Ko, 2012; Ocampo-Sosa et al., 2012; Kim et al., 2016) and it is associated with multidrug resistance. In most of our cases, the loss of oprD was caused by different mutations within the gene causing a premature stop codon as a consequence of a large insertion, a frameshift or a nonsense mutation. The loss of oprD was also associated with either ESBL (blaSHV, blaTEM or blaCTX-M) or AmpC resistance genes. The presence of different mutations and β-lactamase genes indirectly demonstrates the lack of clonal relatedness at least between the P. aeruginosa isolates. The absence of clonal spread in apparent pseudo-outbreaks is very frequent for Gram-negative non-fermenting bacteria and it is correlated to their ability to colonize the medical device and surface (Valdezate et al., 2004) and produce multispecies biofilms (Tan et al., 2017).
Together with Acinetobacter spp. and P. aeruginosa, other carbapenemase-resistant non-fermenter GNB, namely S. maltophilia, were found. S. maltophilia is not an inherently primary pathogen, but it is rather environmental bacteria intrinsically resistant to β-lactams including carbapenems due to inducible β-lactamase (Gould et al., 2006; Okazaki and Avison, 2008). It should be noted that, in S. maltophilia human nosocomial infections, the antibiotic of choice for treatment of infections caused by these bacteria is SXT (Madi et al., 2016). Our isolates actually showed complete resistance to SXT. Both sul1 associated with class 1 integron, and sul2 genes were found in the isolates. Plasmid-mediated sul1 and sul2 are responsible for the worldwide emergence of resistance to sulfonamides reported beginning since 2007 (Toleman et al., 2007). In 2009, a comprehensive meta-analysis carried out on 3872 strains, found 4% of SXT resistant strains (Looney et al., 2009). As far as we know, the only report from southern Europe, namely from Greece, showed that the majority of S. maltophilia clinical isolates were sensitive to SXT (Samonis et al., 2012) while most of the concerns were from Middle east (Turkey, Iran) and Far East (Korea and China) (Liaw et al., 2010). The origin of this resistance at the genetic level was ascribed to sul genes. In particular, the presence of sul2 is reported in isolates showing very high resistance to SXT (Toleman et al., 2007). However, sul genes alone do not confer trimethoprim resistance but certainly, they are frequently associated with dfrA genes in the gene cassettes of the integrons. In our isolates, dfrA genes were not found and the high resistance to SXT was not clarified. Furthermore, class 1 integrons may carry genes coding for tolerance to common disinfectants and facilitate the horizontal spread of resistance to other bacterial species. Overall, these features make these opportunistic pathogens isolated in this study an insidious threat.
On a more general note, the findings of the study show that hospitalization in VTCFs providing cures to pets represents a significant risk factor of colonization with carbapenem resistant GNB embodying the most significant current hazard to public health in VTCFs. These findings are even more worrying if compared with those found in human hospitals in very similar cross-sectional epidemiological survey during non-epidemic outbreaks. The carbapenemase producing GNB was 0.4% compared with the 6% of this study and the overall prevalence of carbapenem-resistant GNB was 4.8% compared with the 11.4% of this study (Pantel et al., 2015). The other side of the coin, thankfully, is that the more pathogenic Enterobacteriaceae carrying carbapenemase resistance genes reported to cause epidemic cases around the world and also recently found in Germany in many different veterinary hospitals (Pulss et al., 2018) were not found in these veterinary healthcare settings. It should be noted that KPC producers (mainly Enterobacteriaceae) may exhibit susceptibility to imipenem and resistance to ertapenem causing an underestimation of their prevalence using MacConkey supplemented with imipenem (1 mg/L) as carried out in this study. However, KPC producers were not shown either using ceftazidime (1 mg/L) for ESBL bacteria nor using multiplex PCR directly on the DNA purified from stools for molecular screening (data not shown).
Non-fermenter GNB may cause nosocomial infections, particularly in immunocompromised critically ill patients, raising the mortality rate (Siempos et al., 2010; Chiu et al., 2015). Furthermore, Acinetobacter spp. and S. maltophilia, but also P. aeruginosa, show great ability to adapt to any environment, including hospitals; the clonal relatedness between the isolates of A. baumannii carried by pets hospitalized in one VTCF over a short period of time demonstrated the colonization of the environment. Non-fermenter GNB are also readily carried on the skin of colonized animals thus enabling the easy transmission to veterinarians, personnel providing care and owners as well as other hospitalized pets. These bacteria represent a rising challenge for healthcare management because they are frequently multidrug resistant as a consequence of a multitude of mechanisms. In addition, non-fermenter GNB may be a source of genes which are carbapenemase resistant for Enterobacteriaceae (Toleman et al., 2012; Bonnin et al., 2014). It should be emphasized that non-fermenter GNB were not isolated in clinical samples, but they colonized the intestines of the pets; no outbreaks sustained by non-fermenter GNB were reported in either of the investigated VTCFs during the period of study and, hence, no awareness of the risk was evident and no specific measures of control were activated.
In VTCFs, the concerns of carbapenem resistance should be even more compelling with respect to human hospitals due to the extensive, systematic use of broad-spectrum antibiotics, the relative lack of antimicrobial susceptibility-based therapy, the objective difficulties in applying and maintaining hygienic practices and the very limited use of systematic monitoring programs. In fact, these findings and considerations give rise to concerns regarding the risk for veterinary personnel veterinary students or trainees and for the pet owners, highlighting the possible role that veterinary facilities play in spreading carbapenemase-resistant genes in the environment, including the human community.
In the VTCFs investigated in this study, wide spectrum β-lactam drugs are used as first line therapy in hospitalized dogs. The consequent high environmental pressure of such antibiotics represents a risk factor for the co-selection of carbapenem-resistant bacteria (Maseda et al., 2017). In fact, almost all the carbapenem-resistant isolates are either Sulbactam resistant or AmpC or ESBL producers (data not shown), i.e., the “restricted use” of a specific class of antibiotics is not per se sufficient to avoid the emergence of the relative resistance. Since it is evident that carbapenem-resistant gram negative non-fermenters represent an actual threat in VTCFs, it appears more reasonable to also extend the public health surveillance system to VTCFs, and to allow the traceable and targeted use of carbapenems or other critical antibiotics, such as colistin, to control infection or to eradicate the carriage of multidrug resistant bacteria in pets. Appropriate antibiotic stewardships as well as active and continuous surveillance of resistance should be implemented in VTCFs which ask to be accredited for the use of carbapenems or last-choice antimicrobials.
Author Contributions
FG and RZ provided substantial contributions to the conception and design of the work and the acquisition, analysis, and interpretation of data and drafting the work. FP and DM performed the acquisition, analysis, and interpretation of data and drafting the work. VS and AP performed the acquisition, analysis, and interpretation of data and critical revising of the work for important intellectual content. MT, NR, EZ, DT, GP, and JP performed the acquisition and analysis of data and critical revising of the work for important intellectual content. All authors have approved the final version to be published. All authors are to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02872/full#supplementary-material
References
- Agodi A., Voulgari E., Barchitta M., Quattrocchi A., Bellocchi P., Poulou A., et al. (2014). Spread of a carbapenem- and colistin-resistant Acinetobacter baumannii ST2 clonal strain causing outbreaks in two Sicilian hospitals. J. Hosp. Infect. 86 260–266. 10.1016/j.jhin.2014.02.001 [DOI] [PubMed] [Google Scholar]
- Ambrosi C., Scribano D., Aleandri M., Zagaglia C., Di Francesco L., Putignani L., et al. (2017). Acinetobacter baumannii virulence traits: a comparative study of a novel sequence type with other Italian endemic international clones. Front. Microbiol. 8:1977. 10.3389/fmicb.2017.01977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2016). Babraham Bioinformatics – FastQC a Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Boerlin P., Eugster S., Gaschen F., Straub R., Schawalder P. (2001). Transmission of opportunistic pathogens in a veterinary teaching hospital. Vet. Microbiol. 82 347–359. 10.1016/S0378-1135(01)00396-0 [DOI] [PubMed] [Google Scholar]
- Bonnin R. A., Poirel L., Naas T., Pirs M., Seme K., Schrenzel J., et al. (2012). Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 18 E362–E365. 10.1111/j.1469-0691.2012.03928.x [DOI] [PubMed] [Google Scholar]
- Bonnin R. A., Poirel L., Nordmann P. (2014). New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol. 9 33–41. 10.2217/fmb.13.69 [DOI] [PubMed] [Google Scholar]
- Brigante G., Migliavacca R., Bramati S., Motta E., Nucleo E., Manenti M., et al. (2012). Emergence and spread of a multidrug-resistant Acinetobacter baumannii clone producing both the carbapenemase OXA-23 and the 16S rRNA methylase ArmA. J. Med. Microbiol. 61 653–661. 10.1099/jmm.0.040980-0 [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. (1978). Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 75 4801–4805. 10.1073/pnas.75.10.4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretto E., Barbarini D., Dijkshoorn L., van der Reijden T. J. K., Brisse S., Passet V., et al. (2011). Widespread carbapenem resistant Acinetobacter baumannii clones in Italian hospitals revealed by a multicenter study. Infect. Genet. Evol. 11 1319–1326. 10.1016/j.meegid.2011.04.024 [DOI] [PubMed] [Google Scholar]
- Cerqueira G. C., Earl A. M., Ernst C. M., Grad Y. H., Dekker J. P., Feldgarden M., et al. (2017). Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc. Natl. Acad. Sci. U.S.A. 114 1135–1140. 10.1073/pnas.1616248114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Datta S., Roy S., Ramanan L., Saha A., Viswanathan R., et al. (2016). Carbapenem resistance in Acinetobacter baumannii and other Acinetobacter spp. causing neonatal sepsis: focus on NDM-1 and its linkage to ISAba125. Front. Microbiol. 7:1126. 10.3389/fmicb.2016.01126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.-W., Li M.-C., Ko W.-C., Li C.-W., Chen P.-L., Chang C.-M., et al. (2015). Clinical impact of Gram-negative nonfermenters on adults with community-onset bacteremia in the emergency department. J. Microbiol. Immunol. Infect. 48 92–100. 10.1016/j.jmii.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Corvec S., Poirel L., Naas T., Drugeon H., Nordmann P. (2007). Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51 1530–1533. 10.1128/AAC.01132-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arezzo S., Principe L., Capone A., Petrosillo N., Petrucca A., Visca P. (2011). Changing carbapenemase gene pattern in an epidemic multidrug-resistant Acinetobacter baumannii lineage causing multiple outbreaks in central Italy. J. Antimicrob. Chemother. 66 54–61. 10.1093/jac/dkq407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L., Nordmann P., Poirel L. (2012). Association of the emerging carbapenemase NDM-1 with a bleomycin resistance pProtein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob. Agents Chemother. 56 1693–1697. 10.1128/AAC.05583-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D., Peirano G., Lascols C., Lloyd T., Church D. L., Pitout J. D. (2012). Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol. 50 3877–3880. 10.1128/JCM.02117-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmaz R., Otlu B., Koksal F., Hosoglu S., Ozturk R., Ersoy Y., et al. (2009). The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn. J. Infect. Dis. 62 372–377. [PubMed] [Google Scholar]
- Endimiani A., Hujer K. M., Hujer A. M., Bertschy I., Rossano A., Koch C., et al. (2011). Acinetobacter baumannii isolates from pets and horses in Switzerland: molecular characterization and clinical data. J. Antimicrob. Chemother. 66 2248–2254. 10.1093/jac/dkr289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Medicines Agency (2015). European Surveillance of Veterinary Antimicrobial Consumption. 5th Report Released Sales of Veterinary Antimicrobial Agents in 26 EU/EEA Countries in 2013’. London: EMA. [Google Scholar]
- Ewers C., Klotz P., Leidner U., Stamm I., Prenger-Berninghoff E., Göttig S., et al. (2017). OXA-23 and ISAba1 –OXA-66 class D β-lactamases in Acinetobacter baumannii isolates from companion animals. Int. J. Antimicrob. Agents 49 37–44. 10.1016/j.ijantimicag.2016.09.033 [DOI] [PubMed] [Google Scholar]
- Ewers C., Klotz P., Scheufen S., Leidner U., Göttig S., Semmler T. (2016). Genome sequence of OXA-23 producing Acinetobacter baumannii IHIT7853, a carbapenem-resistant strain from a cat belonging to international clone IC1. Gut Pathog. 8:37. 10.1186/s13099-016-0119-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Du X., Ji J., Chen Y., Jiang Y., Yu Y. (2012). Epidemiological characteristics and genetic structure of blaNDM-1 in non-baumannii Acinetobacter spp. in China. J. Antimicrob. Chemother. 67 2114–2122. 10.1093/jac/dks192 [DOI] [PubMed] [Google Scholar]
- Gould V. C., Okazaki A., Avison M. B. (2006). β-Lactam resistance and β-lactamase expression in clinical Stenotrophomonas maltophilia isolates having defined phylogenetic relationships. J. Antimicrob. Chemother. 57 199–203. 10.1093/jac/dki453 [DOI] [PubMed] [Google Scholar]
- Guardabassi L., Schwarz S., Lloyd D. H. (2004). Pet animals as reservoirs of antimicrobial-resistant bacteria: review. J. Antimicrob. Chemother. 54 321–332. 10.1093/jac/dkh332 [DOI] [PubMed] [Google Scholar]
- Halachev M. R., Chan J. Z., Constantinidou C. I., Cumley N., Bradley C., Smith-Banks M., et al. (2014). Genomic epidemiology of a protracted hospital outbreak caused by multidrug-resistant Acinetobacter baumannii in Birmingham. Engl. Genome Med. 6 70. 10.1186/s13073-014-0070-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérivaux A., Pailhoriès H., Quinqueneau C., Lemarié C., Joly-Guillou M.-L., Ruvoen N., et al. (2016). First report of carbapenemase-producing Acinetobacter baumannii carriage in pets from the community in France. Int. J. Antimicrob. Agents 48 220–221. 10.1016/j.ijantimicag.2016.03.012 [DOI] [PubMed] [Google Scholar]
- Higgins P. G., Dammhayn C., Hackel M., Seifert H. (2010). Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65 233–238. 10.1093/jac/dkp428 [DOI] [PubMed] [Google Scholar]
- Higgins P. G., Prior K., Harmsen D., Seifert H. (2017). Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One 12:e0179228. 10.1371/journal.pone.0179228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.-F., Chang X., Ye Y., Wang Z.-X., Shao Y.-B., Shi W., et al. (2011). Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int. J. Antimicrob. Agents 37 230–234. 10.1016/j.ijantimicag.2010.10.025 [DOI] [PubMed] [Google Scholar]
- Jia B., Raphenya A. R., Alcock B., Waglechner N., Guo P., Tsang K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45 D566–D573. 10.1093/nar/gkw1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkada U. H., Adamic L. A., Kahn J. M., Iwashyna T. J. (2011). Limiting the spread of highly resistant hospital-acquired microorganisms via critical care transfers: a simulation study. Intensive Care Med. 37 1633–1640. 10.1007/s00134-011-2341-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan K., Thirunarayan M. A., Krishnan P. (2010). Coexistence of blaOXA-23 with blaNDM-1 and ArmA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother. 65 2253–2254. 10.1093/jac/dkq273 [DOI] [PubMed] [Google Scholar]
- Kim C. H., Kang H. Y., Kim B. R., Jeon H., Lee Y. C., Lee S. H., et al. (2016). Mutational inactivation of OprD in carbapenem-resistant Pseudomonas aeruginosa isolates from Korean hospitals. J. Microbiol. 54 44–49. 10.1007/s12275-016-5562-5 [DOI] [PubMed] [Google Scholar]
- Krizova L., Poirel L., Nordmann P., Nemec A. (2013). TEM-1 β-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii. J. Antimicrob. Chemother. 68 2786–2791. 10.1093/jac/dkt275 [DOI] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., Ko K. S. (2012). OprD mutations and inactivation, expression of efflux pumps and AmpC, and metallo-β-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from South Korea. Int. J. Antimicrob. Agents 40 168–172. 10.1016/j.ijantimicag.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Leite-Martins L., Meireles D., Bessa L. J., Mendes Â., de Matos A. J., da Costa P. M. (2014). Spread of multidrug-resistant Enterococcus faecalis within the household setting. Microb. Drug Resist. 20 501–507. 10.1089/mdr.2013.0217 [DOI] [PubMed] [Google Scholar]
- Looney W. J., Narita M., Mühlemann K. (2009). Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect. Dis. 9 312–323. 10.1016/S1473-3099(09)70083-0 [DOI] [PubMed] [Google Scholar]
- Lupo A., Châtre P., Ponsin C., Saras E., Boulouis H.-J., Keck N., et al. (2017). Clonal spread of Acinetobacter baumannii sequence type 25 carrying blaOXA-23 in companion animals in France. Antimicrob. Agents Chemother. 61:e1881-16. 10.1128/AAC.01881-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madi H., Lukić J., Vasiljević Z., Bioèanin M., Kojić M., Jovèić B., et al. (2016). Genotypic and phenotypic characterization of Stenotrophomonas maltophilia strains from a pediatric tertiary care hospital in Serbia. PLoS One 11:e0165660. 10.1371/journal.pone.0165660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A.-P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Maseda E., Salgado P., Anillo V., Ruiz-Carrascoso G., Gómez-Gil R., Martín-Funke C., et al. (2017). Risk factors for colonization by carbapenemase-producing enterobacteria at admission to a surgical ICU: a retrospective study. Enferm. Infecc. Microbiol. Clin. 35 333–337. 10.1016/j.eimc.2016.02.017 [DOI] [PubMed] [Google Scholar]
- Mezzatesta M. L., D’Andrea M. M., Migliavacca R., Giani T., Gona F., Nucleo E., et al. (2012). Epidemiological characterization and distribution of carbapenem-resistant Acinetobacter baumannii clinical isolates in Italy. Clin. Microbiol. Infect. 18 160–166. 10.1111/j.1469-0691.2011.03527.x [DOI] [PubMed] [Google Scholar]
- Ocampo-Sosa A. A., Cabot G., Rodriguez C., Roman E., Tubau F., Macia M. D., et al. (2012). Alterations of OprD in carbapenem-intermediate and -susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a Spanish multicenter study. Antimicrob. Agents Chemother. 56 1703–1713. 10.1128/AAC.05451-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki A., Avison M. B. (2008). Induction of L1 and L2 beta-lactamase production in Stenotrophomonas maltophilia is dependent on an AmpR-type regulator. Antimicrob. Agents Chemother. 52 1525–1528. 10.1128/AAC.01485-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel A., Marchandin H., Prère M. F., Boutet-Dubois A., Brieu-Roche N., Gaschet A.et al. (2015). Faecal carriage of carbapenemase-producing Gram-negative bacilli in hospital settings in southern France. Eur. J. Clin. Microbiol. Infect. Dis. 34 899–904. 10.1007/s10096-014-2298-1 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez F. J., Hanson N. D. (2002). Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40 2153–2162. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli M., Sabatini A., Pontieri E., Celenza G., Segatore B., Bottoni C., et al. (2015). OXA-23 carbapenemase in multidrug-resistant Acinetobacter baumannii ST2 type: first identification in L’Aquila Hospital (Italy). Microb. Drug Resist. 21 97–101. 10.1089/mdr.2014.0056 [DOI] [PubMed] [Google Scholar]
- Pfeifer Y., Wilharm G., Zander E., Wichelhaus T. A., Göttig S., Hunfeld K.-P., et al. (2011). Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66 1998–2001. 10.1093/jac/dkr256 [DOI] [PubMed] [Google Scholar]
- Piana A., Palmieri A., Deidda S., Mura E., Are B. M., Rubino S., et al. (2015). Molecular typing of XDR Acinetobacter baumannii strains in an Italian ICU. Epidemiol. Prev. 39 129–133. [PubMed] [Google Scholar]
- Poirel L., Berçot B., Millemann Y., Bonnin R. A., Pannaux G., Nordmann P. (2012). Carbapenemase-producing Acinetobacter spp. in Cattle, France. Emerg. Infect. Dis. 18 523–532. 10.3201/eid1803.111330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Figueiredo S., Cattoir V., Carattoli A., Nordmann P. (2008). Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52 1252–1256. 10.1128/AAC.01304-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomba C., Endimiani A., Rossano A., Saial D., Couto N., Perreten V. (2014). First report of OXA-23-mediated carbapenem resistance in sequence type 2 multidrug-resistant Acinetobacter baumannii associated with urinary tract infection in a cat. Antimicrob. Agents Chemother. 58 1267–1268. 10.1128/AAC.02527-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principe L., Piazza A., Giani T., Bracco S., Caltagirone M. S., Arena F., et al. (2014). Epidemic diffusion of OXA-23-producing Acinetobacter baumannii isolates in Italy: results of the first cross-sectional countrywide survey. J. Clin. Microbiol. 52 3004–3010. 10.1128/JCM.00291-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulss S., Stolle I., Stamm I., Leidner U., Heydel C., Semmler T., et al. (2018). Multispecies and clonal dissemination of OXA-48 carbapenemase in Enterobacteriaceae from companion animals in Germany, 2009—2016. Front. Microbiol. 9:1265. 10.3389/fmicb.2018.01265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonis G., Karageorgopoulos D. E., Maraki S., Levis P., Dimopoulou D., Spernovasilis N. A., et al. (2012). Stenotrophomonas maltophilia infections in a general hospital: patient characteristics, antimicrobial susceptibility, and treatment outcome. PLoS One 7:e37375. 10.1371/journal.pone.0037375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch A.C., Arredondo-Alonso S., Willems R. J. L., Goering R. V. (2018). Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin. Microbiol. Infect. 24 350–354. 10.1016/j.cmi.2017.12.016 [DOI] [PubMed] [Google Scholar]
- Shaheen B. W., Nayak R., Boothe D. M. (2013). Emergence of a New Delhi metallo-β-lactamase (NDM-1)-encoding gene in clinical Escherichia coli isolates recovered from companion animals in the United States. Antimicrob. Agents Chemother. 57 2902–2903. 10.1128/AAC.02028-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siempos I. I., Vardakas K. Z., Kyriakopoulos C. E., Ntaidou T. K., Falagas M. E. (2010). Predictors of mortality in adult patients with ventilator-associated pneumonia: a meta-analysis. Shock 33 590–601. 10.1097/SHK.0b013e3181cc0418 [DOI] [PubMed] [Google Scholar]
- Smet A., Boyen F., Pasmans F., Butaye P., Martens A., Nemec A., et al. (2012). OXA-23-producing Acinetobacter species from horses: a public health hazard? J. Antimicrob. Chemother. 67 3009–3010. 10.1093/jac/dks311 [DOI] [PubMed] [Google Scholar]
- Stolle I., Prenger-Berninghoff E., Stamm I., Scheufen S., Hassdenteufel E., Guenther S., et al. (2013). Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J. Antimicrob. Chemother. 68 2802–2808. 10.1093/jac/dkt259 [DOI] [PubMed] [Google Scholar]
- Tan C. H., Lee K. W. K., Burmølle M., Kjelleberg S., Rice S. A. (2017). All together now: experimental multispecies biofilm model systems. Environ. Microbiol. 19 42–53. 10.1111/1462-2920.13594 [DOI] [PubMed] [Google Scholar]
- Toleman M. A., Bennett P. M., Bennett D. M., Jones R. N., Walsh T. R. (2007). Global Emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg. Infect. Dis. 13 559–565. 10.3201/eid1304.061378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman M. A., Spencer J., Jones L., Walsh T. R. (2012). BlaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56 2773–2776. 10.1128/AAC.06297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton J. F., Ward M. E., Woodford N., Kaufmann M. E., Pike R., Livermore D. M., et al. (2006). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258 72–77. 10.1111/j.1574-6968.2006.00195.x [DOI] [PubMed] [Google Scholar]
- Valdezate S., Vindel A., Martín-Dávila P., Del Saz B. S., Baquero F., Cantón R. (2004). High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital. J. Clin. Microbiol. 42 693–699. 10.1128/JCM.42.2.693-699.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fu Y., Lan P., Xu Q., Jiang Y., Chen Y., et al. (2018). Molecular epidemiology and mechanism of sulbactam resistance in Acinetobacter baumannii isolates with diverse genetic backgrounds in China. Antimicrob. Agents Chemother. 62:e1947-17. 10.1128/AAC.01947-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Wu D., Lei L., Shen Z., Wang Y., Liao K. (2016). The detection of fosfomycin resistance genes in Enterobacteriaceae from pets and their owners. Vet. Microbiol. 193 67–71. 10.1016/j.vetmic.2016.07.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.