Abstract
The twin-arginine translocation (Tat) system is a specialized secretion pathway required for bacteria to export fully folded proteins through the cytoplasmic membrane. This system is crucial during Salmonella infection of animal hosts. In this study, we show that Salmonella enterica serovar Typhimurium (S. Typhimurium) requires the Tat system to survive and proliferate intracellularly in the social amoeba Dictyostelium discoideum. To achieve this, we developed a new infection assay to assess intracellular bacterial loads in amoeba by direct enumeration of colony forming units (CFU) at different times of infection. Using this assay we observed that a ΔtatABC mutant was internalized in higher numbers than the wild type, and was defective for intracellular survival in the amoeba at all times post infection evaluated. In addition, we assessed the effect of the ΔtatABC mutant in the social development of D. discoideum. In contrast to the wild-type strain, we observed that the mutant was unable to delay the social development of the amoeba at 2 days of co-incubation. This phenotype correlated with defects in intracellular proliferation presented by the ΔtatABC mutant in D. discoideum after 24 h of infection. All phenotypes described for the mutant were reverted by the presence of a plasmid carrying tatABC genes, indicating that abrogation of Tat system attenuates S. Typhimurium in this model organism. Overall, our results indicate that the Tat system is crucial for S. Typhimurium to survive and proliferate intracellularly in D. discoideum and for virulence in this host. To the best of our knowledge, this is the first report on the relevance of the Tat system in the interaction of any bacterial pathogen with the social amoeba D. discoideum.
Keywords: Salmonella, Dictyostelium, Tat system, infection, intracellular survival, social development, virulence
Introduction
The genus Salmonella includes species S. enterica and S. bongori, which can be differentiated into more than 2,500 serovars according to variations of surface antigens. Some serovars within S. enterica, such as Salmonella enterica serovar Typhimurium (S. Typhimurium), are named generalist as they can infect a wide range of hosts, causing illnesses ranging from gastroenteritis to severe systemic disease (Tsolis et al., 1999; Zhang et al., 2003). These pathogens are highly versatile and can adapt to a variety of conditions both in the natural environment and within a host.
Internalization and intracellular survival in host cells are essential processes for Salmonella virulence that depend on the translocation of bacterial effector proteins to eukaryotic cells through the type 3 secretion systems (T3SS) encoded in SPI-1 and SPI-2 (T3SSSPI-1 and T3SSSPI-2, respectively) (Haraga et al., 2008). In addition, it has been reported that other secretion systems, such as the twin-arginine translocation (Tat) system, are required for Salmonella infection (Mickael et al., 2010; Reynolds et al., 2011; Craig et al., 2013).
The Tat system is found in most bacteria and has the unusual property of transporting fully folded proteins from the cytoplasm to the periplasmic space (Palmer and Berks, 2012). In Escherichia coli and Salmonella, the main components of this system are proteins TatA, TatB, and TatC. TatB and TatC form an integral membrane complex that recognizes substrates containing an amino-terminal signal peptide that includes the twin-arginine consensus motif S/T-R-R-x-F-L-K. Once the substrate has been recognized, the TatBC complex induces the polymerization of TatA to form the pore that allows the passage of the fully folded protein through the cytoplasmic membrane (Palmer and Berks, 2012).
It has been described that inactivation of genes encoding Tat components in several bacteria including E. coli, Legionella pneumophila, Yersinia pseudotuberculosis, Mycobacterium smegmatis, Vibrio cholerae, Pseudomonas aeruginosa and S. Typhimurium results in defects in several bacterial properties and processes, including cell morphology, growth and biofilm formation, cell wall integrity, transport of virulence factors, cell division, motility and chemotaxis (Bogsch et al., 1998; Santini et al., 1998; Voulhoux et al., 2001; Ochsner et al., 2002; Ding and Christie, 2003; Pradel et al., 2003; Harrison et al., 2005; McDonough et al., 2005; Rossier and Cianciotto, 2005; Lavander et al., 2006; Posey et al., 2006; Zhang et al., 2009; Reynolds et al., 2011). Regarding the role of the Tat system in Salmonella virulence, it has been reported that a ΔtatC mutant of S. Typhimurium presents colonization defects in mice (Santiviago et al., 2009; Reynolds et al., 2011; Craig et al., 2013) and reduced intracellular replication in J774-A1 murine macrophages (Reynolds et al., 2011). In addition, inactivation of genes tatB and tatC in Salmonella Enteritidis results in impaired Caco-2 cell invasion and reduced colonization in mice and chickens (Mickael et al., 2010; Silva et al., 2012).
During its life cycle outside animal hosts, Salmonella interacts with a wide variety of predatory eukaryotic organisms in the environment, including amoeba. Amoeba are organisms that feed on fungi and bacteria by phagocytosis (Salah et al., 2009; Denoncourt et al., 2014; Hoffmann et al., 2014). Several bacterial pathogens have been described to survive predation by these organisms, including L. pneumophila, Mycobacterium marinum, Bordetella bronchiseptica, P. aeruginosa and V. cholerae, among others (Abd et al., 2005, 2007; Cardenal-Muñoz et al., 2017; Strassmann and Shu, 2017; Taylor-Mulneix et al., 2017; Swart et al., 2018). Recently, we and other researchers have reported that S. Typhimurium can survive within the social amoeba Dictyostelium discoideum (Sillo et al., 2011; Riquelme et al., 2016; Varas et al., 2018) and requires T3SSSPI-1 and T3SSSPI-2 for this process, among other virulence factors (Riquelme et al., 2016).
In this work, we assessed the contribution of the Tat system to the interaction of S. Typhimurium with D. discoideum. To this end, we developed a CFU-based infection assay using axenic D. discoideum to evaluate the intracellular survival of wild-type and ΔtatABC strains in the amoeba. Our results showed that the ΔtatABC mutant presents intracellular survival and proliferation defects during infection in this host. In addition, we observed that the ΔtatABC mutant was unable to delay the social development of the amoeba caused by the wild-type strain. Altogether, our results indicate that the Tat system plays a major role during the interaction of S. Typhimurium with D. discoideum.
Materials and Methods
Bacterial Strains, Media and Culture Conditions
Bacterial strains used in this study are described in Table 1. All S. Typhimurium strains are derivatives of the wild-type, virulent strain 14028s (Fields et al., 1986; Jarvik et al., 2010). Bacteria were routinely grown statically in Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L NaCl and 5 g/L yeast extract) at 37°C. When required, the medium was supplemented with ampicillin (100 mg/L), chloramphenicol (20 mg/L) or kanamycin (75 mg/L). Media were solidified by the addition of agar (15 g/L). All procedures involving the use of pathogenic organisms were conducted following the guidelines in the Biosafety Manual (2018 version) of the National Commission of Scientific and Technological Research (CONICYT, Chile), and were approved by the Biosafety Committee of Universidad de Chile, Campus Norte.
Table 1.
Bacteria and Dictyostelium strains used in this study.
| Strain | Characteristic or genotype | Source or reference |
|---|---|---|
| Salmonella Typhimurium | ||
| WT | Wild-type, virulent strain 14028s | Laboratory collection (Fields et al., 1986; Jarvik et al., 2010) |
| ΔtatABC | ΔtatABC::Kan | This study |
| ΔtatABC/pTAT | ΔtatABC::Kan transformed with plasmid pBAD-TOPO::tatABC | This study |
| ΔtatABC/pBAD | ΔtatABC::Kan transformed with plasmid pBAD-TOPO | This study |
| ΔaroA | ΔaroA::Kan | Laboratory collection (Varas et al., 2017) |
| ΔphoN | ΔphoN::Cam | Laboratory collection (Riquelme et al., 2016) |
| Escherichia coli | ||
| B/r (DBS0348878) | Wild-type strain | Dicty Stock Center (dictyBase) |
| Klebsiella aerogenes | ||
| DBS0305928 | Wild-type strain | Dicty Stock Center (dictyBase) |
| Dictyostelium discoideum | ||
| AX4 (DBS0302402) | axeA1 axeB1 axeC1 | Dicty Stock Center (dictyBase) |
Dictyostelium Strains, Media and Culture Conditions
D. discoideum strain AX4 (DBS0302402) (Table 1) was obtained from Dicty Stock Center (Basu et al., 2013; Fey et al., 2013), and cultured according to standard protocols (Fey et al., 2007). Briefly, amoeba were maintained at 22°C in SM agar (10 g/L glucose, 10 g/L peptone, 1 g/L yeast extract, 1 g/L MgSO4 × 7H2O, 1.9 g/L KH2PO4, 0.6 g/L K2HPO4, 20 g/L agar), growing on a confluent lawn of Klebsiella aerogenes (DBS0305928). For infection assays, amoeba were grown axenically at 22°C in HL5 medium (14 g/L tryptone, 7 g/L yeast extract, 0.35 g/L Na2HPO4, 1.2 g/L KH2PO4, 14 g/L glucose, pH 6.3) with agitation at 180 rpm. When required, HL5 medium was supplemented with a mix of streptomycin (300 mg/L) and ampicillin (100 mg/L). Prior to infection, amoeba were harvested at the early exponential phase (1–2 × 106 cells/mL) and centrifuged at 300 ×g for 5 min at 4°C. The supernatant was discarded and the pellet was washed three times using Soerensen buffer (2 g/L KH2PO4, 0.36 g/L Na2HPO4 × 2H2O, pH 6.0). Trypan blue exclusion and counting in a Neubauer chamber was used to determine the population of viable cells.
Construction of Mutant Strains
A S. Typhimurium ΔtatABC mutant strain was constructed using the Lambda Red recombination method (Datsenko and Wanner, 2000) with modifications (Santiviago et al., 2009), using primers tatABC_(H1+P1) and tatABC_(H2+P2), and plasmid pCLF4 (KanR, GenBank accession number EU629214) as template for PCR amplification. The correct allelic replacement in this mutant was confirmed by PCR amplification using primers tatABC_Out5 and tatABC_Out3, flanking the substitution site. All primers for PCR amplifications are listed in Table 2.
Table 2.
Primers used in this study.
| Primer name | Sequence |
|---|---|
| tatABC_(H1+P1) | AGGAACATGTATGGGTGGTATCAGTATTTGGCAGTTGTTGGTGCAGGCTGGAGCTGCTTC |
| tatABC_(H2+P2) | GCGGTTGTGTTTAGTCTTCAGTGTGCTCGGCCTTTTCGGTCATATGAATATCCTCCTTAG |
| tatABC_Out5 | GAGCGGGTCATTCTTACTCG |
| tatABC_Out3 | TTCGTTCCGGTCAGTAGCAT |
| pBAD_Forward | ATGCCATAGCATTTTTATCC |
| pBAD_Reverse | GATTTAATCTGTATCAGG |
Underlined sequences anneal to the 5′ or 3′ end of the antibiotic-resistance cassette in template vector pCLF4 (KanR, GenBank accession number EU629214).
Construction of Complementing Plasmid pTAT
A DNA fragment containing genes tatABC (including the promoter region) was amplified from the genome of S. Typhimurium strain 14028s using Taq DNA polymerase (Invitrogen) and primers tatABC_Out5 and tatABC_Out3 (Table 2). The PCR product was purified from 1% agarose gels using the “QIAquick Gel Extraction Kit” (QIAGEN) and cloned into pBAD-TOPO using the “pBAD-TOPO TA Expression Kit” (Invitrogen). The presence and orientation of the insert in the recombinant plasmid generated (pTAT) was confirmed by PCR amplification using combinations of primers tatABC_Out5, tatABC_Out3, pBAD_Forward and pBAD_Reverse (Table 2). The S. Typhimurium ΔtatABC mutant was transformed by electroporation with plasmids pTAT or pBAD-TOPO for complementation assays.
Individual Infection Assay
An infection assay was developed (Supplementary Figure S1) based on a method previously described by our group (Riquelme et al., 2016). Each bacterial strain to be evaluated was grown overnight, suspended in LB medium adjusting the OD600nm to 0.2, harvested and washed two times with Soerensen buffer. Next, 150 μL of each bacterial suspension was further diluted by adding 850 μL of Soerensen buffer. In parallel, D. discoideum AX4 cells from axenic cultures grown to exponential phase were washed two times with Soerensen buffer, and titrated by Trypan blue exclusion and counting on a Neubauer chamber. Next, a suspension containing ∼1 × 106 amoeba/mL was prepared using Soerensen buffer. Aliquots of 100 μL from bacteria and amoeba suspensions were mixed in an Eppendorf tube in order to obtain a multiplicity of infection (MOI) of ∼100 bacteria/amoeba. The mixture was centrifuged at 9,200 ×g for 20 s (to promote the interaction of bacteria with amoeba), and incubated at 22°C for 1 h. After the incubation, all mixes were centrifuged at 300 ×g for 5 min, washed once with Soerensen buffer supplemented with gentamicin (10 mg/L) to kill extracellular bacteria, then centrifuged and washed twice with Soerensen buffer to remove the antibiotic, and finally centrifuged and suspended in Soerensen buffer (final volume: 100 μL). The content of two tubes carrying a suspension of infected amoeba after 0, 1, 3, 5, 10 or 24 h post infection was analyzed in parallel to determine viable amoeba and intracellular bacteria. Viable amoeba in the first tube were determined at each time point by Trypan blue exclusion and counting on a Neubauer chamber. In addition, infected amoeba recovered from the second tube were washed once with Soerensen buffer and finally lysed with 0.2% Triton X-100. Titers of intracellular bacteria were determined by serial dilutions and plating on LB agar.
Bacterial Adherence Assay
A procedure similar to our infection assay was implemented to evaluate adherence of different bacterial strains to D. discoideum AX4 cells. Briefly, suspensions of bacteria and amoeba were prepared as indicated above and mixed at a MOI of ∼100 bacteria/amoeba in an Eppendorf tube. Next, each mixture was centrifuged (9,200 ×g for 20 s) to promote the interaction of bacteria with amoeba, and further incubated at 4°C for 30 min. After the incubation, all mixtures were centrifuged (300 ×g for 5 min) at 4°C and the infected amoeba were washed 3 sequential times with Soerensen buffer at 4°C to remove non-adhered bacteria. Finally, amoeba were lysed with 0.2% Triton X-100 and titers of adhered bacteria were determined by serial dilutions and plating on LB agar.
Bacterial Toxicity Assay
A modified version of the virulence assay reported by Champion and colleagues (Champion et al., 2016) was implemented to indirectly evaluate bacterial cytotoxicity toward D. discoideum evidenced by changes in cell morphology of amoeba co-incubated with different S. Typhimurium strains and E. coli B/r. Briefly, bacteria and amoeba were cultured as indicated in the infection assay procedure. Aliquots (100 μL) of a suspension containing ∼1 × 106 amoeba/mL prepared in HL5 medium were added to the wells of a 96-well plate and incubated overnight at 22°C to allow adhesion of amoeba to the bottom of the wells. The next day, the HL5 medium in wells containing adhered amoeba was replaced with aliquots (100 μL) of bacterial suspensions prepared in Soerensen buffer in order to obtain MOIs of 100 or 1,000 bacteria/cell. The plates were incubated at 22°C and the cell shape of infected amoeba in each well was monitored at different co-incubation times (0, 30, 60, 90, 120, 150, and 180 min) using a Motic AE 2000 inverted microscope equipped with a Plan Achromatic Phase LWD PL Ph2 40× objective. Representative images were acquired using a Moticam 580 (5.0 MP) digital camera attached to the trinocular port of the microscope using a C-mount adapter (0.5×).
Social Development Assay
Individual wells of a 24-well plate containing N agar (Soerensen buffer supplemented with 1 g/L peptone, 1 g/L glucose and 20 g/L agar) were inoculated with 30 μL of an overnight culture from each bacterial strain to be evaluated. The plate was incubated overnight at 22°C to obtain bacterial lawns. The next day, 10 μL of a suspension containing ∼1 × 104 axenic D. discoideum AX4 cells in HL5 was spotted in the center of each well on top of the corresponding bacterial lawn and the plate was further incubated at 22°C for 2 days. Representative images of D. discoideum development were obtained at days 1 and 2 using a Motic SMZ-171 stereomicroscope equipped with a Moticam 580 (5.0 MP) digital camera attached to the trinocular port of the stereomicroscope using a C-mount adapter (0.5×).
Results and Discussion
New Infection Assay to Evaluate Internalization and Intracellular Survival in D. discoideum
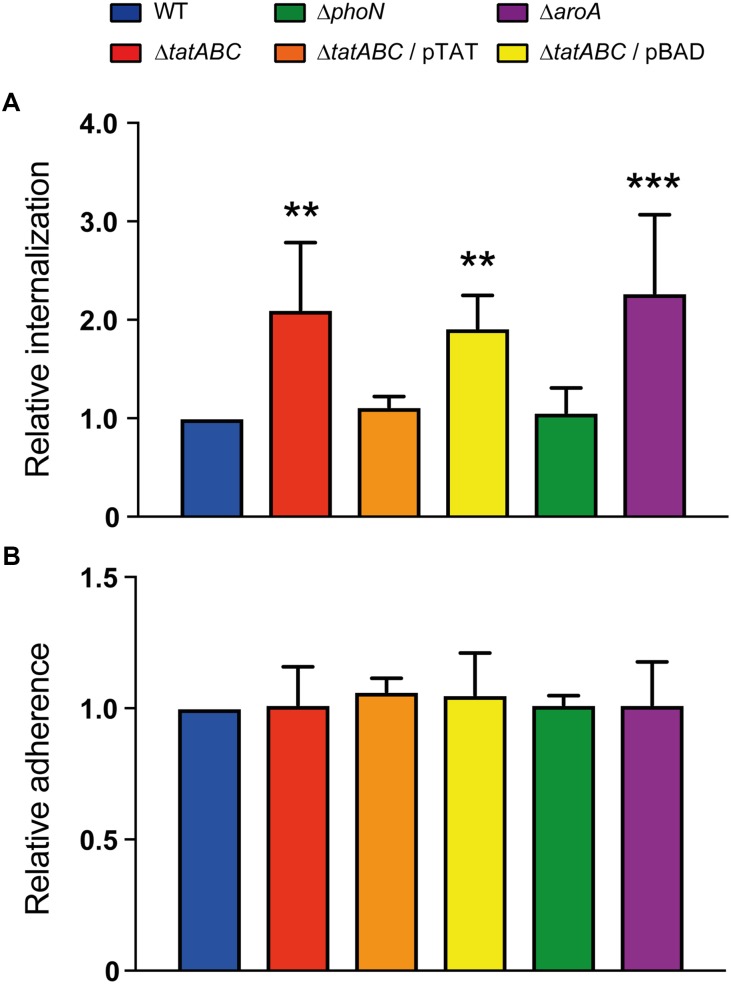
To evaluate the role played by the Tat system in the survival of S. Typhimurium in D. discoideum, we developed a new CFU-based infection assay where amoeba were infected with the wild type and mutant strains, and intracellular bacteria were recovered from infected amoeba and titrated at different times post infection to evaluate internalization and intracellular survival (Supplementary Figure S1). The main advantage of this method is that it uses smaller volumes, and therefore fewer amoeba cells, than assays previously reported by our group (Riquelme et al., 2016; Varas et al., 2018). This simplifies the handling of samples, allowing the analysis of more strains per experiment than assays requiring larger volumes. Although this method is based on a described assay (Riquelme et al., 2016), we used our new infection assay to analyze the wild-type strain and included ΔaroA and ΔphoN mutants as controls in order to validate our observations. The internalization of each strain was evaluated after 1 h of infection. We observed that the ΔaroA mutant was internalized in higher numbers than the wild type, and the ΔphoN mutant was internalized at wild-type levels (Figure 1A). In addition, the ΔaroA mutant presented defects in intracellular survival in the amoeba while the ΔphoN mutant presented wild-type levels of intracellular survival (Figure 3A). These observations are consistent with results from different infection assays showing that ΔaroA and ΔphoN mutants are attenuated and not attenuated in this model organism, respectively (Riquelme et al., 2016; Varas et al., 2018).
FIGURE 1.
Adherence and internalization of S. Typhimurium strains in D. discoideum. Individual infection assays were conducted to evaluate the adherence and internalization of different S. Typhimurium strains in D. discoideum AX4. (A) Relative internalization after 1 h of infection at 22°C. (B) Relative adherence after 30 min of co-incubation at 4°C. All values (internalization and adherence) were calculated as CFUt =0/CFUinoculum and further normalized to the value of the wild-type strain. Graphs show mean values ± SD from at least 7 independent assays. Statistical significance of differences in internalization or adherence between each strain and the wild type was determined using a one-way ANOVA with Dunnett’s test (∗∗P < 0.01; ∗∗∗P < 0.001).
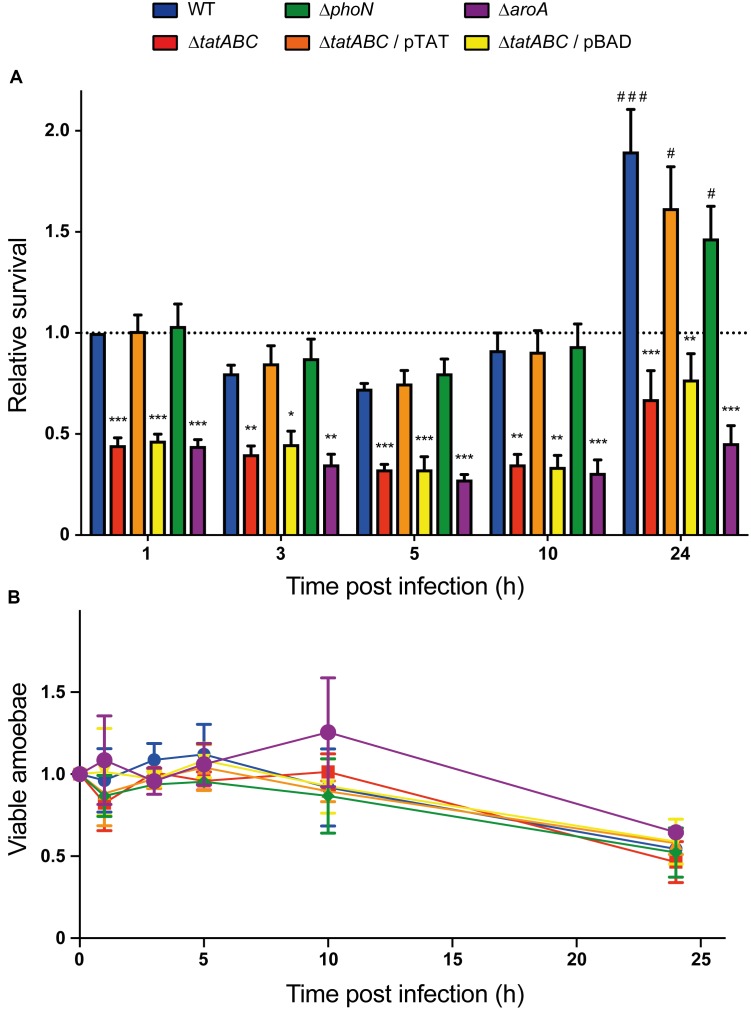
FIGURE 3.
Intracellular survival of S. Typhimurium strains in D. discoideum. Individual infection assays were conducted to evaluate the intracellular survival of different S. Typhimurium strains in D. discoideum AX4. (A) Relative intracellular survival at different times post infection. All values were calculated as CFUt =x/CFUt =0 and further normalized to the value of the wild-type strain at t = 1. (B) Variation in the population of viable amoeba during the infection assay. All values were calculated as cells/mL and further normalized to the value of amoeba infected with the wild-type strain at t = 0. Graphs show mean values ± SD from at least 7 independent assays. Statistical significance of differences in intracellular survival between each strain and the wild type at a given time was determined using a one-way ANOVA with Dunnett’s test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Statistical significance of differences in intracellular survival of a given strain at t = 1 versus t = 24 was determined using a one-way ANOVA with Dunnett’s test (#P < 0.05; ###P < 0.001).
A S. Typhimurium ΔtatABC Mutant Is Defective for Intracellular Survival in D. discoideum
Once validated, we used our infection assay to evaluate the internalization and intracellular survival of a ΔtatABC mutant in D. discoideum. We observed that this mutant was internalized at higher levels than the wild-type strain, as in the case of the ΔaroA mutant (Figure 1A). To gain further insight into the causes of the observed phenotype, we conducted an assay to evaluate the adherence of our strains to D. discoideum cells. We observed that all strains presented identical adherence levels (Figure 1B), indicating that the elevated internalization levels presented by ΔtatABC (and other mutants under study) are not attributable to increased adherence to amoeba cells resulting in higher uptake.
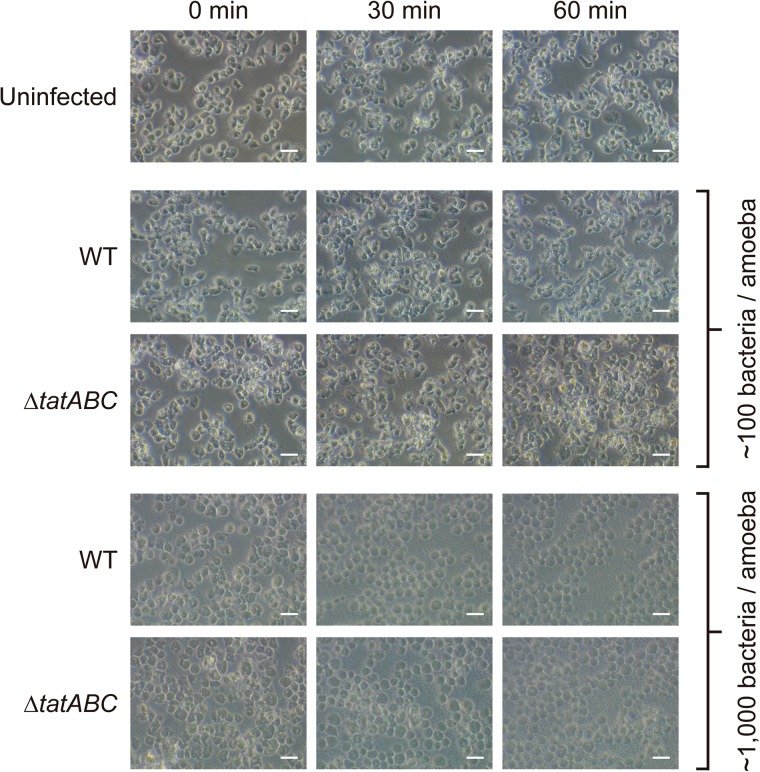
Furthermore, we conducted a modified version of an assay originally developed to evaluate virulence of Pseudomonas aeruginosa strains during co-incubation with D. discoideum in buffer (Champion et al., 2016). The authors of the method reported that virulent strains of P. aeruginosa caused cell rounding and cytoplasmic shrinkage of amoeba, that were interpreted as a cytotoxic effect exerted by these strains (Champion et al., 2016). In our case, co-incubations of amoeba with each bacterial strain at a MOI of ∼100 bacteria/amoeba for up to 180 min produced no changes in D. discoideum cell shape (Supplementary Figure S2). Figure 2 shows representative results of amoeba co-incubated for up to 60 min with the wild-type strain or the ΔtatABC mutant. These results suggest that all bacterial strains evaluated produced no cytotoxic effects to amoeba cells under the experimental conditions evaluated. In contrast, during co-incubations conducted at a MOI of ∼1,000 bacteria/amoeba all strains caused detachment of amoeba from the wells and a rounded cell morphology at all times evaluated (Figure 2 and Supplementary Figure S2), suggesting the existence of a cytotoxic effect toward D. discoideum under these experimental conditions. Therefore, most probably the differences in internalization levels presented by each strain under study are not caused by differential cytotoxic effects exerted by these bacteria on amoeba cells under the conditions routinely used in our infection assay (MOI of ∼100 bacteria/amoeba).
FIGURE 2.
Qualitative evaluation of cytotoxicity caused by S. Typhimurium strains on D. discoideum. The shape of D. discoideum AX4 cells infected with different strains of S. Typhimurium was monitored at 0, 30, and 60 min of co-incubation at 22°C in Soerensen buffer. Representative images from 3 independent assays carried out at MOIs of ∼100 or ∼1,000 bacteria/amoeba are shown. Scale bar, 20 μm.
In addition to the internalization phenotype revealed by our infection assay, we observed that the ΔtatABC mutant was impaired for intracellular survival in D. discoideum at all times evaluated, as in the case of the ΔaroA mutant (Figure 3A). No effect in amoeba viability was observed during the course of the infections (Figure 3B), indicating that the phenotypes shown by the different strains are not attributable to changes in the number of viable amoeba all through the assay. Of note, all phenotypes presented by the ΔtatABC mutant were reverted by the presence of a plasmid carrying genes tatABC (pTAT), and the presence of the empty vector (pBAD) did not affect the phenotypes shown by this mutant (Figures 1, 3). During the course of the infection assay we noticed that all strains slightly decreased their intracellular titers during the first 5 h post infection. We also observed intracellular proliferation in the case of the wild-type strain at 24 h post infection. The proliferation level shown by the wild-type strain at this time point was comparable to that shown by strains ΔphoN and ΔtatABC harboring plasmid pTAT, roughly reaching twice the amount of intracellular bacteria present at 1 h post infection. In contrast, in the case of strains ΔtatABC, ΔtatABC harboring plasmid pBAD and ΔaroA the amount of intracellular bacteria at 24 h post infection was comparable to the levels shown by each strain at 1 h post infection (Figure 3A).
Taken together, our results indicate that S. Typhimurium requires the Tat system to survive and proliferate in D. discoideum. This is the first report on the relevance of the Tat system in Salmonella survival within this amoeba. These observations are consistent with the role reported for Tat in the intracellular survival of S. Typhimurium in J774-A1 murine macrophages (Reynolds et al., 2011). Regarding the role played by Tat in the survival of other bacterial pathogens in protozoa, it has been reported that inactivation of genes encoding components of this system in L. pneumophila results in intracellular survival defects in Acanthamoeba castellanii and Hartmannella vermiformis (De Buck et al., 2005; Rossier and Cianciotto, 2005). These studies also revealed that L. pneumophila requires Tat for intracellular survival in U937 human monocytes differentiated into macrophage-like cells (De Buck et al., 2005; Rossier and Cianciotto, 2005).
A S. Typhimurium ΔtatABC Mutant Is Unable to Delay the Social Development of D. discoideum
It has been reported that virulent pathogenic bacteria delay the social development of D. discoideum, while attenuated or non-pathogenic bacteria allow its rapid progression (Bravo-Toncio et al., 2016; Ouertatani-Sakouhi et al., 2017; Marcoleta et al., 2018). In fact, wild-type S. Typhimurium causes a significant delay in the development of this amoeba (Sillo et al., 2011; Varas et al., 2018) and requires a fully functional T3SSSPI-2 for this process (Sillo et al., 2011). Thus, to evaluate the role played by the Tat system in the virulence of S. Typhimurium in D. discoideum, we compared the effect of feeding the amoeba with the wild-type strain or its ΔtatABC derivative during the development cycle, which mainly involves three sequential stages: aggregation, elevation and culmination (Figure 4A). E. coli B/r, routinely used to feed the amoeba during growth in the laboratory (Fey et al., 2007), and the attenuated mutant ΔaroA were included as controls in our assay.
FIGURE 4.
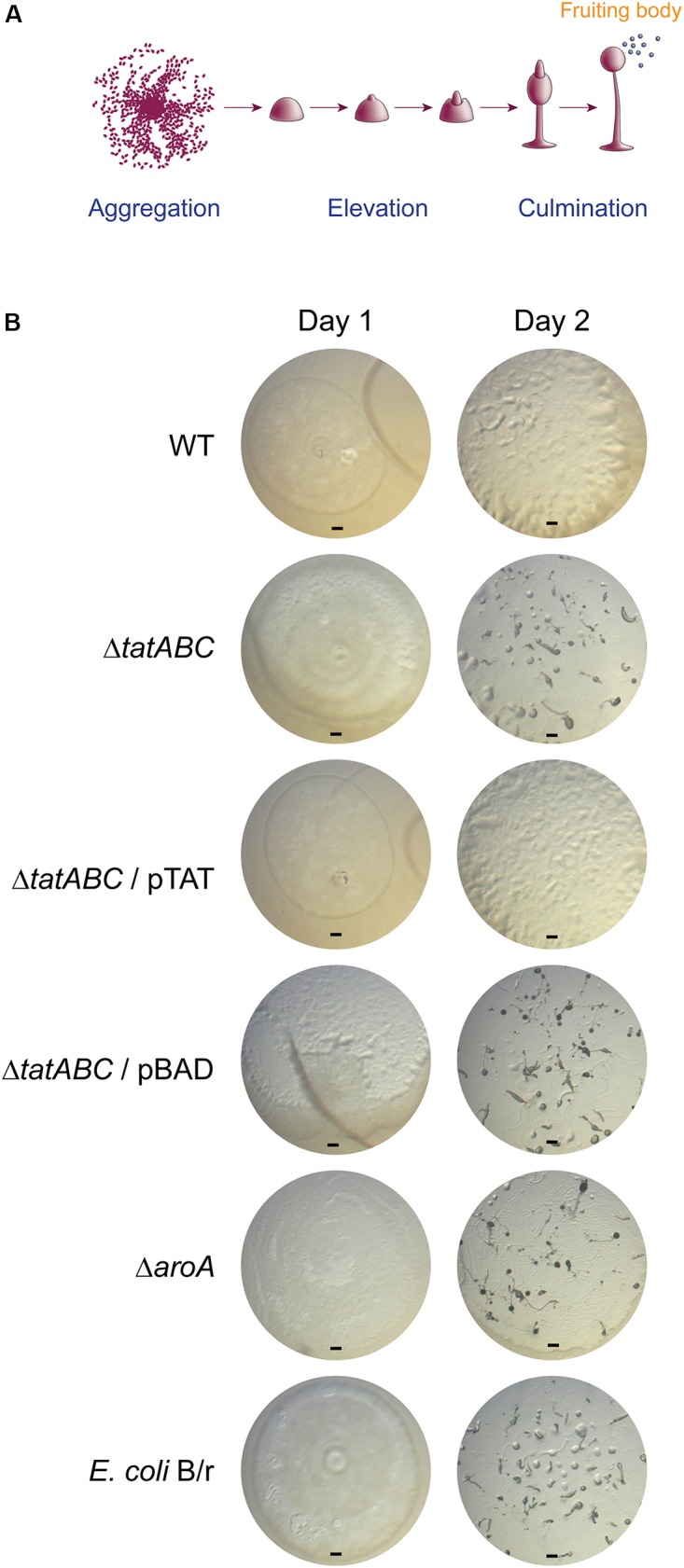
Social development of D. discoideum co-incubated with S. Typhimurium 14028s derivatives and E. coli B/r. (A) Main phases of the D. discoideum development cycle, including aggregation, elevation and culmination to generate fruiting bodies (scheme adapted from Fey et al., 2007). (B) D. discoideum development after 1 and 2 days of co-incubation with S. Typhimurium strains or E. coli B/r. Representative images from 3 independent assays are shown. Scale bar, 100 μm.
As reported (Varas et al., 2018), the wild-type strain caused a delay in the social development of the amoeba where only the aggregation phase was reached after 2 days of co-incubation (Figure 4B). In contrast, the ΔtatABC mutant allowed the development of the amoeba until reaching the elevation phase (and in some cases the culmination phase) after 2 days of co-incubation. The same phenotype was observed in the case of the ΔaroA mutant and E. coli B/r. Noteworthy, the ΔtatABC mutant harboring plasmid pTAT caused a delayed social development of the amoeba similar to that caused by the wild-type strain, and the presence of the empty vector did not affect the phenotype shown by the ΔtatABC mutant (Figure 4B). It is important to mention that the ability of each bacterial strain to delay the social development of the amoeba correlated with its ability to proliferate intracellularly in this organism after 24 h of infection (compare Figures 3A, 4B).
Given that monitoring the progress of D. discoideum social development is used to evaluate the virulence of pathogenic bacteria (Sillo et al., 2011; Bravo-Toncio et al., 2016; Ouertatani-Sakouhi et al., 2017; Marcoleta et al., 2018; Varas et al., 2018), our results indicate that abrogation of Tat system attenuates S. Typhimurium in this model organism. Therefore, the Tat system contributes to S. Typhimurium virulence in D. discoideum. This is consistent with the attenuation showed by a ΔtatC mutant of S. Typhimurium in mice and murine macrophages (Santiviago et al., 2009; Reynolds et al., 2011; Craig et al., 2013), and by tatB and tatC mutants of S. Enteritidis in mice, chickens and Caco-2 epithelial cells (Mickael et al., 2010; Silva et al., 2012).
According to our results, the attenuation showed by the ΔtatABC mutant in D. discoideum (as revealed by its inability to delay the social development of the amoeba) cannot be explained by a diminished toxic effect exerted by this strain (for instance, caused by defective secretion of a particular exotoxin via the Tat system), as no differences in cytotoxicity were detected when D. discoideum was co-incubated with the wild-type strain or the ΔtatABC mutant under the same experimental conditions (MOI and time of infection) (Figure 2 and Supplementary Figure S2). On the other hand, the attenuation phenotype can be explained by the inability of the ΔtatABC mutant to survive and proliferate intracellularly in this amoeba, as revealed by our infection assay (Figure 3A). In a recent study, the role played by Tat substrates in S. Typhimurium virulence was studied (Craig et al., 2013). To this end, the authors inactivated every gene encoding a Tat-exported protein and evaluated the virulence of each mutant strain in a mouse model of infection. Noteworthy, no single Tat-exported substrate accounted for the strong attenuation showed by a Tat-defective mutant (ΔtatC) in this model. However, this attenuation was attributed to failure translocating three Tat substrates: AmiA, AmiC, and SufI. AmiA and AmiC are N-acetylmuramoyl-L-alanine amidases, and SufI (also named FtsP) is involved in bacterial division. Therefore, the attenuation of a Tat-defective mutant of S. Typhimurium in the mouse model of infection was associated with failure to translocate Tat substrates linked to the cell division machinery, resulting in envelope defects (Craig et al., 2013). Most probably, the attenuation and intracellular survival and proliferation defects shown by the ΔtatABC mutant in D. discoideum can be explained by these envelope defects. Further studies are required to confirm this hypothesis.
Conclusion
Overall, our results indicate that the Tat system is essential for S. Typhimurium to survive and proliferate intracellularly in D. discoideum and for virulence in this host. To the best of our knowledge, this is the first report on the relevance of the Tat system in the interaction of any bacterial pathogen with the social amoeba D. discoideum.
Author Contributions
ÍU, AS, CV, and CS conceived and designed the experiments, and analyzed the data. ÍU, AS, BL, and CV performed the experiments. ÍU, CV, SÁ, and CS contributed reagents, materials, and analysis tools, and wrote the paper. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Macarena Varas for technical guidance during the implementation of the social development assay.
Footnotes
Funding. This work was supported by FONDECYT grants 1140754, 1171844 (to CS), and 1130225 (to SÁ). ÍU, AS, BL, and CV were supported by CONICYT fellowships 21150005, 22140758, 21180946, and 21140615, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03001/full#supplementary-material
References
- Abd H., Saeed A., Weintraub A., Nair G. B., Sandström G. (2007). Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol. Ecol. 60 33–39. 10.1111/j.1574-6941.2006.00254.x [DOI] [PubMed] [Google Scholar]
- Abd H., Weintraub A., Sandström G. (2005). Intracellular survival and replication of Vibrio cholerae O139 in aquatic free-living amoebae. Environ. Microbiol. 7 1003–1008. 10.1111/j.1462-2920.2005.00771.x [DOI] [PubMed] [Google Scholar]
- Basu S., Fey P., Pandit Y., Dodson R., Kibbe W. A., Chisholm R. L. (2013). DictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 41 D676–D683. 10.1093/nar/gks1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogsch E. G., Sargent F., Stanley N. R., Berks B. C., Robinson C., Palmer T. (1998). An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273 18003–18006. 10.1074/jbc.273.29.18003 [DOI] [PubMed] [Google Scholar]
- Bravo-Toncio C., Álvarez J. A., Campos F., Ortiz-Severín J., Varas M., Cabrera R., et al. (2016). Dictyostelium discoideum as a surrogate host-microbe model for antivirulence screening in Pseudomonas aeruginosa PAO1. Int. J. Antimicrob. Agents 47 403–409. 10.1016/j.ijantimicag.2016.02.005 [DOI] [PubMed] [Google Scholar]
- Cardenal-Muñoz E., Barisch C., Lefrancois L. H., López-Jiménez A. T., Soldati T. (2017). When Dicty met Myco, a (not so) romantic story about one amoeba and its intracellular pathogen. Front. Cell. Infect. Microbiol. 7:529. 10.3389/fcimb.2017.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion A. C., Houston N. K., Bradbury R. S., Reid D. W. (2016). Preliminary feasibility and modelling of a liquid matrix Dictyostelium discoideum virulence assay for Pseudomonas aeruginosa. Br. J. Biomed. Sci. 73 51–55. 10.1080/09674845.2016.1157249 [DOI] [PubMed] [Google Scholar]
- Craig M., Sadik A. Y., Golubeva Y. A., Tidhar A., Slauch J. M. (2013). Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol. Microbiol. 89 887–902. 10.1111/mmi.12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck E., Maes L., Meyen E., Van Mellaert L., Geukens N., Anné J., et al. (2005). Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. Biochem. Biophys. Res. Commun. 331 1413–1420. 10.1016/j.bbrc.2005.04.060 [DOI] [PubMed] [Google Scholar]
- Denoncourt A. M., Paquet V. E., Charette S. J. (2014). Potential role of bacteria packaging by protozoa in the persistence and transmission of pathogenic bacteria. Front. Microbiol. 5:240. 10.3389/fmicb.2014.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Christie P. J. (2003). Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 185 760–771. 10.1128/JB.185.3.760-771.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P., Dodson R. J., Basu S., Chisholm R. L. (2013). One stop shop for everything Dictyostelium: dictyBase and the Dicty Stock Center in 2012. Methods Mol. Biol. 983 59–92. 10.1007/978-1-62703-302-2_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P., Kowal A. S., Gaudet P., Pilcher K. E., Chisholm R. L. (2007). Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2 1307–1316. 10.1038/nprot.2007.178 [DOI] [PubMed] [Google Scholar]
- Fields P. I., Swanson R. V., Haidaris C. G., Heffron F. (1986). Mutants of Salmonella typhimurium that cannot survive within the macrophage are a virulent. Proc. Natl. Acad. Sci. U.S.A. 83 5189–5193. 10.1073/pnas.83.14.5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A., Ohlson M. B., Miller S. I. (2008). Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6 53–66. 10.1038/nrmicro1788 [DOI] [PubMed] [Google Scholar]
- Harrison J. J., Ceri H., Badry E. A., Roper N. J., Tomlin K. L., Turner R. J. (2005). Effects of the twin-arginine translocase on the structure and antimicrobial susceptibility of Escherichia coli biofilms. Can. J. Microbiol. 51 671–683. 10.1139/w05-048 [DOI] [PubMed] [Google Scholar]
- Hoffmann C., Harrison C. F., Hilbi H. (2014). The natural alternative: protozoa as cellular models for Legionella infection. Cell. Microbiol. 16 15–26. 10.1111/cmi.12235 [DOI] [PubMed] [Google Scholar]
- Jarvik T., Smillie C., Groisman E. A., Ochman H. (2010). Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J. Bacteriol. 192 560–567. 10.1128/JB.01233-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavander M., Ericsson S. K., Bröms J. E., Forsberg A. (2006). The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis. Infect. Immun. 74 1768–1776. 10.1128/IAI.74.3.1768-1776.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoleta A. E., Varas M. A., Ortiz-Severín J., Vásquez L., Berríos-Pastén C., Sabag A. V., et al. (2018). Evaluating different virulence traits of Klebsiella pneumoniae using Dictyostelium discoideum and zebrafish larvae as host models. Front. Cell. Infect. Microbiol. 8:30. 10.3389/fcimb.2018.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough J. A., Hacker K. E., Flores A. R., Pavelka M. S., Jr., Braunstein M. (2005). The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J. Bacteriol. 187 7667–7679. 10.1128/JB.187.22.7667-7679.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickael C. S., Lam P. K., Berberov E. M., Allan B., Potter A. A., Köster W. (2010). Salmonella enterica serovar Enteritidis tatB and tatC mutants are impaired in Caco-2 cell invasion in vitro and show reduced systemic spread in chickens. Infect. Immun. 78 3493–3505. 10.1128/IAI.00090-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A., Snyder A., Vasil A. I., Vasil M. L. (2002). Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99 8312–8317. 10.1073/pnas.082238299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouertatani-Sakouhi H., Kicka S., Chiriano G., Harrison C. F., Hilbi H., Scapozza L., et al. (2017). Inhibitors of Mycobacterium marinum virulence identified in a Dictyostelium discoideum host model. PLoS One 12:e0181121. 10.1371/journal.pone.0181121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer T., Berks B. C. (2012). The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10 483–496. 10.1038/nrmicro2814 [DOI] [PubMed] [Google Scholar]
- Posey J. E., Shinnick T. M., Quinn F. D. (2006). Characterization of the twin-arginine translocase secretion system of Mycobacterium smegmatis. J. Bacteriol. 188 1332–1340. 10.1128/JB.188.4.1332-1340.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel N., Ye C., Livrelli V., Xu J., Joly B., Wu L. F. (2003). Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71 4908–4916. 10.1128/IAI.71.9.4908-4916.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds M. M., Bogomolnaya L., Guo J. B., Aldrich L., Bokhari D., Santiviago C. A., et al. (2011). Abrogation of the twin arginine transport system in Salmonella enterica serovar Typhimurium leads to colonization defects during infection. PLoS One 6:e15800. 10.1371/journal.pone.0015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme S., Varas M., Valenzuela C., Velozo P., Chahin N., Aguilera P., et al. (2016). Relevant genes linked to virulence are required for Salmonella Typhimurium to survive intracellularly in the social amoeba Dictyostelium discoideum. Front. Microbiol. 7:1305. 10.3389/fmicb.2016.01305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier O., Cianciotto N. P. (2005). The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 73 2020–2032. 10.1128/IAI.73.4.2020-2032.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah I. B., Ghigo E., Drancourt M. (2009). Free-living amoebae, a training field for macrophage resistance of mycobacteria. Clin. Microbiol. Infect. 15 894–905. 10.1111/j.1469-0691.2009.03011.x [DOI] [PubMed] [Google Scholar]
- Santini C. L., Ize B., Chanal A., Müller M., Giordano G., Wu L. F. (1998). A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 17 101–112. 10.1093/emboj/17.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiviago C. A., Reynolds M. M., Porwollik S., Choi S. H., Long F., Andrews-Polymenis H. L., et al. (2009). Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. 10.1371/journal.ppat.1000477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillo A., Matthias J., Konertz R., Bozzaro S., Eichinger L. (2011). Salmonella typhimurium is pathogenic for Dictyostelium cells and subverts the starvation response. Cell. Microbiol. 13 1793–1811. 10.1111/j.1462-5822.2011.01662.x [DOI] [PubMed] [Google Scholar]
- Silva C. A., Blondel C. J., Quezada C. P., Porwollik S., Andrews-Polymenis H. L., Toro C. S., et al. (2012). Infection of mice by Salmonella enterica serovar Enteritidis involves additional genes that are absent in the genome of serovar Typhimurium. Infect. Immun. 80 839–849. 10.1128/iai.05497-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassmann J. E., Shu L. (2017). Ancient bacteria-amoeba relationships and pathogenic animal bacteria. PLoS Biol. 15:e2002460. 10.1371/journal.pbio.2002460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart A. L., Harrison C. F., Eichinger L., Steinert M., Hilbi H. (2018). Acanthamoeba and Dictyostelium as cellular models for Legionella infection. Front. Cell. Infect. Microbiol. 8:61. 10.3389/fcimb.2018.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Mulneix D. L., Hamidou Soumana I., Linz B., Harvill E. T. (2017). Evolution of Bordetella from environmental microbes to human respiratory pathogens: amoebae as a missing link. Front. Cell. Infect. Microbiol. 7:510. 10.3389/fcimb.2017.00510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis R. M., Adams L. G., Ficht T. A., Bäumler A. J. (1999). Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67 4879–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas M., Ortiz-Severín J., Marcoleta A. E., Díaz-Pascual F., Allende M. L., Santiviago C. A., et al. (2017). Salmonella Typhimurium induces cloacitis-like symptomsin zebrafish larvae. Microb. Pathog. 107 317–320. 10.1016/j.micpath.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Varas M. A., Riquelme-Barrios S., Valenzuela C., Marcoleta A. E., Berríos-Pastén C., Santiviago C. A., et al. (2018). Inorganic polyphosphate is essential for Salmonella Typhimurium virulence and survival in Dictyostelium discoideum. Front. Cell. Infect. Microbiol. 8:8. 10.3389/fcimb.2018.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R., Ball G., Ize B., Vasil M. L., Lazdunski A., Wu L. F., et al. (2001). Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20 6735–6741. 10.1093/emboj/20.23.6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhu Z., Jing H., Zhang J., Xiong Y., Yan M., et al. (2009). Pleiotropic effects of the twin-arginine translocation system on biofilm formation, colonization, and virulence in Vibrio cholerae. BMC Microbiol. 9:114. 10.1186/1471-2180-9-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Kingsley R. A., Santos R. L., Andrews-Polymenis H., Raffatellu M., Figueiredo J., et al. (2003). Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 71 1–12. 10.1128/IAI.71.1.1-12.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.