Abstract
GRAS proteins belong to a plant-specific transcription factor family and play roles in diverse physiological processes and environmental signals. In this study, we identified and characterized a GRAS transcription factor gene in Brassica rapa, BrLAS, an ortholog of Arabidopsis AtLAS. BrLAS was primarily expressed in the roots and axillary meristems, and localized exclusively in the nucleus of B. rapa protoplast cells. qRT-PCR analysis indicated that BrLAS was upregulated by exogenous abscisic acid (ABA) and abiotic stress treatment [polyethylene glycol (PEG), NaCl, and H2O2]. BrLAS-overexpressing Arabidopsis plants exhibited pleiotropic characteristics, including morphological changes, delayed bolting and flowering time, reduced fertility and delayed senescence. Transgenic plants also displayed significantly enhanced drought resistance with decreased accumulation of ROS and increased antioxidant enzyme activity under drought treatment compared with the wild-type. Increased sensitivity to exogenous ABA was also observed in the transgenic plants. qRT-PCR analysis further showed that expression of several genes involved in stress responses and associated with leaf senescence were also modified. These findings suggest that BrLAS encodes a stress-responsive GRASs transcription factor that positively regulates drought stress tolerance, suggesting a role in breeding programs aimed at improving drought tolerance in plants.
Keywords: Brassica rapa, GRAS transcription factor, BrLAS, drought tolerance, ABA sensitivity
Introduction
Transcription factors are known to participate in various growth and development processes in higher plants. GRAS proteins belong to a plant-specific protein family, named after the first three members identified, GAI, RGA, and SCR (Peng et al., 1997; Pysh et al., 1999; Bolle, 2004). GRAS proteins typically consist of 400–770 amino acids and possess a highly conserved homologous sequence at the C-terminus, which includes five conserved motifs, LHR I, VHIID, LHR II, PFYRE, and SAW (Pysh et al., 1999; Bolle, 2004; Lee et al., 2008). In contrast, the N-terminus is highly variable in both length and sequence. Studies have shown that the N-terminus of GRAS proteins can form a unique stretch region, perhaps contributing to the functional specificity of each member (Tian et al., 2004; Sun et al., 2011).
Members of the GRAS family reportedly possess a variety of functions, playing important roles in diverse physiological processes, such as gibberellin signal transduction, root radial patterning, axillary meristems development, phytochrome signaling, male gametogenesis and detoxification (Dill et al., 2001; Greb et al., 2003; Bolle, 2004; Lee et al., 2008). Two rice GRAS genes, OsCIGR1 and OsCIGR2, show sensitivity to N-acetylchitooligosaccharide elicitor and are induced by phytoactive gibberellins (Day et al., 2004), while AtRGA and AtGAI negatively regulate a series of GA signal transduction pathways (Peng et al., 1997; Silverstone et al., 1998) and are major inhibitors of Arabidopsis vegetative growth (Tyler et al., 2004). Meanwhile, AtSCR and AtSHR participate in radial organization of the roots by forming a SCR/SHR complex (Cui et al., 2007), while AtSCL3 plays an essential role in gibberellin-promoted cell elongation during root development via the formation of a regulation network with the DELLA protein and SHR/SCR complex (Heo et al., 2011). The PAT1 sub-branch in Arabidopsis, which consists of four GRAS proteins, PAT1, SCL5, SCL13, and SCL21, mediates phytochrome signaling pathways (Bolle et al., 2000; Torres-Galea et al., 2006, 2013). LiSCl, a GRAS gene isolated from Lilium longiflorum, participates in microsporogenesis of lily pollen by affecting the transcriptional activity of meiosis-related genes (Morohashi et al., 2003). The LS subfamily of GRAS proteins plays important regulatory roles in axillary meristem development. Analysis of the lateral suppressor (Lels) mutant in tomato revealed that the Ls gene is involved in the development of axillary meristems and formation of lateral buds (Schumacher et al., 1999). OsMOC1 in rice and AtLAS/SCL18 in Arabidopsis, both of which are homologous to Lels, were also found to play an important role in axillary meristem formation (Schumacher et al., 1999; Greb et al., 2003). OsMOC1 is mainly expressed in the axillary buds and is required for axillary meristem initiation and tiller bud formation. AtLAS/SCL18 is expressed in the leaf axils and its mutant does not develop axillary buds during vegetative growth in Arabidopsis (Li et al., 2003).
A large number of GRAS genes are also involved in response to biotic and abiotic stress. For example, NtGRAS1 participates in signal transduction pathways in tobacco under stress conditions (Czikkel and Maxwell, 2007). PeSCL7, a GRAS family gene in Poplar, improves salt and drought resistance in transgenic Arabidopsis plants (Ma et al., 2010). Meanwhile, SlGRAS6-silenced tomato plants exhibited impaired tolerance to drought stress (Mayrose et al., 2006). Overexpression of BrLAS, a GRAS gene from Brassica napus, increased the chlorophyll content and improved drought resistance in Arabidopsis (Yang et al., 2010). Rice OsGRAS23 is also involved in drought and oxidative stress tolerance via regulation of stress-related gene expression (Kai et al., 2015). Overexpression of the SlGRAS40 gene in tomato was also found to enhance tolerance to abiotic stress and influence auxin and gibberellin signaling (Liu et al., 2017).
Phytohormones play pivotal roles in regulating the protective responses in plants under stress conditions. Amongst these, abscisic acid (ABA) is the one of the crucial phytohormones regulating plant growth and development under stress conditions by involving in H2O2 induced signaling (Saxena et al., 2016). H2O2, the by-product of oxidative plant aerobic metabolism, induces oxidative stress under abiotic or biotic stress conditions and acts as a secondary messenger in signal transduction networks (Jubany-Mari et al., 2009; Chen et al., 2012). The cross-talk between ABA signals and H2O2 production in plant tissue has been discussed in several reports (Pei et al., 2000; Saxena et al., 2016). ABA can induce stomatal closure by reducing the turgor of guard cells under drought stress, and H2O2, one of the major reactive oxygen species (ROS), plays a vital role as a second messenger in the ABA-induced stomatal closure (Pei et al., 2000; Miao et al., 2006). Water deficit and ABA treatment triggers H2O2 accumulation, together with major changes in gene expression of stress-responsive genes and higher antioxidant enzyme activities (Jiang and Zhang, 2002a,b). Furthermore, the increase of H2O2 content caused by water deficit was more pronounced than that response to exogenous ABA (Phillips and Ludidi, 2017). Abiotic stress condition enhances ABA/gibberellic acid (GA) ratio supporting DELLA protein accumulation which in turn lowers H2O2 levels (Considine and Foyer, 2014). Several evidences show that MAPK (mitogen activated protein kinase) cascades is implicated in ABA signaling, that is activated by the exogenous H2O2 which in turn mediated by the hormones like jasmonic acid (JA) and ABA (Rodriguez et al., 2010).
Brassica rapa, a member of the Brassica genus, is an important vegetable in China and is cultivated and consumed worldwide. However, B. rapa plants are frequently exposed to adverse environmental conditions, including drought, extreme temperatures, and high salinity stress. Amongst these, drought is the major stress factors resulting in reduced agricultural productivity (Abe et al., 2011; Ahmed et al., 2012). Therefore, understanding the molecular adaptation mechanisms of stress is necessary for genetic improvement of stress resistance in B. rapa. In this study, we isolated and characterized a GRAS transcription factor gene in B. rapa, BrLAS, which is an ortholog of Arabidopsis AtLAS (Greb et al., 2003). Expression pattern analysis revealed that BrLAS is expressed primarily in the roots and axillary meristems and is up-regulated under NaCl, PEG, H2O2 and exogenous ABA treatment. Overexpression of BrLAS also conferred pleiotropic phenotypes and enhanced drought tolerance in transgenic Arabidopsis plants. Collectively, these findings provide details of the GRAS gene family in B. rapa, revealing the potential function of BrLAS in development and drought stress tolerance.
Materials and Methods
Plant Materials, Growth Conditions, and Treatments
Brassica rapa var. Japonica, which shows multiple shoot branching during the vegetative stage, was used for BrLAS gene isolation and expression pattern analysis in different tissues and under various treatments. Seeds were germinated in plastic petri dishes containing two layers of filter paper saturated with distilled water at 23°C for 2 days in the dark then transferred to pots containing soil growth medium under artificial growth conditions at 23–25°C, a photoperiod of 16/8 h light/dark and relative humidity of 60 %. Hoagland nutrient solution was added once a week. Roots, stems, leaves, axillary buds and shoot tips were sampled from 4-week-old plants. Flowers were harvested after vernalization and shoot apical meristems (SAMs) were sampled every 7 days during shoot branching development (from 14 to 42 days after germination). Five well-grown plants were thoroughly mixed to form one sample. Three biological replicates of each tissue and organ were sampled.
For hormone treatment, 2-week-old seedlings were transferred to 4 L hydroponic containers containing half-strength Murashige and Skoog (1/2 MS) liquid medium (pH = 5.8) renewed once every 3 days. At the five-leaf-stage, seedlings were exposed to 1/2 MS solution with 100 μM ABA. For stress treatment, 4-week-old plants grown in pots were irrigated with 200 mM NaCl, 15 % (w/v) polyethylene glycol (PEG) and 100 mM hydrogen peroxide (H2O2), respectively. Leaves were sampled 0.5, 1, 2, 4, 8, 12, and 24 h after treatment and seedlings without treatment were used as a control. In addition, the leaves of seedlings grown under solution without any treatment were also sampled at the designated time (8:00 a.m. to 8:00 a.m. the next day). Five well-grown plants were thoroughly mixed to form one sample. Three biological replicates of each tissue and organ were sampled for RNA extraction.
A. thaliana ecotype Columbia (Col-0) and BrLAS-overexpressing Arabidopsis plants were used in this study. Seeds of transformed Arabidopsis were surface sterilized with 1% sodium hypochlorite and selected on 1/2 MS medium containing appropriate antibiotics. Homozygous lines of T3 generations were used for further analysis.
Cloning of the BrLAS Gene
To clone BrLAS from B. rapa, a specific pair of primers, TL-F and TL-R, were designed based on sequence information in the Brassica genome database (BRAD1) (Wang et al., 2011) and sequence information of Arabidopsis LAS. The full-length cDNA of BrLAS was obtained via PCR using B. rapa total RNA, specific primers and Tks Gflex DNA Polymerase. The PCR products were then cloned into the pMD 18-T Vector (Takara, Japan) for sequencing.
BrLAS homologous genes were obtained by BLAST searches of the National Center for Biotechnology Information2 and Brassica database (BRAD, see footnote 1) using the DNA or protein sequences of BrLAS as queries. BrLAS-related proteins were aligned using DNAMAN software and phylogenetic relationships were analyzed using MEGA 6.0 by the neighbor-joining method with 1,000 bootstrap replicates.
RNA Isolation and Real-Time Quantitative PCR Analysis
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. First-strand cDNA was synthesized from total RNA using a PrimeScript®RT reagent Kit (Takara). The real-time PCR reaction was performed using a Roche thermocycler (LightCycler 480, Roche, Germany). Each reverse transcription-PCR procedure was carried out with three biological and three technical replicates. The Chinese cabbage GAPDH gene was used as an internal control to normalize expression of the BrLAS gene and the relative quantification method was used to evaluate quantitative variation. ACTIN2 (At3g18780) was used as an internal control to normalize expression levels of abiotic stress-responsive genes in transgenic Arabidopsis plants.
Constructs and Generation of Transgenic Plants
To construct the overexpression vector 35S: BrLAS, the entire 1,332-bp coding sequence of BrLAS was amplified and subcloned into the pCR8/GW/TOPO entry vector (Invitrogen) according to the manufacturer’s instructions. The CDS fragment was then inserted into the pMDC32 Gateway-compatible binary vector through LR recombination reactions (Curtis and Grossniklaus, 2003). The 35S: BrLAS plasmid was then introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis Col-0 plants using the floral dip method (Clough and Bent, 1998).
Subcellular Localization
To observe subcellular localization of the BrLAS protein, a p35S: BrLAS-GFP vector containing the open reading frame (ORF) of BrLAS (without the termination codon) fused in-frame with green fluorescent protein (GFP) and driven by the 35S promoter was constructed. The p35S: BrLAS-GFP and 35S: GFP control plasmids were then introduced into GV3101 and the transformed Agrobacterium were infiltrated into leaf cells of Nicotiana benthamiana as described previously (Sparkes et al., 2006). GFP signals in leaf epidermal cells were subsequently observed under a Leica fluorescence microscope (Leica Microsystem, Heidelberg, Germany).
Measurements of Chlorophyll Content
To determine the chlorophyll content, 16-day-old seedlings of wild-type (WT) and transgenic Arabidopsis grown on 1/2 MS medium were collected in centrifuge tubes and weighed then immersed in 1 mL pre-chilled 80% acetone and fully ground. After incubation at 4°C for 4 h and centrifugation at 5,000 × g for 10 min at 4°C, the supernatants were transferred to a 10 mL graduated tube and diluted to 3 mL with 80% acetone. A spectrophotometer was then used to measure absorbance at 645 and 663 nm, and chlorophyll a, chlorophyll b, and total chlorophyll contents were calculated as follows:
| (1) |
| (2) |
| (3) |
Germination and Root Growth Assay
Arabidopsis seeds of WT (Col-0) and BrLAS-overexpressing lines were surface sterilized with 1% sodium hypochlorite and kept at 4°C in the dark for 72 h. To conduct a germination rate assay showing the response to ABA, sterilized seeds of WT and transgenic lines were sown on 1/2 MS plates supplemented with 0 or 2 μM ABA and grown at 22°C in a growth chamber with a photoperiod of 16/8 h light/dark. Germination rates (seedling with cotyledon) were then examined after 6-day growth. For the root growth assay, seeds of transgenic and WT lines were firstly sown on 1/2 MS plates then grown at 22°C in a growth chamber with a photoperiod of 16/8 h light/dark for 5 days. Five-day-old seedlings were then transferred to fresh 1/2 MS medium supplemented with 0 or 10 μM ABA. Seedling root lengths were then measured after 8 days growth on vertical plates.
Drought Treatment and Measurements of ABA Content
To examine drought tolerance, transgenic and WT Arabidopsis plants were germinated on 1/2 MS plates under a 16/8 h light/dark cycle at 22°C. Seven-day-old seedlings were then transferred to pots containing an identical soil mix (1:1 vermiculite:humus) under normal growth conditions. Twenty-five-day-old seedlings were then subjected to water stress by withholding watering until distinct phenotypic differences between transgenic and WT plants were observed. Plants were then re-watered and survival rates surveyed after re-watering for 3 days. To measure ABA content, leaves of WT and transgenic plants at an identical rosette stage were collected under normal and drought stress, and the procedure for extraction, purification and quantification of ABA was performed as described by Cornish and Zeevaart (1984) with some modifications. Each experiment was performed in triplicate.
Water Loss Measurements
For water loss measurements, leaves of transgenic and WT plants at an identical rosette stage were removed and weighed immediately. The samples were then weighed at designated time intervals under normal light conditions. Leaves from five plants were mixed to form one sample. Three biological replicates were obtained for each transgenic line. Water loss was calculated as follows: (Leaf initial fresh weight - leaf weight at each time point)/leaf initial fresh weight.
Scanning Electron Microscope (SEM)
For SEM, intact fully expanded rosette leaves of WT and transgenic plants were collected in parallel then freshly excised into thin pieces 3 mm wide and immediately fixed in 2.5% (v/v) glutaraldehyde in PBS buffer at 4°C overnight. After washing in the same phosphate buffer, the samples were post-fixed with 1% OsO4 and dehydrated with a graded ethanol series. After trough-critical point drying, the samples were mounted and sputter-coated with gold then observed under a SEM (HITACHI S-3400N).
3,3′-Diaminobenzidine (DAB) Staining
To examine H2O2 accumulation, 4-week-old rosette leaves of WT and transgenic plants under drought stress were collected and immersed in freshly prepared 1 mg mL-1 DAB solution for 12 h in the dark. Stained leaves were then discolored with a bleaching solution (ethanol/acetic acid/glycerol = 3:1:1) and photographed.
Reactive Oxygen Species (ROS)-Related Biochemical Assay
To detect the changes in physiological indicators under drought stress, leaves of WT and T3 transgenic plants at an identical rosette stage were harvested. For extraction of H2O2, O2-, superoxide dismutase (SOD), peroxidase (POD) and chloramphenicol acetyltransferase (CAT), 0.2 g samples were placed in 1.8 mL 0.1 M PBS (pH 7.4) on ice. After centrifugation at 10,000 × g for 10 min at 4°C, the supernatants were then used to measure H2O2 and O2- contents, and SOD, POD, and CAT enzyme activity using corresponding detection kits. MDA assay was performed using the thiobarbituric acid method (TBA) (Heath and Packer, 1968).
Primers
All primers used in this study are listed in Supplementary Table S1.
Statistical Analysis
All experiments included three independent repeats, and significant differences analyses were performed using the t-test at significance levels of ∗P < 0.05 and ∗∗P < 0.01.
Results
Cloning and Sequence Analysis of BrLAS
The full-length cDNA sequence of BrLAS was 1,866-bp long and contained a 1,332-bp ORF with a 5′-untranslated region (UTR) of 151-bp and a 3′-UTR of 383-bp. It encoded a protein of 443 amino acid residues with a predicted molecular mass of 49.278 kDa. Amino acid sequence alignment analysis revealed that BrLAS is highly homologous to GRAS genes in other species except for its two progenitor species (BnLAS in B. napus and BoLAS in B. oleracea). For example, Arabidopsis AtLAS shared 89% homology, tomato LeLs shared 81% homology and rice OsMOC1 shared 71% homology (Figure 1A). The deduced amino acid sequence of BrLAS contains a typical GRAS domain consisting of the LHRI, VHIID, LHRII, PFYRE, and SAW motifs in its C-terminus (Bolle, 2004) (Figure 1A). Here, phylogenetic analysis indicated that BrLAS groups with the AtLAS branch contained LeLS and OsMOC1 (Figure 1B). These results strongly suggest that BrLAS is a member of the GRAS gene family in B. rapa and an ortholog of LAS in Arabidopsis.
FIGURE 1.
Sequence alignment and phylogenetic relationship between BrLAS and its homologous genes. (A) Sequence alignment of the deduced amino acid sequences of BrLAS protein, Arabidopsis AtLAS, tomato SlLS, and rice OsMOC1 as determined using DNAMAN. Identical residues are displayed by color. Conserved leucine residues in heptad repeats are identified by asterisks and conserved motifs are underlined. (B) Phylogenetic tree of BrLAS and its homologous proteins in Arabidopsis thaliana, Brassica napus (BnLAS) and B. oleracea (BoLAS) (GenBank accession numbers: HQ324233 and HQ324234, respectively).
Expression Patterns of BrLAS
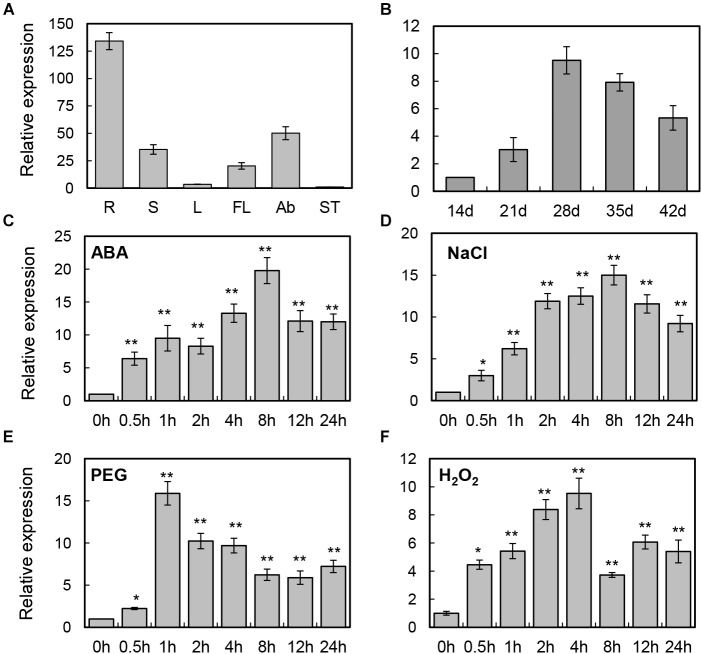
To examine the potential functions of the BrLAS gene, quantitative real-time analysis was performed to investigate expression patterns in the roots, stems, leaves, flowers, axillary buds, and shoot tips. As shown in Figure 2A, BrLAS showed relatively high levels of expression in the roots, axillary buds, and flowers. Since relatively higher expression levels were observed in the axillary buds, expression levels of BrLAS at five stages of axillary bud development (14–42 days after germination) were examined further (Figure 2B). The results revealed that transcript levels of BrLAS underwent a dramatic change during axillary bus development, suggesting a role in the formation of shoot branching.
FIGURE 2.
Expression patterns of BrLAS in B. rapa. (A) Relative expression of BrLAS in different tissues (R, roots; S, stem; L, leaves; FL, flowers; Ab, axillary buds; ST, shoot tips) under normal conditions. Expression data in the shoot tips were normalized to 1. (B) Relative expression profiles of BrLAS during axillary bud development (14–42 days after germination). Expression data 21 days after germination were normalized to 1. (C–F) Expression patterns of BrLAS in response to 100 μM ABA, 250 mM NaCl, 15% (w/v) polyethylene glycol (PEG), and 100 mM H2O2. Total RNA was isolated from 4-week-old seedlings exposed to the various stress treatments. Expression data of the control sample were normalized to 1. Error bars show the standard error between three replicates, asterisks indicate significant differences using Student’s t-test ∗P < 0.05, ∗∗P < 0.01.
To examine the detailed expression profiles of BrLAS under various abiotic stresses and phytohormone treatment, 4-week-old B. rapa seedlings were treated with exogenous 100 μM ABA, and NaCl, PEG, and H2O2 treatment. Expression levels of BrLAS were significantly induced by ABA, reaching a maximum at 8 h after treatment and up-regulated more than 20-fold compared with the control, and then decreasing sharply (Figure 2C). BrLAS expression was also induced by NaCl, reaching a peak of more than 15-fold at 8 h after treatment (Figure 2D). Similarly, expression was markedly induced by PEG, reaching a peak of more than 15-fold at 1 h after treatment then declining gradually (Figure 2E). Under H2O2 treatment, expression reached a peak of more than 10-fold at 4 h after treatment then decreased (Figure 2F). Moreover, the expression of BrLAS under normal conditions using solution without any treatment was also examined, and the result showed that the expression of BrLAS was affected by mechanical stress, reaching a peak of approximately fivefold at 0.5 h after tissues cut then increasing gradually, and showed the circadian rhythm, the expression levels of which at 12 and 16 h (8:00 p.m. and 00:00 a.m.) were higher than other time points during the day (Supplementary Figure S1). These data suggest that expression of BrLAS is sensitive to NaCl, PEG, H2O2 and exogenous ABA treatment, affected by mechanical stress and showed circadian rhythm.
BrLAS Is a Nuclear Protein
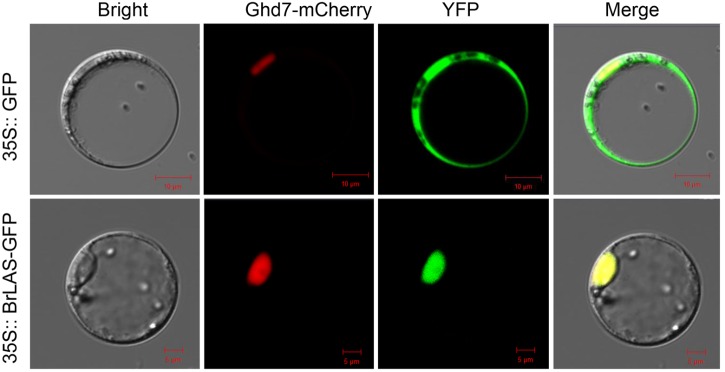
To determine subcellular localization of the BrLAS protein, we constructed a p35S: BrLAS-GFP construct containing the ORF of BrLAS (without the termination codon) fused in-frame with GFP driven by the 35S promoter and transformed it into B. rapa protoplast cells by means of modified PEG treatment (Yoo et al., 2007). As shown in Figure 3, fluorescent signals of 35S: GFP were uniformly distributed throughout the cells, while BrLAS-GFP was exclusively localized in the nucleus. Consistent with the subcellular localization of other members of the GRAS gene family and possible function as a transcription factor, these findings suggest that BrLAS is a nuclear protein.
FIGURE 3.
Subcellular localization of BrLAS. Fluorescence microscopy of 35S: BrLAS-GFP and 35S: GFP (control) plasmids transiently expressed in B. rapa protoplast cells.
35S: BrLAS Transgenic Arabidopsis Plants Exhibit Pleiotropic Characteristics
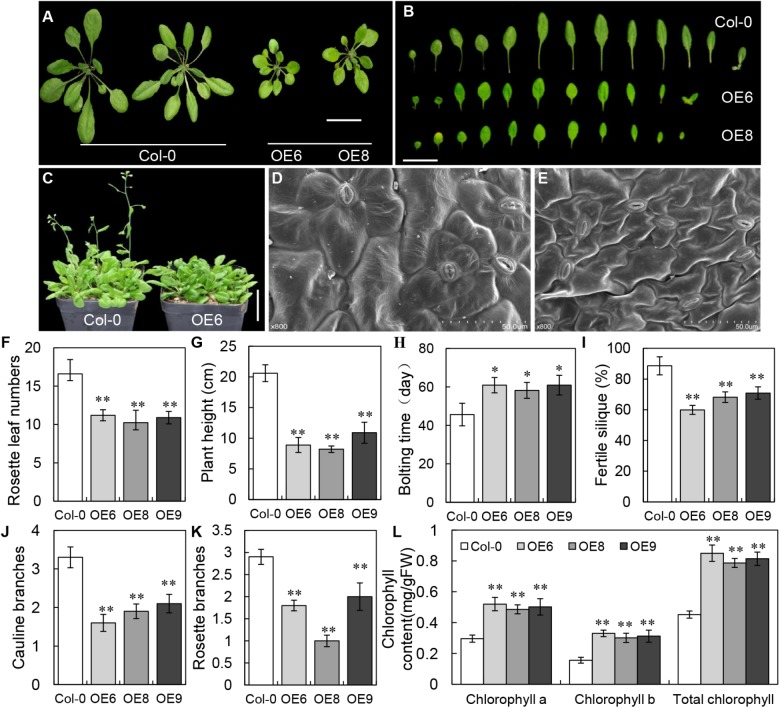
To determine the potential function of BrLAS, we constructed a 35S: BrLAS construct to transform Arabidopsis Col-0 plants. Approximately 40 transgenic plants were identified through hygromycin B resistance analysis and PCR verification with primers of BrLAS and a selection marker gene (hygromycin, HYG). Phenotypes of T1 transgenic plants were then observed and representative individuals were propagated for further experiments. Compared with WT Arabidopsis, the morphology and developmental characteristics of the overexpression lines showed obvious changes (Figure 4 and Supplementary Table S2). During the same developmental period, BrLAS-overexpressing plants grew at a slower rate and had fewer leaves, which were darker in color than the WT plants (Figures 4A,B). Some overexpressing lines exhibited delayed bolting and flowering, to varying degrees, at the reproductive stage (Figures 4C,H), with fewer senesced rosette leaves. All changes were stably inherited by the following generations; therefore, three representative T3 transgenic lines (OE6, OE8, and OE9) were chosen for further quantitative analysis. Rosette leaf numbers in the three overexpressing lines were significantly reduced compared to the WT plants before bolting (Figure 4F). Moreover, plant height at day 65 was markedly reduced (Figure 4G). The bolting times of all three lines were delayed by about 14 days compared with the WT (Figure 4H), and fertility was also reduced, resulting in fewer fertile siliques on the main stem (Figure 4I).
FIGURE 4.
Phenotypes of BrLAS-overexpressing transgenic plants and wild-type (WT) plants. (A) Comparison of transgenic T3 generation OE6 and OE8 and wild-type plants. The transgenic plants were smaller and had less rosette leaves. Bar = 1 cm. (B) Comparison of 30-day-old rosettes leaves from WT and transgenic plants. Bar = 1 cm. (C) Bolting and flowering in transgenic compared with WT plants. Both were delayed in the transgenic plants. Bar = 1 cm. (D,E) The size of leaf epidermis cells in transgenic (E) and WT plants (D). Cell size was significantly smaller in the transgenic plants. (F) Numbers of rosette leaves in the transgenic and WT plants before bolting. There were significantly less rosette leaves in the transgenic plants. (G) Plant height in the transgenic and WT plants at 65 days. A marked reduction was observed in the transgenic plants. (H) Bolting and flowering in the transgenic and WT plants. Both were delayed in the transgenic plants. (I) Fertility in the transgenic compared with WT plants. Reduced fertility was observed in the transgenic plants. (J,K) The number of rosette and cauline branches in the transgenic compared with WT plants. Both were reduced in the transgenic plants. (L) Chlorophyll contents in transgenic plants were increased compared with WT plants. Error bars represent the standard deviation of three independent experiments, and asterisks indicate a significant difference from the WT at ∗P < 0.05 and ∗∗P < 0.01.
Since the majority of BrLAS-overexpressing plants grown both on plates and in soil showed smaller leaves with a darker green color than the WT, we also examined cell morphology and chlorophyll content. SEM showed that the leaf epidermis cells were significantly smaller in the transgenic lines than the WT (Figures 4D,E), suggesting that this was the cause of the smaller leaves in BrLAS-overexpressing plants. The total chlorophyll content of the three transgenic lines increased by 88.1, 74.1, and 80.1%, respectively, with increases in both chlorophyll a and chlorophyll b contents (Figure 4L), suggesting that this was the major cause of the darker green leaves in the BrLAS-overexpressing lines. Since the AtLAS gene in Arabidopsis is required for the initiation of axillary meristems, we also examined branching numbers. As shown in Figures 4J,K, numbers of rosette branches and cauline branches both decreased in the 35S: BrLAS transgenic plants compared with the WT.
Overexpression of BrLAS Confers Drought Tolerance in Transgenic Arabidopsis
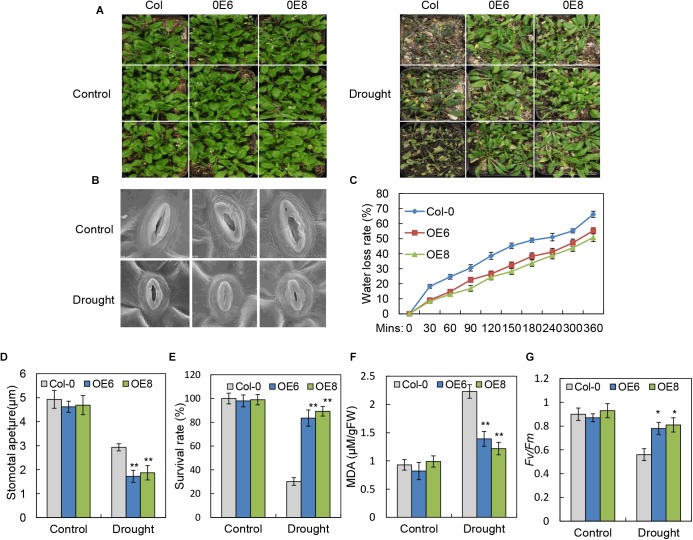
Since expression of BrLAS was up-regulated with ABA, PEG, and H2O2 treatment (Figure 2), and considering the altered leaf morphology and delayed leaf senescence in the 35S: BrLAS transgenic plants, it is plausible that BrLAS is involved in drought stress tolerance in BrLAS-overexpressing Arabidopsis plants. To verify this hypothesis, 3-week-old transgenic and WT seedlings were subjected to drought stress. Survival rates were then surveyed after re-watering for 3 days. As shown in Figure 5A, the leaves of WT plants displayed severe wilting and yellowing, while the overexpression lines maintained good growth after drought stress. Moreover, only 30% of WT plants were able to recover from stress, while the survival rates of the OE6 and OE8 transgenic plants were 86 and 93%, respectively (Figure 5E). Consistent with these findings, water loss rates of detached leaves from the overexpression lines decreased at each time point compared with the WT plants (Figure 5C). Stomatal apertures of WT and transgenic lines under normal and drought stress conditions were also examined. Under normal conditions, no significant differences were observed; however, at 6 h after drought stress treatment, the stomatal apertures of OE6 and OE8 leaves decreased by 65.0 and 68.4%, respectively, while those of WT plants decreased by only 35.3% (Figures 5B,D). The MDA content, a sign of oxidative damage, was also measured, revealing a significant increase in the WT plants, but only a slight increase in the transgenic plants (Figure 5F). The overexpression lines also maintained a higher Fv/Fm ratio than the WT plants after drought treatment (Figure 5G). These findings confirm that overexpression of BrLAS confers drought resistance in the transgenic plants.
FIGURE 5.
Improved drought resistance in 35S: BrLAS transgenic Arabidopsis. (A) Drought resistance in 35S: BrLAS transgenic (OE6 and OE8) compared with wild-type (WT) plants. Twenty-five-day-old seedlings grown under normal conditions were subjected to drought stress by withholding watering for up to 14 days followed by re-watering. (B) The stomatal apertures of 35S: BrLAS transgenic lines compared with WT plants under drought stress. A significant decrease in size was observed in the transgenic lines. (C) Water loss from detached leaves of 35S: BrLAS compared with the WT plants. A significant decrease in water loss was observed in the transgenic plants. Data represent the means of 20 leaves each per three independent experiments. (D) Stomatal aperture size in the 35S: BrLAS transgenic and WT plants under drought stress. A significant decrease in size was observed in the transgenic lines. (E) Survival rates of the transgenic and WT plants under drought stress. (F,G) Comparisons of the MDA content (F) and Fv/Fm ratios (G) of the transgenic and WT plants. Arabidopsis seedlings treated with water were used as a control. Error bars represent the standard deviation of three independent experiments, and asterisks indicate a significant difference from the WT at ∗P < 0.05 and ∗∗P < 0.01.
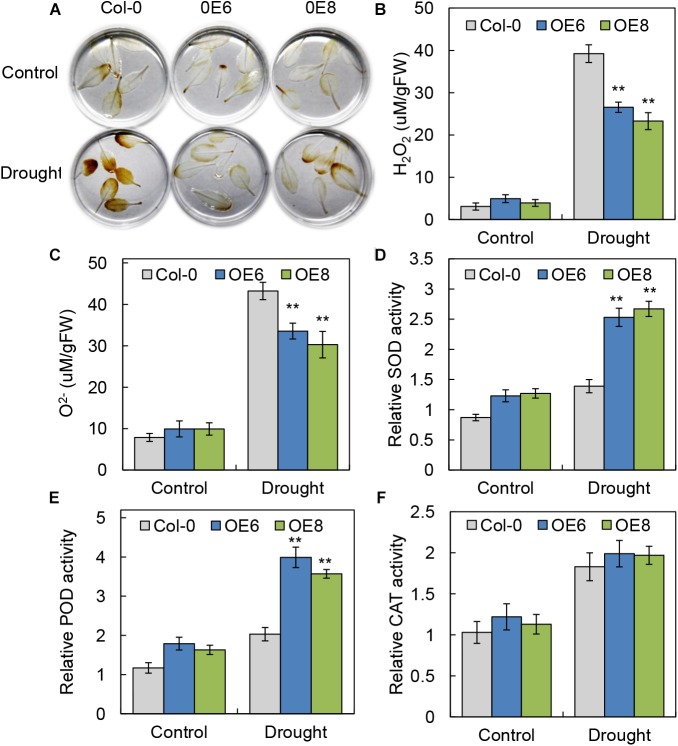
Overexpression of BrLAS Is Involved in the Regulation of ROS in Transgenic Arabidopsis
Abiotic stress can lead to increased accumulation of ROS, having damaging effects on plant development (Wang et al., 2012). To investigate the levels of active oxygen accumulation in the transgenic and WT plants, we performed DAB staining to determine H2O2 accumulation. Compared with WT plants, the overexpression lines showed a visible decrease in DAB staining intensity under drought stress treatment (Figure 6A), consistent with the lower accumulation of H2O2 in detached leaves of the BrLAS-overexpressing plants. Moreover, the WT plants exhibit higher contents of H2O2 and O2- than the OE lines after drought treatment (Figures 6B,C). These findings indicate that the transgenic lines experienced less damage than the WT plants under stress conditions.
FIGURE 6.
BrLAS-overexpressing plants exhibited increased oxidative stress tolerance. (A) ROS accumulation in detached leaves using DAB staining to visualize H2O2 under normal and drought stress conditions. (B,C) Contents of H2O2 and O2- in transgenic plants and WT plants under normal and drought stress conditions. (D–F) Antioxidant enzyme activity in the transgenic and WT plants under normal and drought conditions. (D) SOD activity, (E) POD activity, and (F) CAT activity. Arabidopsis seedlings treated with water were used as a mock control. Error bars show the standard error between three replicates, and asterisks indicate significant differences from the WT at ∗∗P < 0.01.
Antioxidant enzyme activity is another important indicator of a plant’s ability to scavenge ROS. We therefore measured relative activities of SOD, POD, and catalase CAT (Hu et al., 2012) in WT and BrLAS-overexpressing lines. Activities of SOD and POD in the transgenic plants were both higher than in the WT plants under normal conditions. Meanwhile, under drought stress, levels of SOD and POD activity increased significantly in the OE lines, but only slightly in the WT (Figures 6D,E). CAT activity was up-regulated in both the OE lines and WT following drought stress treatment, with no obvious differences between the two (Figure 6F). These findings suggest that BrLAS-overexpression confers increased drought stress resistance in transgenic plants through enhanced ROS scavenging ability.
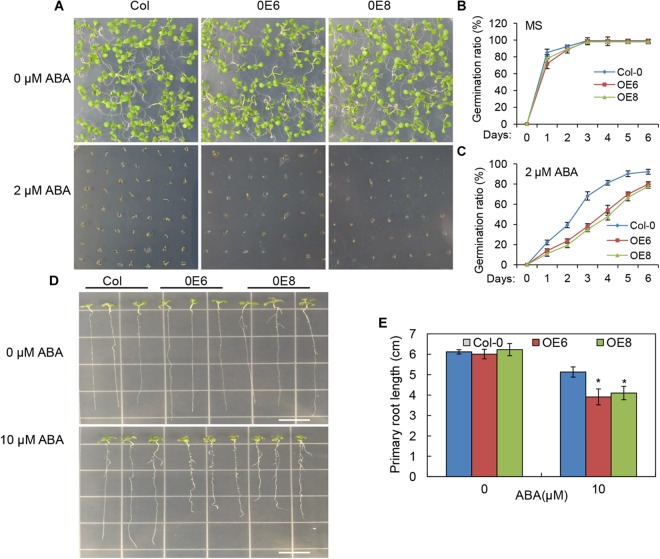
BrLAS-Overexpressing Plants Exhibit an ABA-Hypersensitive Response
To determine whether the enhanced drought resistance in the BrLAS-overexpressing lines was associated with ABA, seeds of transgenic and WT plants were germinated on 1/2 MS medium with or without ABA. The germination rates were then compared at designated time points. No significant differences in germination rates were observed between the transgenic and WT seeds when grown without ABA (Figures 7A,B). However, on 1/2 MS medium containing 2 μM ABA, germination rates of the transgenic seeds were lower than those of WT seeds at each time point (Figures 7A,C). It is well known that ABA inhibits plant growth (Luo et al., 2014), therefore, the differences in ABA-induced inhibition of root growth were also observed. Five-day-old seedlings were transferred to 1/2 MS plates supplemented with ABA and the primary root lengths were measured after vertical growth for 8 days. Exogenous ABA-induced inhibition of root growth was more severe in the transgenic than the WT seedlings when grown on 1/2 MS medium containing 10 μM ABA (Figures 7D,E). These findings suggest that overexpression of BrLAS in Arabidopsis enhances plant sensitivity to exogenous ABA.
FIGURE 7.
ABA hypersensitivity in the BrLAS-overexpressing (OE) lines. (A) Seed germination in wild-type (WT) and BrLAS-overexpressing lines 6 days after germination on 1/2 MS medium supplemented with 0 or 2 μM ABA and quantitative analysis of seed germination rates (B,C). Values represent means ± SE (n = 60–90 seeds). (D) Phenotypes of WT and OE lines grown for 8 days on 1/2 MS medium supplemented with 0 or 10 μM ABA and primary root lengths (E). Values represent means ± SE (n = 20–25 plants). Error bars show the standard error of three replicates, and the asterisk indicates significant differences from the WT plants at ∗P < 0.05.
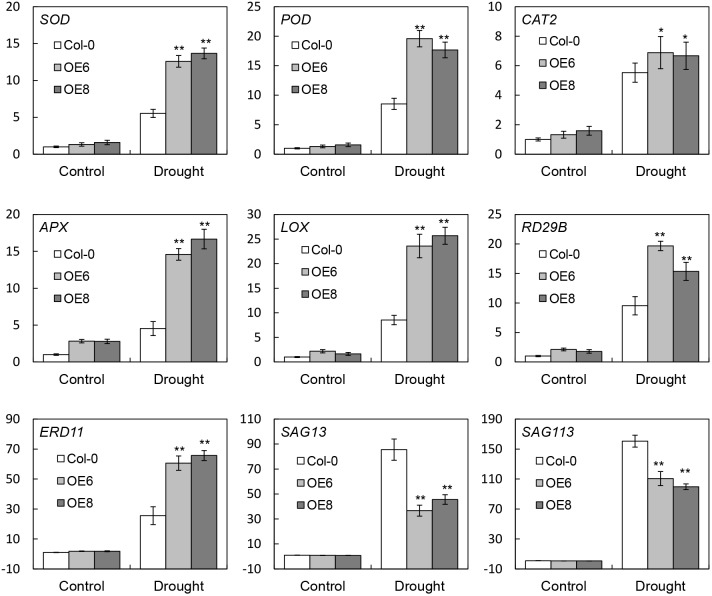
Expression Patterns of Stress-Related Genes Under Drought Stress in Transgenic Arabidopsis
To further investigate the molecular mechanism of BrLAS-enhanced drought stress tolerance, expression patterns of plant stress-related marker genes were examined by qRT-PCR (Figure 8). Abundant transcripts of several genes involved in ROS generation and scavenging; namely, SOD, POD, CAT2, ascorbate peroxidase (APX), and LOX, a lipoxygenase gene (Jin et al., 2013), were detected in BrLAS-overexpressing and WT plans under both normal and drought conditions. Moreover, while expression levels increased in both transgenic and WT plants, transcript levels were significantly higher in transgenic compared with WT plants under drought stress conditions, except for CAT. CAT was up-regulated in both OE and WT plants after stress treatment, with no obvious differences in expression levels, which is consistent with the change in catalase activity in both plants under drought stress (Figure 6F). Accumulation of ERD11 (Jensen et al., 2010) and RD29B (Yang et al., 2011), two drought-responsive genes, was also higher in the OE than the WT plants under drought stress treatment. Meanwhile, the two leaf senescence-related genes SAG13 (senescence-associated gene 13) (Garapati et al., 2015) and SAG113 (senescence-associated gene 113) (Zhang et al., 2011) were both down-regulated in BrLAS overexpressing compared to WT plants both under normal and drought stress conditions (Figure 8), suggesting that BrLAS overexpression delays leaf senescence in OE plants. Taken together, these findings suggest that BrLAS is involved in drought stress via modulation of key stress- and senescence-related genes.
FIGURE 8.
Transcript levels of stress-related marker genes in wild-type (WT) and transgenic plants under normal and drought stress conditions. Error bars show the standard error of three replicates, and asterisks indicate significant differences from the WT at ∗P < 0.05 and ∗∗P < 0.01.
BrLAS-Overexpressing Plants Had Altered Endogenous ABA Content
The high-level expression patterns of ABA signaling related genes in 35S: BrLAS plants, under normal and drought stress conditions, suggested that the endogenous ABA content may be elevated in transgenic plants. To verify this hypothesis, we measured endogenous ABA contents of transgenic and WT plants with and without drought stress. As shown in Figure 9, the ABA contents in all lines were increased under drought stress conditions. As expected, the level of ABA in 35S: BrLAS plants were both higher than WT plants under normal and drought stress conditions. Drought stress treatment induced a dramatically increase in endogenous ABA content of 35S: BrLAS plants, which is up to approximately fourfold higher than ABA increase in WT plants. These data demonstrate that overexpression of BrLAS may affect endogenous ABA content.
FIGURE 9.
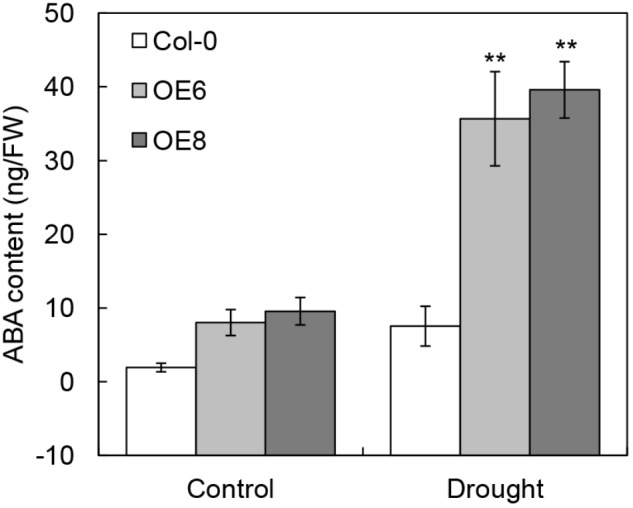
Contents of endogenous ABA in 35S: BrLAS transgenic and wild-type (Col-0) plants under normal and drought stress conditions. Endogenous ABA contents of WT and transgenic plants were measured under normal and drought stress conditions. Error bars show the standard error of three replicates, and the asterisk indicates significant differences from the WT plants at ∗∗P < 0.01.
Discussion
To date, available information on GRAS proteins in B. rapa was relatively limited, despite their important roles in plant growth and development. Song et al. (2014) analyzed the GRAS transcription factor family in Chinese cabbage and identified 48 BraGRAS genes in the entire genome through comparative genomic analysis. In this study, we characterized a GRAS transcription factor gene, BrLAS, from B. rapa and found that it was an ortholog of Arabidopsis AtLAS. BrLAS showed relatively high expression levels in the roots and axillary buds, and was regulated by salt, PEG and H2O2 stress as well as exogenous ABA (Figure 2). Meanwhile, transgenic BrLAS overexpressing Arabidopsis plants displayed significantly enhanced drought resistance with decreased ROS accumulation and increased antioxidant enzyme activity under drought treatment compared with WT plants.
BrLAS-overexpressing plants showed pleiotropic characteristics, including morphological changes, delayed bolting and flowering time, reduced fertility and altered chlorophyll content (Figure 4). These findings suggest that BrLAS is involved in various aspects of growth and development in B. rapa. A previous study revealed that LAS controls axillary meristem formation in Arabidopsis (Greb et al., 2003). Considering the high sequence identity and similar expression pattern in axillary meristems between LAS and BrLAS, it is speculated that BrLAS also plays a potential role in shoot branching development in B. rapa. Interestingly, 35S: LAS overexpressing Arabidopsis plants not only exhibited increased shoot branches, but also presented reduced numbers of rosette and cauline branches, suggesting that relative expression levels of BrLAS are vital, with overexpression disrupting normal branch morphogenesis. Expression levels of BrLAS were also high in the roots, similar to its homologous gene AtLAS (Greb et al., 2003), and some 35S: BrLAS transgenic plants showed a relatively larger number of lateral roots (Supplementary Figure S2). However, whether or not this increase in transcript abundance contributed to the increase in lateral roots in transgenic plants needs to be studied further. The darker green leaves and increased chlorophyll content of the transgenic plants was thought to be the result of smaller leaf epidermis cells, leading to more cells within a given leaf area. However, this also requires further confirmation in the future.
BrLAS overexpressing plants also displayed significantly enhanced drought resistance compared with WT plants (Figures 5A–G). To date, few reports have documented a role of GRAS proteins in drought stress. For example, NtGRAS1 in tobacco (Czikkel and Maxwell, 2007), BnLAS in B. napus (Yang et al., 2010), PeSCL7 in Poplar (Ma et al., 2010), OsGRAS23 in rice (Kai et al., 2015), and SlGRAS40 in tomato (Liu et al., 2017) were found to be involved in drought stress. In this study, the increased drought tolerance in BrLAS overexpressing plants provides novel evidence of the role of GRAS proteins in drought stress responses. Leaf water loss rate is an important indicator of drought resistance in plants (Hu et al., 2006). Here, the BrLAS-overexpressing plants displayed a reduced water loss rate and smaller stomatal apertures than the WT plants (Figures 5B–D), suggesting that BrLAS improves water retentivity in transgenic plants by regulating stomatal closure to reduce water loss. Drought stress can also cause a decrease in photosynthetic efficiency, oxidative damage to cells, and limited metabolic reactions (Farooq et al., 2009). As an indicator of photosynthetic efficiency, Fv/Fm values significantly decreased in WT plants following drought treatment; however, in OE plants levels remained high, suggesting that the PSII reaction center was less damaged (Figure 5G). In addition, an increase in MDA content was observed in the WT plants, with only a slight increase in transgenic plants, suggesting that the overexpression plants suffered less oxidative damage than the WT plants (Figure 5F). On the whole, these findings suggest that BrLAS is involved in the regulation of drought resistance, with overexpression effectively improving drought tolerance.
Abiotic stress can trigger oxidative stress in plants (Hu et al., 2012; Wang et al., 2012), with the accumulation of ROS causing damage to plant growth. Maintaining a steady-state level of ROS within cells is important, representing balance between the amount of total reactive oxygen produced and the removal ability of the cellular antioxidant system (Foyer and Noctor, 2005). In our study, the BrLAS-overexpressing plants showed a significant decrease in H2O2 and O2- contents compared to the WT following drought stress (Figures 6A–C). Antioxidant enzymes such as CAT, SOD, and POD also play a crucial role in scavenging ROS and enhancing stress tolerance in plants (Hu et al., 2012; Wang et al., 2012). In our study, activities of SOD and POD were both higher in the transgenic compared to WT plants under normal and drought conditions. Meanwhile, CAT activity was up-regulated in both the OE lines and WT after drought treatment (Figures 6D–F). Taken together, these findings suggest that BrLAS alleviated oxidative damage in the OE lines by regulating activities of antioxidant enzymes, thereby enhancing drought tolerance.
Numerous genes are reportedly up-regulated under stress conditions, with overexpression of these genes enhancing tolerance to abiotic stresses (Seki et al., 2002; Zhu, 2002). Expression levels of certain genes involved in ROS scavenging and the drought response were therefore monitored in OE and WT plants (Figure 8). Transcript levels of SOD, POD, CAT2, APX, and LOX, a lipoxygenase gene (Jin et al., 2013), were all significantly higher in OE compared to WT plants under drought treatment. Meanwhile, accumulation of ERD11 and RD29B, both of which are involved in ABA-independent stress signaling pathways, was also higher in OE compared to WT plants under drought stress treatment. These findings suggest that BrLAS confers enhanced drought tolerance in OE plants through regulation of certain marker genes involved in abiotic stress and ROS scavenging. Moreover, expression of SAG13 (Garapati et al., 2015) and SAG113 (Zhang et al., 2011), senescence-related marker genes, was down-regulated in OE compared to WT plants under both control and drought stress treatment (Figure 8), suggesting that BrLAS leads to down-regulation of SAG13 and SAG113, further contributing to enhanced drought stresses tolerance in OE plants.
Abscisic acid is a phytohormone that plays critical roles in plant stress response. It accumulates as a response to stressful environmental conditions and regulates the expression of various stress-responsive genes by a complex regulatory network (Zhu, 2002; Cutler et al., 2010; Raghavendra et al., 2010). Consistent with this function, the endogenous ABA content and expression levels of some ABA signaling related genes were changed in 35S: BrLAS plants compared with WT plants after drought stress. It is reported that overproduction of ABA induces H2O2 accumulation, which in turn, results in the up-regulation of antioxidant enzyme activities in ABA signaling through activating MAPK cascade (Pei et al., 2000; Jiang and Zhang, 2002a,b; Zhang et al., 2007). In our study, the contents of endogenous ABA and H2O2 were both higher in 35S: BrLAS plant (Figure 6) than WT plant without water deficit. This result indicated that overexpression of the BrLAS increases the cellular ABA content, which leads to the increased generation of H2O2. In contrast, the content of H2O2 in transgenic plants was lower than WT plants with the increase of ABA content under drought stress conditions. This observation may be explained at least in part by increased antioxidant enzyme activities triggered by ABA accumulation (Fath et al., 2001). Thus, our findings illustrate the complex interaction of H2O2 and ABA signaling in the regulation of drought stress response in plants and a clear integration and crosstalk between ROS and hormone signaling pathways remain to be characterized in the further research.
Conclusion
In conclusion, this study revealed, for the first time, that BrLAS encodes a drought-responsive GRAS transcription factor in B. rapa, which was also the first identified GRAS gene positively regulating drought tolerance in leafy vegetable crops that consuming a large amount of fresh water resources in China. Compared with GRAS members in other crops, BrLAS is primarily involved in ABA-mediated signaling pathways, and BrLAS-overexpressing Arabidopsis plants exhibited hypersensitive to exogenous ABA and pleiotropic characteristics in vegetative and reproductive growth stages. These findings suggest that BrLAS is an ideal candidate gene for genetic manipulation of B. rapa drought stress tolerance breeding. The drought-resistant BrLAS may be used as genetic resources in drought-tolerant breeding via transgenic approaches; however, it needs huge cost and breeding time. The genome editing, a new technology, have the potential to generate varieties with improved drought tolerance by precise introduction of drought-resistant BrLAS into drought-sensitive, locally adapted elite varieties at a lower cost. In addition, the resistant alleles or haplotypes detected in BrLAS can be used as molecular markers for marker-assisted selection (MAS) in genetic improvement of B. rapa drought resistance.
Author Contributions
SY, PL, and FZ designed the experiments. PL, TS, BZ, XX, WW, XZ, YY, and DZ carried out the experiments. PL, PRL, TS, SY, and FZ wrote the paper. All authors discussed the results and commented on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This research was financially supported by the National Key Research and Development Program of China (2017YFD0101801), Key Program of Beijing Municipal Science and Technology Committee (D171100007617001), the Technology Innovation Program, BAAFS (KJCX20180204), the National Natural Science Foundation of China (Nos. 31801873 and 31772307), Beijing Postdoctoral Research Foundation (2017-ZZ-085) and the earmarked fund for China Agriculture Research System (CARS-23-A-05).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01792/full#supplementary-material
References
- Abe H., Narusaka Y., Sasaki I., Hatakeyama K., Shin-I S., Narusaka M., et al. (2011). Development of full-length cdnas from chinese cabbage (Brassica rapa Subsp. pekinensis) and identification of markergenes for defence response. DNA Res. 18 277–289. 10.1093/dnares/dsr018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N. U., Park J. I., Seo M. S., Kumar T. S., Lee I. H., Park B. S., et al. (2012). Identification and expression analysis of chitinase genes related to biotic stress resistance in Brassica. Mol. Biol. Rep. 39 3649–3657. 10.1007/s11033-011-1139-x [DOI] [PubMed] [Google Scholar]
- Bolle C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218 683–692. 10.1007/s00425-004-1203-z [DOI] [PubMed] [Google Scholar]
- Bolle C., Koncz C., Chua N. H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome a signal transduction. Genes Dev. 14 1269–1278. 10.1101/gad.14.10.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. J., Wu S. D., Huang G. J., Shen C. Y., Afiyanti M., Li W. J., et al. (2012). Expression of a cloned sweet potato catalase SPCAT1 alleviates ethephon- mediated leaf senescence and H2O2 elevation. J. Plant Pysiol. 169 86–97. 10.1016/j.jplph.2011.08.002 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Considine M. J., Foyer C. H. (2014). Redox regulation of plant development. Antioxid. Redox Signal. 21 1305–1326. 10.1089/ars.2013.5665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish K., Zeevaart J. A. D. (1984). Abscisic acid metabolism in relation to water stress and leaf age in Xanthium strumarium. Plant Physiol. 76 1029–1035. 10.1104/pp.76.4.1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Levesque M. P., Vernoux T., Jung J. W., Paquette A. J., Gallagher K. L., et al. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316 421–425. 10.1126/science.1139531 [DOI] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61 651–679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Czikkel B. E., Maxwell D. P. (2007). NTGRAS1, a novel stress-induced member of the GRAS family in tobacco, localizes to the nucleus. J. Plant Physiol. 164 1220–1230. 10.1016/j.jplph.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Day R. B., Tanabe S., Koshioka M., Mitsui T., Itoh H., Ueguchi-Tanaka M., et al. (2004). Two rice GRAS family genes responsive to N-acetylchitooligosaccharide elicitor are induced by phytoactive gibberellins: evidence for cross-talk between elicitor and gibberellin signaling in rice cells. Plant Mol. Biol. 54 261–272. 10.1023/B:PLAN.0000028792.72343.ee [DOI] [PubMed] [Google Scholar]
- Dill A., Jung H. S., Sun T. P. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. U.S.A. 98 14162–14167. 10.1073/pnas.251534098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M., Wahid A., Dongjin L., Ito O., Siddique K. H. M. (2009). Advances in drought resistance of rice. Crit. Rev. Plant Sci. 28 199–217. 10.1080/07352680902952173 [DOI] [Google Scholar]
- Fath A., Bethke P. C., Jones R. L. (2001). Enzymes that scavenge reactive oxygen species are down-regulated prior to gibberellic acid-induced programmed cell death in barley aleurone. Plant Physiol. 126 156–166. 10.1104/pp.126.1.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C. H., Noctor G. (2005). Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 28 1056–1071. 10.1111/j.1365-3040.2005.01327.x [DOI] [Google Scholar]
- Garapati P., Xue G.-P., Munné-Bosch S., Balazadeh S. (2015). Transcription Factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168 1122–1139. 10.1104/pp.15.00567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schäfer E., Müller D., Herrero R., Schmitz G., et al. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17 1175–1187. 10.1101/gad.260703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath R. L., Packer L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125 189–198. 10.1016/0003-9861(68)90654-1 [DOI] [PubMed] [Google Scholar]
- Heo J. O., Chang K. S., Kim I. A., Lee M. H., Lee S. A., Song S. K., et al. (2011). Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 108 2166–2171. 10.1073/pnas.1012215108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., et al. (2006). Overexpressing a NAM, ATAF, AND CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103 12987–12992. 10.1073/pnas.0604882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Li H., Pang H., Fu J. (2012). Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass (Lolium perenne) differing in salt tolerance. J. Plant Physiol. 169 146–156. 10.1016/j.jplph.2011.08.020 [DOI] [PubMed] [Google Scholar]
- Jensen M. K., Kjaersgaard T., Nielsen M. M., Galberg P., Petersen K., O’Shea C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426 183–196. 10.1042/BJ20091234 [DOI] [PubMed] [Google Scholar]
- Jiang M., Zhang J. (2002a). Involvement of plasma-membrane NADPH oxidase in abscisic acid-and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 215 1022–1030. 10.1007/s00425-002-0829-y [DOI] [PubMed] [Google Scholar]
- Jiang M., Zhang J. (2002b). Waters tress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 53 2401–2410. 10.1093/jxb/erf090 [DOI] [PubMed] [Google Scholar]
- Jin J., Zheng Y., Boeglin W. E., Brash A. R. (2013). Biosynthesis, isolation, and NMR analysis of leukotriene a epoxides: substrate chirality as a determinant of the cis or trans epoxide configuration. J. Lipid Res. 54 754–761. 10.1194/jlr.M033746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubany-Mari T., Munne-Bosch S., Lopez-Carbonell M., Alegre L. (2009). Hydrogen peroxide is involved in the acclimation of the mediterranean shrub, cistus albidus l., to summer drought. J. Exp. Bot. 60 107–120. 10.1093/jxb/ern274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai X., Chen S., Li T., Ma X., Liang X., Ding X., et al. (2015). OSGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 15:141. 10.1186/s12870-015-0532-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Kim B., Song S. K., Heo J. O., Yu N. I., Lee S. A., et al. (2008). Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 67 659–670. 10.1007/s11103-008-9345-1 [DOI] [PubMed] [Google Scholar]
- Li X., Qian Q., Fu Z., Wang Y., Xiong G., Zeng D., et al. (2003). Control of tillering in rice. Nature 422 618–621. 10.1038/nature01518 [DOI] [PubMed] [Google Scholar]
- Liu Y., Huang W., Xian Z., Hu N., Lin D., Ren H., et al. (2017). Overexpression of SlGRAS40 in tomato enhances tolerance to abiotic stresses and influences auxin and gibberellin signaling. Front. Plant Sci. 8:1659. 10.3389/fpls.2017.01659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Chen Z., Gao J., Gong Z. (2014). Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J. 79 44–55. 10.1111/tpj.12534 [DOI] [PubMed] [Google Scholar]
- Ma H. S., Liang D., Shuai P., Xia X. L., Yin W. L. (2010). The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 61 4011–4019. 10.1093/jxb/erq217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose M., Ekengren S. K., Melech-Bonfil S., Martin G. B., Sessa G. (2006). A novel link between tomato GRAS genes, plant disease resistance and mechanical stress response. Mol. Plant Pathol. 7 593–604. 10.1111/j.1364-3703.2006.00364.x [DOI] [PubMed] [Google Scholar]
- Miao Y., Lv D., Wang P., Wang X. C., Chen J., Miao C., et al. (2006). An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18 2749–2766. 10.1105/tpc.106.044230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Minami M., Takase H., Hotta Y., Hiratsuka K. (2003). Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 278 20865–20873. 10.1074/jbc.M301712200 [DOI] [PubMed] [Google Scholar]
- Pei Z. M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406 731–734. 10.1038/35021067 [DOI] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D. E., King K. E., Cowling R. J., Murphy G. P., et al. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. 10.1101/gad.11.23.3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips K., Ludidi N. (2017). Drought and exogenous abscisic acid alter hydrogen peroxide accumulation and differentially regulate the expression of two maize RD22-like genes. Sci. Rep. 7:8821. 10.1038/s41598-017-08976-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh L. D., Wysocka-Diller J. W., Camilleri C., Bouchez D., Benfey P. N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18 111–119. 10.1046/j.1365-313X.1999.00431.x [DOI] [PubMed] [Google Scholar]
- Raghavendra A. S., Gonugunta V. K., Christmann A., Grill E. (2010). ABA perception and signalling. Trends Plant Sci. 15 395–401. 10.1016/j.tplants.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Rodriguez M. C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61 621–649. 10.1146/annurev-arplant-042809-112252 [DOI] [PubMed] [Google Scholar]
- Saxena I., Srikanth S., Chen Z. (2016). Cross talk between H2O2 and interacting signal molecules under plant stress response. Front. Plant Sci. 7:570. 10.3389/fpls.2016.00570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G., Theres K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. U.S.A. 96 290–295. 10.1073/pnas.96.1.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31 279–292. 10.1046/j.1365-313X.2002.01359.x [DOI] [PubMed] [Google Scholar]
- Silverstone A. L., Ciampaglio C. N., Sun T. P. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. 10.1016/S1369-5266(98)80052-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X. M., Liu T. K., Duan W. K., Ma Q. H., Ren J., Wang Z., et al. (2014). Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp. pekinensis). Genomics 103 135–146. 10.1016/j.ygeno.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Sparkes I., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1 2019–2025. 10.1038/nprot.2006.286 [DOI] [PubMed] [Google Scholar]
- Sun X., Xue B., Jones W. T., Rikkerink E., Dunker A. K., Uversky V. N. (2011). A functionally required unfoldome from the plant kingdom: intrinsically disordered n-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 77 205–223. 10.1007/s11103-011-9803-z [DOI] [PubMed] [Google Scholar]
- Tian C., Wan P., Sun S., Li J., Chen M. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54 519–532. 10.1023/B:PLAN.0000038256.89809.57 [DOI] [PubMed] [Google Scholar]
- Torres-Galea P., Hirtreiter B., Bolle C. (2013). Two GRAS proteins, SCARECROW-LIKE21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, function cooperatively in phytochrome a signal transduction. Plant Physiol. 161 291–304. 10.1104/pp.112.206607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Galea P., Huang L. F., Chua N. H., Bolle C. (2006). The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome a responses. Mol. Genet. Genomics 276 13–30. 10.1007/s00438-006-0123-y [DOI] [PubMed] [Google Scholar]
- Tyler L., Thomas S. G., Hu J., Dill A., Alonso J. M., Ecker J. R., et al. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. 10.1104/pp.104.039578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu C., Xie B., Liu Y., Cui J., Chen G., et al. (2012). Model analysing the antioxidant responses of leaves and roots of switchgrass to NACL-salinity stress. Plant Physiol. Biochem. 58 288–296. 10.1016/j.plaphy.2012.06.021 [DOI] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43 1035–1039. 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- Yang M., Yang Q., Fu T., Zhou Y. (2010). Overexpression of the Brassica napus BnLAS gene in Arabidopsis affects plant development and increases drought tolerance. Plant Cell Rep. 30 373–388. 10.1007/s00299-010-0940-7 [DOI] [PubMed] [Google Scholar]
- Yang S. D., Seo P. J., Yoon H. K., Park C. M. (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23 2155–2168. 10.1105/tpc.111.084913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Zhang A., Jiang M., Zhang J., Ding H., Xu S., Hu X., et al. (2007). Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol. 175 36–50. 10.1111/j.1469-8137.2007.02071.x [DOI] [PubMed] [Google Scholar]
- Zhang Z. L., Ogawa M., Fleet C. M., Zentella R., Hu J., Heo J. O., et al. (2011). SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 2160–2165. 10.1073/pnas.1012232108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.