Abstract
ANRIL (CDKN2B antisense RNA 1, CDKN2B-AS1) is involved in the progression of various cancers. However, its role in head and neck squamous cell carcinoma (HNSCC) remains unclear. In this study, we found that ANRIL expression was upregulated in HNSCC and correlated with tumor progression. Further functional analysis showed that knockdown of ANRIL significantly inhibited proliferation in vivo and in vitro. ANRIL functioned as a ceRNA (competing endogenous RNAs) for miR-125a-3p and upregulated FGFR1 (fibroblast growth factor receptor-1), which could promote tumor growth. Moreover, we confirmed that ANRIL promoted HNSCC activity via FGFR1 with a FGFR1 inhibitor in vivo and in vitro. Thus, it could be concluded that ANRIL promoted the progression of HNSCC via miR-125a-3p/FGFR1/MAPK signaling, which might provide a new target for the diagnosis and treatment of HNSCC.
Keywords: Head and neck squamous cell carcinoma, ANRIL, miR-125a-3p, FGFR1, proliferation
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignant tumor worldwide [1]. Despite recent treatment advances, the overall 5-year survival rate of HNSCC remains less than 50%, and many HNSCC patients will eventually develop metastatic recurrence [2]. Although a number of risk factors, such as smoking, alcohol consumption, human papilloma virus (HPV) and Epstein-Barr virus (EBV) infection, adverse work environment and poor nutrition, have been identified, the mechanism of HNSCC carcinogenesis remains elusive. Clearly, a better understanding of the mechanism that facilitates HNSCC progression is essential to improve the treatment of HNSCC. Long non-coding RNAs (lncRNAs), a newly discovered class of ncRNAs, are non-protein coding molecules of more than 200 nucleotides (nt) in length. However, although more than 3000 human lncRNAs have been identified, less than 1% of them have been characterized [3]. There is increasing evidence that lncRNAs play an essential role in various physiological cellular processes, such as decoys, scaffolds for interacting proteins, and functions in chromatin remodelling and post-transcriptional modifications [4]. Upregulated lncRNA expression has been implicated in the pathogenesis of many types of cancers. For example, MALAT1 is upregulated in lung [5] and colorectal cancers [6], and overexpression of MALAT1 is also associated with increased cell proliferation, cell migration and tumor metastasis [6-8]. HOTAIR can function as an oncogene for predicting the prognosis of non-small cell lung cancer (NSCLC) and determining whether a patient can benefit from chemotherapy [9,10].
The lncRNA ANRIL (CDKN2B antisense RNA 1, CDKN2B-AS1) is transcribed as a 3.8-kb lncRNA that binds to the Polycomb Repressor Complex (PRC) [11]. Recent studies have shown that dysregulation of ANRIL could participate in the progression of many human diseases [11-13]. Zhang et al. found that ANRIL knockdown inhibited the proliferation and invasion and promoted the apoptosis of cervical cancer cells by sponging miR-186 [14]. ANRIL was overexpressed in pancreatic cancer, which could promote epithelial-mesenchymal transition by activating the ATM-E2F1 signaling pathway both in vitro and in vivo [15]. Chai et al. showed that ANRIL could modulate the carcinogenesis of oral cancer and the relationship between ANRIL and miR-125a in vitro, and it might sponge miR-125a in OSCC [16]. However, the biological function of ANRIL in vivo and the mechanism of downstream regulation through miR-125a in OSCC remain unclear.
In the present study, we investigated the expression of ANRIL and its biological function in HNSCC in vitro and in vivo. Then, we explored the ceRNA network potentially involved in regulating tumorgenesis in HPV negative HNSCC. The results indicated that ANRIL could be novel biomarkers and therapeutic strategies for HNSCC.
Materials and mathods
Patients
Fresh HNSCC tissues and adjacent normal tissues were collected postoperatively from 49 HPV negative patients treated in the Department of Oral and Maxillofacial Head and Neck Oncology from April 2011 to January 2015. Tumors were classified according to the tumor-node-metastasis (TNM) system of classification (2010 version). This study was approved by the Human Research Ethics Committee of the Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China). All patients signed informed consent forms for sampling and molecular analysis. The clinical data of patients are shown in Table 1.
Table 1.
Relationship between lncRNA ANRIL expression and their clinicopathologic parameters in 49 of HNSCC patients
| Clinicopathologic parameters | Number of cases | Median expression of lncRNA ANRIL | |
|---|---|---|---|
|
| |||
| Mean ± s.d. | P-value | ||
| Gender | |||
| Male | 28 | 1.6992±0.3211 | 0.7 |
| Female | 21 | 1.4165±0.3091 | |
| Ages, years | |||
| ≤ 59 | 24 | 1.6667±0.3402 | 0.336 |
| > 59 | 25 | 1.4753±0.2951 | |
| T stage, TNM | |||
| T1+T2 | 37 | 1.3665±0.2247 | 0.005 |
| T3+T4 | 12 | 1.7262±0.4983 | |
| Clinical stage | |||
| I-II | 18 | 1.2038±0.2692 | 0.058 |
| III-IV | 31 | 1.7124±0.3181 | |
| Lymph node metastasis | |||
| Absent | 26 | 1.1078±0.2173 | 0.001 |
| Present | 23 | 1.6925±0.3529 | |
| Alcohol drinking | |||
| Absent | 27 | 1.4706±0.2830 | 0.427 |
| Present | 22 | 1.7131±0.3652 | |
| Smoking | |||
| Absent | 30 | 1.6199±0.2958 | 0.335 |
| Present | 19 | 1.4911±0.3421 | |
| Location | |||
| Lips | 12 | 1.8201±0.5256 | |
| Cheek | 5 | 1.0918±0.4883 | 0.304 |
| Buccal mucosa | 13 | 1.2690±0.3520 | |
| Tongue | 19 | 1.6466±0.3778 | |
Abbreviations: HNSCC, head and neck squamous cell carcinoma; TNM, tumor node metastasis. P-value represents the probability from a Student’s t-test for lncRNA ANRIL expression between variable subgroups.
Cell culture
CAL-27 and SCC-9 cell lines were obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). HN4, HN6 and HN30 cell lines were obtained from the US National Institutes of Health. Normal oral epithelial cells (Normal) were isolated from adjacent normal tissues of HNSCC patients by primary culture. All cells except SCC-9 were cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO-BRL, USA) supplemented with 10% heat-inactivated FBS (GIBCO-BRL), penicillin (100 units/mL), and streptomycin (100 μg/mL) at 37°C in a humidified 5% CO2 atmosphere, while SCC-9 cells were maintained in DMEM/F12 medium containing 10% FBS. In addition, normal primary head and neck epithelial cells were cultured in keratinocyte serum-free medium (KSF; GIBCO-BRL, USA) with 0.2 ng/mL recombinant epidermal growth factor (rEGF; Invitrogen, USA).
Cell transfection
ANRIL siRNA, control siRNA, ANRIL overexpression (ANRIL OE), control ANRIL overexpression (ANRIL OE NC), miR-125a-3p mimic, control mimic (NC-mimic), miR-125a-3p inhibitor, control inhibitor (NC-inhibitor), and FGFR1 siRNA were synthesized by GenePharma Co. (Shanghai, China). Stably transfected cells were grown in 6-well plates and transfected using Lipofectamine 2000 according to the manufacturer’s instructions. Cells were collected for real-time PCR or western blot analyses 48 h after transfection. The final concentrations of miRNAs and plasmids used in this study were as follows: ANRIL siRNA/control siRNA 30 nM/ml, ANRIL OE/ANRIL OE NC 30 nM/ml, miR-125a-3p mimic/NC mimic 120 nM/ml, miR-125a-3p inhibitor/NC-inhibitor 200 nM/ml, and FGFR1 siRNA 30 nM/ml. The lentiviral shRNA clones targeting human ANRIL, GGUCAUCUCAUUGCUCUAU, was purchased from GenePharma (Shanghai, China). Transfected cell lines were screened for stable expression of shANRIL using puromycin in the culture medium for 10 d.
Luciferase reporter assay
PmirGLO, pmirGLO-NC-wt, pmirGLO-ANRIL-wt and pmirGLO-ANRIL-mut were co-transfected with miRNA-125a-3p mimics or miRNA NC into shANRIL CAL27 cells in a 6-well dish using Lipofectamine 2000 reagent (Invitrogen, USA) according to the manufacturer’s instructions, respectively. PmirGLO, pmirGLO-FGFR1-wt and pmirGLO-FGFR1-mut were transfected into shANRIL CAL27 cells using Lipofectamine 2000 reagent (Invitrogen, USA) according to the manufacturer’s instructions, respectively. The relative luciferase activity was normalized to the Renilla luciferase activity 48 h after transfection. Cells were co-transfected with the pPenilla Renilla luciferase reporter to normalize for transfection efficiency. The transfection medium was replaced with fresh medium 6 h later, and cells were cultured for another 24 h. Cells were pre-treated with external stimulus for 12 h and harvested in passive lysis buffer. Finally, the luciferase activity was measured using the Dual Luciferase System (Promega, USA).
RNA preparation, RT and qPCR
The total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen) and then reverse transcribed into cDNAs using the Primer-Script One Step RT-PCR kit (TaKaRa, Dalian, China). The cDNA template was amplified by real-time RT-PCR using the SYBR Premix Dimmer Eraser kit (TaKaRa). Gene expression in each sample was normalized to the GADPH expression. The primer sequences used were as follows: for GAPDH, 5’-GTCAACGGATTTGGTCTGTATT-3’ (forward) and 5’-AGTCTTCTGGGTGGCAGTGAT-3’ (reverse); for ANRIL, 5’-CTTATTTTATTCCTGGCTCCCCT-3’ (forward) and 5’-ATCATTCTCCTCAAATTACAGAG-3’ (reverse). Real-time PCR was performed in triplicate on the ABI7300 system (Applied Biosystems, Carlsbad, CA, USA). The relative expression fold change of mRNAs was calculated by the 2-ΔΔCt method, and the primers used in this study were listed in Supplementary Table 1.
Cell fractionation
Cells were grown in 15-cm dishes (NUNC). One million cells were harvested for each sample and nuclear/cytoplasmic fractionation was performed using Nuclei EZ Lysis Buffer (Sigma) according to the manufacturer’s instructions. In the ANRIL siRNA knockdown experiments, CAL27 cells were harvested 48 h after transfection.
Real-time qPCR of mature miRNAs
Primers were designed on the basis of mature miRNA sequences. The total RNA was isolated from cells by TRIzol extraction (Invitrogen, Shanghai, China) on ice in an RNA-free environment and then subjected to reverse transcription using the miRcute miRNA first-stand cDNA Kit (TIANGEN, Shanghai, China) according to the manufacturer’s instructions. Poly (A) polymerase was used to add a poly (A) tail to the total RNA before reverse transcription. Real-time PCR was performed in triplicate under the following conditions: degeneration at 95°C for 5 min, 40 cycles of annealing at 60°C for 30 s, and extension at 72°C for 30 s. miRNA expression was analyzed using SYBR Green methods and a miRcute miRNA qPCR kit according to the manufacturer’s instructions (TIANGEN, Shanghai, China). Small nuclear RNA U6 was used as the internal control. The relative expression fold change of mRNAs was calculated by the 2-ΔΔCt method.
Western blotting
Cells from each group were detached with trypsin, centrifuged, and washed twice with pre-chilled PBS. Cell lysis buffer was added and incubated on ice for protein extraction. The protein concentration was determined using a BCA Protein Assay Kit (Beyotime, China). Equal amounts of proteins were separated via 8% SDS-PAGE and then transferred to a PVDF membrane (Millipore Corporation, Billerica, MA, USA). The membrane was soaked in 10% skim milk in PBS containing 0.1% Tween-20 (pH 7.2) for 2 h and incubated with an appropriate amount of primary antibody at 4°C overnight. Bound antibodies were detected by peroxidase-conjugated secondary antibodies (KPL, Gaithersburg, MD, USA) and chemiluminescence (Millipore Corporation). GAPDH was used as a control. Antibodies (1:1000) for FGFR1, AKT, p-AKT, ERK, and p-ERK were purchased from Cell Signaling Technology (Boston, MA, USA).
Cell viability assay
Cell proliferation was determined 24 h after transfection by a Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. ANRIL siRNA-transfected CAL27 and HN6 cells were seeded into 96-well plates at a density of 1 × 103 cells per well with 100 μl of medium and incubated at 37°C. At the indicated time points, 10 μl of CCK-8 solution was added to each well. After incubation at 37°C for 1 h, the absorbance was measured with a plate reader at 450 nm, and the growth curves were examined to determine the growth rates.
Colony forming assay
A total of 1000 ANRIL siRNA-transfected/ANRIL OE-transfected CAL27 and HN6 cells were placed in fresh 6-well plates and maintained in medium containing 10% FBS, and the medium was replaced every 3 days. After 14 days, cells were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA). Visible colonies were manually counted. Wells were assessed in triplicate for each group.
Flow cytometric analysis
Cells transfected with the desired plasmid and negative control were plated in 6-well plates, respectively. After 48 h of incubation, cultures were incubated with propidium iodide for 30 min in dark, and then they were collected for the analysis of cell cycle using a flow cytometer (FACSCalibur, BD Biosciences, San Jose, CA, USA) after propidium iodide staining. The results were expressed as the percentages of cells in the G0/G1, S and G2/M phases of the cell cycle.
Gene ontology (GO) analysis
GO analysis was performed to determine the biological implications of unique genes in the significant or representative profiles or the target genes of miR-125a-3p. We downloaded the GO annotations from NCBI (http://www.ncbi.nlm.nih.gov/), UniProt (http://www.uniprot.org/) and Gene Ontology (http://www.geneontology.org/) database. Significant GO categories were identified by Fisher’s exact test, and FDR was used to correct the P-values.
RNA immunoprecipitation (RIP) assay
RIP assay was performed by the EZ-Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA). Cells were lysed in complete RIP lysis buffer, and then 100 μl of cell lysate was incubated with RIP buffer containing magnetic beads conjugated with human anti-Ago2 antibody (1:50 dilution, Millipore) and negative control normal mouse IgG. Samples were incubated with Proteinase K buffer, and then target RNA was extracted for future use.
Immunohistochemistry (IHC)
IHC was performed on paraformaldehyde-fixed paraffin sections using Ki-67 antibody (Abcam) by the streptavidin peroxidase-conjugated method. The percentage of positive tumor cells was graded according to the following criteria: 0, < 10%; 1, 10-30%; 2, 31-50%; 3, > 50%, and then patients were divided into low- (0 or 1) and high-Ki-67 expression group (2 or 3).
Xenograft mouse model
CAL27 cells (1 × 106) stably expressing LV-NC or LV-shANRIL were subcutaneously injected into either side of the flank area of 6-week-old female athymic nude mice (n = 8 per group). Tumor volumes were measured (0.5 × length × width2) on a weekly basis. After 3 weeks, mice were killed and tumors were excised and subjected to immunohistochemical analysis of ki67 and western blot analysis of FGFR1, AKT, p-AKT, ERK and p-ERK. All animal experiments were performed in the Animal Laboratory Centre of the Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China) in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication number 85-23, revised 1996). The study protocol was approved by the Animal Care and Use Committee of the Ninth People’s Hospital.
Statistical statistics
All statistical analyses were performed using SPSS 20.0 (SPSS, Chicago, IL, USA). The expression differences between HNSCC and matched normal tissues were analysed using paired samples t-tests. Pearson correlation analysis was also performed. The expression differences between high/low stages, expression changes after transfection, S-phase fraction and cell proliferation assay results were analysed using independent samples t-tests. A two-sided P value less than 0.05 was considered statistically significant.
Results
ANRIL expression in HNSCC and its relationship with prognosis
The ANRIL levels in primary HNSCC and adjacent normal tissues were detected by qRT-PCR. ANRIL was significantly upregulated in HNSCC tissues compared with that in their normal counterparts (Figure 1A), and it was also elevated in samples with a high tumor stage (T stage) and lymph node dissemination stage (N stage) (N1/2, T3+T4) compared with that with a low T/N stage (N0, T1+T2) in HNSCC (Figure 1B, 1C). To further elucidate the clinical significance of ANRIL in HNSCC, we investigated the relationships between ANRIL expression and patients’ clinicopathological features, as shown in Table 1. Patients with an ANRIL expression higher than 50% in HNSCC tissues showed significantly poorer recurrence-free survival and overall survival (Figure 1D, 1E). Univariate Cox proportional hazards regression analyses revealed that the poor overall survival in HNSCC patients was associated with high ANRIL expression in HNSCC tissues (P < 0.001), high T stage (T3+T4; P < 0.001), and lymph node metastasis (P < 0.001) (Table 2). Multivariate analysis identified the high ANRIL expression in HNSCC tissues as an independent prognostic marker for the poor overall survival of HNSCC patients (Table 2). Then, we examined ANRIL expression in HNSCC cell lines, and the results showed that the ANRIL expression levels were increased in HNSCC cell lines compared with that in normal oral epithelial cells (Titled Normal), which was consistent with the biopsy results (Figure 1F).
Figure 1.
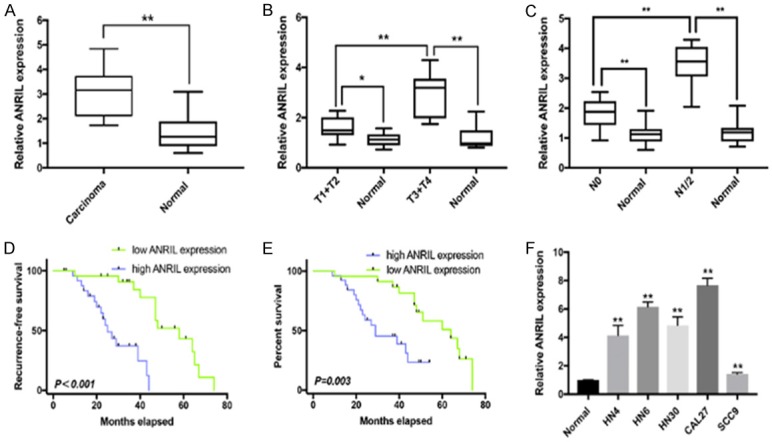
ANRIL expression levels in HNSCC and its clinical significance. A. Difference in ANRIL expression levels between HNSCC tissues and matched normal tissues. The ANRIL expression was normalized to GADPH. Significant differences between samples were analysed with paired samples t tests (n = 49, P < 0.0001). B. Relationship between ANRIL expression and primary tumor growth (P < 0.0001). The ANRIL expression in T1+T2/T3+T4 stage tumors was normalized to corresponding paired normal tissues. C. Relationship between ANRIL expression and lymph node metastasis (P < 0.0001). The ANRIL expression in N1+2/N0 stage tumors was normalized to the corresponding paired normal tissues. D, E. HNSCC patients with high ANRIL expression in tumor tissues showed significantly poorer recurrence-free survival and overall survival than those with low ANRIL expression. F. The ANRIL expression levels in five HNSCC cell lines (HN4, HNz6, HN30, CAL27 and SCC9) and normal oral epithelial cells (titled Normal). The ANRIL expression was normalized to that in Normal. The relative expression fold change of mRNAs was calculated by the 2-ΔΔCt method. Horizontal lines in the box plots represent the medians; the boxes represent the interquartile range; and the whiskers represent the 2.5th and 97.5th percentiles. *P < 0.05; **P < 0.01; n.s., not significant.
Table 2.
Univariate and multivariate analysis of factors for predicting poor prognosis in HNSCC patients (Cox proportional hazards regression model)
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% | P value | HR | 95% | P value | |
| Gender (Male vs. Female) | 0.987 | 0.465-2.085 | 0.972 | - | - | - |
| Ages, years (≤ 59 vs. ≥ 59) | 2.001 | 0.932-4299 | 0.075 | - | - | - |
| T stage, TNM (T1+T2 vs. T3+T4) | 7.018 | 2.721-18.104 | < 0.001 | 6.549 | 2.092-20.500 | 0.001 |
| Clinical stage (Grade I-II vs. Grade III-IV) | 2.123 | 0.935-4.822 | 0.072 | - | - | - |
| Lymph node metastasis (Absent vs. Present) | 7.917 | 3.206-19.552 | < 0.001 | 14.554 | 4.686-45.207 | < 0.001 |
| Alcohol drinking (Absent vs. Present) | 0.696 | 0.323-1.502 | 0.356 | - | - | - |
| Smoking (Absent vs. Present) | 1.388 | 0.647-2.978 | 0.400 | - | - | - |
| lncRNA ANRIL expression (Low vs. High) | 2.257 | 1.553-3.280 | < 0.001 | 1.939 | 1.278-2.944 | 0.002 |
HR, hazard ratio; CI, confidence interval.
ANRIL was required for tumor proliferation of HNSCC cells
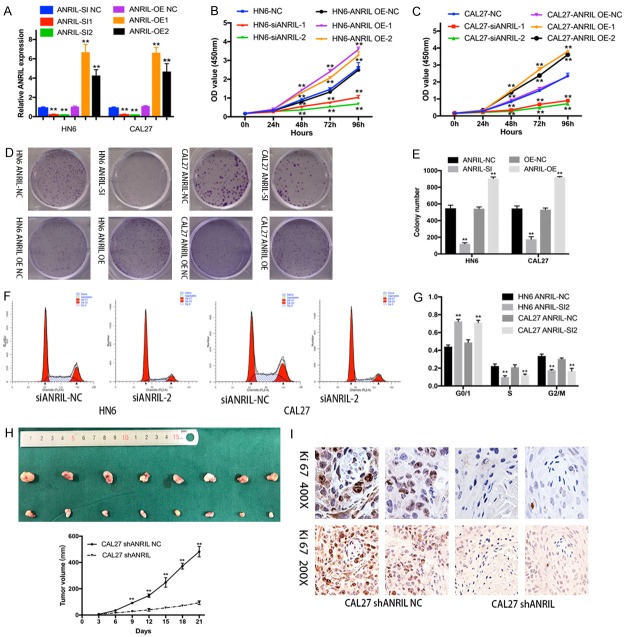
The functional role of ANRIL was evaluated using CAL27 and HN6 cells as they showed the highest ANRIL levels in HNSCC cells (Figure 1F). The inhibitory and overexpression efficiency was verified using qRT-PCR (Figure 2A). Knockdown of ANRIL significantly inhibited cell viability and proliferation in both HN6 and CAL27 cell lines (Figure 2B, 2C). Similarly, colony formation assays revealed that the clonogenic survival was significantly decreased by the inhibition of ANRIL (Figure 2D, 2E). In contrast, lncRNA ANRIL overexpresson (OE) in HNSCC cells increased cell viability and proliferation (Figure 2B-E). We further examined whether the effect of ANRIL on the proliferation of HNSCC cells was mediated by cell cycle progression. The results revealed that cell cycle progression was stalled at the G1-G0 phase (Figure 2F, 2G). Thus, we concluded that ANRIL could promote HNSCC proliferation in vitro.
Figure 2.
ANRIL promoted the growth of HNSCC in vitro and in vivo. A. ANRIL-specific siRNA 1 and 2 reduced endogenous ANRIL mRNA levels in HN6 and CAL27 cells. ANRIL OE1 and OE2 increased endogenous ANRIL mRNA levels. B, C. Cell growth viability was assayed in ANRIL siRNA NC-, siRNA 1/2-transfected and ANRIL OE NC, OE 1/2 transfected HN6 and CAL27 cells by CCK-8 assays. D, E. Representative results of colony formation assays of HN6 and CAL27 cells transfected with ANRIL siRNA NC, siRNA or ANRIL OE NC, OE. F, G. The cell cycle profile was examined in ANRIL siRNA NC- or siRNA 2-transfected HN6 and CAL27 cells by flow cytometry. H. Depletion of ANRIL inhibited the growth of subcutaneous tumors. Tumor volumes were measured every 3 d. Mice were killed at 21 d after injection for the excision of tumors. Scale bars, 1 mm. The results were represented by the mean ± s.d. of n = 8 mice in each group by the Mann-Whitney test. I. Representative images of Ki-67 IHC staining in HNSCC and normal tissues. Scale bar, 100 μm. Significant differences between groups were analysed using independent samples t test. Error bars represent the mean ± S.D. of triplicate experiments. *P < 0.05; **P < 0.01; n.s., not significant.
CAL27 cells stably transfected with LV-shANRIL and LV-NC were injected subcutaneously into nude mice. Strikingly, depletion of ANRIL inhibited tumorigenesis of these cells in vivo (Figure 2H). The staining intensity of Ki-67 was significantly decreased in the ANRIL-depleted group (Figure 2I). These results identified ANRIL as a potential therapeutic target for HNSCC.
Identification of potential ANRIL-targeted miRNAs
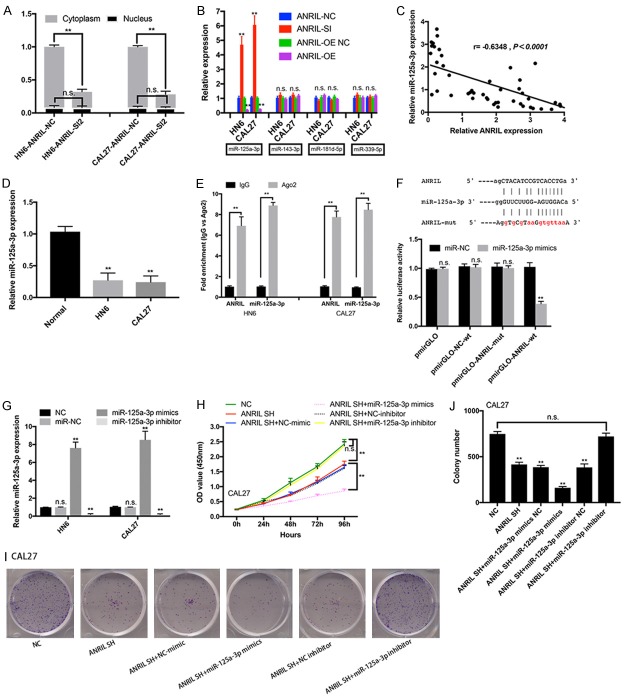
Previous studies have suggested that lncRNAs could serve as a ceRNAs to sponge miRNAs and thus regulated gene expression in cancers [17,18]. Here, we hypothesized that ANRIL might also act as a ceRNA to sponge miRNA. To test this hypothesis, we first examined expression levels and locations of ANRIL in HN6 and CAL27 cells. As shown in Figure 3A, a higher expression level of ANRIL was found in cytoplasm than in nucleus, suggesting that ANRIL interacted with miRNAs in cytoplasm and functioned as an endogenous sponge for miRNAs.
Figure 3.
Identification of miRNA-125a-3p as a target of ANRIL. A. Changes in the expression of ANRIL in cytoplasm and nucleus of ANRIL siRNA NC- or siRNA 1/2-transfected HN6 and CAL27 cells. B. Relative expression (miR-125a-3p, miR-145-3p, miR-181d-5p, and miR-339-5p) in ANRIL siRNA- or OE-transfected HN6 and CAL27 cells. C. The correlation between ANRIL transcript level and miR-125a-3p mRNA level was measured in 49 HNSCC tissues. The ΔCt values (normalized to GADPH) were subjected to Pearson correlation analysis. D. The miR-125a-3p expression levels in normal oral epithelial cells (titled Normal), HN6 and CAL27 cells. E. Associations of miR-125a-3p and ANRIL with Ago2. HN6 and CAL27 cell lysates were collected for RIP using an anti-Ago2 antibody. miR-125a-3p and ANRIL were detected by qRT-PCR. F. Alignment of potential ANRIL base pairing with miR-125a-3p as identified by Starbase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php). Mutant ANRIL at the putative binding site. Luciferase activity in CAL27 cells co-transfected with miR-NC or miR-125a-3p mimics and luciferase reporters containing nothing, ANRIL, NC mutant transcripts or ANRIL mutant transcripts. The results are presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. G. Change in miR-125a-3p expression levels in miR-NC-, miR-125a-3p mimic-, and miR-125a-3p inhibitor-transfected HN6 and CAL27 cells. Cell growth and viability were assayed in shANRIL NC-, shANRIL, shANRIL+miR-125a-3p mimic-NC-, shANRIL+miR-125a-3p mimic-, shANRIL+miR-125a-3p inhibitor-NC-, and shANRIL+miR-125a-3p inhibitor-transfected HN6 and CAL27 cells by CCK-8 and colony formation assays. H-J. *P < 0.05; **P < 0.01; n.s., not significant.
Several miRNAs with ANRIL binding sites were identified using Starbase (http://starbase.sysu.edu.cn/mirLncRNA.php). miR-125a-3p was significantly increased after knockdown of ANRIL (Figure 3B); while lncRNA ANRIL overexpression decreased the expression of miR-125a-3p in cells (Figure 3B). Furthermore, the ANRIL transcript level was significantly negatively correlated with the miRNA-125a-3p expression level in HNSCC tissues (Figure 3C). The miR-125a-3p expression levels were also determined in HNSCC cells (Figure 3D). To test the hypothesis, we performed RIP analysis with antibodies against AGO2, which was a vital component of the RNA-induced silencing complex (RISC). Figure 3E showed that ANRIL and miR-125a-3p were increased by 6.5-9.4-fold following immunoprecipitation using the anti-Ago2 antibody compared to IgG. Thus, ANRIL might be recruited to AGO2-related RISCs and functionally interacted with miR-125a-3p in HNSCC cells. We also investigated whether ANRIL bound to miR-125a-3p to exert its function. It was found that miR-125a-3p mimics reduced the luciferase activities of wild-type (WT) ANRIL reporter vectors, but the luciferase activities in cells transfected with NC or ANRIL mutant and miR-125a-3p mimics were comparable to that of control cells (Figure 3F). The CCK-8 assay and colony formation assay showed that miR-125-3p mimic significantly impaired proliferation of CAL27 cells under ANRIL knockdown, which, however, could be recovered by miR-125a-3p inhibitor (Figure 3H-J). Thus, ANRIL might act as a sponge of miR-125a-3p to inhibit its expression by direct binding to miR-125a-3p.
MiR-125a-3p regulated ANRIL-induced proliferation in HNSCC cells by targeting FGFR1
To elucidate the molecular mechanism underlying the role of miR-125a-3p in tumorigenesis, we performed transcriptome sequencing of CAL27 cells with stable depletion of miR-125a-3p, and identified 104 genes with significant expression changes compared with normal CAL27 cells. GO analysis showed that these genes were associated with the mitogen-activated protein kinases (MAPK) signaling pathway, supporting the role of miR-125a-3p in HNSCC proliferation (Figure 4A). Among these genes, 7 mRNAs were chosen as they had vital functions in cell proliferation (Figure 4B). qRT-PCR analysis showed that FGF1 and FGFR1 were downregulated in ANRIL depleted cells. However, only FGFR1 was further downregulated in ANRIL depleted cells by miR-125a-3p mimic, and its expression was upregulated when miR-125a-3p was inhibited (Figure 4C).
Figure 4.
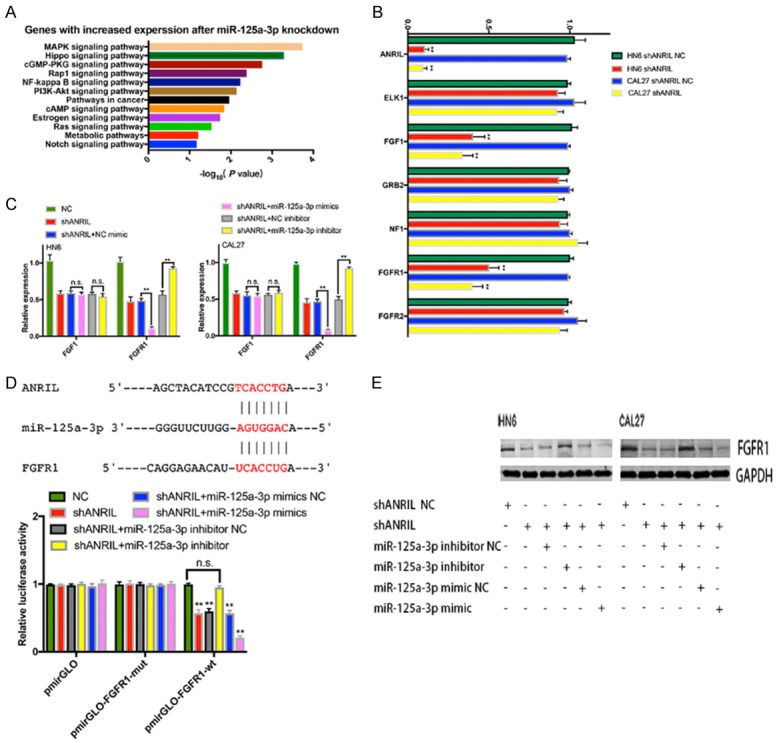
FGFR1 was the target gene of miR-125a-3p in HNSCC. A. GO analysis of miR-125a-3p-regulated gene expression. Fisher’s exact P values for each enriched functional category were shown. B. Changes in the expression of target genes (ELK1, FGF1, GRB2, NF1, FGFR1 and FGFR2) after knockdown of ANRIL. C. Identification of the target genes of miR-125a-3p in HN6 or CAL27 cells by transfecting miR-125a-3p mimics or inhibitor. D. Luciferase activity in CAL27 cells transfected with luciferase reporters containing or not containing FGFR1 3’-UTR. The results were represented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. E. The protein levels of FGFR1 were confirmed by western blot analysis of shANRIL NC-, shANRIL-, shANRIL+miR-125a-3p mimic-NC-, shANRIL+miR-125a-3p mimic-, shANRIL+miR-125a-3p inhibitor-NC-, shANRIL+miR-125a-3p inhibitor-transfected HN6 and CAL27 cells. Error bars represent the mean ± S.D. of triplicate experiments. *P < 0.05; **P < 0.01; n.s., not significant.
Our results revealed that ANRIL and FGFR1 shared the same miRNA-responsive element in their sequences and showed the same miR-125a-3p-dependent regulation pattern (Figure 4D). To explore whether the observed effects depended on the regulation of the 3’ untranslated regions (3’-UTR) of FGFR1, we cloned the sequence of the FGFR1 3’-UTR region with the miR-125a-3p predicted binding sites downstream of a luciferase reporter and co-transfected it into CAL27 cells with miR-125a-3p mimic, miR-125a-3p inhibitor or the respective scrambled control. Figure 4D showed that ANRIL knockdown decreased the luciferase activity of WT FGFR1 reporter vectors transfected CAL27 cells, which was recovered by the miR-125a-3p inhibitor but amplified by the miR-125a-3p mimics. The luciferase activities in cells transfected with FGFR1 mutant transfected CAL27 cells were comparable to that of control cells. We also found that FGFR1 protein levels were decreased by miR-125a-3p mimic but increased by miR-125a-3p inhibitor in CAL27, HN6, shANRIL-CAL27 and shANRIL-HN6 cells (Figure 4E). Thus, FGFR1 could be a potential target of miR-125a-3p.
ANRIL enhanced HNSCC proliferation by regulating the miR-125a-3p/FGFR1/MAPK signaling pathway
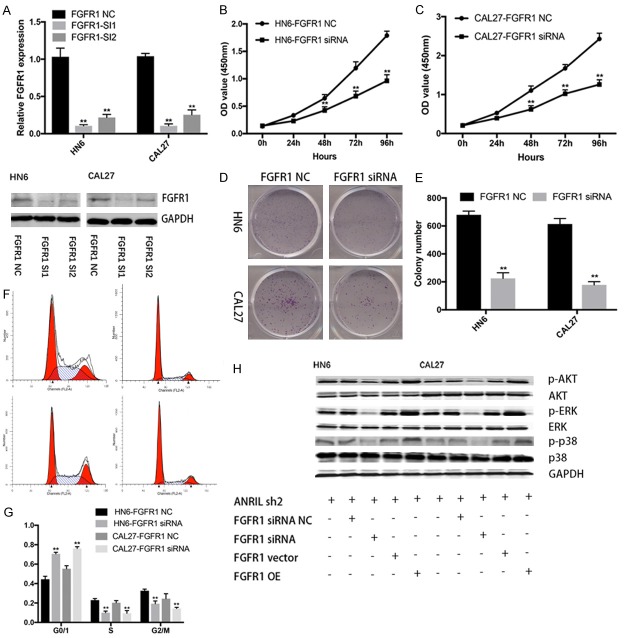
Previous studies showed that FGFR-mediated reactivation of MAPK signaling pathway attenuated the antitumor effects of imatinib in gastrointestinal stromal tumors [19]. We found that knockdown of FGFR1 significantly inhibited viability and proliferation of both HN6 and CAL27 cell lines compared with that of control cells (Figure 5A-C). The percentage of S-phase cells and the number of colonies were also reduced after FGFR1 knockdown (Figure 5D-G). Then, we investigated the expression of MAPK-related proteins (P-p38, P-ERK and P-AKT) in ANRIL depleted HNSCC cells. We found that FGFR1 knockdown decreased MAPK-related protein expression levels in ANRIL depleted HNSCC cells, while the opposite was true in FGFR1 overexpressed cells (Figure 5H). All these results implied that ANRIL promoted HNSCC proliferation by regulating the miR-125a-3p/FGFR1/MAPK signaling pathway in vitro.
Figure 5.
FGFR1 promoted the growth of HNSCC cells by activating MAPK signaling. A. FGFR1-specific siRNA 1 and 2 reduced endogenous FGFR1 mRNA levels in HN6 and CAL27 cells. B, C. Cell viability was assayed in FGFR1 siRNA NC- or siRNA 1-transfected HN6 and CAL27 cells by CCK-8 assays. D, E. Representative results of colony formation assays of HN6 and CAL27 cells transfected with FGFR1 siRNA NC or siRNA 1. F, G. Cell cycle profile was examined in FGFR1 siRNA NC- or siRNA 1-transfected HN6 and CAL27 cells by flow cytometry. H. The protein levels of a panel of core factors in MAPK signaling were evaluated in FGFR1 knockdown or overexpression shANRIL HN6 and CAL27 cells. *P < 0.05; **P < 0.01; n.s., not significant.
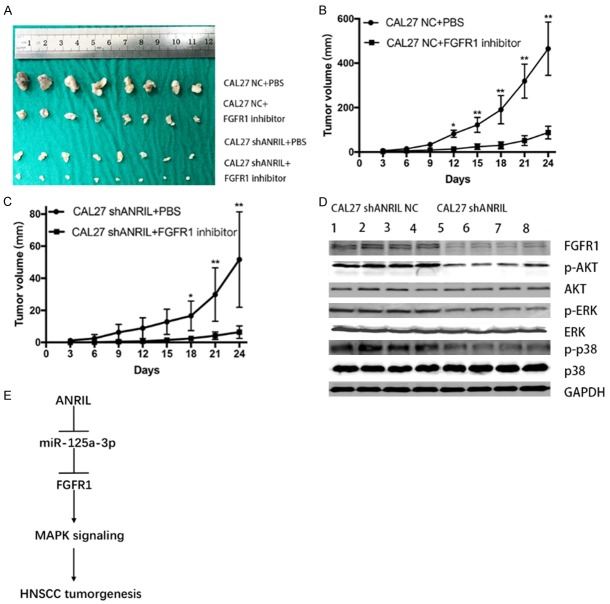
FGFR1 inhibitor inhibited HNSCC tumor growth in vivo
To further determine the tumorigenic potential of FGFR1 in vivo, we inoculated CAL27 cells transfected with LV-NC or LV-shANRIL into nude mice, and then injected PBS or FGFR1 inhibitors (PD173074, MCE; 25 mg/kg, i.p.). A reduction in tumor volume was found at the end of the experiment in mice injected with inhibitors compared with those with PBS (Figure 6A-C). Western blot analysis of tumor tissues resected from PBS-injected mice showed that ANRIL expression was positively correlated with FGFR1, p-p38, p-ERK and p-AKT; but negatively correlated with p38, ERK and AKT (Figure 6D). These results further verified that ANRIL promoted HNSCC tumorigenesis by activating the FGFR1/MAPK signaling pathway in vivo (Figure 6E).
Figure 6.
FGFR1 inhibitor inhibited the growth of subcutaneous tumors. Tumor volumes were measured every 3. Mice were killed at 24 d after injection of cells and FGFR1 inhibitor, and the tumors were excised (A-C). Scale bars, 1 mm. The results were represented as the mean ± s.d. of n = 8 mice in each group by the Mann-Whitney test. (D) The protein levels of a panel of core factors in MAPK signaling were reduced in shANRIL NC or shANRIL tumor tissues. (E) A schematic of the proposed mechanism of ANRIL in HNSCC. ANRIL functioned as a ceRNA to sponge miR-125a-3p and upregulated the expression of FGFR1 as well as FGFR1 downstream effector that promoted HNSCC progression. *P < 0.05; **P < 0.01; n.s., not significant.
Discussion
lncRNAs have been reported to be involved in various human diseases, especially cancers [20-22]. Dysregulated lncRNAs are significantly related to tumorigenesis, metastasis, prognosis and diagnosis [23,24]. Thus, more efforts should be made to clarify the biological and molecular mechanisms of lncRNAs in cancers. lncRNA ANRIL has been reported to promote proliferation of numerous solid tumors [25,26]. It also indicated a poor prognosis of cervical cancers, and the ANRIL/miR-186 axis might advance carcinogenesis via the PI3K/Akt pathways [14]. Moreover, ANRIL overexpression enhanced cell proliferation and migration through the let-7a/TGF-β1/Smad cascade [27] and inhibited apoptosis by interacting with KLF2 and P21 [28]. Our study showed that ANRIL was highly expressed in HNSCC biopsies, and K-M analysis showed that ANRIL expression was positively correlated with poor prognosis. Knockdown of ANRIL significantly inhibited cell proliferation and stalled cell cycle progression at the G1-G0 phase in HN6 and CAL27 cells. Thus, ANRIL may play an oncogenic role in HNSCC and holds great promise as a novel diagnostic and prognostic biomarker for HNSCC.
There is evidence that a widespread interaction network of ceRNAs is involved in carcinogenesis, in which ncRNAs could regulate modulatory RNAs by binding and titrating them off their binding sites on protein-coding mRNAs [1,29]. Previous studies have confirmed that ANRIL could act as a molecular sponge by binding miR-125a to modulate tumorigenesis, metastasis and invasion of hepatocellular carcinoma cells [30]. MiR-125a-3p, a tumor-suppressor microRNA, has been reported to be involved in several solid tumors, such as non-small cell lung carcinoma [31], glioma [32] and gastric cancer [33]. Reduced miR-125a-3p could increase the expression of endogenous MTA1 and initiate docetaxel acquired resistance in prostate cancer cells [34]. MiR-125a-3p also played a vital role in mediating colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via the PI3K-Akt pathway [35]. In our study, we identified several miRNA binding sites of ANRIL through a public database, and RIP assays and luciferase assays confirmed that ANRIL might act as a sponge of miR-125a-3p to inhibit the expression of certain miRNAs by direct binding in HNSCC cells. Thus, ANRIL may regulate oncogenesis of HNSCC through a ceRNA network via the ANRIL/miR-125a-3p axis.
FGFR1 is expected to be a potential prognostic biomarker and therapeutic target in HNSCC [36]. FGFR1 expression level was associated with sensitivity to a FGFR inhibitor (NVP-BGJ398) and predicted tyrosine kinase inhibitor sensitivity of PDX models in FGFR-specific inhibitor clinical trials [37]. FGFR1 could be regulated by miRNAs in some malignancies, including NSCLC [38], gastric carcinoma [39] and colorectal carcinoma [40]. We observed opposite correlations between mRNA or protein expression of FGFR1 and the miR-125a-3p levels in HNSCC cell lines, and luciferase assays confirmed that FGFR1 was a direct target of miR-125a-3p. Since MAPKs are the primary signaling cascade activated downstream of FGFR signaling [18,19], the expression levels of FGFR1 and MAPK-related proteins were elevated and miR-125a-3p was downregulated in ANRIL-overexpressed HNSCC cell lines. These findings indicated that ANRIL could regulate HNSCC growth by miR-125a-3p/FGFR1/MAPK signaling.
In summary, we first showed that ANRIL might serve as an oncogene by promoting proliferation of human HNSCC. Notably, we revealed a novel ANRIL/miR-125a-3p/FGFR1/MAPK signaling pathway regulatory network in HNSCC. These results may lead to the development of novel biomarkers and therapeutic strategies for HNSCC.
Acknowledgements
This study was supported by grants from the National Program on Key Research Project of China (2016YFC0902700), National Natural Science Foundation of China (No. 31140007, 81472516) and the project of Science and Technology Commission of Shanghai Municipality (Grant #14DZ1941402).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Datta J, Islam M, Dutta S, Roy S, Pan Q, Teknos TN. Suberoylanilide hydroxamic acid inhibits growth of head and neck cancer cell lines by reactivation of tumor suppressor microRNAs. Oral Oncol. 2016;56:32–39. doi: 10.1016/j.oraloncology.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 6.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 7.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, De W, Wang Z, Wang R. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao JJ, Hao S, Wang LL, Hu CY, Zhang S, Guo LJ, Zhang G, Gao B, Jiang Y, Tian WG, Luo DL. Long non-coding RNA ANRIL promotes the invasion and metastasis of thyroid cancer cells through TGF-beta/Smad signaling pathway. Oncotarget. 2016;7:57903–57918. doi: 10.18632/oncotarget.11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D, Sun G, Zhang H, Tian J, Li Y. Long non-coding RNA ANRIL indicates a poor prognosis of cervical cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed Pharmacother. 2017;85:511–516. doi: 10.1016/j.biopha.2016.11.058. [DOI] [PubMed] [Google Scholar]
- 13.Wei X, Wang C, Ma C, Sun W, Li H, Cai Z. Retraction note: long noncoding RNA ANRIL is activated by hypoxia-inducible factor-1alpha and promotes osteosarcoma cell invasion and suppresses cell apoptosis upon hypoxia. Cancer Cell Int. 2017;17:60. doi: 10.1186/s12935-017-0425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JJ, Wang DD, Du CX, Wang Y. Long noncoding RNA ANRIL promotes cervical cancer development by acting as a sponge of miR-186. Oncol Res. 2018;26:345–352. doi: 10.3727/096504017X14953948675449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, Zhang JQ, Chen JZ, Chen HX, Qiu FN, Yan ML, Chen YL, Peng CH, Tian YF, Wang YD. The over expression of long non-coding RNA ANRIL promotes epithelial-mesenchymal transition by activating the ATM-E2F1 signaling pathway in pancreatic cancer: an in vivo and in vitro study. Int J Biol Macromol. 2017;102:718–728. doi: 10.1016/j.ijbiomac.2017.03.123. [DOI] [PubMed] [Google Scholar]
- 16.Chai L, Yuan Y, Chen C, Zhou J, Wu Y. The role of long non-coding RNA ANRIL in the carcinogenesis of oral cancer by targeting miR-125a. Biomed Pharmacother. 2018;103:38–45. doi: 10.1016/j.biopha.2018.01.105. [DOI] [PubMed] [Google Scholar]
- 17.Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun X, Du P, Yuan W, Du Z, Yu M, Yu X, Hu T. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015;6:e1907. doi: 10.1038/cddis.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Huynh H, Li X, Ruddy DA, Wang Y, Ong R, Chow P, Qiu S, Tam A, Rakiec DP, Schlegel R, Monahan JE, Huang A. FGFR-mediated reactivation of MAPK signaling attenuates antitumor effects of imatinib in gastrointestinal stromal tumors. Cancer Discov. 2015;5:438–451. doi: 10.1158/2159-8290.CD-14-0763. [DOI] [PubMed] [Google Scholar]
- 20.Hong Q, Li O, Zheng W, Xiao WZ, Zhang L, Wu D, Cai GY, He JC, Chen XM. LncRNA HOTAIR regulates HIF-1alpha/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017;8:e2772. doi: 10.1038/cddis.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Peng Z, Zhang C, Duan C. Functions and mechanisms of long noncoding RNAs in lung cancer. Onco Targets Ther. 2016;9:4411–4424. doi: 10.2147/OTT.S109549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 25.Deng W, Wang J, Zhang J, Cai J, Bai Z, Zhang Z. TET2 regulates LncRNA-ANRIL expression and inhibits the growth of human gastric cancer cells. IUBMB Life. 2016;68:355–364. doi: 10.1002/iub.1490. [DOI] [PubMed] [Google Scholar]
- 26.Meseure D, Vacher S, Alsibai KD, Nicolas A, Chemlali W, Caly M, Lidereau R, Pasmant E, Callens C, Bieche I. Expression of ANRIL-polycomb complexes-CDKN2A/B/ARF genes in breast tumors: identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol Cancer Res. 2016;14:623–633. doi: 10.1158/1541-7786.MCR-15-0418. [DOI] [PubMed] [Google Scholar]
- 27.Zhao B, Lu YL, Yang Y, Hu LB, Bai Y, Li RQ, Zhang GY, Li J, Bi CW, Yang LB, Hu C, Lei YH, Wang QL, Liu ZM. Overexpression of lncRNA ANRIL promoted the proliferation and migration of prostate cancer cells via regulating let-7a/TGF-beta1/Smad signaling pathway. Cancer Biomark. 2018;21:613–620. doi: 10.3233/CBM-170683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 29.Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, Li D. Long non-coding RNA NEAT1 promotes nonsmall cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma J, Li T, Han X, Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144:205–214. doi: 10.1007/s00432-017-2543-y. [DOI] [PubMed] [Google Scholar]
- 31.Hou L, Luo P, Ma Y, Jia C, Yu F, Lv Z, Wu C, Fu D. MicroRNA-125a-3p downregulation correlates with tumorigenesis and poor prognosis in patients with non-small cell lung cancer. Oncol Lett. 2017;14:4441–4448. doi: 10.3892/ol.2017.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Xu T, Jiang Y, Xu H, Yan Y, Fu D, Chen J. The challenges and the promise of molecular targeted therapy in malignant gliomas. Neoplasia. 2015;17:239–255. doi: 10.1016/j.neo.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashiguchi Y, Nishida N, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Mochizuki H, Hase K, Doki Y, Mori M. Down-regulation of miR-125a-3p in human gastric cancer and its clinicopathological significance. Int J Oncol. 2012;40:1477–1482. doi: 10.3892/ijo.2012.1363. [DOI] [PubMed] [Google Scholar]
- 34.Liu JZ, Yin FY, Yan CY, Wang H, Luo XH. Regulation of docetaxel sensitivity in prostate cancer cells by hsa-miR-125a-3p via modulation of metastasis-associated protein 1 signaling. Urology. 2017;105:208, e211–208, e217. doi: 10.1016/j.urology.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Liang L, Gao C, Li Y, Sun M, Xu J, Li H, Jia L, Zhao Y. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017;8:e2968. doi: 10.1038/cddis.2017.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koole K, Brunen D, van Kempen PM, Noorlag R, de Bree R, Lieftink C, van Es RJ, Bernards R, Willems SM. FGFR1 is a potential prognostic biomarker and therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res. 2016;22:3884–3893. doi: 10.1158/1078-0432.CCR-15-1874. [DOI] [PubMed] [Google Scholar]
- 37.Göke F, Franzen A, Hinz TK, Marek LA, Yoon P, Sharma R, Bode M, von Maessenhausen A, Lankat-Buttgereit B, Göke A, Golletz C, Kirsten R, Boehm D, Vogel W, Kleczko EK, Eagles JR, Hirsch FR, Van Bremen T, Bootz F, Schroeck A, Kim J, Tan AC, Jimeno A, Heasley LE, Perner S. FGFR1 expression levels predict BGJ398 sensitivity of FGFR1-dependent head and neck squamous cell cancers. Clin Cancer Res. 2015;21:4356–4364. doi: 10.1158/1078-0432.CCR-14-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schultheis AM, Bos M, Schmitz K, Wilsberg L, Binot E, Wolf J, Büttner R, Schildhaus HU. Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod Pathol. 2014;27:214–221. doi: 10.1038/modpathol.2013.141. [DOI] [PubMed] [Google Scholar]
- 39.Murase H, Inokuchi M, Takagi Y, Kato K, Kojima K, Sugihara K. Prognostic significance of the co-overexpression of fibroblast growth factor receptors 1, 2 and 4 in gastric cancer. Mol Clin Oncol. 2014;2:509–517. doi: 10.3892/mco.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C, Wang FH, Chiao LJ, Pelicano H, Huang P, Li YH, Xu RH. Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology. 2014;60:598–609. doi: 10.1002/hep.27118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.