Abstract
In recent years, changes in microRNA (miRNA) expression have been detected in almost all human cancer types, including glioblastoma (GBM). Dysregulation of miRNAs may play tumor-suppressing or oncogenic roles in the initiation and progression of GBM, and may be involved in the regulation of multiple pathological behaviors. Therefore, identifying the clinical value and functional role of GBM-related miRNAs may provide effective therapeutic targets for the treatment of patients with this fatal malignancy. Dysregulation of miR-744 has been identified in several human cancer types. However, to the best of our knowledge, little is known concerning the expression pattern and biological roles of miR-744 in GBM. In this study, we found that miR-744 was significantly downregulated in GBM tissues and cell lines. Decreased miR-744 expression was significantly correlated with the Karnofsky Performance Scale (KPS) and World Health Organization (WHO) grade in GBM patients. miR-744 upregulation inhibited the proliferation, colony formation, migration, and invasion, in addition to inducing apoptosis of GBM cells in vitro. Inhibition of miR-744 had the opposite effect on these behaviors in GBM cells. Additionally, miR-744 attenuated the tumor growth of GBM cells in vivo. Furthermore, NIN1/RPN12 binding protein1 homolog (NOB1) was identified as a direct target gene of miR-744 in GBM cells. NOB1 was confirmed to be upregulated in GBM tissues, and this was inversely correlated with upregulation of miR-744 expression. Moreover, NOB1 knockdown exhibited similar inhibitory effects as miR-744 overexpression in GBM cells. Notably, recovered NOB1 expression counteracted the tumor-suppressing roles of miR-744 in the malignant phenotypes of GBM cells. Taken together, these results demonstrate that miR-744 directly targets NOB1 to inhibit the aggressive behaviors of GBM cells. Hence, the miR-744/NOB1 axis may be useful in the identification of novel therapies for GBM patients.
Keywords: Glioblastoma, microRNA-744, NIN1/RPN12 binding protein1 homolog
Introduction
Glioma is the most common and prevalent malignancy in the human central nervous system [1]. Glioblastoma (GBM), also known as World Health Organization (WHO) grade IV glioma, is the most common and deadly glioma [2]. The therapeutic strategies for GBM, including surgical resection, chemoradiotherapy, gene therapy, immunotherapy, have developed rapidly in recent years [3]. Unfortunately, the prognosis of GBM patients remains unsatisfactory, with a median survival duration of only 9-12 months [4]. Metastasis, recurrence, unlimited proliferation, fast diffuse infiltration, and significant apoptosis resistance are considered to be the major factors that cause poor prognosis of patients with GBM [5,6]. The mechanism underlying the occurrence and development of GBM has not been fully elucidated, and this explains the lack of improvement in therapeutic outcomes [7]. Thus, it is urgent to further elucidate the molecular pathogenesis of GBM and identify effective therapeutic methods to improve the survival of patients with this fatal disease.
The discovery of microRNAs (miRNAs) has provided novel insight and their subsequent study has provided in-depth understanding of the carcinogenesis and cancer progression in recent years [8]. miRNAs are a series of endogenous, non-coding, and short RNAs 17-24 nucleotides in length [9]. miRNAs are considered to be a novel group of gene regulators, and they modulate gene expression by directly binding to partially complimentary sequences in the 3’-untranslated regions (3’-UTRs) of their target genes, thereby causing translation suppression and/or the degradation of mRNAs [10]. In total, >1,500 mature miRNAs have been validated in the human genome, and these miRNAs participate in the regulation of ~30% of human protein-coding genes [11]. Dysregulation of miRNAs has been identified in nearly all human cancer types, including GBM [12], colorectal cancer [13], thyroid cancer [14], bladder cancer [15], and osteosarcoma [16]. A variety of miRNAs have been reported to be aberrantly expressed in GBM, which is closely related to the aggressive behaviors of GBM cells [17-19]. Highly expressed miRNAs in GBM may play oncogenic roles by inhibiting tumor suppressors [20]. By contrast, low-expressed miRNAs may serve as tumor suppressors by blocking oncogenes [21]. Accordingly, miRNAs may be promising biomarkers for the diagnosis, treatment, and prognosis of patients with this deadly malignancy.
Dysregulation of miR-744 has been identified in several human cancer types [22-24]. However, to the best of our knowledge, little is known concerning the expression pattern and biological roles of miR-744 in GBM. In this study, the expression level of miR-744 in GBM was detected and its clinical significance was also evaluated. Functional experiments were performed to determine the specific roles of miR-744 in the progression and development of GBM. Moreover, the molecular mechanisms underlying the activity of miR-744 in GBM cells were illustrated. Our study may offer a novel therapeutic target for the management of patients with this malignancy.
Materials and methods
Human tissue specimens
A total of 47 paired GBM tissues and adjacent non-tumor tissues were obtained from Huizhou Municipal Central Hospital between August 2014 and June 2017. None of the patients had been treated with pre-operative radiotherapy or chemotherapy. All tissues were obtained immediately after surgical resection and stored at -80°C until further use. This study was approved by the Ethics Committee of Huizhou Municipal Central Hospital. Written informed consent was obtained from all participations enrolled in the current research.
Cell culture
Normal human astrocytes (NHAs) were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA), and grown in astrocyte medium (ScienCell Research Laboratories) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Waltham, MA, USA). In total, 4 GBM cell lines, including T98, LN229, U138, and U251, were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% FBS and 1% penicillin/streptomycin mixture (both from Gibco; Thermo Fisher Scientific). All cells were maintained at 37°C in a humidified chamber supplied with 5% CO2.
Transfection experiments
Synthetic miR-744 mimics, miR-744 inhibitor, miRNA mimic negative control (miR-NC), and negative control RNA inhibitor (NC inhibitor) were chemically synthesized by Genepharma (Shanghai, China). miR-NC represents as a control for the miR-744 mimics, and NC inhibitor was used as the corresponding control for miR-744 inhibitor. NOB1 siRNA and negative control siRNA (NC siRNA) were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The NOB1 overexpression plasmid pcDNA3.1-NOB1 (pc-NOB1) and empty pcDNA3.1 plasmid were constructed by the Chinese Academy of Sciences (Changchun, China). Cells were plated into 6-well plates at an initial density of 5 × 105 cells per well. Cell transfection was performed using Lipofectamine 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific) in accordance with the manufacturer’s protocol. The transfected cells were incubated at 37°C with 5% CO2 for 6 h, and the transfection mixture was replaced with fresh DMEM containing 10% FBS.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA of cell lines or tissue specimens was isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific), according to the manufacturer’s instructions. The concentration and quality of total RNA was determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). For miR-744 detection, total RNA was converted into cDNA using a TaqManTM MicroRNA Reverse Transcription Kit (Applied Biosystems; Thermo Fisher Scientific). The synthesized cDNA was then subjected to quantitative PCR (qPCR) with a TaqMan microRNA assay kit (Applied Biosystems; Thermo Fisher Scientific). To quantify NOB1 mRNA expression, reverse transcription was conducted using a Prime-Script RT Reagent Kit (Takara Bio, Dalian, China), followed by qPCR using the SYBR Premix Ex Taq TM Kit (Takara Bio). U6 snRNA and β-actin were used as internal controls to normalize the relative expression levels of miR-744 and NOB1, respectively. Relative gene expression was analyzed by the 2-ΔΔCq method [25].
Cell counting kit-8 (CCK-8) assay
After incubation at 37°C with 5% CO2 for 24 h, cells were collected and inoculated into 96-well plates at 3 × 103 cells/well, and the volume of the culture medium was 100 μL each well. CCK-8 assay (Dojindo, Kumamoto, Japan) was carried out to evaluate cell proliferation at 4 time points: 0, 24, 48, and 72 h after inoculation. In brief, a total of 10 μL CCK-8 solution was added into each well and the culture plates were incubated at 37°C with 5% CO2 for an additional 2 h. The absorbance was detected at 450 nm by using a microplate reader (Bio-Rad Laboratories, Richmond, CA, USA).
Colony formation assay
Transfected cells were harvested after 24 h of incubation and seeded into 6-well plates at an initial density of 1000 cells/well. After culturing for 2 weeks at 37°C and 5% CO2, cells were fixed with 100% methanol and stained with methyl violet. The number of colonies formed was evaluated under an inverted light microscope (Olympus, Tokyo, Japan).
Flow cytometry analysis of cell apoptosis
The percentage of apoptosis cells was determined by an Annexin V fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (BioLegend, San Diego, CA, USA), based on the manufacturer’s protocol. In detail, transfected cells were harvested at 48 h post-transfection, washed thrice with PBS (Gibco; Thermo Fisher Scientific) and resuspended in 100 µL of binding buffer. The cells were then stained with 5 µL annexin V-FITC and 5 µL propidium iodide at room temperature for 30 min in the dark. Finally, stained cells were analyzed by a flow cytometer (FACScanTM; BD Biosciences, Franklin Lakes, NJ, USA).
Transwell migration and invasion assays
The migratory and invasiveness abilities of GBM cells were assessed using an 8-µm pore polycarbonate membrane Boyden chamber insert in a Corning Transwell apparatus (Corning, NY, USA). The chambers used for invasion assays were coated with Matrigel (BD Biosciences, Bedford, MA, USA), while no Matrigel was used for migration assays. For both assays, transfected cells were collected at 48 h post-transfection, washed with FBS-free medium and mechanically dissociated into a single cell suspension. A total of 5 × 104 cells in 200 µL FBS-free DMEM were plated into the upper compartment of chamber insets. The lower compartments were filled with 500 µL DMEM containing 20% FBS to serve as a chemoattractant. After 24 h of incubation, a cotton swab was used to gently remove the cells that remained on the upper surface of chamber inserts. The migrated and invaded cells were fixed with 100% methanol, stained with 0.05% crystal violet, washed with PBS, and air-dried. The migratory and invasiveness capacities were analyzed by counting the number of migrated and invaded cells in 5 randomly chosen microscopic fields as observed under an inverted light microscope.
Xenograft tumor assay
BALB/c nude mice aged 4-6 weeks old were acquired from the Shanghai Laboratory Animal Center (Shanghai, China). Cells transfected with miR-744 mimics or miR-NC was subcutaneously injected into both flanks of nude mice (4 nude mice per group). The nude mice were maintained in pathogen-free conditions for 1 month. The width and length of the tumor xenografts formed was detected every 2 days using a Vernier caliper. On day 31, all mice were sacrificed, and the weights of the tumor xenografts were measured. The tumor volumes were calculated by the following formula: volume (mm3) = width2 (mm2) * length (mm)/2. The animal experiments were approved by the Ethics Review Committee of Huizhou Municipal Central Hospital.
Bioinformatics analysis and luciferase reporter assay
Two miRNA target prediction websites, including TargetScan (http://targetscan.org/) and miRDB (http://www.mirdb.org/), were used to search for the putative targets of miR-744. NOB1 was predicted as the potential target of miR-744, and this association was then evaluated using a luciferase reporter assay. The 3’-UTR of NOB1 containing wild type (wt) and mutant (mut) miR-744 binding site was chemically constructed by Genepharma, cloned into the pmirGLO luciferase reporter vector (Promega Corporation, Madison, WI, USA) to generate pmirGLO-NOB1-3’-UTR wt and pmirGLO-NOB1-3’-UTR mut, respectively. For the reporter assay, cells were plated into 24-well plates at 1.0 × 105 cells per well. Lipofectamine 2000 was employed to co-transfect cells with miR-744 mimics/inhibitor or miR-NC/NC inhibitor and pmirGLO-NOB1-3’-UTR wt or pmirGLO-NOB1-3’-UTR mut, according to the manufacturer’s protocol. A total of 48 h after transfection, luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega Corporation). The Renilla luciferase activity was normalized to that of the firefly luciferase activity.
Protein extraction and western blot analysis
A total protein extraction kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) was used to isolate total protein from tissue specimens or cells according to the manufacturer’s instructions. The concentration of total protein was determined with a Bicinchoninic Acid Assay Kit (Pierce Biotechnology Inc., Rockford, IL, USA). Equal amounts of protein were loaded onto 10% SDS-PAGE gels for electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore, Burlington, MA, USA). After blocking for 2 h with 5% fat-free milk, the membranes were incubated overnight at 4°C with primary antibodies against NOB1 (cat. no. ab224619; 1:1,000 dilution) or GAPDH (cat. no. ab201822; 1:1,000 dilution; both from Abcam, Cambridge, UK). After that, the membranes were washed thrice with Tris-buffered saline and 0.05% Tween-20 (TBST) followed by incubation with a goat anti-mouse horseradish peroxidase-conjugated secondary antibody (cat. no. ab6721; 1:5,000 dilution; Abcam) at room temperature for 2 h. Finally, the protein signals were developed using an ECL Protein Detection Kit (Pierce Biotechnology, Inc.). GAPDH was used as a loading control.
Statistical analysis
SPSS software (version 18.0; SPSS, Inc., Chicago, IL, USA) was used for all statistical analysis. All data are presented as the mean ± SD from 3 independent experiments. The comparison of data was performed using Student’s t-test or one-way analysis of variance (ANOVA). Dunnett’s test was applied for post hoc analysis following ANOVA. χ2 test was adopted to analyze the relation between miR-744 expression and clinicopathological variables of GBM patients. The correlation between miR-744 and NOB1 mRNA levels was determined using Spearman’s correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-744 is downregulated in GBM tissues and cell lines
To examine the expression status of miR-744 in GBM, we performed RT-qPCR analysis in 47 paired GBM tissues and adjacent non-tumor tissues. The data revealed that miR-744 was significantly downregulated in GBM tissues compared with that in adjacent non-tumor tissues (Figure 1A, P<0.05). We then explored the relationship between miR-744 and clinicopathological characteristics of GBM patients. All patients were divided into either an miR-744-low or miR-744-high expression group based on the median value of miR-744. Statistical analysis indicated that expression level of miR-744 was correlated with the Karnofsky Performance Scale (KPS; P = 0.013) and World Health Organization (WHO) grade (P = 0.005) and GBM patients (Table 1). However, there was no significant association with gender (P = 0.474), age (P = 0.423) or extension of resection (P = 0.679) in GBM patients.
Figure 1.
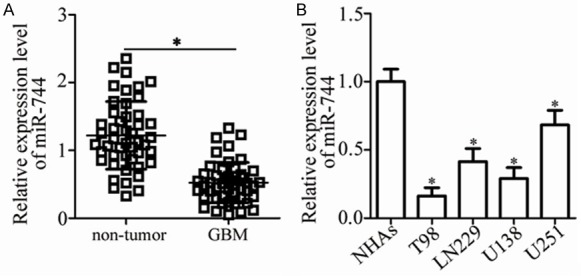
miR-744 is downregulated in GBM tissues and cell lines. A. Relative miR-744 expression was determined in 47 paired GBM tissues and adjacent non-tumor tissues. *P<0.05 vs. non-tumor tissues. B. RT-qPCR analysis was used to detect miR-744 expression in 4 GBM cell lines (T98, LN229, U138, and U251) and normal human astrocytes (NHAs). *P<0.05 vs. NHAs.
Table 1.
The relationship between of miR-744 and clinicopathological characteristics of GBM patients
| Characteristics | miR-744 expression | p | |
|---|---|---|---|
|
| |||
| Low | High | ||
| Gender | 0.474 | ||
| Male | 15 | 12 | |
| Female | 9 | 11 | |
| Age | 0.423 | ||
| <55 years | 10 | 7 | |
| ≥55 years | 14 | 16 | |
| Extension of resection | 0.679 | ||
| Subtotal | 8 | 9 | |
| Total | 16 | 14 | |
| KPS | |||
| ≥80 | 7 | 15 | 0.013* |
| <80 | 17 | 8 | |
| WHO grade | |||
| I-II | 8 | 17 | 0.005* |
| III | 16 | 6 | |
P<0.05.
KPS, Karnofsky performance score; WHO, World Health Organization.
We next detected the expression levels of miR-744 in 4 GBM cell lines (T98, LN229, U138, and U251) and normal human astrocytes (NHAs). The data obtained from RT-qPCR analysis showed that expression level of miR-744 was lower in all tested GBM cell lines compared to NHAs (Figure 1B, P<0.05). Taken together, these results imply that miR-744 expression is decreased in GBM, suggesting that this miRNA may have tumor-suppressor activity in GBM progression.
miR-744 inhibits the proliferation and colony formation, and induces apoptosis of GBM cells
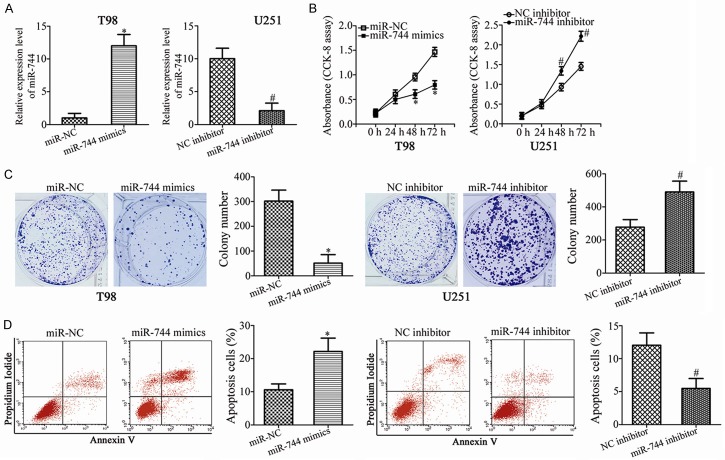
We performed gain- and loss-of-function assays to investigate the functional roles of miR-744 in GBM cells. T98 and U251 exhibited the relatively lowest and highest miR-744 expression among the 4 GBM cell lines, respectively; thus, T98 and U251 cells were transfected with miR-744 mimics and miR-744 inhibitor, respectively. miR-744 expression in the transfected cells was evaluated by RT-qPCR and was demonstrated to be markedly upregulated in T98 cells transfected with miR-744 mimics but significantly decreased in U251 cells treated with the miR-744 inhibitor (Figure 2A, P<0.05). The proliferation of GBM cells, measured by CCK-8 assay, was shown to be markedly decreased in the miR-744 overexpressing-T98 cells, and increased in the U251 cells after transfection with miR-744 inhibitor (Figure 2B, P<0.05). Additionally, colony formation assays revealed that the colony formation ability in T98 cells transfected with miR-744 mimics was suppressed compared to cells transfected with miR-NC; the colony formation ability in U251 cells transfected with the miR-744 inhibitor was elevated relative to that in NC inhibitor groups (Figure 2C, P<0.05). Furthermore, flow cytometry analysis was utilized to examine the effect of miR-744 expression in GBM cell apoptosis. As shown in Figure 2D, in miR-744 mimics-transfected T98 cells, the apoptosis rate was noticeably elevated. On the contrary, the percentage of apoptotic cells was significantly restricted in U251 cells following miR-744 inhibitor transfection (Figure 2D, P<0.05). These results suggest that miR-744 may play inhibitory roles in the growth of GBM cells in vitro.
Figure 2.
Effects of miR-744 on cell proliferation, colony formation, and apoptosis in GBM cells. T98 cells were transfected with miR-744 mimics or miR-NC, while U251 cells were treated with miR-744 inhibitor or NC inhibitor. A. Expression level of miR-744 in T98 cells was significantly increased by miR-744 mimics but was decreased by miR-744 inhibitor in U251 cells. *P<0.05 vs. miR-NC. #P<0.05 vs. NC inhibitor. B. The proliferation ability in the abovementioned cells was evaluated using CCK-8 assay. *P<0.05 vs. miR-NC. #P<0.05 vs. NC inhibitor. C. Colony formation assay was applied to examine the colony formation ability in T98 and U251 cells treated as described above. *P<0.05 vs. miR-NC. #P<0.05 vs. NC inhibitor. D. Following transfection 48 h, flow cytometry analysis was carried out to measure the apoptosis rate. *P<0.05 vs. miR-NC. #P<0.05 vs. NC inhibitor.
miR-744 attenuates the migration and invasion of GBM cells
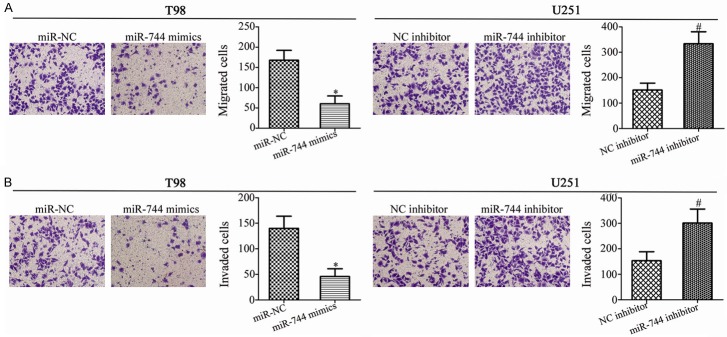
Next, transwell migration and invasion assays were carried out in order to investigate the effects of miR-744 in GBM cell metastasis. The results showed that miR-744 mimics transfection prohibited the migration (Figure 3A, P<0.05) and invasion (Figure 3B, P<0.05) of T98 cells, while miR-744 inhibitor promoted these abilities compared with NC inhibitor groups in U251 cells. These results suggest that miR-744 may markedly decrease in vitro metastasis of GBM cells.
Figure 3.
miR-744 plays an inhibitory role in the migratory and invasive abilities of GBM cells. T98 and U251 cells were transfected with miR-744 mimics and miR-744 inhibitor for 48 h, respectively. Transwell migration and invasion assays were employed to assess the migration (A) and invasion (B). *P<0.05 vs. miR-NC. #P<0.05 vs. NC inhibitor.
NOB1 is a direct target gene of miR-744 in GBM cells
To illustrate the mechanism underlying the tumor-suppressor activity of miR-744 in GBM cells, we predicted the putative target genes of miR-744 by bioinformatics analysis. The seed sequences of miR-744 matched the 3’-UTR of NOB1 (Figure 4A). NOB1 was chosen for further identification because this gene was previously reported to be involved in the genesis and development of GBM [26,27]. The luciferase reporter assay was carried out to confirm that NOB1 is the target gene of miR-744 as predicted. The results revealed that the luciferase activity of plasmid carrying wt NOB1 3’-UTR was significantly decreased by miR-744 overexpression in T98 cells or increased by miR-744 inhibition in U251 cells (Figure 4B, P<0.05). However, these effects were abolished when the miR-744 binding sequences in the 3’-UTR of NOB1 were mutated. Furthermore, results of RT-qPCR showed that miR-744 upregulation reduced NOB1 expression in T98 cells, while miR-744 knockdown increased NOB1 expression in U251 cells, at both the mRNA (Figure 4C, P<0.05) and protein (Figure 4D, P<0.05) levels. These results suggest that NOB1 is a direct target gene of miR-744 in GBM cells.
Figure 4.
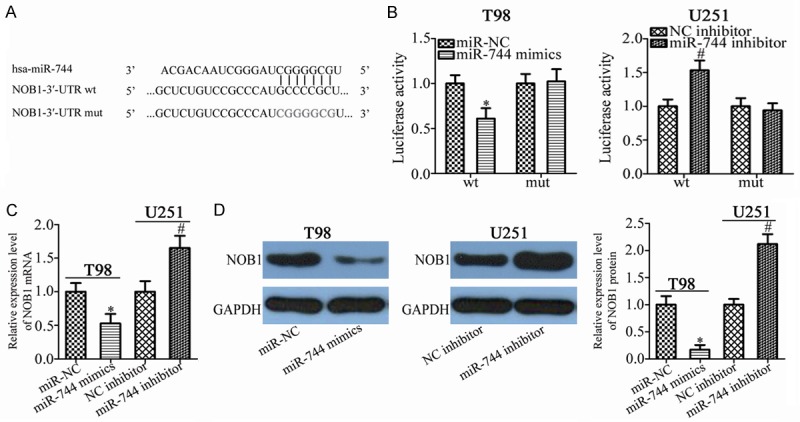
NOB1 is a direct target gene of miR-744 in GBM cells. A. The NOB1 3’-UTR contains the predicted miR-744 binding site. The mut miR-744 binding sequences are also shown. B. T98 and U251 cells were transfected with luciferase reporter plasmid harboring wt or mut NOB1 3’-UTR and miR-744 mimics or miR-744 inhibitor. Luciferase reporter assay was performed after 48 h of incubation. *P<0.05 vs. miR-NC. #P<0.05 vs. NC inhibitor. C, D. The effects of miR-744 on the expression of NOB1 mRNA and protein levels in T98 and U251 cells using RT-qPCR and western blot analysis, respectively. *P<0.05 vs. miR-NC. #P<0.05 vs. NC inhibitor.
miR-744 expression is inversely correlated with NOB1 expression in GBM tissues
To clarify the association between miR-744 and NOB1 in GBM, we detected NOB1 expression in 47 paired GBM tissues and adjacent non-tumor tissues. RT-qPCR analysis showed that the expression level of NOB1 was higher in GBM tissues than that in adjacent non-tumor tissues (Figure 5A, P<0.05). Meanwhile, western blot analysis was utilized to measure NOB1 protein expression in several pairs of GBM tissues and adjacent non-tumor tissues. It was observed that the NOB1 protein was upregulated in GBM tissues compared with that in adjacent non-tumor tissues (Figure 5B and 5C, P<0.05). Furthermore, an inverse correlation was identified between miR-744 and NOB1 mRNA levels in GBM tissues (Figure 5D; r = -0.5150, P = 0.0002). These results suggest that the upregulation of NOB1 in GBM tissues was, at least partly, induced by miR-744 downregulation.
Figure 5.
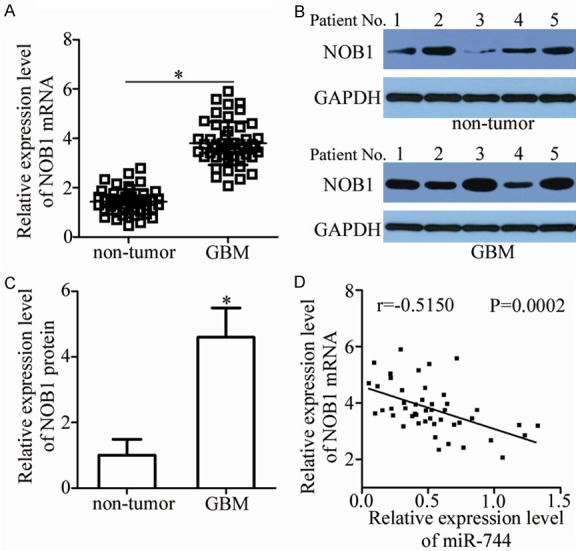
Upregulation of NOB1 is inversely correlated with miR-744 expression in GBM tissues. A. Expression levels of NOB1 mRNA in 47 paired GBM tissues and adjacent non-tumor tissues were detected by RT-qPCR. *P<0.05 vs. non-tumor tissues. B, C. NOB1 protein expression in several paired GBM tissues and adjacent non-tumor tissues was evaluated using western blot analysis. *P<0.05 vs. non-tumor tissues. D. Spearman’s correlation analysis of the relationship between miR-744 and NOB1 mRNA levels in GBM tissues. r = -0.5150, P = 0.0002.
Inhibition of NOB1 phenocopies the tumor suppressive effects of miR-744 overexpression in GBM cells
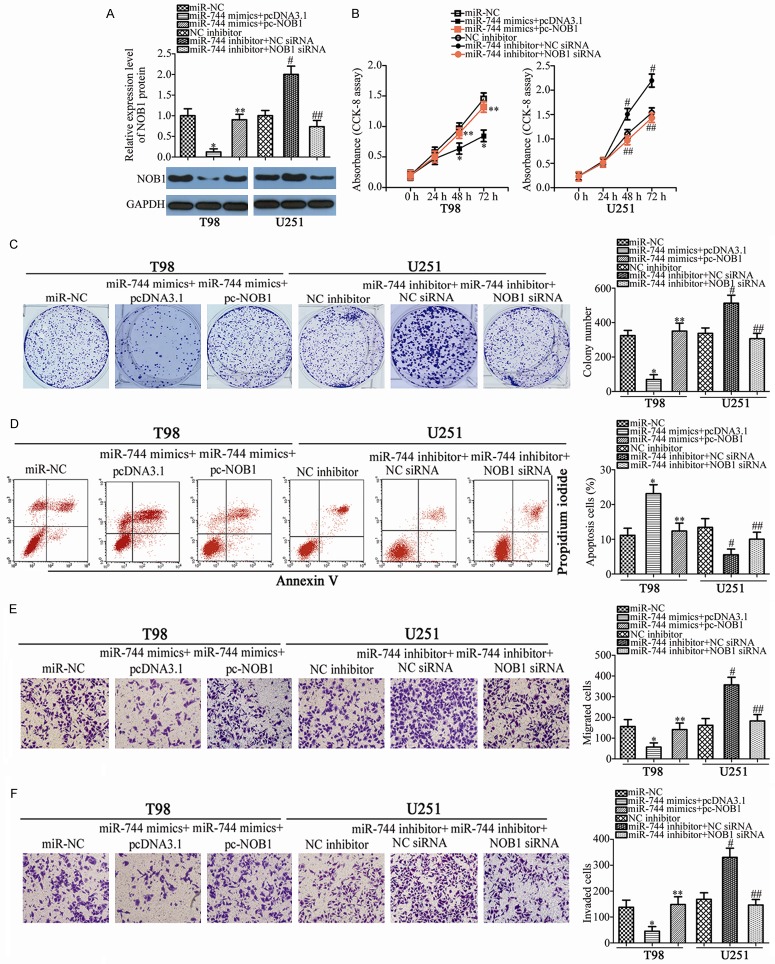
NOB1 was validated as a direct target gene of miR-744 in GBM cells; hence, we subsequently explored the functional roles of NOB1 in GBM cells. To this end, siRNA targeting the expression of NOB1 (NOB1 siRNA) was applied to knock down endogenous NOB1 expression in T98 and U251 cells. Western blot analysis confirmed that NOB1 siRNA transfection effectively decreased NOB1 protein expression in T98 and U251 cells (Figure 6A, P<0.05). As expected, NOB1 knockdown restricted the proliferation (Figure 6B, P<0.05) and colony formation (Figure 6C, P<0.05), promoted apoptosis (Figure 6D, P<0.05), and decreased migration (Figure 6E, P<0.05) and invasion (Figure 6F, P<0.05) of T98 and U251 cells. These results demonstrate that the biological roles of NOB1 knockdown are similar with those induced by miR-744 overexpression in GBM cells, further suggesting NOB1 as a functional downstream target of miR-744 in GBM cells.
Figure 6.
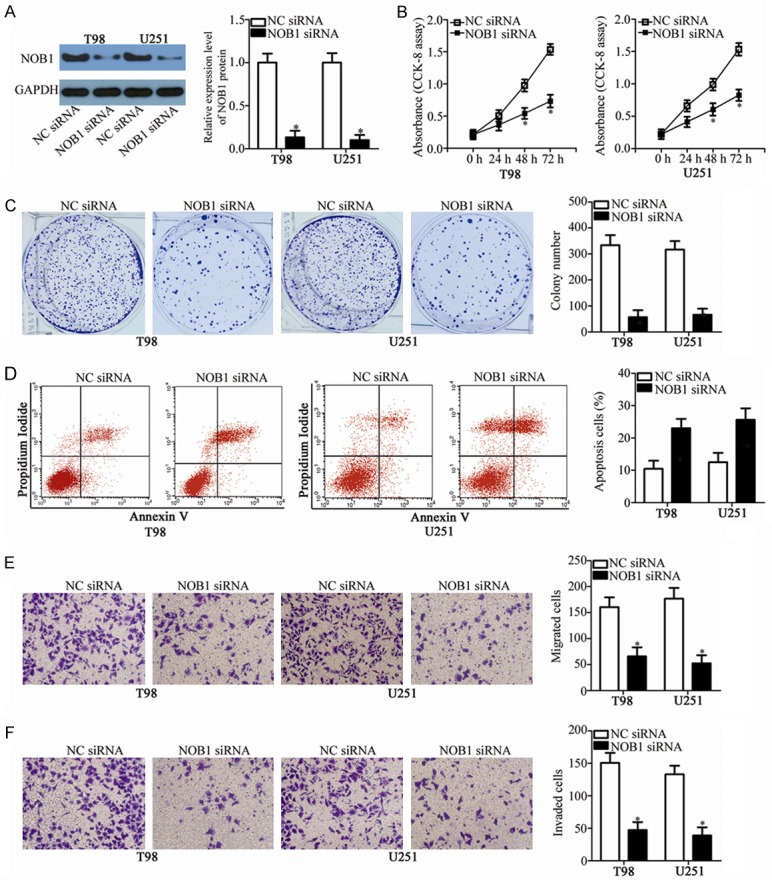
Downregulation of NOB1 suppresses GBM cell proliferation and colony formation, promotes cell apoptosis, and impedes cell migration and invasion. T98 and U251 cells were treated with NOB1 siRNA or NC siRNA. After incubation for different times, transfected cells were used in the following assays. A. Western blot analysis was conducted to determine NOB1 protein expression in T98 and U251 cells transfected with NOB1 siRNA or NC siRNA. *P<0.05 vs. NC siRNA. B, C. Proliferation and colony formation capacities in the abovementioned cells were examined using CCK-8 and colony formation assays, respectively. *P<0.05 vs. NC siRNA. D. The percentage of apoptotic cells was evaluated in NOB1 siRNA or NC siRNA transfected- T98 and U251 cells via flow cytometry analysis. *P<0.05 vs. NC siRNA. E, F. Transwell migration and invasion assays were used to determine the effects of NOB1 knockdown on the migration and invasion in T98 and U251 cells. *P<0.05 vs. NC siRNA.
Recovered NOB1 expression abolishes the effects of miR-744 in the malignant phenotypes of GBM cells
Rescue experiments were conducted to address the fact that the effects of miR-744 in malignant phenotypes of GBM cells were mediated by regulation of NOB1. miR-744-overexpressing T98 cells were transfected with a NOB1 overexpression plasmid (pc-NOB1), while miR-744 inhibitor-transfected U251 cells were co-transfected with NOB1 siRNA, in order to recover NOB1 expression. Following transfection, western blot analysis revealed that the downregulation of NOB1 caused by miR-744 overexpression was restored in T98 cells after co-transfection with pc-NOB1 (Figure 7A, P<0.05). In addition, NOB1 siRNA co-transfection recovered NOB1 expression in U251 cells that was increased by miR-744 underexpression (Figure 7A, P<0.05). Functional experiments demonstrated that recovered NOB1 expression abolished the effects of miR-744 in the proliferation (Figure 7B, P<0.05), colony formation (Figure 7C, P<0.05), apoptosis (Figure 7D, P<0.05), migration (Figure 7E, P<0.05), and invasion (Figure 7F, P<0.05) of T98 and U251 cells. These results suggest that miR-744 may serve a suppressive role in GBM cells, at least partly, via directly targeting NOB1.
Figure 7.
NOB1 reintroduction reverses the effects of miR-744 on GBM cells in vitro. T98 cells were co-transfected with miR-744 mimics and pc-NOB1 or pcDNA3.1, while miR-744 inhibitor along with NC siRNA or NOB1 siRNA was transfected into U251 cells. A. Western blot analysis was carried out to measure NOB1 protein expression. *P<0.05 vs. miR-NC. **P<0.05 vs. miR-744 mimics+pcDNA3.1. #P<0.05 vs. NC inhibitor. **P<0.05 vs. miR-744 inhibitor+NC siRNA. B-F. Proliferation, colony formation, apoptosis, migration and invasion in T98 and U251 cells treated as above were examined using CCK-8 assay, colony formation assay, flow cytometry analysis, and transwell migration and invasion assays, respectively. *P<0.05 vs. miR-NC. **P<0.05 vs. miR-744 mimics+pcDNA3.1. #P<0.05 vs. NC inhibitor. **P<0.05 vs. miR-744 inhibitor+NC siRNA.
miR-744 inhibits tumor growth of GBM in vivo
To further investigate the effect of miR-744 in the GBM in in vivo tumor growth, a xenograft tumor assay was performed. Firstly, T98 cells transfected with miR-744 mimics or miR-NC were subcutaneously implanted into nude mice. The tumor volume was determined, demonstrating that the volume of tumor xenografts in miR-744 group were lower than those of the miR-NC group (Figure 8A and 8B, P<0.05). The maximum diameter of a single tumor was 1.8 cm, while the minimum was 0.5 cm. The weights of tumor xenografts formed were also measured on day 31, after all mice were sacrificed. It was observed that the tumor weights in the miR-744 group were lower than those of the miR-NC-treated group (Figure 8C, P<0.05). In addition, the expression level of miR-744 in the tumor xenografts was detected by RT-qPCR. The results showed that miR-744 was markedly upregulated in the miR-744 mimics-treated tumor xenografts (Figure 8D, P<0.05). Furthermore, the tumor xenografts of the miR-744 group expressed lower levels of NOB1 protein compared to the miR-NC group (Figure 8E). These results demonstrate that miR-744 directly targets NOB1 to exert a significant inhibitory effect on the growth of GBM cells in vivo.
Figure 8.
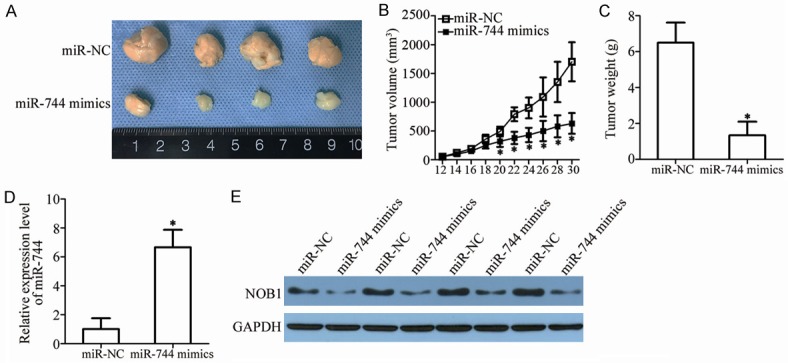
miR-744 hinders GBM tumor growth in vivo. A. Tumor xenografts derived from miR-744 mimics or miR-NC-transfected T98 cells. B. The volume of tumor xenografts was detected every 2 days until mice were sacrificed. The tumor volumes were calculated by the following formula: volume (mm3) = width2 (mm2) * length (mm)/2. *P<0.05 vs. miR-NC. C. The weight of excised tumor xenografts in the miR-744 or miR-NC group. *P<0.05 vs. miR-NC. D. miR-744 expression in tumor xenografts derived from miR-744 mimics or miR-NC-transfected T98 cells was detected using RT-qPCR. *P<0.05 vs. miR-NC. E. Western blot analysis was used to measure protein expression in tumor xenografts.
Discussion
In recent years, miRNAs have garnered increasing attention for their crucial roles in tumorigenesis and tumor development [28,29]. Changes in miRNA expression have been detected in almost all human cancer types, including GBM [12,13,30]. Dysregulation of miRNAs may play tumor-suppressing or oncogenic roles in GBM initiation and progression, and may be involved in the regulation of multiple pathological behaviors. Therefore, identifying the clinical value and functional role of GBM-related miRNAs may provide effective therapeutic targets for the treatment of patients with this fatal malignancy. In this study, we detected miR-744 expression in GBM for the first time and determined its clinical significance. Additionally, the specific roles and underlying molecular mechanisms of miR-744 in GBM progression were investigated. This study helps to illustrate the role of miR-744 in GBM and may be an effective therapeutic target for GBM therapy.
miR-744 is downregulated in colorectal cancer, and the downregulation of miR-744 is significantly correlated with lymphatic metastasis and TNM stage [22]. miR-744 is also low-expressed in cervical cancer [23] and hepatocellular carcinoma [24]. By contrast, miR-744 is upregulated in nasopharyngeal carcinoma. Highly expressed miR-744 is strongly associated with the tumor node stage, and clinical stage and grade in nasopharyngeal carcinoma patients. Nasopharyngeal carcinoma patients with high miR-744 expression exhibit lower five-year overall and relapse-free survival rates compared to patients with high miR-744 expression [31]. Notably, miR-744 expression level is identified as an independent biomarker for predicting the clinical outcomes of nasopharyngeal carcinoma patients [31]. Furthermore, miR-744 is overexpressed in prostate cancer [32], laryngeal squamous cell carcinoma [33], and pancreatic cancer patients [34,35]. However, little is known about the expression pattern of miR-744 in GBM. To address this, RT-qPCR analysis was used to measure miR-744 expression in GBM tissues and cell lines. More importantly, the clinical significance of miR-744 in GBM patients was determined. The data revealed that miR-744 was decreased in GBM, and it exhibited a significant relationship with KPS and WHO grade. These studies suggest that miR-744 may be a biomarker for the diagnosis of patients with these specific cancer types.
miR-744 overexpression inhibits the proliferation and invasion of colorectal cancer cells [22]. In cervical cancer, ectopic miR-744 expression attenuates cell proliferation and promotes cell apoptosis [23]. In hepatocellular carcinoma, resumption of miR-744 expression restricts cell proliferation and induces cell cycle arrest [36]. By contrast, miR-744 plays an oncogenic role in prostate cancer by regulating cell growth and metastasis in vitro as well tumor growth in vivo [32]. In addition, miR-744 serves as an oncogene in laryngeal squamous cell carcinoma [33], pancreatic cancer [35], and nasopharyngeal carcinoma [37]. However, the detailed roles of miR-744 in GBM progression and development have not been elucidated as of yet. Herein, a series of experiments were performed to explore the effects of miR-744 in GBM cells. The results showed that restoration of miR-744 expression prohibited GBM cell proliferation and colony formation, increased cell apoptosis, decreased migration and invasion in vitro, and inhibited tumor growth in vivo. miR-744 knockdown exerted an opposite effect on these behaviors of GBM cells. These findings suggest miR-744 as a novel promising therapeutic target for treating patients with these human cancer types.
Identification of the direct targets of miR-744 in GBM is essential for understanding their functional roles in the GBM genesis and development, and may be useful in developing effective therapeutic techniques. In our current study, NOB1, an essential gene encoding the Nin one binding protein, was demonstrated to be a novel and downstream direct target gene of miR-744 in GBM cells. High expression of NOB1 has been reported in several human malignant tumor types, such as prostate cancer [38], ovarian cancer [39], gastric cancer [40], and non-small cell lung cancer [41]. It is upregulated in GBM tissues and significantly correlates with tumor grade [26]. GBM patients with high NOB1 levels possess unfavorable clinical outcomes compared to patients with low NOB1 levels [27]. NOB1 is implicated in the regulation of GBM genesis and development by affecting cell proliferation, colony formation, cell cycle, apoptosis, and metastasis [26,27]. In our current study, miR-744 directly targeted NOB1 to inhibit the aggressive behaviors of GBM cells. Therefore, the miR-744/NOB1 axis may be a therapeutic target to block the rapid growth and metastasis of GBM.
In conclusion, we found that miR-744 was downregulated in GBM tissues and cell lines. Low miR-744 expression was significantly correlated with KPS and WHO grade in GBM patients. Further functional investigation revealed that miR-744 directly targets NOB1 to exert tumor suppressor activity in GBM cell proliferation, colony formation, apoptosis, migration, invasion in vitro and tumor growth in vivo. This study provides new clues to guide therapeutic intervention for GBM patients.
Acknowledgements
This study was supported by The Medical Scientific Research Foundation of Guangdong Province (No. B2014417).
Written informed consent was obtained from all patients for the use of their clinical tissues.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Thorne AH, Zanca C, Furnari F. Epidermal growth factor receptor targeting and challenges in glioblastoma. Neuro Oncol. 2016;18:914–918. doi: 10.1093/neuonc/nov319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 6.Zhuang W, Qin Z, Liang Z. The role of autophagy in sensitizing malignant glioma cells to radiation therapy. Acta Biochim Biophys Sin (Shanghai) 2009;41:341–351. doi: 10.1093/abbs/gmp028. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Jiang T. Understanding high grade glioma: molecular mechanism, therapy and comprehensive management. Cancer Lett. 2013;331:139–146. doi: 10.1016/j.canlet.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 8.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De Paoli E, Lu C, Schroth G, Meyers BC, Green PJ. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang SW, Ali ND, Zhong L, Shi J. MicroRNAs as biomarkers for human glioblastoma: progress and potential. Acta Pharmacol Sin. 2018;39:1405–1413. doi: 10.1038/aps.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.To KK, Tong CW, Wu M, Cho WC. MicroRNAs in the prognosis and therapy of colorectal cancer: from bench to bedside. World J Gastroenterol. 2018;24:2949–2973. doi: 10.3748/wjg.v24.i27.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celano M, Rosignolo F, Maggisano V, Pecce V, Iannone M, Russo D, Bulotta S. MicroRNAs as biomarkers in thyroid carcinoma. Int J Genomics. 2017;2017:6496570. doi: 10.1155/2017/6496570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wieczorek E, Reszka E. mRNA, microRNA and lncRNA as novel bladder tumor markers. Clin Chim Acta. 2018;477:141–153. doi: 10.1016/j.cca.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Kim YH, Goh TS, Lee CS, Oh SO, Kim JI, Jeung SH, Pak K. Prognostic value of microRNAs in osteosarcoma: a meta-analysis. Oncotarget. 2017;8:8726–8737. doi: 10.18632/oncotarget.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang DZ, Wang YP, Liu J, Hui XB, Wang XD, Chen X, Liu D. MicroRNA-129-3p suppresses tumor growth by targeting E2F5 in glioblastoma. Eur Rev Med Pharmacol Sci. 2018;22:1044–1050. doi: 10.26355/eurrev_201802_14387. [DOI] [PubMed] [Google Scholar]
- 18.Wang N, Zhang Y, Liang H. microRNA-598 inhibits cell proliferation and invasion of glioblastoma by directly targeting metastasis associated in colon cancer-1. Oncol Res. 2018;26:1275–1283. doi: 10.3727/096504018X15185735627746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahir BK, Ozer H, Engelhard HH, Lakka SS. MicroRNAs in glioblastoma pathogenesis and therapy: a comprehensive review. Crit Rev Oncol Hematol. 2017;120:22–33. doi: 10.1016/j.critrevonc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Mao K, Lei D, Zhang H, You C. MicroRNA-485 inhibits malignant biological behaviour of glioblastoma cells by directly targeting PAK4. Int J Oncol. 2017;51:1521–1532. doi: 10.3892/ijo.2017.4122. [DOI] [PubMed] [Google Scholar]
- 21.Yin J, Weng C, Ma J, Chen F, Huang Y, Feng M. MicroRNA1288 promotes cell proliferation of human glioblastoma cells by repressing ubiquitin carboxylterminal hydrolase CYLD expression. Mol Med Rep. 2017;16:6764–6770. doi: 10.3892/mmr.2017.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen J, Li M. MicroRNA-744 inhibits cellular proliferation and invasion of colorectal cancer by directly targeting oncogene Notch1. Oncol Res. 2018 doi: 10.3727/096504018X15188747585738. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen XF, Liu Y. MicroRNA-744 inhibited cervical cancer growth and progression through apoptosis induction by regulating Bcl-2. Biomed Pharmacother. 2016;81:379–387. doi: 10.1016/j.biopha.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Tan YL, Bai ZG, Zou WL, Ma XM, Wang TT, Guo W, Liu J, Li JS, Jie Y, Zang YJ, Zhang ZT. miR-744 is a potential prognostic marker in patients with hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2015;39:359–365. doi: 10.1016/j.clinre.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Li P, Zhao B. Knockdown of NOB1 expression by RNAi inhibits cellular proliferation and migration in human gliomas. Gene. 2013;528:146–153. doi: 10.1016/j.gene.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Xu T, Yan Y, Qin R, Wang H, Zhang X, Huang Y, Wang Y, Lu Y, Fu D, Chen J. MicroRNA-326 functions as a tumor suppressor in glioma by targeting the Nin one binding protein (NOB1) PLoS One. 2013;8:e68469. doi: 10.1371/journal.pone.0068469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. 2018 doi: 10.1016/j.mam.2018.07.003. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Alderman C, Sehlaoui A, Xiao Y, Wang W. MicroRNAs as immunotherapy targets for treating gastroenterological cancers. Can J Gastroenterol Hepatol. 2018;2018:9740357. doi: 10.1155/2018/9740357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan HL, Wang T, Zhang KH. MicroRNAs as potential biomarkers for diagnosis, therapy and prognosis of gastric cancer. Onco Targets Ther. 2018;11:3891–3900. doi: 10.2147/OTT.S156921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Q, Zhang F, Du Z, Xiang Y. Up-regulation of serum miR-744 predicts poor prognosis in patients with nasopharyngeal carcinoma. Int J Clin Exp Med. 2015;8:13296–13302. [PMC free article] [PubMed] [Google Scholar]
- 32.Guan H, Liu C, Fang F, Huang Y, Tao T, Ling Z, You Z, Han X, Chen S, Xu B, Chen M. MicroRNA-744 promotes prostate cancer progression through aberrantly activating Wnt/beta-catenin signaling. Oncotarget. 2017;8:14693–14707. doi: 10.18632/oncotarget.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li JZ, Gao W, Lei WB, Zhao J, Chan JY, Wei WI, Ho WK, Wong TS. MicroRNA 744-3p promotes MMP-9-mediated metastasis by simultaneously suppressing PDCD4 and PTEN in laryngeal squamous cell carcinoma. Oncotarget. 2016;7:58218–58233. doi: 10.18632/oncotarget.11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyamae M, Komatsu S, Ichikawa D, Kawaguchi T, Hirajima S, Okajima W, Ohashi T, Imamura T, Konishi H, Shiozaki A, Morimura R, Ikoma H, Ochiai T, Okamoto K, Taniguchi H, Otsuji E. Plasma microRNA profiles: identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br J Cancer. 2015;113:1467–1476. doi: 10.1038/bjc.2015.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W, Li Y, Gou S, Xiong J, Wu H, Wang C, Yan H, Liu T. MiR-744 increases tumorigenicity of pancreatic cancer by activating Wnt/betacatenin pathway. Oncotarget. 2015;6:37557–37569. doi: 10.18632/oncotarget.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin F, Ding R, Zheng S, Xing D, Hong W, Zhou Z, Shen J. Decrease expression of microRNA-744 promotes cell proliferation by targeting c-Myc in human hepatocellular carcinoma. Cancer Cell Int. 2014;14:58. doi: 10.1186/1475-2867-14-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang Y, Zhu X, Wang J, Li N, Li D, Sakib N, Sha Z, Song W. MiR-744 functions as a protooncogene in nasopharyngeal carcinoma progression and metastasis via transcriptional control of ARHGAP5. Oncotarget. 2015;6:13164–13175. doi: 10.18632/oncotarget.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Zhang D, Qu F, Hong Y, Cao J, Pan X, Li L, Huang Y, Huang H, Yin L, Chen L, Ren J, Wang Z, Xu D, Cui X. Knockdown of NOB1 expression inhibits the malignant transformation of human prostate cancer cells. Mol Cell Biochem. 2014;396:1–8. doi: 10.1007/s11010-014-2126-z. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y, Xu T, Teng H, Cui M. Anticancer activity of NOB1-targeted shRNA combination with TRAIL in epithelial ovarian cancer cells. Int J Clin Exp Pathol. 2015;8:10061–10071. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Zhou WP, Liu X, Yang Y, Liu YF. Expression mechanism and clinical significance of Nob1 in gastric cancer tissue and adjacent normal tissue. J Biol Regul Homeost Agents. 2015;29:423–430. [PubMed] [Google Scholar]
- 41.Liu K, Chen HL, Wang S, Gu MM, Chen XM, Zhang SL, Yu KJ, You QS. High expression of RIOK2 and NOB1 predict human non-small cell lung cancer outcomes. Sci Rep. 2016;6:28666. doi: 10.1038/srep28666. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]