Abstract
Inhibitor of DNA binding/differentiation (Id2) is an important regulator involved in the initiation and progression of cancer. However, the function and mechanism of the regulation of Id2 in hepatocellular carcinoma (HCC) was unclear. In the present study, we found that the overexpression of Id2 increased HCC cell proliferation in vitro and in vivo. Knockdown of Id2 inhibited HCC cell proliferation in vitro and in vivo. Furthermore, knockdown of Id2 enhanced sorafenib-induced apoptosis in HCC. Conversely, overexpression of Id2 weakened sorafenib-induced apoptosis in HCC. In addition, the transcription factor CCAAT/enhancer-binding protein alpha (C/EBPα) bound to the Id2 promoter and decreased its expression in HCC cells. Therefore, all results suggest that Id2 promotes the proliferation of HCC cells by inhibiting cell apoptosis. Id2 may serve as a potential target in HCC therapy.
Keywords: Id2, hepatocellular carcinoma, proliferation, C/EBPα, sorafenib
Introduction
Hepatocellular carcinoma (HCC) has a poor prognosis and high mortality rate worldwide [1]. Surgery is the main means of treating HCC, while frequent recurrence and metastasis have become critical issues. It has been widely accepted that the occurrence, progression and prognosis of HCC may be accompanied by complex molecular alterations, including genetic and epigenetic changes [2]. Therefore, it is necessary to explore the function and mechanisms that are involved in the progression of these tumors.
Inhibitor of DNA binding/differentiation (Id) proteins act as a negative regulators of the basic helix-loop-helix (bHLH) type of transcription factors by forming heterodimers [3]. Studies have shown that bHLH factors are critically involved in diverse types of tissue-specific differentiation and cellular functions [4]. Therefore, Id proteins play a wide array of biological roles in cell proliferation, differentiation and development, as well as oncogenesis. Four members of the Id protein family (Id1-Id4) have been identified in humans [3,5]. Recent studies have shown that Id2 expression is correlated with the prognosis of patients [6]. Id2 may be a potential target for cancer diagnosis and treatment [7,8]. Id2 is involved in cell-cycle progression through interacting with the retinoblastoma protein (RB) [9,10]. In Rb mutant mice, the tumor formation rate of Id2-/- mice was significantly lower than that of the control group [11]. Id2 regulates invasion and migration by interacting with Snail in TNF-α-induced oral squamous cell carcinoma cells [12]. However, the function of Id2 remains controversial in HCC. Studies have shown that Id2 promotes the proliferation and metastasis of HCC cells [13,14]. Nevertheless, Id2 has been correlated with less portal vein invasion and a better prognosis in HCC patients. Therefore, the function of Id2 needs to be reassessed in HCC.
Although abnormal Id2 expression is found in many human tumors, such as breast cancer, bladder cancer, pancreatic cancer, colorectal cancer and HCC, suggesting that it might be a possible prognostic marker for tumor patients, little is known regarding the regulatory mechanism of Id2 in HCC [7,15-17]. Previous studies have shown that hypoxia-inducible factor-1 (HIF-1), p53 and CCAAT/enhancer-binding protein beta (C/EBPβ) regulate Id2 expression in neuroblastoma cells, neural progenitor cells and NIH3T3 cells, respectively [18-20]. However, the molecular mechanism of the regulation of Id2 in HCC remains unclear. For this reason, it is important to elucidate the mechanism(s) of the regulation of Id2 in HCC.
Materials and methods
Cell lines and cell culture
The human HCC cell line Huh7 was purchased from Riken BRC Cell Bank (Japan). The SMMC-7721 cell line was provided by the Cell Bank of the Institute of Biochemistry and Cell Biology (Shanghai, China). The Li-7 cell line was purchased from Shanghai Sixin Biotechnology Co. (Shanghai, China), and the HEK 293T, SK-Hep-1 and PLC/PRF/5 cell lines were purchased from the American Type Culture Collection (ATCC, USA). All of these cell lines were continuously cultured in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, USA) containing 10% fetal bovine serum (FBS) (Gibco, New York, USA), 100 IU/ml penicillin G and 100 μg/ml streptomycin (Sigma-Aldrich, USA). The cell lines were incubated at 37°C under a humidified atmosphere with 5% CO2.
Quantitative real-time RT-PCR
The total RNA of cells or tissues was extracted using the TRIzol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA) and was reversed transcribed to cDNA using a PrimeScriptTM RT Reagent (TaKaRa, Japan). Quantitative real-time RT-PCR (qRT-PCR) analyses were carried out as previously described using an ABI Prism 7500 System (Applied Biosystems, Carlsbad, CA, USA) with SYBR Premix Ex Taq (Takara, Dalian, China) [21]. The primers used for the qPCR of the referenced genes are shown in Supplementary Table 1.
Western blot analysis
Total protein was extracted from fresh cells that were lysed in RIPA buffer (Thermo Scientific, MA, USA) supplemented with protease inhibitor and phosphatase inhibitor (Roche, Welwyn Garden, Switzerland), and the protein was quantified using the BCA reagent. Then, the protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and electroblotted onto 0.2 μm polyvinylidene difluoride (PVDF) membranes (Merck Millipore, USA). Antibody information is listed in Supplementary Table 2.
Plasmid construction
The full-length human Id2 open reading frame cDNA sequence was generated from cDNA derived from cells using specific primers (forward: 5’-ATGAAAGCCTTCAGTCCCG-3’ and reverse: 5’-TCAGCCACACAGTGCTTT-3’) and was cloned into the lentiviral vector pWPXL (Addgene, Cambridge, UK). A C/EBPα plasmid tagged with FLAG was purchased from Genechem (Shanghai, China). All promoter sequences were cloned into the pGL3-basic vector (Promega, Madison, WI, USA). The primer information is listed in Supplementary Table 3.
RNA interference
Short hairpin RNA (shRNA) for Id2 and a general negative control (NC) were synthesized by Genechem (Shanghai, China). The fragments were designed to target Id2 transcripts. Two target sequences of Id2 were selected: 5’-GAAGGTGAGCAAGATGGAA-3’ and 5’-GACGACTACATTGAACAA-3’.
Lentivirus production and cell transduction
Viral packaging was performed in HEK 293T cells after the cotransfection of the Id2 vector or the shId2 vector with the packaging plasmid psPAX2 and the envelope plasmid pMD2.G (Addgene) using Lipofectamine 2000 (Invitrogen) according to a previously described protocol [22]. SMMC-7721, li-7, Sk-Hep1, PLC/PRF/5 and Huh7 cells were infected with 1×106 recombinant lentivirus-transducing units in the presence of 6 μg/ml polybrene (Sigma).
Cell viability assay
One thousand cells per well were seeded into a 96-well plate. On the second day, 10 μl MTT reagent (5 mg/ml, Sigma-Aldrich, USA) per well was added to medium of the cells. After incubating for 4 h at 37°C, the medium was discarded, and the formazan was dissolved in 100 μl DMSO (Sigma-Aldrich, USA). The absorbance at 490 nm was read by means of a Thermo Scientific Microplate Reader. Each experiment was performed in triplicate.
Colony formation assay
Six-well plates were seeded with 1×103 cells per well, and the medium was renewed every three days. After 2 weeks, the cells in the plates were washed twice with PBS and fixed with formaldehyde solution for 30 minutes. Then, the cells were stained with Giemsa (Sigma-Aldrich, USA). Each experiment was performed in triplicate.
Apoptosis assay
HCC cells were washed twice with PBS and centrifuged. The cells were then suspended in 100 μl binding buffer, and 5 μl PE-conjugated Annexin-V and 5 μl 7-AAD were added. The cells were incubated for 30 min on ice in the dark. Flow cytometric analysis was used to detect apoptotic cells. The percentage of apoptotic cells was determined by using the FlowJo software 7.6 (Treestar, USA). Each experiment was performed in triplicate.
Luciferase assay
Cells were seeded into 96-well plates and grown to approximately 80% confluency on the second day. Next, the relevant reporter plasmids and the PRL-TK reporter were transiently cotransfected into the cells. After forty-eight hours, firefly luciferase activity and Renilla activity were determined. The ratio of firefly luciferase activity to Renilla activity was regarded as indicating the activity of the gene promoters. Each experiment was performed in triplicate.
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation assay assays were performed using a ChIP assay kit according to a previously described protocol [23]. Cells were cross-linked with 10% formaldehyde and reversed with 1 M glycine. Cells were collected and lysed. The cells were then suspended in buffer to lyse the nuclei, and the nucleic acid was fragmented using an ultrasonic cell disruptor. Rabbit anti-FLAG or rabbit IgG was added along with protein A/G-agarose beads (Sigma), and the samples were incubated overnight at 4°C to immunoprecipitate the DNA-containing complexes. After washing, the DNA was isolated from the agarose beads and used for PCR analysis. The primer information is listed in Supplementary Table 1.
Tumor xenograft models
Six- to 8-week-old male nude mice were randomly divided into two groups. To establish the liver orthotropic model, 1×106 experimental or control cells for each mouse were suspended in serum-free DMEM mixed with an equal volume of Matrigel (BD Biosciences) and orthotopically injected into the left hepatic lobe. All the mice were sacrificed after 6-8 weeks. Xenograft tumors were collected and weighed. Tumor tissues were analyzed by H&E staining and western blot analysis. All of the animal experiments were approved by the Institutional Animal Care and Use Committee of the Shanghai Jiao Tong University School of Medicine.
Statistical analysis
Statistical analyses were performed using the SPSS 19.0 software. The results are presented as the mean ± S.D., and two-group comparisons were evaluated using Student’s t-test. Multiple group comparisons were evaluated using a one-way ANOVA. P<0.05 was considered significant.
Results
Overexpression of Id2 promotes the proliferation of HCC cells in vitro and in vivo
To verify a role for Id2 in HCC cells, HCC cell lines stably overexpressing Id2 (SMMC-7721, SK-Hep-1 and Li-7) were established using lentiviral infection according to the endogenous expression levels of Id2 in HCC cells (Figure 1A-C). MTT and colony formation assays showed that the overexpression of Id2 promoted HCC cell proliferation in vitro (Figure 1D and 1E).
Figure 1.
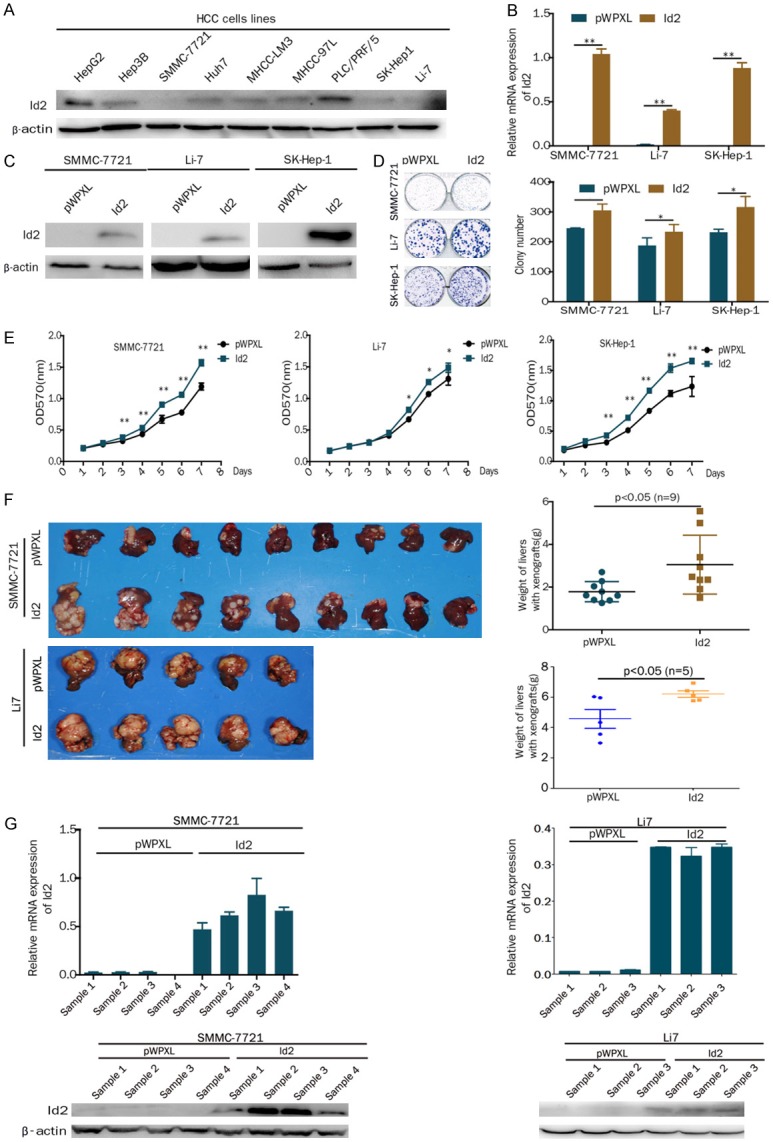
Overexpression of Id2 promotes the proliferation of HCC cells in vitro and in vivo. A. Western blot analysis of Id2 expression in HCC cell lines. B. Expression of Id2 was detected by real-time PCR assays in Id2-overexpressing HCC cells. C. Expression of Id2 was detected by western blot analysis in Id2-overexpressing HCC cells. D. A colony formation assay was performed with Id2-overexpressing or control HCC cells. E. A cell growth assay was performed with Id2-overexpressing or control HCC cells. F. The overexpression of Id2 promoted SMMC-7721 and Li7 cell proliferation in vivo. The weight of the liver is shown in the right panel. G. qRT-PCR and western blot analysis of Id2 expression in the SMMC-7721 and Li7 cell-derived xenograft samples. *P<0.05; **P<0.01.
To confirm the function of Id2 in vivo, SMMC-7721 and Li7 cells that overexpressed Id2 or control cells were inoculated into the hepatic lobes of nude mice. After 8 or 4 weeks, the weight of the liver was significantly increased in the Id2-overexpressing group compared with the control group (Figure 1F). Furthermore, the expression of Id2 in the tumor tissues separated from 4 mice was still higher than that in the control group based on real-time PCR and western blot assays (Figure 1G). Therefore, our results suggest that the overexpression of Id2 promotes the proliferation of HCC cells in vitro and in vivo.
Knockdown of Id2 inhibits the proliferation of HCC in vitro and in vivo
To analyze the effect of Id2 knockdown on cell proliferation, the stable knockdown of Id2 was established in HCC cell lines (PLC/PRF/5 and Huh7) using lentivirus shRNA constructs targeting Id2 (Figure 2A and 2B). The Results showed that the knockdown of Id2 inhibited cell proliferation and colony formation capacity (Figure 2C and 2D).
Figure 2.
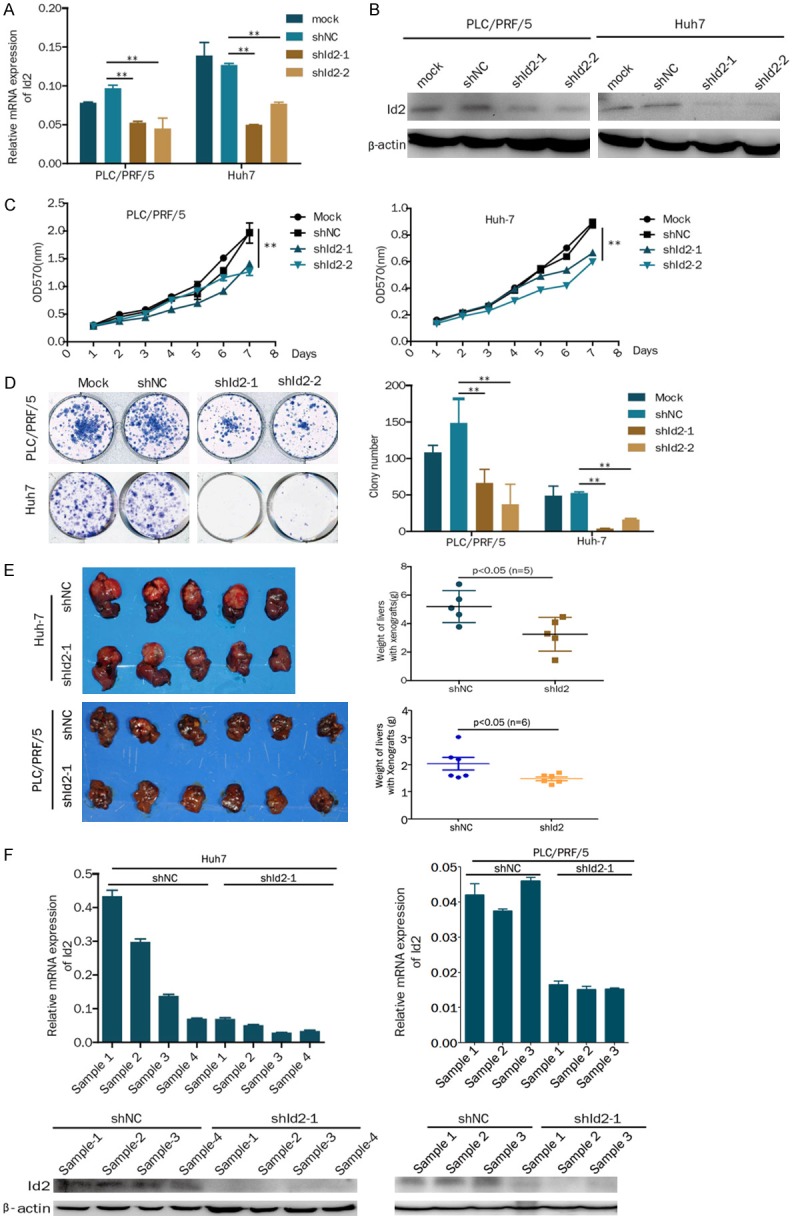
Knockdown of Id2 inhibits the proliferation of HCC cells in vitro and in vivo. A. Expression of Id2 as detected by real-time PCR in Id2-knockdown HCC cells. B. Expression of Id2 detected by western blot analysis in Id2-knockdown HCC cells. C. A cell growth assay was performed in Id2-knockdown or control HCC cells. D. A colony formation assay was performed in Id2-knockdown or control HCC cells. E. The knockdown of Id2 inhibited Huh7 and PLC/PRF/5 cell proliferation in vivo. The weight of the liver is shown in the right panel. F. qRT-PCR and western blot analysis of Id2 expression in the Huh7 and PLC/PRF/5 cell-derived xenograft samples. *P<0.05; **P<0.01.
To confirm the function of Id2 in vivo, Id2-knockdown Huh7 and PLC/PRF/5 cells or control cells were inoculated into the hepatic lobes of nude mice. After 8 or 5 weeks, the weight of the liver was significantly decreased in the Id2-knockdown group compared with the control group (Figure 2E). Furthermore, the expression of Id2 in the tumor tissues separated from 4 mice was still lower than that in the control group based on real-time PCR and western blot assays (Figure 2F). Therefore, our results indicate that the knockdown of Id2 inhibits the proliferation of HCC cells in vitro and in vivo.
Knockdown of Id2 enhances sorafenib-induced apoptosis
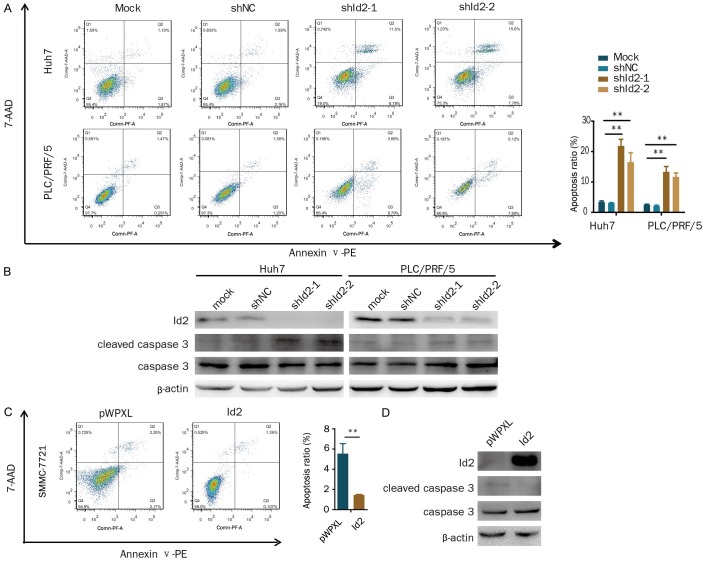
The above results showed that the knockdown of Id2 inhibited HCC cell proliferation. Therefore, we determined the effect of Id2 on cell apoptosis. Our results showed that the knockdown of Id2 increased cell apoptosis compared with the control (Figure 3A). Meanwhile, the levels of the apoptosis-associated protein cleaved caspase 3 were markedly increased in the Id2-knockdown cells (Figure 3B). Conversely, the overexpression of Id2 decreased cell apoptosis in HCC cells (Figure 3C), and cleaved caspase 3 levels were markedly decreased in the Id2-overexpressing cells (Figure 3D). Therefore, these results suggest that Id2 increases cell proliferation by inhibiting apoptosis.
Figure 3.
Id2 inhibits apoptosis in HCC cells. A. Apoptotic cells were assessed by flow cytometry following Annexin V/7-AAD staining in Id2-knockdown HCC cells. B. Cleaved caspase 3 and Id2 were detected by western blotting in Id2-knockdown HCC cells. C. Apoptotic cells were assessed by flow cytometry following Annexin V/7-AAD staining in Id2-overexpressing SMMC-7721 cells. D. Cleaved caspase 3 and Id2 were detected by western blotting in Id2-overexpressing SMMC-7721 cells. **P<0.01.
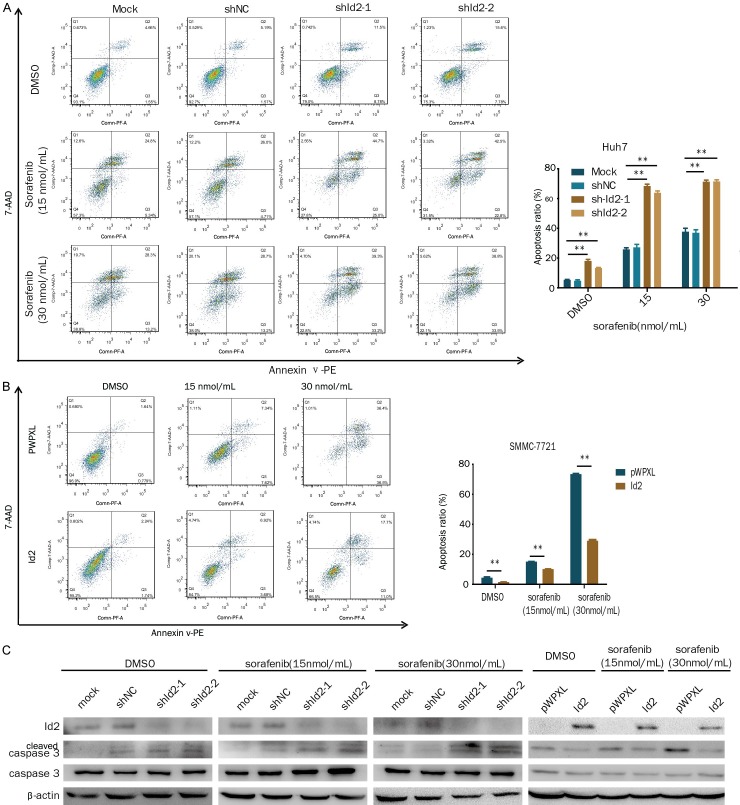
The multikinase inhibitor sorafenib is the only FDA-approved treatment for HCC. Therefore, we asked if the inhibition of Id2 would sensitize HCC cells to sorafenib treatment. The results showed that the knockdown of Id2 significantly enhanced sorafenib-induced cell apoptosis (Figure 4A). Conversely, the overexpression of Id2 decreased sorafenib-induced cell apoptosis (Figure 4B). Meanwhile, the knockdown of Id2 enhanced cleaved caspase 3 levels in the sorafenib-treated cells, while the overexpression of Id2 decreased cleaved caspase 3 levels in the sorafenib-treated cells (Figure 4C). Therefore, these results suggest that the knockdown of Id2 sensitizes HCC cells to sorafenib treatment.
Figure 4.
Id2 influences sorafenib-induced HCC cell apoptosis. A. Id2-knockdown Huh7 cells or control cells were treated with sorafenib (0, 15, 30 µM) for 24 h, and apoptotic cells were assessed by flow cytometry. B. Id2-overexpressing SMMC-7721 cells or control cells were treated with sorafenib (0, 15, 30 µM) for 24 h, and apoptotic cells were assessed by flow cytometry. C. Cleaved caspase 3 and Id2 were detected by western blotting in HCC cells with the overexpression or knockdown of Id2. **P<0.01.
C/EBPα binds to the Id2 promoter and inhibits Id2 expression in HCC cells
To analyze the regulatory mechanism of Id2 in HCC, we performed a reporter assay to identify the regulatory elements that control Id2 transcription. The results of assays with deletion constructs showed that luciferase activity was decreased in the -102/+360-bp construct compared with the -1044/+360-bp, -747/+360-bp and -164/+360-bp constructs, suggesting that the promoter activity was restricted to -164/+360 bp (Figure 5A). We analyzed the Id2 promoter via three websites that predicted transcription factor binding, including the JASPAR database (http://jaspar.genereg.net/), Alggen-promo (http://alggen.lsi.upc.es) and Gene regulation (http://gene-regulation.com). We found that the transcription factor CCAAT/enhancer binding protein alpha (C/EBPα) can bind directly to the promoter of Id2. A luciferase assay showed that C/EBPα inhibited Id2 promoter activity (-164/+360-bp construct) compared with the control. However, C/EBPα did not inhibit Id2 promoter activity (-102/+360-bp construct) compared with the control, suggesting that the region from -164 bp to -156 bp may be the binding site of C/EBPα (Figure 5B). A conserved C/EBPα binding motif was found at position -164 to -156 bp (Figure 5C). However, C/EBPα did not decrease the activity of an Id2 promoter containing a mutation in the putative C/EBPα binding site (Figure 5D). Moreover, the binding of C/EBPα to Id2 promoters was further confirmed by means of a ChIP assay (Figure 5E). In addition, the overexpression of C/EBPα inhibited the expression of Id2 in HCC cells (Figure 5F and 5G). Therefore, all of these results indicate that C/EBPα binds to the Id2 promoter and inhibits Id2 expression in HCC cells.
Figure 5.
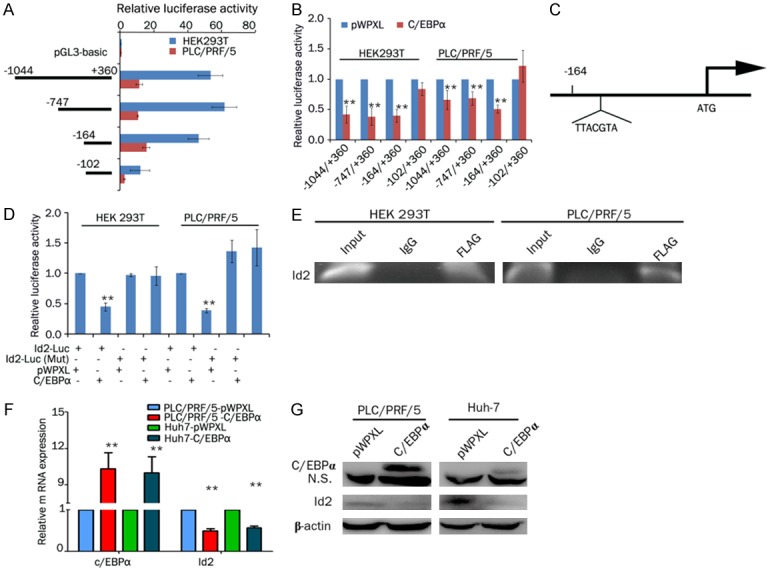
C/EBPα directly modulates Id2 transcriptional activity. A. HEK 293T and PLC/PRF/5 cells were transfected with different truncations of Id2 luciferase reporter vectors (-1044/+360, -747/+360, -164/+360 or -102/+360). The corresponding relative luciferase activities were determined by a reporter gene assay. B. HEK 293T and PLC/PRF/5 cells were cotransfected with Id2 luciferase reporter vectors (-1044/+360, -747/+360, -164/+360 or -102/+360) and C/EBPα or pWPXL. The corresponding relative luciferase activities were determined by a reporter gene assay. C. Potential binding site for C/EBPα in the Id2 promoter identified with the JASPAR database. D. C/EBPα or pWPXL and Id2 luciferase reporter vectors (wild-type or mutant C/EBPα-binding sites, -164/+360) were cotransfected into HEK 293T and PLC/PRF/5 cells. The corresponding relative luciferase activities were determined by a reporter gene assay. **P<0.01. E. ChIP analysis of C/EBPα (tagged with FLAG) binding to the Id2 promoter. F. PLC/PRF/5 and Huh7 cells were transfected with C/EBPα or pWPXL, and the expression levels of C/EBPα and Id2 were detected by real-time PCR. G. PLC/PRF/5 and Huh7 cells were transfected with C/EBPα or pWPXL, and the expression levels of C/EBPα and Id2 were detected by western blot analysis.
Discussion
Id2 not only plays an important role in embryonic development and histological differentiation but is also implicated in tumor proliferation [15]. Id2 is overexpressed in breast cancer, bladder cancer, pancreatic cancer and colorectal cancer [6,17,24,25]. In this study, we found that Id2 promoted HCC cell proliferation in vitro and in vivo. Previous studies have shown that Id2 promotes the proliferation of cancer cells by activating the NF-kappaB/cyclin D1 pathway in squamous cell carcinoma [26]. Id2 influences the potential for tumor metastasis by regulating EMT of cancer cells [27]. Our results showed that the silencing of Id2 with RNA interference induces HCC cell apoptosis. Sorafenib is the only FDA-approved treatment for advanced HCC, but its use is hampered by the occurrence of drug resistance [28]. Therefore, the combination of sorafenib with other targeted or chemotherapeutic reagents can improve the management of HCC. In this study, we found that the combination of sorafenib with the silencing of Id2 via RNA interference significantly promoted HCC cell apoptosis, indicating that Id2 may be a promising target for the treatment of HCC.
Although abnormal Id2 expression is found in many human tumors, the regulatory mechanism of Id2 is unclear in HCC. The p53 tumor suppressor gene product was the first transcription factor that was found to bind to the promoter of Id2 [19]. Subsequent studies found more transcription factors involved in the transcription of Id2. C/EBPβ negatively regulates the expression of Id2 at the transcriptional level [20]. In this study, we found that C/EBPα could bind to the promoter of Id2 and inhibit its expression. C/EBPα is the first transcription factor identified in the C/EBP transcription factor family. Previous studies have suggested that C/EBPα is a potential tumor suppressor. A C/EBPα short-activating RNA suppresses the growth and metastasis of HCC and is currently in a phase I clinical trial for patients with liver cancer [29,30]. However, a report by Lu et al. has shown that C/EBPα is elevated in some patients with HCC and increases HCC cell growth [31]. Recent studies have shown that C/EBPα at Ser193 (Ser190 in the human protein) or the mutation of Ser193 to Ala results in a modified protein with oncogenic activities [32]. Therefore, further studies are needed to investigate the role of C/EBPα in HCC.
In conclusion, our study demonstrates that Id2 promotes HCC cell proliferation by inhibiting apoptosis. The knockdown of Id2 enhances sorafenib-induced apoptosis in HCC. C/EBPα binds to the Id2 promoter and inhibits its expression in HCC cells. Thus, Id2 may serve as a potential target in HCC therapy.
Acknowledgements
This work was supported in part by grants from the National Natural Science Foundation of China (81773152, 81472570, 81472726), the SKLORG Research foundation (91-17-29, 91-17-22) and the Research Project of Shanghai Municipal Health and Family Planning Commission (201540107).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Kanda M, Sugimoto H, Kodera Y. Genetic and epigenetic aspects of initiation and progression of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10584–10597. doi: 10.3748/wjg.v21.i37.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 4.Ohashi-Ito K, Fukuda H. Functional mechanism of bHLH complexes during early vascular development. Curr Opin Plant Biol. 2016;33:42–47. doi: 10.1016/j.pbi.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. Id-related genes encoding helix-loop-helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Biol Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- 6.Li K, Yao L, Chen L, Cao ZG, Yu SJ, Kuang XY, Hu X, Shao ZM. ID2 predicts poor prognosis in breast cancer, especially in triplenegative breast cancer, and inhibits E-cadherin expression. Onco Targets Ther. 2014;7:1083–1094. doi: 10.2147/OTT.S64759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppe JP, Itahana Y, Moore DH, Bennington JL, Desprez PY. Id-1 and Id-2 proteins as molecular markers for human prostate cancer progression. Clin Cancer Res. 2004;10:2044–2051. doi: 10.1158/1078-0432.ccr-03-0933. [DOI] [PubMed] [Google Scholar]
- 8.Lasorella A, Boldrini R, Dominici C, Donfrancesco A, Yokota Y, Inserra A, Iavarone A. Id2 is critical for cellular proliferation and is the oncogenic effector of N-myc in human neuroblastoma. Cancer Res. 2002;62:301–306. [PubMed] [Google Scholar]
- 9.Lasorella A, Iavarone A, Israel MA. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 11.Lasorella A, Rothschild G, Yokota Y, Russell RG, Iavarone A. Id2 mediates tumor initiation, proliferation, and angiogenesis in Rb mutant mice. Mol Cell Biol. 2005;25:3563–3574. doi: 10.1128/MCB.25.9.3563-3574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C, Yang X, Pursell B, Mercurio AM. Id2 complexes with the SNAG domain of Snai1 inhibiting Snai1-mediated repression of integrin beta4. Mol Cell Biol. 2013;33:3795–3804. doi: 10.1128/MCB.00434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng S, Zhang Y, Ma J, Deng G, Qu Y, Guo C, Han Y, Yin L, Cai C, Li Y, Wang G, Bonkovsky HL, Shen H. BMP4 promotes metastasis of hepatocellular carcinoma by an induction of epithelial-mesenchymal transition via upregulating ID2. Cancer Lett. 2017;390:67–76. doi: 10.1016/j.canlet.2016.12.042. [DOI] [PubMed] [Google Scholar]
- 14.Ma J, Zeng S, Zhang Y, Deng G, Qu Y, Guo C, Yin L, Han Y, Shen H. BMP4 enhances hepatocellular carcinoma proliferation by promoting cell cycle progression via ID2/CDKN1B signaling. Mol Carcinog. 2017;56:2279–2289. doi: 10.1002/mc.22681. [DOI] [PubMed] [Google Scholar]
- 15.Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–8333. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- 16.Perk J, Gil-Bazo I, Chin Y, de Candia P, Chen JJ, Zhao Y, Chao S, Cheong W, Ke Y, Al-Ahmadie H, Gerald WL, Brogi E, Benezra R. Reassessment of id1 protein expression in human mammary, prostate, and bladder cancers using a monospecific rabbit monoclonal anti-id1 antibody. Cancer Res. 2006;66:10870–10877. doi: 10.1158/0008-5472.CAN-06-2643. [DOI] [PubMed] [Google Scholar]
- 17.Wilson JW, Deed RW, Inoue T, Balzi M, Becciolini A, Faraoni P, Potten CS, Norton JD. Expression of Id helix-loop-helix proteins in colorectal adenocarcinoma correlates with p53 expression and mitotic index. Cancer Res. 2001;61:8803–8810. [PubMed] [Google Scholar]
- 18.Lofstedt T, Jogi A, Sigvardsson M, Gradin K, Poellinger L, Pahlman S, Axelson H. Induction of ID2 expression by hypoxia-inducible factor-1: a role in dedifferentiation of hypoxic neuroblastoma cells. J Biol Chem. 2004;279:39223–39231. doi: 10.1074/jbc.M402904200. [DOI] [PubMed] [Google Scholar]
- 19.Paolella BR, Havrda MC, Mantani A, Wray CM, Zhang Z, Israel MA. p53 directly represses Id2 to inhibit the proliferation of neural progenitor cells. Stem Cells. 2011;29:1090–1101. doi: 10.1002/stem.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karaya K, Mori S, Kimoto H, Shima Y, Tsuji Y, Kurooka H, Akira S, Yokota Y. Regulation of Id2 expression by CCAAT/enhancer binding protein beta. Nucleic Acids Res. 2005;33:1924–1934. doi: 10.1093/nar/gki339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou H, Ge C, Sun H, Li H, Li J, Tian H. Tunicamycin inhibits cell proliferation and migration in hepatocellular carcinoma through suppression of CD44s and the ERK1/2 pathway. Cancer Sci. 2018;109:1088–1100. doi: 10.1111/cas.13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian H, Ge C, Li H, Zhao F, Hou H, Chen T, Jiang G, Xie H, Cui Y, Yao M, Li J. Ribonucleotide reductase M2B inhibits cell migration and spreading by early growth response protein 1-mediated phosphatase and tensin homolog/Akt1 pathway in hepatocellular carcinoma. Hepatology. 2014;59:1459–1470. doi: 10.1002/hep.26929. [DOI] [PubMed] [Google Scholar]
- 23.Tian H, Ge C, Zhao F, Zhu M, Zhang L, Huo Q, Li H, Chen T, Xie H, Cui Y, Yao M, Li J. Downregulation of AZGP1 by Ikaros and histone deacetylase promotes tumor progression through the PTEN/Akt and CD44s pathways in hepatocellular carcinoma. Carcinogenesis. 2017;38:207–217. doi: 10.1093/carcin/bgw125. [DOI] [PubMed] [Google Scholar]
- 24.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013;280:1709–1716. doi: 10.1111/febs.12185. [DOI] [PubMed] [Google Scholar]
- 25.Kleeff J, Ishiwata T, Friess H, Buchler MW, Israel MA, Korc M. The helix-loop-helix protein Id2 is overexpressed in human pancreatic cancer. Cancer Res. 1998;58:3769–3772. [PubMed] [Google Scholar]
- 26.Wang C, Chen Q, Hamajima Y, Sun W, Zheng YQ, Hu XH, Ondrey FG, Lin JZ. Id2 regulates the proliferation of squamous cell carcinoma in vitro via the NF-kappaB/Cyclin D1 pathway. Chin J Cancer. 2012;31:430–439. doi: 10.5732/cjc.011.10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou JP, Gao ZL, Zhou ML, He MY, Xu XH, Tao DT, Yang CC, Liu LK. Snail interacts with Id2 in the regulation of TNF-alpha-induced cancer cell invasion and migration in OSCC. Am J Cancer Res. 2015;5:1680–1691. [PMC free article] [PubMed] [Google Scholar]
- 28.Niu L, Liu L, Yang S, Ren J, Lai PBS, Chen GG. New insights into sorafenib resistance in hepatocellular carcinoma: Responsible mechanisms and promising strategies. Biochim Biophys Acta. 2017;1868:564–570. doi: 10.1016/j.bbcan.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Reebye V, Saetrom P, Mintz PJ, Huang KW, Swiderski P, Peng L, Liu C, Liu X, Lindkaer-Jensen S, Zacharoulis D, Kostomitsopoulos N, Kasahara N, Nicholls JP, Jiao LR, Pai M, Spalding DR, Mizandari M, Chikovani T, Emara MM, Haoudi A, Tomalia DA, Rossi JJ, Habib NA. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59:216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huan H, Wen X, Chen X, Wu L, Liu W, Habib NA, Bie P, Xia F. C/EBPalpha short-activating RNA suppresses metastasis of hepatocellular carcinoma through inhibiting EGFR/betacatenin signaling mediated EMT. PLoS One. 2016;11:e0153117. doi: 10.1371/journal.pone.0153117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu GD, Leung CH, Yan B, Tan CM, Low SY, Aung MO, Salto-Tellez M, Lim SG, Hooi SC. C/EBPalpha is up-regulated in a subset of hepatocellular carcinomas and plays a role in cell growth and proliferation. Gastroenterology. 2010;139:632–43. 643.e1–4. doi: 10.1053/j.gastro.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 32.Cast A, Valanejad L, Wright M, Nguyen P, Gupta A, Zhu L, Shin S, Timchenko N. C/EBPalpha-dependent preneoplastic tumor foci are the origin of hepatocellular carcinoma and aggressive pediatric liver cancer. Hepatology. 2018;67:1857–1871. doi: 10.1002/hep.29677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.