Abstract
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the world. RING finger-related E3 ubiquitin ligases play a role in tumorigenesis and can function either as oncogenes or tumor suppressors based on their target proteins. Here, we show that the expression of RNF152, a ring finger protein, in CRC tissues was significantly reduced compared with adjacent non-cancerous tissues. High expression levels of RNF152 correlated with better prognosis in patients with colorectal cancer. Low expression of RNF152 correlated with lymphatic metastasis. Overexpression of RNF152 inhibited CRC cell proliferation both in vitro and in vivo by inactivating the mechanistic target of rapamycin complex 1 (mTORC1) and inducing autophagy and apoptotic cell death. This strong inhibition was dependent on the E3 ligase activity of RNF152. Ectopic expression of the RNF152-CS-mutant, which lacks E3 ligase activity, significantly restored the proliferation ability of CRC cells. Our findings showed that RNF152 inhibits colorectal cancer growth and may be a novel prognostic biomarker for the treatment of CRC.
Keywords: Colorectal cancer, RNF152, prognosis, proliferation, E3 ligase
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer, accounting for 9.7% of all cancer cases worldwide. It accounts for approximately 1.4 million new cases and 697,000 deaths per year [1]. The incidence of CRC increases with age: the median age for diagnosis of CRC is 68 years in men and 72 years in women [2]. The 5-year relative survival rate of CRC has risen to 50% in developing countries and 65% in developed countries [3]. The treatment of CRC depends on the site of the tumor and the stage of diagnosis. For early stages of CRC, surgery alone can eliminate cancer [4]. Advanced CRC is treated with either chemotherapy alone or radiotherapy combined with surgery for metastases [5]. If the tumor has metastasized to distant organs, the 5-year relative survival rate is only 11.7% [2]. Clinical outcomes in patients with CRC are far from satisfactory, especially in patients with advanced cancer (stages III and IV).
Really interesting new gene (RING) finger proteins (RNFs) are characterized by a canonical RING finger domain that has a consensus sequence of C-X2-C-X[9-39]-C-X[1-3]-H-X[2-3]-C-X2-C-X[4-48]-C-X2-C, where C and H are cysteine and histidine residues that participate in zinc coordination, and X represents any amino acid [6,7]. This domain is important for the E3 ubiquitin ligase activity of RNFs [8]. Ubiquitination involves the sequential action of three major enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) and is crucial for many cellular processes [9]. RNFs are important for growth, differentiation, gene transcription, signal transduction, and apoptosis [7]. In tumorigenesis, RNFs have shown conflicting roles of suppression or progression of cancer [10]. The RING finger E3 ligase, MDM2, regulates p53 tumor suppressor activity by targeting it for ubiquitination and proteasomal degradation [24]. Overexpression of MDM2 results in unrestricted tumor growth [25]. BRCA1, a noted tumor suppressor gene with an amino-terminal RING finger, is often mutated in ovarian and familial breast cancer [26]. Overexpression of RNF139 inhibits the growth of renal cancer cells [11]. In colorectal cancer, increased RNF183 expression is associated with tumor size and metastasis and promotes CRC cell growth through the NF-κB signaling pathway [12]. RNF43 is a frequently mutated gene in colorectal and endometrial cancers [13].
RNF152 is a new member of the RNF protein family and is polyubiquitinated via its RING finger domain [14]. The Ras-related small G protein, RagA GTPase, is an ubiquitinated substrate of RNF152 [15]. Decreased RNF152 expression has been observed in breast cancer cell lines [16]. Using expression profile and bioinformatics analyses, we have previously shown that RNF152 expression is significantly downregulated in CRC tissues compared with normal colorectal mucosa [17]. Here, we used real-time quantitative polymerase chain reaction (qPCR) to confirm that RNF152 was significantly downregulated in colorectal cancer tissue. Overexpression of RNF152 in CRC cells in vitro and in vivo inhibited their proliferation. Retrospective analysis showed that high RNF152 expression was associated with better prognosis in patients with CRC. Our results suggested an important role for RNF152 in the development of CRC.
Methods
CRC specimen collection
We collected a total of 169 human CRC and paired normal tissue samples from patients at the Department of Colorectal Surgery at Xinhua Hospital, Shanghai Jiaotong University School of Medicine. We obtained informed consent from all study participants, and the study was approved by the Institutional Review Board. These experiments were conducted in accordance with the ethical standards laid down in the Declaration of Helsinki 1964 and its subsequent amendments or comparable ethical standards. We analyzed 20 CRC samples using qPCR. We prepared tissue arrays using the 169 CRC and normal tissue specimens to perform immunohistochemical analysis.
Cell culture
Six human colorectal cancer cell lines, SW480, SW620, HT29, HCT116, LOVO, and DLD1, and 293T cells used for virus infection were maintained in Dulbecco’s Modified Eagle’s medium (Corning, USA) containing 10% fetal bovine serum (Gibco, USA). Cells were incubated in humid atmospheric 5% CO2 and passaged every 3-4 days.
RNA isolation and qPCR
Total RNA was extracted using RNAiso Plus (TaKaRa, Dalian, China) and reverse transcribed into cDNA using PrimeScript® RT-PCR kit (TaKaRa). SYBR® Premix Ex TaqTM (TaKaRa) and ABI 7500 cycler (Fischer Scientific, USA) were used for qPCR. Beta actin (ACTB) served as an internal control. The primers used in this study were as follows: RNF152-F, 5’-TGTGTCTTTGCAGTGCAGGT-3’ and RNF152-R, 5’-GGGTGAAAGAGACTCGGTGA-3’; and ACTB-F, 5’-GTTGTCGACGACGAGCG-3’ and ACTB-R, 5’-GCACAGAGCCTCGCCTT-3’. The PCR was performed in triplicate and gene expression was analyzed using the 2-ΔΔCT method [18].
Immunohistochemistry
Paraffin sections of CRC tissues were dewaxed, hydrated, and processed according to standard protocols. Sections were incubated with rabbit polyclonal antibody against RNF152 (A gift from Dr. Lu Deng of East China Normal University) [15] or phosphate buffer (PBS; negative control) overnight, washed three times with PBS, and incubated with horseradish peroxidase-conjugated secondary antibody (GK500710; Gene Company Ltd., Shanghai, China) for 30 minutes at room temperature. After rinsing in PBS for 3 minutes, sections were stained with 3,3’-diaminobenzidine solution and subsequently counterstained with 0.1% hematoxylin using a coverslip. RNF152 staining was scored according to the following criteria: staining intensity was classified as 0 (no staining), 1 (mild staining), 2 (moderate staining), or 3 (strong positive); staining rate was defined as 1 (0-25% positive cells), 2 (50% or more), 3 (75%), or 4 (>75%) [19]. For each section, a product of staining intensity and staining rate values (from 0 to 12) was used to calculate a semi-quantitative score. Two histopathologists were blindly assigned slides to review and score.
Immunoblotting
Western blotting was performed as described [20]. Proteins were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. The following antibodies were used: anti-FLAG antibody (1:2000; F1804; Sigma-Aldrich, USA); anti-PS6K (1:1000; #9234; Cell Signaling Technology, USA); anti-S6K1 (1:5000; ab32529; Abcam, USA); anti-LC3B (1:1000; #3868; Cell Signaling Technology); anti-cleaved PARP (1:5000; ab32064; Abcam); and anti-b-ACTIN (1:10,000; A3854; Sigma-Aldrich). The membrane was incubated with horseradish peroxidase-labeled secondary antibody (1:1000; Beyotime, China), and antibody-bound protein was visualized using chemiluminescence (Millipore, USA).
RNF152 overexpression
The entire coding sequence of RNF152 and RNF152-CS-mutant cDNAs was amplified and cloned into the pLVX vector (containing FLAG tag) obtained from Addgene (http://www.addgene.org). The pCDNA3.1-RNF152 and pCDNA3.1-RNF152-CS-mutant vectors were gifts from Dr. Lu Deng of East China Normal University [15]. Lentivirus production and transduction were performed according to the instructions provided by Addgene.
RNA interference
For doxycycline (DOX)-inducible small hairpin RNA (shRNA)-mediated knockdown of RNF152, two sets of single-stranded oligonucleotides encoding RNF152 target shRNAs (RNF152-sh1: sense, 5’-CCGGATGTCAGATCTGTTTCAATTCTCGAGTAATTGAAACAGATCTGACATTTTTTG-3’; RNF152-sh2: sense, 5’-CCGGCTTCACAACATGTCTTGCATTCTCGAGAATGCAAGACATGTTGTGAAGTTTTTG-3’) were synthesized and cloned into the pLKO-TET-ON lentiviral expression system (Addgene) [21]. Cells stably expressing DOX-inducible shRNA were cultured in medium containing puromycin (1 μg/mL). Gene knockdown was induced by incubating cells in 500 ng/mL DOX for 48 hours.
Cell proliferation assay
Cell growth was measured using the CCK-8 kit (Dojindo, Japan) to detect the dehydrogenase activity of living cells. Briefly, cells (2 × 103 cells/well) were seeded in 96-well plates and incubated for 24 hours at 37°C in a humidified incubator (5% CO2). CCK-8 solution (10 μL) was added to each cell incubator for 1 hour. The absorbance at 450 nm was measured using a microplate reader. For the colony formation assay, 1000 cells per well were inoculated in a 6-well plate and cultured at 37°C for 2 weeks. They were then fixed with 100% methanol and stained with 0.1% crystal violet. Experiments were performed in triplicate.
Apoptosis detection
Cell apoptosis was measured using the FITC Annexin V Apoptosis Detection Kit (BD Pharmingen, USA). Cells were trypsinized, washed with cold PBS, and then resuspended in 1 × Binding Buffer at a concentration of 1 × 106 cells/mL. A cell suspension volume of 100 μL (1 × 105 cells) was transferred to a 5-mL culture tube and 5 μL of FITC Annexin V and 5 μL of propidium iodide were added to the tube. The cells were gently vortexed and incubated for an additional 15 minutes at room temperature (25°C) in the dark and resuspended in 400 μL 1 × Binding Buffer. Apoptosis was analyzed using flow cytometry (Beckman Coulter) within 1 hour.
Xenograft tumor formation
Nude mice (4-6 weeks old, male) were obtained from SLAC Laboratory Animals LLC, Shanghai, China. All mouse procedures were approved by Xinhua Hospital Animal Care and Use Committee. Xenograft tumors were established in nude mice. HCT116-PLVX and HCT116-PLVX-RNF152 cells (1 × 106) were injected into the bilateral axillary fat pad. The mice were randomly divided into two groups (5 mice/group). Tumor weight was measured 4 weeks after surgery. The following antibodies were used for immunohistochemistry: anti-Ki-67 (1:1 ready to use, M7240; Dako, Denmark); anti-cleaved PARP (1:100; ab32064; Abcam); and anti-LC3B (1:200; #3868; Cell Signaling Technology).
Statistical analysis
Statistical analysis was performed using SPSS 18 software. Student’s t-test was used to compare continuous variables. Pearson’s χ2 test was used to evaluate the correlation between expression and clinicopathological parameters of RNF152. The survival curve was plotted using the Kaplan-Meier method and compared with a log-rank test. Bilateral P values <0.05 were considered statistically significant.
Results
Downregulation of RNF152 in CRC cells and its prognostic value for patients with CRC
We had previously used mRNA microarrays to identify RNF152 as a downregulated gene in CRC [17]. Here, we confirmed our microarray results by using qPCR to detect mRNA expression levels of RNF152 in 20 paired CRC tissues and adjacent normal colorectal tissues. We found that RNF152 mRNA expression was significantly reduced in CRC tissues compared with normal tissues (P<0.0001; Figure 1A). These data were consistent with that in The Cancer Genome Atlas database (Figure 1B, downloaded from http://gepia.cancer-pku.cn/).
Figure 1.
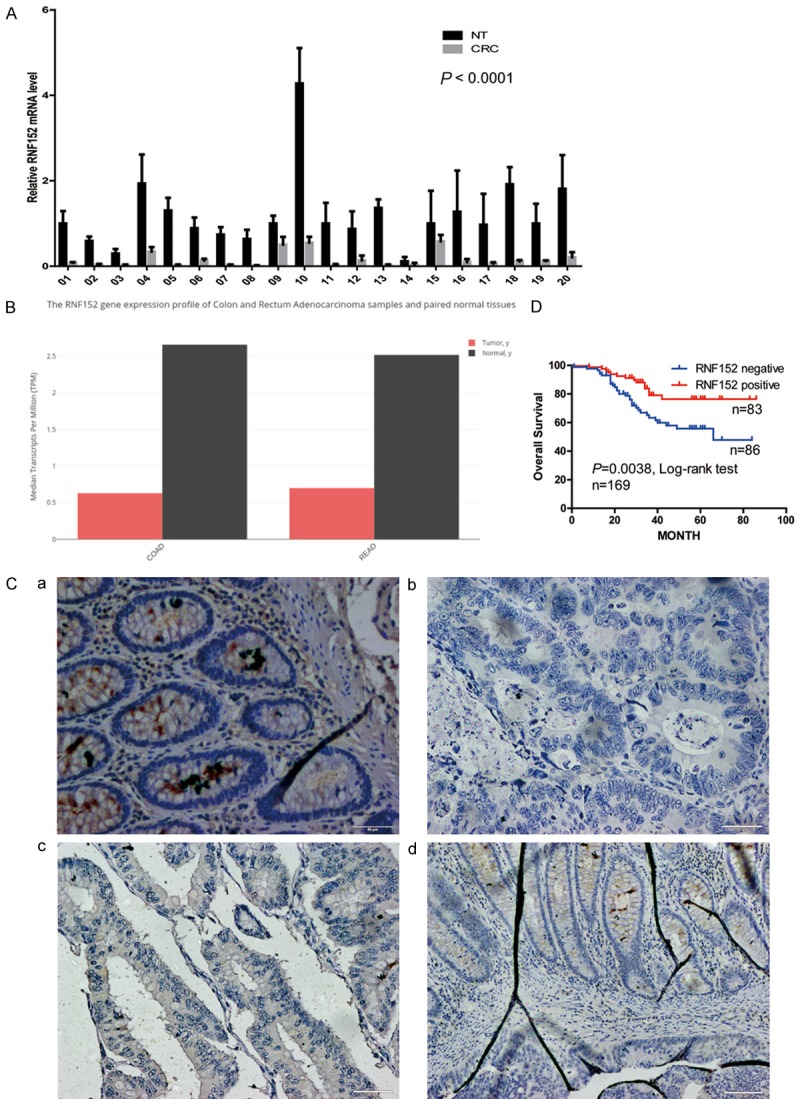
Reduced expression of RNF152 in CRC. (A) QPCR detection of RNF152 expression in 20 CRC tumor tissue samples and corresponding normal mucosa. RNF152 mRNA expression was significantly declined in CRC tissue compared with normal tissue. Student’s paired t-test; P<0.0001. (B) Expression of RNF152 in Colon Cancer and Rectal Cancer in TCGA Database (http://gepia.cancer-pku.cn). RNF152 mRNA expression was significantly declined in CRC tissue compared with normal tissue. The y-axis stands for the median RNF152 transcripts per million (TPM). (C) Immunohistochemical analysis of RNF152 in CRC and normal mucosa tissue samples. Representative images of (a) positive RNF152 expression of normal mucosa tissue, (b) negative expression of RNF152 in CRC cells, (c) positive expression of RNF152 in CRC cells, and (d) RNF152 expression in CRC cells in the invasive margin. In the invasive margin of CRC tissue, CRC cells did not express RNF152. But the normal colonic gland, which has not been invaded by CRC cells, expressed high level of RNF152. (Ca-Cc) Magnification, 200 ×. Scale bar, 50 μm; (Cd) Magnification, 100 ×. Scale bar, 100 μm. (D) RNF152 expression-stratified Kaplan-Meier plots for overall survival in CRC patients, negative RNF152 expression was significantly associated with shortened patient survival. Statistical significance was determined by a log-rank test. P = 0.0038, n = 169. RNF152: Ring Finger Proteins 152; CRC: colorectal cancer; TCGA: The Cancer Genome Atlas; QPCR: quantitative-polymerase chain reaction.
To assess the association between tumor protein levels of RNF152 and the clinicopathological features of CRC patients (Table 1), we performed immunohistochemistry using tissue microarrays of 169 CRC and adjacent normal colon tissues. Representative images are shown in Figure 1C. RNF152 staining intensities of 2 and 3 were marked as high expression levels and 0 and 1 were marked as low expression levels (Table 2). We detected high levels of RNF152 expression in the epithelial cells of normal tissue (Figure 1Ca). High RNF152 staining was found in 155/169 (91.7%) normal tissue samples (Table 2). In tumor epithelial cells, high levels of RNF152 staining were found only in 83/169 (49.1%) tissue samples (Figure 1Cb, 1Cc; Table 2), which again confirmed the reduced expression of RNF152 in CRC tissues. Distribution of RNF152 expression was also different between normal and CRC tissues. In the invasive margin of CRC tissue, CRC cells did not express RNF152. However, the normal colonic gland, which was untouched by CRC cells, expressed high levels of RNF152 (Figure 1C, 1D).
Table 1.
Case information
| Clinical parameters | Case (%) | |
|---|---|---|
| Gender | Male | 91 (53.8) |
| Female | 78 (46.2) | |
| Age | ≤60 | 79 (46.7) |
| >60 | 90 (53.3) | |
| Stage | I | 13 (7.7) |
| II | 66 (39.1) | |
| III | 71 (42.0) | |
| IV | 19 (11.2) | |
| T | 1 | 3 (1.8) |
| 2 | 15 (8.9) | |
| 3 | 57 (33.7) | |
| 4 | 94 (55.6) | |
| N | 0 | 85 (50.3) |
| 1 | 55 (32.5) | |
| 2 | 29 (17.2) | |
| M | 0 | 148 (87.6) |
| 1 | 21 (12.4) | |
| Pathology | I | 31 (18.3) |
| II | 125 (74.0) | |
| III | 13 (7.7) | |
Table 2.
IHC expression of RNF152
| Tissues | Cases (N) | Score of RNF152 expression | P | |||
|---|---|---|---|---|---|---|
|
| ||||||
| 0 | 1 | 2 | 3 | |||
| CRC | 169 | 19 | 67 | 57 | 26 | <0.0001 |
| NT | 169 | 4 | 10 | 67 | 88 | |
| Stage | ||||||
| I | 13 | 3 | 2 | 7 | 1 | |
| II | 66 | 4 | 24 | 26 | 12 | |
| III | 71 | 11 | 30 | 18 | 12 | |
| IV | 19 | 1 | 11 | 6 | 1 | |
| Stage N | ||||||
| N0 | 85 | 8 | 28 | 33 | 16 | = 0.041 |
| N1/2 | 84 | 11 | 39 | 24 | 10 | |
Statistical analysis of the relationship between RNF152 protein expression and CRC pathological features showed that low expression levels of RNF152 were significantly associated with stage N of the American joint committee on cancer (AJCC) tumor, node, metastasis (TNM) staging system (P = 0.041, Table 2). Kaplan-Meier survival analysis (Figure 1D) showed that patients with low RNF152 expression had shorter overall survival (P = 0.0038). Together, our results suggested that RNF152 played an important role in CRC and is a potential prognostic marker for patients with CRC.
RNF152 overexpression inhibits CRC cell proliferation in vitro
We examined RNF152 mRNA levels in six CRC cell lines and normal colonic mucosa from two patients (Figure 2A) to study the role of RNF152 in CRC cell proliferation. We generated RNF152 knockdown cells by infecting HCT116 with two pLKO-TET-ON lentivirus vectors encoding RNF-152-targeting shRNAs. The knockdown efficiency of RNF152 inhibition was confirmed using qPCR (Figure 2B). We evaluated the effects of RNF152 knockdown on cell proliferation in the stable cell lines using CCK-8 assays. We found that knockdown of RNF152 in HCT116 cells did not have an effect on CRC cell growth (Figure 2C).
Figure 2.
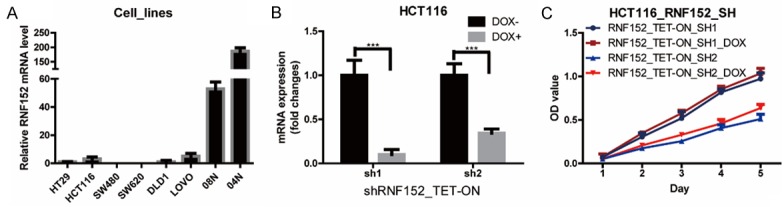
The low expression of RNF152 in CRC cell lines and the effect of RNF152 knockdown in CRC cells. A. QPCR analysis of RNF152 mRNA levels in 6 CRC cell lines and 2 patients’ normal mucosa. ACTB mRNA was used as an internal control. B. QPCR analysis of RNF152 expression level in HCT116 cells transduced with 2 shRNA of RNF152 TET-ON virus in the absence and presence of doxycycline. Student’s t-test; ***P<0.001. C. The proliferation of shRNF152 TET-ON HCT116 cells in the absence and presence of doxycycline was analyzed by a CCK-8 assay. Knocking down of RNF152 in HCT116 showed no difference of CRC cell growth. The cell numbers were analyzed every day for 5 days. RNF152: Ring Finger Proteins 152; CRC: colorectal cancer; QPCR: quantitative-polymerase chain reaction; sh: short hairpin RNA.
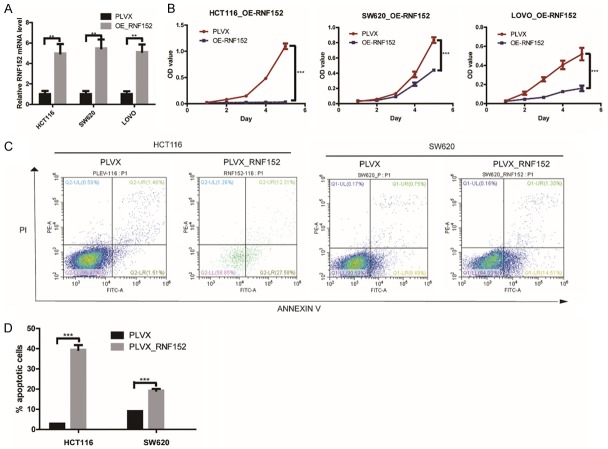
For RNF152 overexpression, we infected HCT-116, SW620, and LOVO cell lines with an RNF-152-expressing PLVX lentivirus vector. RNF152 overexpression was confirmed using qPCR (Figure 3A; P<0.01). CCK-8 assays showed that overexpression of RNF152 in HCT116, SW620, and LOVO cells significantly decreased CRC cell growth in vitro (Figure 3B; P<0.001 for all three cell lines).
Figure 3.
The effect of RNF152 ectopic expression in CRC cells in vitro. A. QPCR analysis of RNF152 expression level in HCT116, SW620 and LOVO cells transduced with RNF152-expressing PLVX lentivirus (containing the FLAG tag). The overexpression efficiency of RNF152 was significant. Student’s t-test; **P<0.01. B. The proliferation of RNF152-ectopic expression HCT116, SW620 and LOVO cells was analyzed by CCK-8 assays. Overexpression of RNF152 in HCT116, SW620 and LOVO cells significantly decreased CRC cell growth in vitro. The cell numbers were analyzed every day for 5 days. Student’s t-test; ***P<0.001. C and D. The proportion of apoptotic cells as determined by flow cytometry analysis in RNF152-ectopic expression HCT116 and SW620 cells. RNF152 could promote cell apoptosis. Student’s t-test; ***P<0.001. RNF152: Ring Finger Proteins 152; CRC: colorectal cancer; QPCR: quantitative-polymerase chain reaction.
RNF152 overexpression induces cell apoptosis in CRC cells
To test whether the anti-proliferative effect of RNF152 overexpression in CRC cells was mediated by apoptotic cell death, we performed flow cytometry analysis on HCT116 and SW620 cells that overexpressed RNF152. We found that 39.9% of HCT116 and 15.8% of SW620 cells showed Annexin V positive staining, which indicated apoptosis (Figure 3C and 3D; P<0.001 for both cell lines). However, only 3.0% of HCT116 and 9.2% of SW620 control-vector-expressing cells were Annexin V positive. Our results suggested that RNF152 inhibited CRC cell proliferation by inducing apoptosis.
RNF152 overexpression impairs the growth of tumor xenografts in vivo
We studied the effect of RNF152 overexpression on colorectal cancer cells in vivo. Immunodeficient nude mice were subcutaneously injected with control or RNF152-overexpressing HCT116 cells. The mice were monitored for tumor xenograft growth for 4 weeks post-injection. We found that RNF152 overexpression significantly reduced tumor growth in mice (Figure 4A). Tumors in mice that overexpressed RNF152 had a lower weight compared with tumors in control mice (P<0.0001; Figure 4B). Immunohistochemical staining of xenograft tumor tissue (Figure 4C) confirmed RNF152 overexpression. Xenograft tumor tissues that ectopically expressed RNF152 showed reduced expression of Ki-67, which is a marker for cell proliferation. However, they showed high levels of expression of cleaved PARP and LC3B, which are markers of apoptosis and autophagy, respectively (Figure 4C). Our results showed that overexpression of RNF152 reduced CRC growth in vivo.
Figure 4.
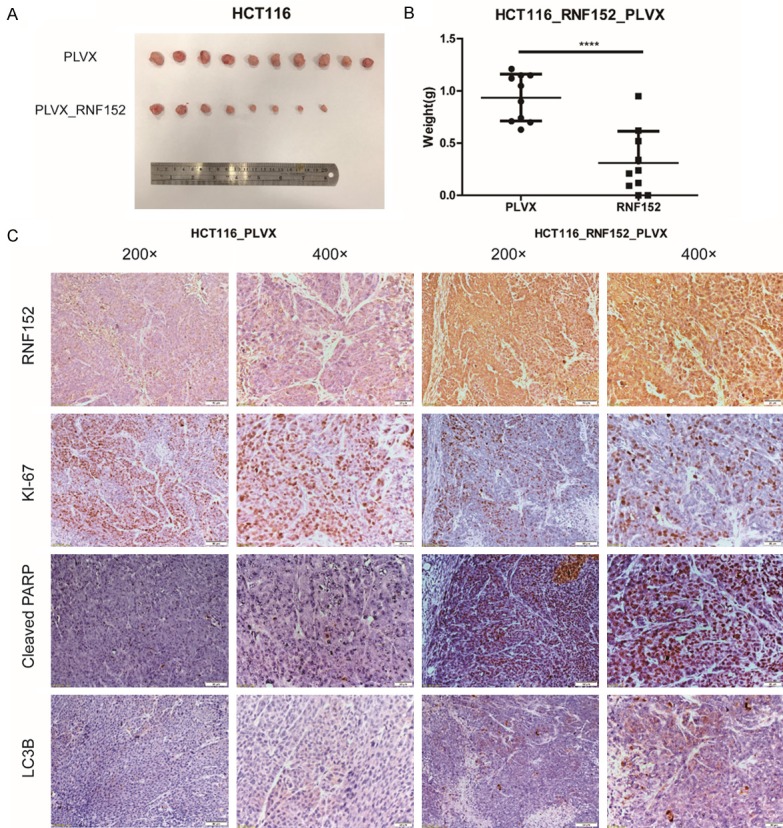
The effect of RNF152 ectopic expression in CRC cells in vivo. A. 1 million PLVX-HCT116 cells or RNF152-ectopic expression HCT116 cells were inoculated into mice to create a xenograft tumor model. Mice were randomized into two groups (5 mice/group). Images of tumors were taken from both groups after 4 weeks of treatment. B. The tumor weights in both groups after 4 weeks of treatment. Tumors derived from the RNF152-overexpression group were significantly smaller compared with tumors in the control group. Student’s t-test; ****P<0.0001. C. Immunohistochemical analysis of RNF152, KI-67, Cleaved PARP and LC3B in xenograft tumor of PLVX-HCT116 cells and RNF152-ectopic expression HCT116 cells. RNF152-ectopic expression HCT116 xenograft tumor revealed low expression of Ki-67, but high expression of Cleaved PARP and LC3B. Left column of HCT116_PLVX and HCT116_RNF152: Magnification, 200 ×. Scale bar, 50 μm; Righ column of HCT116_PLVX and HCT116_RNF152: Magnification, 400 ×. Scale bar, 20 μm. RNF152: Ring Finger Proteins 152; CRC: colorectal cancer; QPCR: quantitative-polymerase chain reaction.
The E3 ligase activity of RNF152 is important for inhibiting CRC cell proliferation
RNF152 is a RING family E3 ligase and its activity depends on the RING domain [14]. To test whether RNF152 inhibits the proliferation of CRC cells via its E3 ligase activity, we generated the RNF152-CS mutant. This mutant lacked E3 ubiquitin ligase activity because four cysteine residues are replaced with serines in the RING domain of RNF152. We used qPCR and western blotting to confirm RNF152 expression in HCT116 cells (Figure 5A and 5B). Stable ectopic expression of wild-type RNF152 resulted in a significant decrease in the proliferation of HCT116 cells, but proliferation capacity was restored in cells expressing the CS mutant (P<0.0001; Figure 5C). We found an approximately 36% decrease in apoptosis in RNF152-CS mutant cells compared with cells overexpressing RNF152 (P<0.0001; Figure 5D). Colony formation assays confirmed the restored proliferative ability of RNF152-CS mutant cells (P<0.0001; Figure 5E and 5F). Our results suggested that the E3 ligase activity of RNF152 was important for its inhibition of CRC cell proliferation.
Figure 5.
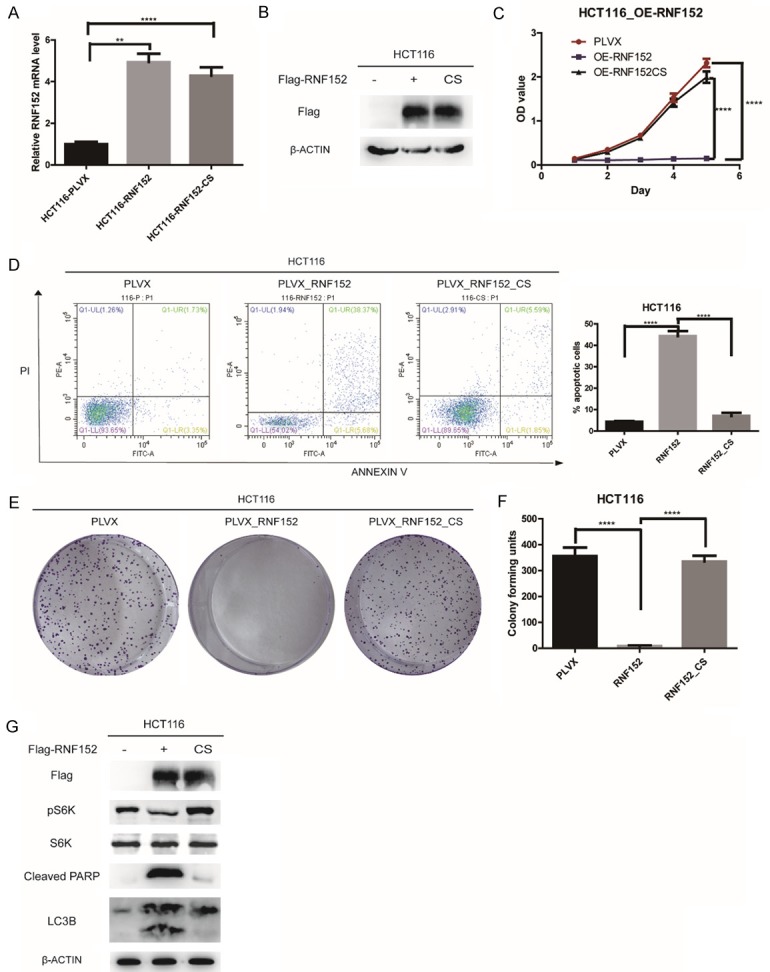
RNF152 regulates mTORC1 inactivation, induces cell autophagy and apoptotic cell death in CRC through its E3 ligase activity. A. QPCR analysis of RNF152 expression level in PLVX-HCT116 cells, RNF152-ectopic expression HCT116 cells (containing the FLAG tag) and RNF152-CS mutant-ectopic expression HCT116 cells (containing the FLAG tag). ACTB was used as control. B. Western blot analysis of FLAG expression level in PLVX-HCT116 cells, RNF152-ectopic expression HCT116 cells (containing the FLAG tag) and RNF152-CS mutant-ectopic expression HCT116 cells (containing the FLAG tag). ACTB was used as control. C. The proliferation of WT of CS mutant RNF152-ectopic expression HCT116 cells was analyzed by CCK-8 assays. Stable expression of the RNF152 wild-type construct results in a significant decrease in the proliferation of HCT116 cells, but cells expressing the CS mutant show a restored proliferative capacity. The cell numbers were analyzed every day for 5 days. D. The proportion of apoptotic cells as determined by flow cytometry analysis in WT of CS mutant RNF152-ectopic expression HCT116 cells. Over 36% of HCT116 cells avoided apoptosis by CS mutation. E, F. Colony-forming assay in WT of CS mutant RNF152-ectopic expression HCT116 cells. Stable expression of the RNF152 wild-type construct results in a significant decrease in the proliferation of HCT116 cells, but cells expressing the CS mutant show a restored proliferative capacity. G. WB analysis in WT of CS mutant RNF152-ectopic expression HCT116 cells. pS6K1 was significantly inhibited by ectopic expression of RNF152. Overexpression of RNF152 could significantly promote PARP degradation and LC3B expression. ACTB was used as the loading control. Student’s t-test; **P<0.01, ***P<0.001, ****P<0.0001. RNF152: Ring Finger Proteins 152; CRC: colorectal cancer; QPCR: quantitative-polymerase chain reaction.
RNF152 regulates mTORC1 inactivation and induces cell autophagy and apoptotic cell death in CRC
To assess the mechanism by which RNF152 impairs CRC proliferation, we examined the levels of several signaling pathway markers. Ectopic expression of wild-type RNF152 significantly inhibited mTORC1-dependent phosphorylation of Thr389 in S6K1 (Figure 5G). This inhibition relied on the E3 ligase activity of RNF152 because the CS mutant showed restored levels of phosphorylated S6K1 (Figure 5G). Our results confirmed that the E3 ligase activity of RNF152 negatively regulated mTORC1 activation. Inhibition of mTORC1 activity is known to induce cell autophagy [22]. We found that RNF152 overexpression resulted in increased expression of the autophagy marker protein, LC3B. However, the RNF152-CS mutant showed no difference in LC3B expression compared with control cells (Figure 5G). Our flow cytometry analysis showed that RNF152 overexpression induced cell apoptosis (Figure 3C and 3D). We analyzed the degradation of the nuclear protein, poly (ADP-ribose) polymerase (PARP), which serves as a molecular biomarker for apoptotic activity. We found that RNF152 overexpression resulted in significant degradation of PARP. In the RNF152-CS mutant, cleavage of PARP was markedly reduced (Figure 5G). Taken together, our results suggested that RNF152 inhibited proliferation of CRC cells by negatively regulating mTORC1 to induce autophagy and cell death.
Discussion
RNFs are E3 ubiquitin ligases [23] that regulate key cellular functions, such as cell signaling, DNA repair, and responses to hypoxia [10]. RNFs also play an important role in the process of tumorigenesis. Interestingly, RNFs have been shown to function both as oncogenes or tumor suppressors based on their ubiquitination targets [10].
RNF152 is a lysosomal E3 ligase, which is polyubiquitinated in a RING finger domain-dependent manner [14]. RNF152 negatively regulates mTORC1 activation by targeting RagA for K63-linked ubiquitination based on amino acid availability [15]. However, the role of RNF152 in tumorigenesis remains largely unknown.
In this study, we found that RNF152 expression in CRC tissues was significantly reduced compared with adjacent non-cancerous tissues. These results were consistent with our previous microarray data [17]. We performed a retrospective analysis of patient samples and data and showed that high expression levels of RNF152 correlated with better prognosis in patients with CRC. Also, low expression levels of RNF152 were significantly associated with stage N of the AJCC TNM staging system in patients with CRC. These results indicated that RNF152 could serve as a prognostic marker for CRC.
We also explored the molecular mechanisms underlying RNF152 function in CRC. We found that RNF152 overexpression inhibited the proliferation of CRC cells both in vitro and in vivo by inactivating mTORC1 to induce autophagy and apoptotic cell death. We also showed that the strong inhibitory effect of RNF152 on CRC cell proliferation depended on its E3 ligase activity. Interestingly, we found that knockdown of RNF152 did not increase CRC cell proliferation. This may be due to the extremely low pre-existing levels of RNF152 in CRC cell lines (Figure 2A).
In conclusion, we are the first to show that RNF152 functions as a tumor suppresser gene in CRC. Low levels of RNF152 mRNA and protein expression was associated with impaired overall survival in patients with CRC. RNF152 negatively regulated mTORC1 activation and induced autophagy and apoptotic cell death in CRC cells via its E3 ligase activity. The results of our study provide new insights into the biology of CRC and show RNF152 as a potential molecular target for future tumor therapy.
Acknowledgements
This study was funded by the National High Technology Research and Development Program of China (863 Program; Grant No. SQ2014SFOZD00314), the National Natural Science Foundation of China (Grant Nos. 81372636, 81572378, and 81302089) and the Natural Science Foundation of Shanghai Municipal Commission of Health and Family Planning (Grant Nos. 201740122 and 20154Y0203). We are very grateful to Dr. Lu Deng and Prof. Ping Wang for their help.
The patient or their parent, guardian or next of kin provided written informed consent for the publication of any associated data and accompanying images.
Disclosure of conflict of interest
None.
References
- 1.International Agency for Research on Cance. 2017. (http://gco.iarc.fr/today)
- 2.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, Lin C, Leach C, Cannady RS, Cho H, Scoppa S, Hachey M, Kirch R, Jemal A, Ward E. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Sankaranarayanan R, Swaminathan R, Brenner H, Chen K, Chia KS, Chen JG, Law SCK, Ahn Y, Xiang YB, Yeole BB, Shin HR, Shanta V, Woo ZH, Martin N, Sumitsawan Y, Sriplung H, Barboza AO, Eser S, Nene BM, Suwanrungruang K, Jayalekshmi P, Dikshit R, Wabinga H, Esteban DB, Laudico A, Bhurgri Y, Bah E, Al-Hamdan N. Cancer survival in Africa, Asia, and Central America: a population-based study. Lancet Oncol. 2010;11:165–173. doi: 10.1016/S1470-2045(09)70335-3. [DOI] [PubMed] [Google Scholar]
- 4.Heald RJ, Ryall R. Recurrence and survival after total mesorectal excision for rectal-cancer. Lancet. 1986;1:1479–1482. doi: 10.1016/s0140-6736(86)91510-2. [DOI] [PubMed] [Google Scholar]
- 5.van Gijn W, Marijnen CA, Nagtegaal ID, Kranenbarg EM, Putter H, Wiggers T, Rutten HJ, Påhlman L, Glimelius B, van de Velde CJ Dutch Colorectal Cancer Group. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol. 2011;12:575–582. doi: 10.1016/S1470-2045(11)70097-3. [DOI] [PubMed] [Google Scholar]
- 6.Borden KL, Freemont PS. The ring finger domain: a recent example of a sequencestructure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 7.Joazeiro CA, Weissman AM. Ring finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 8.Deshaies RJ, Joazeiro CA. Ring domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 9.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Bio. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 10.Lipkowitz S, Weissman AM. Rings of good and evil: ring finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JP, Brauweiler A, Rudolph M, Hooper JE, Drabkin HA, Gemmill RM. The TRC8 ubiquitin ligase is sterol regulated and interacts with lipid and protein biosynthetic pathways. Mol Cancer Res. 2010;8:93–106. doi: 10.1158/1541-7786.MCR-08-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng R, Tan X, Wu J, Pan Z, Yi M, Shi W, Liu R, Yao C, Wang G, Lin J, Qiu L, Huang W, Chen S. RNF183 promotes proliferation and metastasis of colorectal cancer cells via activation of NF-kappaB-IL-8 axis. Cell Death Dis. 2017;8:e2994. doi: 10.1038/cddis.2017.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Planas-Paz L, Orsini V, Boulter L, Calabrese D, Pikiolek M, Nigsch F, Xie Y, Roma G, Donovan A, Marti P, Beckmann N, Dill MT, Carbone W, Bergling S, Isken A, Mueller M, Kinzel B, Yang Y, Mao X, Nicholson TB, Zamponi R, Capodieci P, Valdez R, Rivera D, Loew A, Ukomadu C, Terracciano LM, Bouwmeester T, Cong F, Heim MH, Forbes SJ, Ruffner H, Tchorz JS. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat Cell Biol. 2016;18:467–479. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Wu W, Wu Y, Zheng J, Suo T, Tang H, Tang J. RNF152, a novel lysosome localized E3 ligase with pro-apoptotic activities. Protein Cell. 2010;1:656–663. doi: 10.1007/s13238-010-0083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng L, Jiang C, Chen L, Jin J, Wei J, Zhao L, Chen M, Pan W, Xu Y, Chu H, Wang X, Ge X, Li D, Liao L, Liu M, Li L, Wang P. The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol Cell. 2015;58:804–818. doi: 10.1016/j.molcel.2015.03.033. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto F, Yamamoto M. Scanning copy number and gene expression on the 18q21-qter chromosomal region by the systematic multiplex PCR and reverse transcription-PCR methods. Electrophoresis. 2007;28:1882–1895. doi: 10.1002/elps.200700093. [DOI] [PubMed] [Google Scholar]
- 17.Fu J, Tang W, Du P, Wang G, Chen W, Li J, Zhu Y, Gao J, Cui L. Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst Biol. 2012;6:68. doi: 10.1186/1752-0509-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui X, Liu B, Zheng S, Dong K, Dong R. Genome-wide analysis of DNA methylation in hepatoblastoma tissues. Oncol Lett. 2016;12:1529–1534. doi: 10.3892/ol.2016.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G, Shen W, Liu CY, Liu Y, Wu T, Cui X, Yu T, Zhu Y, Song J, Du P, Cui L. Phosphorylase kinase beta affects colorectal cancer cell growth and represents a novel prognostic biomarker. J Cancer Res Clin Oncol. 2017;143:971–980. doi: 10.1007/s00432-017-2362-1. [DOI] [PubMed] [Google Scholar]
- 20.Cui X, Yang Y, Jia D, Jing Y, Zhang S, Zheng S, Cui L, Dong R, Dong K. Downregulation of bone morphogenetic protein receptor 2 promotes the development of neuroblastoma. Biochem Biophys Res Commun. 2017;483:609–616. doi: 10.1016/j.bbrc.2016.12.095. [DOI] [PubMed] [Google Scholar]
- 21.Wiederschain D, Wee S, Chen L, Loo A, Yang G, Huang A, Chen Y, Caponigro G, Yao YM, Lengauer C, Sellers WR, Benson JD. Singlevector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorick KL, Jensen JP, Fang SY, Ong AM, Hatakeyama S, Weissman AM. Ring fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang SY, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a ring finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 25.Marine J, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 26.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]