Abstract
Long noncoding RNA Zinc Finger E-box-binding homeobox 1 antisense 1 (ZEB1-AS1) reportedly participates in the tumorigenesis of various cancers. However, the clinical significance and biological functions of ZEB1-AS1 in glioma remain virtually unknown. Here, we show that ZEB1-AS1 expression was higher in glioma tissues and cell lines than in corresponding noncancerous samples and primary normal human astrocytes, respectively. The positive correlation of ZEB1-AS1 expression with the poor prognosis and progressive histological stages of glioma patients was clinically proven. In vitro assays revealed that silencing ZEB1-AS1 inhibited glioma cancer-cell growth and motility. Xenograft experiments confirmed that ZEB1-AS1 depletion attenuated tumor growth and metastasis. Dual-luciferase report assay showed that ZEB1-AS1 directly regulated microRNA-200c/141 (miR-200c/141) in glioma cells, which was confirmed by RNA immunoprecipitation assay. Furthermore, the inhibition of miR-200c/141 partially balanced the inhibition effects of cell proliferation and motility induced by ZEB1-AS1 depletion on U87 cells. Additionally, ZEB1-AS1 can regulate ZEB1 through miR-200c/141. Hence, ZEB1-AS1 directly regulated miR-200c/141 in glioma cells and relieved the inhibition of ZEB1 caused by miR-200c/141. Overall, this study revealed a novel regulatory mechanism between ZEB1-AS1 and the miR-200c/141-ZEB1 axis. The interaction between ZEB1-AS1 and miR-200c/141-ZEB1 axis was involved in the progression of glioma cells. Therefore, targeting this interaction was a promising strategy for glioma treatment.
Keywords: ZEB1-AS1, miR-200c/141, ZEB1, glioma, tumorigenesis
Introduction
Brain cancer is a very common public health problem around the world and the leading cause of death among cancers [1]. Gliomas, which are derived from glial or precursor cells, account for nearly 26.5% of all primary brain and other CNS tumors and 80.7% of malignant tumors [2]. Despite considerable improvements in combinational treatments, including surgical resection, chemotherapy, radiation therapy, and other therapeutic strategies, the prognosis and 5-year overall survival of glioblastoma multiforme is only approximately 15 months [3-5]. Therefore, identifying new molecular abnormalities concerning the progression of gliomas is necessary for establishing a specific target for the individual therapeutic strategies of gliomas.
Long noncoding RNAs (lncRNAs) are a class of ncRNAs that is approximately 200 nucleotides in length [6]. With no protein-coding capability, lncRNAs have mechanistically diverse functions in regulating gene expression levels through genomic interactions, protein concentrations, miRNA competition, and chromatin modification [7,8]. LncRNAs have been known to participate in various biological pathways and disease processes, especially in cancers [9-11]. Accumulating evidence has indicated that lncRNAs may function as either tumor suppressors or classical oncogenes in regulating tumor growth and metastasis in various malignant cancers [12-15]. Additionally, lncRNA Zinc Finger E-box-binding homeobox 1 antisense 1 (ZEB1-AS1) has been reportedly implicated in various cancers [16-23], such as hepatocellular cancer (HCC) [17], esophageal squamous cell carcinoma (ESCC) [18], prostate cancer [19], colorectal cancers [20], gastric cancer [21], glioma cancer [22], and osteosarcoma [23]. However, the functions and mechanisms of ZEB1-AS1 in glioma are still largely unknown.
MicroRNAs (miRs) reportedly contribute to important stages of tumor initiation and progression. The miR-200 family (miR-200s), including miR-200a, miR-200b, miR-200c, miR-141, and miR-429, is dysregulated and functions as critical tumor suppressors in many tumors [24-26]. Various studies have reported that miR-200s have a markedly suppressive effect on cell transformation, cancer cell migration, invasion, cell cycle, apoptosis-resistance, tumor growth, and metastasis [27]. Several studies suggested that the expressions of few miR-200 members, such as miR-200c and miR-141, are low in glioma tissues and exhibit a potential suppressive effect on glioma cell proliferation, migration, and invasion [28-30]. Recent studies have revealed that the interplay between lncRNAs and miRNAs plays a critical role in the processes of cancers [21,23,31-33]. Hansen et al. reported that lncRNAs are functional miRNA targets [34]. However, the relationship between ZEB1-AS1 and miR-200c/141 in glioma cancers has not been studied.
In this study, we indicated that ZEB1-AS1 up-regulation was positively correlated with tumor size, advanced histological grades, metastases, and poor prognosis for patients. In vitro and in vivo studies have shown that ZEB1-AS1 silencing suppresses the proliferation, migration, and invasion of glioma cells. Functional experiments have shown that the inhibition of miR-200c/141 partially abolished the effects of ZEB1-AS1 depletion on the proliferation and migration of these cells. Moreover, ZEB1-AS1 depletion and miR-200c/141 overexpression significantly inhibited glioma cell proliferation and migration. ZEB1 overexpression partially reversed the effects of ZEB1-AS1 depletion or miR-200/141 overexpression on glioma cell proliferation and migration. Hence, our study demonstrated that ZEB1-AS1/miR-200c/141-ZEB1 may act as a potential target of glioma cancer treatment.
Material and methods
Patients and tissue samples
Samples were collected from the Department of Neurosurgery of First Affiliated Hospital of Xinxiang Medical University from July 2010 to December 2012. This study was approved and documented by the Medical Ethics Committee of Xinxiang Medical University. A total of 100 patients with primary gliomas and underwent surgical treatment were selected for this study. Glioma tissues (n = 100) and normal tissue samples (n = 16) were collected during surgical treatment, according to the national regulation of clinical sampling in China. Written informed consent was obtained from all patients before surgical treatment. All patients did not receive preoperative radio- or chemotherapy, neither did they receive other forms of therapy before surgical treatment. The patients were all followed-up until 2017 or until death. The clinicopathological features of the patients are summarized in Table 1.
Table 1.
Correlation between ZEB1-AS1 expression and clinicopathological characteristics in 100 glioma patients
| Characteristics | All (n = 100) | ZEB1-AS1 expression | ||
|---|---|---|---|---|
|
| ||||
| High expression (%) | Low expression (%) | P value | ||
| Age (year) | ||||
| < 50 | 45 | 15 (33.3) | 30 (66.7) | 0.384 |
| ≥ 50 | 55 | 23 (41.8) | 32 (58.2) | |
| Gender | ||||
| Male | 58 | 24 (41.4) | 34 (58.6) | 0.741 |
| Female | 42 | 16 (38.1) | 26 (61.9) | |
| Tumor size | ||||
| ≤ 30 mm | 65 | 26 (40.0) | 39 (60.0) | 0.006* |
| > 30 mm | 35 | 24 (68.6) | 11 (31.4) | |
| TNM stage | ||||
| I/II | 47 | 12 (25.5) | 35 (74.5) | 0.015* |
| III/IV | 53 | 26 (49.1) | 27 (50.9) | |
| Tumor location | ||||
| Frontal | 28 | 7 (25.0) | 21 (75.0) | 0.252 |
| Parietal | 10 | 3 (30.0) | 7 (70.0) | |
| Occipital | 16 | 6 (37.5) | 10 (62.5) | |
| Temporal | 20 | 7 (35.0) | 13 (65.0) | |
| Others | 26 | 12 (46.2) | 14 (53.8) | |
The values had statistically significant differences.
Cell culture
The glioma cell lines U87, U251, LN18, U118, and T98G and primary normal human astrocytes (NHA) were purchased from American Type Culture Collection (Manassas, VA, USA). The cell lines were cultured in humidified atmosphere containing 5% CO2 at 37°C. The cells were routinely maintained in complete Dulbecco’s modified Eagle medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 units/mL penicillin, and 100 mg/mL streptomycin (Sigma, St. Louis, MO, USA).
Plasmid construction and transfection
The siRNA sequences targeting human ZEB1-AS1 (siZEB1-AS1: forward 5’-UCAAUGAGAUUGAACUUCAGCUGGA-3’ and reverse 5’-UUUAGGAAGG AAUUCAUGGCCUGUG-3’), negative control RNA (siNC) or double-stranded miRNA mimics, including miR-200c, miR-141, and their respective negative controls (miR-NC), were constructed and purchased from GenePharma (GenePharma Co., Ltd., Shanghai, China). U87 cells were transfected with 50 nM of siZEB1-AS1 and siNC using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. The silence efficiency was analyzed by quantitative real-time PCR (qRT-PCR) assay 48 h after transfection. MiR-200c, miR-141, the mixtures of miR-200c and miR-141 mimics (miR-200s), or miR-NC were transfected into U87 cells using Lipofectamine 2000 for 72 h. Then, 200 μL of miR-NC and miR-200c, miR-141, or miR-200s were transfected into U87 cells, which have been transfected with siZEB1-AS1 or siNC for 72 h. ZEB1 and the negative control vector were transfected into U87 cells, which have been transfected with siZEB1-AS1 or miR-200s for 48 h.
Quantitative real-time reverse transcription PCR (qRT-PCR)
The total RNA was extracted from tissues and treated cells with Trizol Reagent (Invitrogen) according to the manufacturer’s protocols. The RevertAid First Strand cDNA Synthesis kit (Thermo Fisher, Shanghai, China) was used to synthesize the cDNAs. The SYBR-Green PCR Master Mix Kit (Takara) was applied to measure the mRNA expression on an ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, USA). For miRNAs analysis, qPCR was performed using TaqMan microRNA assays (Applied Biosystems) in the StepOne Plus system following the manufacturer’s protocols. The relative gene expression was calculated using the 2-ΔΔCt method. The expressions of ZEB1-AS1 and ZEB1 were normalized against Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). miRNA expression was normalized against U6. The primers for PCR amplification are listed in Table 2.
Table 2.
The primers used for qRT-PCR analyses
| Genes | Primer sequences (5’ to 3’) | |
|---|---|---|
| ZEB1-AS1 | Forward | CCGTGGGCACTGCTGAAT |
| Reverse | CTGCTGGCAAGCGGAACT | |
| ZEB1 | Forward | ACTCTGATTCTACA CCGC |
| Reverse | TGTCACATTGATAGGGCTT | |
| GAPDH | Forward | ACCACAGTCCATGCCATCAC |
| Reverse | TCACCACCCTGTTGCTGT A | |
Protein extraction and western blot
The transfected cells were lysed with RIPA buffer (Beyotime Biotechnology, Shanghai, China) on ice. The supernatants for Western blot analysis were collected from the lysates centrifuged at 12000 × g for 15 min. The protein concentration was detected using a BCA Protein Assay Kit (Pierce Chemical Co., Rockford, IL, USA). Equal amounts of proteins were separated by electrophoresis on sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and transferred onto nitrocellulose membranes (Millipore Corp., Bedford, MA, USA). Subsequently, the membranes were blocked with 5% non-fat milk in Tris-buffered saline and Tween (TBST) 20 for 1 h. After washing with TBST, the blots were incubated with primary antibodies (E-cadherin, MMP2, Vimentin, Snail, ZEB1, Cyclin D1, CDK2, Rb, Bax, Bcl-2 and GAPDH) purchased from Cell Signaling Technology (Beverly, MA, USA) at 4°C overnight. Finally, the membranes were incubated in horseradish peroxidase (HRP)-conjugated secondary antibodies (Sigma) for 1 h at room temperature. The blot signals were visualized using an enhanced chemiluminescence (ECL) kit (Santa Cruz, Dallas, TX, USA) according to the manufacturer’s protocols.
Dual-luciferase reporter assay
For the luciferase assay, U87 cells (5 × 104 cells/well) were seeded in a 24-well plate. Wild-type pGL3-ZEB1-AS1-WT and mutant pGL3-ZEB1-AS1-Mut putative miR-200c/141 seed matching site vectors were purchased from Genechem (Shanghai, China). The cells were transfected with miR-200c or miR-141 using Lipofectamine 2000 (Invitrogen) for 48 h and subsequently co-transfected with ZEB1-AS1-WT or ZEB1-AS1-Mut. The luciferase activities of the cells were measured within 48 h after transfection by the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer’s instructions. The Renilla luciferase activity was used as the internal control.
RNA immunoprecipitation assay
Following the manufacturer’s instructions, the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) was utilized to perform RNA immunoprecipitation (RIP) assay. QRT-PCR assay detected the co-precipitated RNAs using respective primers.
Cell cycle and apoptosis determination
The cell cycle and the apoptosis were detected using flow cytometry as previously described [28]. The U87 cells (1 × 105 cells/well) seeded into 6-well plates were transfected with RNA oligonucleotides. Briefly, the cells were harvested and fixed with 70% ice-cold ethanol. After treatment with 1 mg/mL of RNase at 4°C overnight, the cells were centrifuged and stained with 10 μL of propidium iodide (PI, 50 μg/mL; Sigma) at 37°C for 30 min in the dark. The cells in different phases were analyzed using a BD FACSCaliburflow cytometer (BD Technologies, Carlsbad, CA, USA), and the distribution of cells in each phase was calculated by the ModFit software (Verity Software House Inc., Topsham, ME, USA). For apoptosis assays, the cells were harvested, centrifuged, and washed with PBS. Then, the cells were re-suspended in 100 μL of binding buffer and stained with PI staining (BD Bioscience, San Diego, CA, USA) in the dark for 30 min. Images of cell apoptosis were obtained from the BD FACSCalibur flow cytometer (BD Technologies). The percentage of apoptotic cells was calculated by the CellQuest software (BD Bioscience). Each of the detected samples was prepared three times.
Cell viability assay
Cell viability was determined by Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan). In brief, after the indicated oligonucleotides transfection for 48 h, the cells (1 × 103 cells/well) were seeded into 96-well plates (n = 5 for each time point), supplemented with 100 μL of DMEM, and added with 10% FBS. At 1, 2, 3, or 4 days, 10 μL of CCK-8 solution was added to each well and cultured for 2 h. Optical density values (OD = 450 nm) were determined by a microplate reader (Molecular Devices, Sunnyvale, CA, USA), and the cell viability was calculated.
Colony formation assay
Cells were harvested 24 h after transfection. The transfected cells (1 × 103 cells/well) were seeded in 30 mm dishes and cultured for approximately 2 weeks. After 4% of paraformaldehyde fixation, the colonies in each dish were then stained with 0.1% crystal violet (Beyotime Institute of Biotechnology, Shanghai, China). The plates were washed with phosphate-buffered saline (PBS) three times before air-drying. Finally, images of the stained colonies were captured with a high-resolution camera, and the colonies were counted to calculate the colony formation rate.
Migration and invasion assays
The cell motility was determined by transwell assays. The migration ability of cells was evaluated by migration assay. U87 cells (2 × 104 cells/well in 150 μL of serum-free medium) with different treatments were seeded into the upper chamber of the transwell inserts (8-μm pore size; Corning Costar, NY, USA), and 600 μL of complete medium was added to the lower chamber. For invasion assay, the upper side of the polycarbonate membrane was pre-coated by the diluted Matrigel (BD Biosciences) for 1 h at 37°C. After being cultured for 24 h, the upper chamber cells of the transwell inserts were removed, and the bottom chamber cells were fixed with 4% paraformaldehyde and subsequently stained with 0.1% crystal violet. The cells on the upper side were then removed with cotton swabs. The migrated and invaded cells were counted and photographed under a fluorescence microscope (Olympus, Tokyo, Japan), and the average was calculated.
Animal experiments
All animal experiments were approved by the Animal Research Committee of Xinxiang Medical University and were carried out in accordance with the International Guiding Principles for Animal Research. U87 cells (5 × 106) were stably transfected with siZEB1-AS1, and the controls were subcutaneously injected into the right hind flank of 6-week-old female mice suffering from severe combined immune deficiency (SCID; Institute of Zoology, Chinese Academy of Sciences, Beijing, China) to perform tumor growth assay. Each group had 6 mice. After 0, 4, 8, 12, 16, 20, 24, and 28 days, the tumor volumes (tumor volume = width2 × length/2) were monitored and calculated. All mice were sacrificed 28 days post-inoculation, and the tumors in the right hind flank of the mice were removed and photographed. The tumor weights were quantified.
Moreover, 12 additional 6-week-old SCID female mice were injected with U87 cells (1 × 107) and were stably transfected with siZEB1-AS1 or control through the tail vein for in vivo metastasis assay. The general health status of the mice was monitored, and the incidence associated with primary tumor or metastasis was calculated. All mice were sacrificed 28 days post-injection and anatomized. The metastasis in the lung of each of mouse was examined and calculated by lung section. The sections of lungs with visible tumor colonies were fixed and embedded in paraffin and stained with hematoxylin/eosin (Maixin Biotech). Three non-sequential sections per mice were obtained. The presence of metastasis was observed and photographed by microscopy. The total number of metastases in every lung section was obtained and averaged. Six mice were used for each group.
Identification the correlation by bioinformatics analysis
We used GEPIA (http://gepia.cancer-pku.cn/index.html) to test the ZEB1-AS1 expression level and outcome in glioma cancer [35]. GEPIA is a newly developed interactive web application for gene expression analysis based on 9736 tumors and 8587 normal samples from the TCGA and GTEx databases. It provides customizable functions such as tumor and normal differential expression analysis, survival analysis and correlation based on the gene expression levels. We could demonstrate the expression of ZEB1-AS1 in glioma cancer tissues and normal tissues, correlation between ZEB1 and ZEB1-AS1 in glioma cancer, and the overall survival of patients. The boxplot was performed to visualize the relationship.
Statistical analysis
All data were expressed as mean ± SD. The results of Student’s t-test or one-way analysis of variance (ANOVA) were analyzed by the Graph Prism 5.0 software (GraphPad Prism, San Diego, CA) with P < 0.05 is statistically significant. Chi-squared tests were used to evaluate the frequencies. The five-year survival curves were plotted with the Kaplan-Meier method and analyzed by the log-rank test. All assays were performed independently three times.
Results
LncRNA ZEB1-AS1 was upregulated in glioma cancer
The ZEB1-AS1 level in glioma cancer tissues from 100 patients and 16 normal brain tissues was determined using qPCR assay. Results confirmed that ZEB1-AS1 expression was significantly higher in glioma cancer tissues (n = 100) than in normal brain tissues (n = 16) (Figure 1A). Furthermore, the level of ZEB1-AS1 was much higher in patients with advanced histological grades (III/IV) (Figure 1B; Table 1). ZEB1-AS1 expression was also associated with tumor size but exhibited no correlation with age and gender (Table 1). Meanwhile, the patients with low ZEB1-AS1 levels had higher five-year survival rates than those with high expressions of ZEB1-AS1 (Figure 1C). Additionally, ZEB1-AS1 expression in human glioma cancer cell lines (U87, U251, LN18, U118, and T98G) and the normal human astrocyte (NHA) cell line was detected by qRT-PCR assay. We showed that the ZEB1-AS1 expression was higher in glioma cancer cell lines than in NHA cells (Figure 1D).
Figure 1.
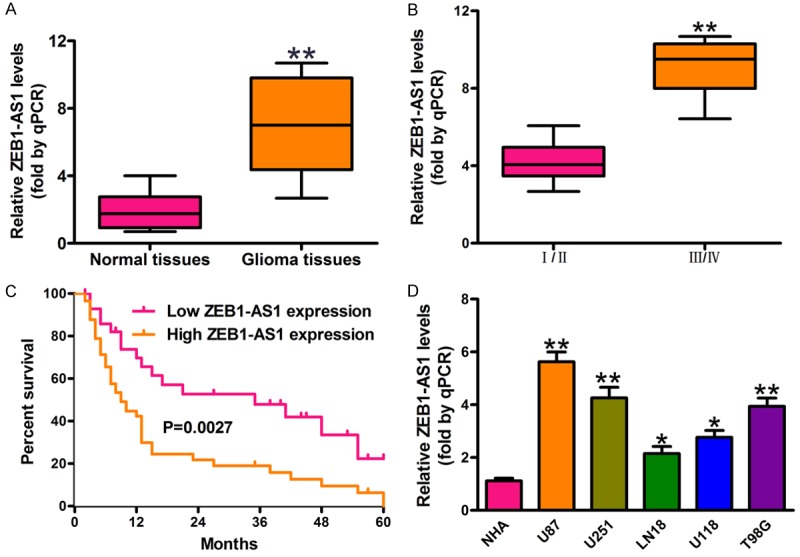
Expression levels of ZEB1-AS1 in glioma cancer tissues and cell lines and its clinical significance. A. Relative expression of ZEB1-AS1 in glioma samples (n = 100) and normal brain tissues (n = 16) was measured by qRT-PCR and normalized to GAPDH. **P < 0.01, Glioma samples versus Normal tissues. B. Comparisons of the levels of ZEB1-AS1 in glioma cancer patients with different tumor stages (I/II, n = 47; III/IV, n = 53). **P < 0.01, III/IV stages versus I/II stages. C. The five-year survival rate of the patients with high (n = 59) and low (n = 41) levels of ZEB1-AS1 was plotted by Kaplan-Meier method (P = 0.0027). D. The expression of ZEB1-AS1 in five glioma cancer cell lines (U87, U251, LN18, U118, and T98G) and in normal human astrocyte (NHA) cell line. *P < 0.05, **P < 0.01, glioma cell lines versus NHA cells. All values are represented as mean ± SD of three replicates.
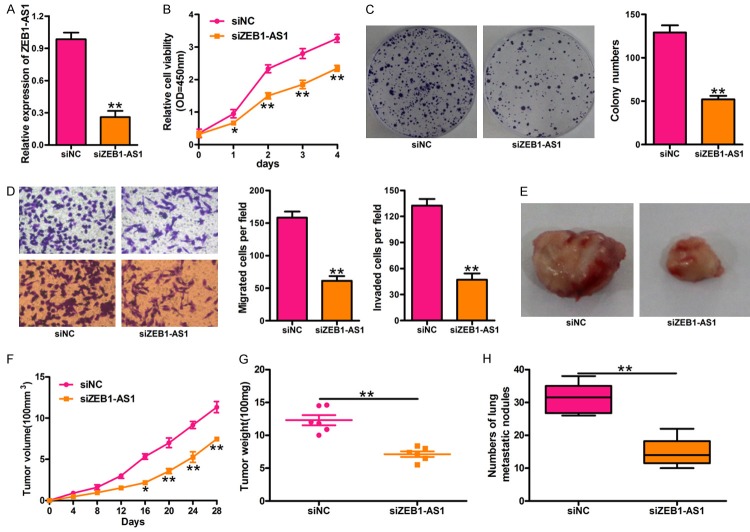
Silencing ZEB1-AS1 expression inhibited glioma cancer progression in vitro and in vivo
To understand the functions of ZEB1-AS1 in glioma cancer, U87 cells were transfected with siZEB1-AS1. qRT-PCR was performed to check the effects of siZEB1-AS1 in U87 cells. Our results indicated that the ZEB1-AS1 expression sharply decreased in the U87 cells transfected with siZEB1-AS1 compared with the control (Figure 2A). CCK-8 assays showed that ZEB1-AS1 deletion significantly suppressed the proliferation of U87 (Figure 2B). The colony formation assay results indicated that silencing ZEB1-AS1 obviously inhibited the glioma cancer cell proliferation (Figure 2C). Moreover, ZEB1-AS1 deletion significantly inhibited the motility of U87 cells. Representative migration and invasion images are shown in Figure 2D. We also explored the effect of ZEB1-AS1 on glioma cancer tumorigenesis in vivo. SCID mice were injected subcutaneously with U87 cells stably transfected with siZEB1-AS1 or the control, and the mice were sacrificed and anatomized at 28 days (Figure 2E). The volume of tumors in the siZEB1-AS1-U87 group was smaller than those in the control group (Figure 2F). The tumor weight of the siZEB1-AS1-U87 group followed the same pattern and was smaller than that of the control group (Figure 2G). The numbers of metastatic nodules were significantly fewer in the siZEB1-AS1-U87 group than in the control group (Figure 2H).
Figure 2.
Silencing ZEB1-AS1 expression suppresses glioma cancer cell proliferation in vitro and tumor growth in vivo. A. The inhibitory efficiency of siZEB1-AS1 transfection on the expression of ZEB1-AS1 was measured by qRT-PCR assay. B. Silencing ZEB1-AS1 by siZEB1-AS1 significantly inhibited proliferation of U87 cells at 2 d, 3 d, and 4 d. Cell proliferation was detected by CCK-8 assay. C. Cell proliferation was detected by colony formation assay in U87 cells transfected with siZEB1-AS1 or siNC. D. Cell migration and invasion was determined by transwell assays. Silencing ZEB1-AS1 by siZEB1-AS1 significantly inhibited the migratory (left, upper panel) and invasive (left, lower panel) ability of U87 cells after transfected with siZEB1-AS1. E. U87 cells were transfected with siZEB1-AS1 or siNC and tumors were collected from SCID mice subcutaneously injected into the right hind flank with the treated cells. Representative images of tumors 4 weeks after subcutaneous xenografting were showed. F. The tumor volume was analyzed once every four days. G. The quantification of tumor weights (n = 6). H. The quantification of pulmonary nodules in each group within 8 weeks after tail vein injection. siNC: siRNA negative control; siZEB1-AS1: ZEB1-AS1 stably deleted cells. *P < 0.05, **P < 0.01, siZEB1-AS1 versus siNC group. All values are expressed as mean ± SD of three replicates.
ZEB1-AS1 negatively regulated miR-200c/141 in glioma cancer cells
To explore the interaction between ZEB1-AS1 and miRNAs, we predicted potential miRNAs binding sites on ZEB1-AS1 using the TargetScan algorithm. Interestingly, we found two binding sites for miR-200s, which are well known for their tumor-suppressing roles and direct targeting and inhibition of ZEB1 [28] (Figure 3A). To investigate the interaction between ZEB1-AS1 and miR-200c/141, we constructed a luciferase reporter containing ZEB1-AS1 or the miR-200c/141 binding sites on mutated ZEB1-AS1. Luciferase reporter assays showed that the ectopic expression of miR-200c or miR-141 significantly inhibited the luciferase activities of wild type ZEB1-AS1, but not the mutant (Figure 3B). RIP assay was performed to analyze the interaction between ZEB1-AS1 and miR-200c/141. Endogenous miR-200c/141 was highly enhanced by ZEB1-AS1 compared with the control, illustrating that ZEB1-AS1 might function as a miR-200c/141 sponge in glioma cancer cells (Figure 3C). To elucidate the molecular mechanisms of ZEB1-AS1 in glioma cancer, U87 cells were transfected with the ZEB1-AS1 or control vector. qRT-PCR results indicated that the expressions of miR-200c and miR-141 (miR-200c/141) were significantly lower in the U87 cells transfected with the ZEB1-AS1 overexpression vector than those in the control (Figure 3D). In contrast, miR-200c/141 was significantly higher in the U87 cells transfected with siZEB1-AS1 than that in the control (Figure 3E). Additionally, the ectopic expression of miR-200c or miR-141 or the mixtures of miR-200c and miR-141 (miR-200s) significantly inhibited ZEB1-AS1 expression in U87 cells (Figure 3F), implying that ZEB1-AS1 is a target of miR-200c/141.
Figure 3.
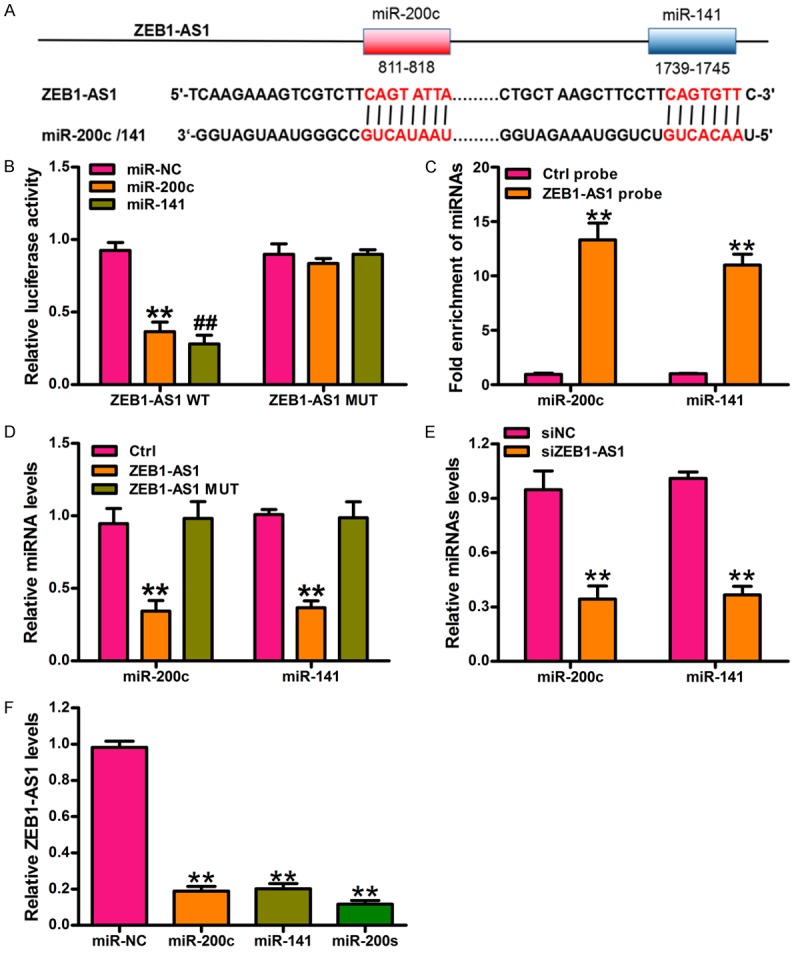
ZEB1-AS1 negatively regulates miR-200s in glioma cancer cells. A. The potential binding sites of miR-200c/141 and ZEB1-AS1 is predicted by using the TargetScan algorithm. B. Luciferase activity in U87 cells cotransfected with miRNA mimics or the miRNA negative control and luciferase reporter containing ZEB1-AS1 wild type (ZEB1-AS1-WT) or miR-200c/141 binding sites mutated ZEB1-AS1 (ZEB1-AS1-Mut). Results are shown as the relative ratio of firefly luciferase activity to renilla luciferase activity. **P < 0.01, miR-200c versus miR-NC group, ##P < 0.01, miR-141 versus miR-NC group. C. miR-200c and miR-141 were significantly enhanced by RNA immunoprecipitation (RIP) assay in the ZEB1-AS1 group compared with control. **P < 0.01, ZEB1-AS1 probe versus Ctrl probe. D. The expression level of miR-200c and miR-141 was measured by qRT-PCR in U87 cells transfected with ZEB1-AS1-WT or ZEB1-AS1-Mut or controls. **P < 0.01, ZEB1-AS1-WT versus Ctrl. E. The expression level of miR-200c and miR-141 was measured by qRT-PCR in U87 cells transfected with siZEB1-AS1 or siNC. **P < 0.01, siZEB1-AS1 versus siNC. F. ZEB1-AS1 expression was measured by qRT-PCR in U87 cells transfected with miR-200c or miR-141 or miR-200s. **P < 0.01, miRNA mimics versus miR-NC group. Ctrl: control; siNC: siRNA negative control; miR-200s: the mixtures of miR-200c and miR-141. All values are expressed as mean ± SD of three replicates.
ZEB1-AS1 promoted glioma cancer-cell growth and motility by miR-200c/141 inhibition
As ZEB1-AS1 is molecularly associated with and inhibits miR-200c/141 expression, we next explored whether miR-200c/141 mediates the facilitative effects of ZEB1-AS1 on glioma cell proliferation and migration. U87 cells were transfected with siZEB1-AS1 and miR-200s inhibitor, siZEB1-AS1 and miR-NC inhibitor, or siNC and miR-NC inhibitor. The ZEB1-AS1 expression was significantly down-regulated by siZEB1-AS1 and up-regulated by the co-transfection with miR-200s inhibitor compared with the control (Figure 4A). CCK-8 assay results indicated that siZEB1-AS1 inhibited the cell growth of U87, and that this effect can be reversed by a miR-200s inhibitor (Figure 4B). Colony formation assay data further revealed that ZEB1-AS1 silencing suppressed the glioma cancer-cell growth through the up-regulation of miR-200s (Figure 4C). The transwell assay results also indicated that ZEB1-AS1 promoted glioma cancer cell migration and invasion by the inhibition of miR-200s. Representative migration and invasion images and the quantification of migrated or invaded cells are presented in Figure 4D and 4E. These results indicated that ZEB1-AS1 promoted glioma cancer-cell growth and motility by miR-200s inhibition.
Figure 4.
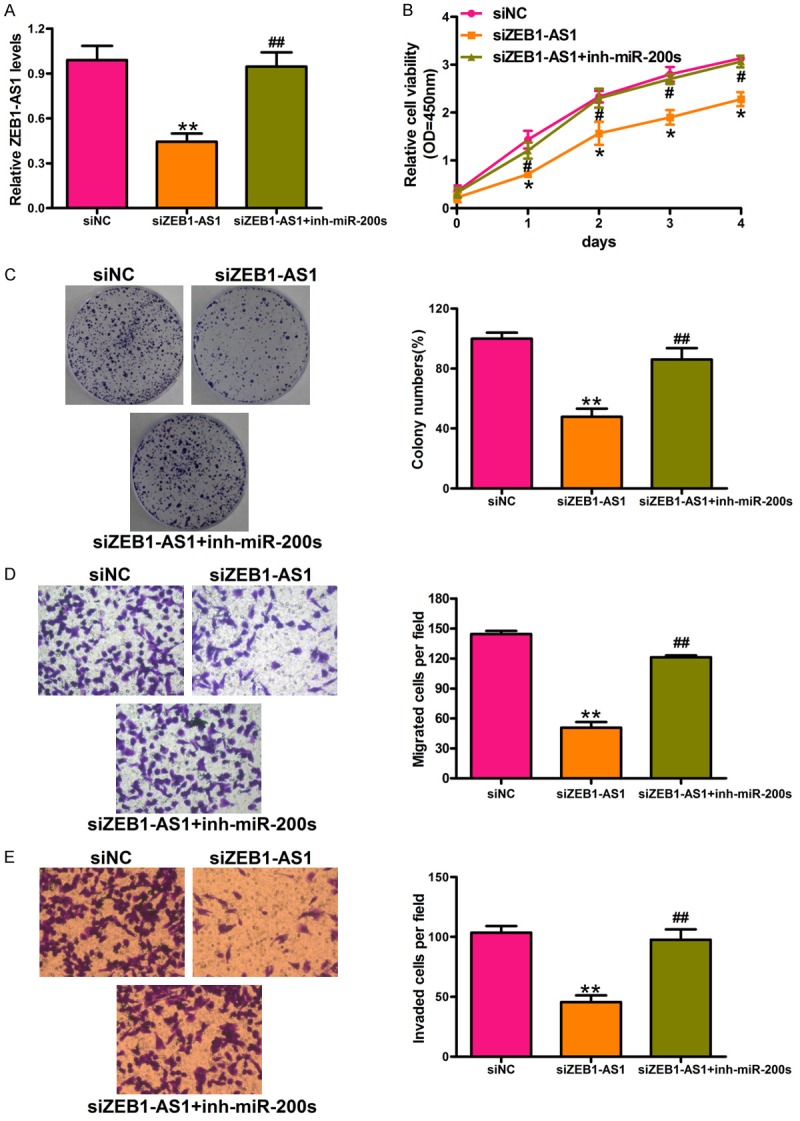
ZEB1-AS1 promotes glioma cancer cell viability and motility through inhibition of miRNA-200s. A. qRT-PCR assay was used to detect the expression level of ZEB1-AS1 in U87 cells transfected with siNC or siZEB1-AS1 or co-transfected with siZEB1-AS1 and miR-200s inhibitors. B and C. Cell proliferation was detected by CCK-8 assay and colony formation assay in treated U87 cells. D. The inhibitory effect of ZEB1-AS1 on glioma cancer cell migration through miRNA-200s by transwell assays. Representative images and quantification of migrated cells were presented. E. The inhibition effect of invasive ability of U87 cells caused by ZEB1-AS1 silencing was reversed by miRNA-200s inhibitor. Representative images and quantification of invaded cells were presented. Total magnification of all images is 200 ×. *P < 0.05, **P < 0.01, siZEB1-AS1 versus siNC; #P < 0.05, ##P < 0.01, siZEB1-AS1 + inh-miR-200s versus siZEB1-AS1. siNC: siRNA negative control; siZEB1-AS1: ZEB1-AS1 stably deleted cell; miR-200s: the mixtures of miR-200c and miR-141; siZEB1-AS1 + inh-miR-200s: ZEB1-AS1 stably deleted cells transfected with miR-200s. All values are expressed as mean ± SD of three replicates.
ZEB1-AS1 promoted the processes of glioma cancer cell tumorigenesis through the miR-200c/141-ZEB1 axis
Previous studies have reported that miR-200s act as a tumor inhibitor by targeting ZEB1 directly in glioma cancers [28]. In this study, we demonstrated the relationships among ZEB1-AS1, miR-200s, and ZEB1. First, U87 cells were transfected with siNC, siZEB1-AS1 + vector, siZEB1-AS1 + ZEB1, miR-200s + vector, and miR-200s + ZEB1. The ZEB1-AS1 and ZEB1 mRNA levels were downregulated by siZEB1-AS1 and miR-200s in U87 cells and restored by ZEB1 overexpression (Figure 5A and 5B). Subsequently, we detected the effects of the miR-200c/141-ZEB1 axis and ZEB1-AS1 on the development and progression of glioma cancer. CCK-8 assay showed that silencing ZEB1-AS1 or introducing miR-200s inhibited the growth of U87 cells, and that the inhibition effect induced by siZEB1-AS1 or miR-200s can be restored by ZEB1 overexpression (Figure 5C and 5D). The transwell assay results showed that ZEB1-AS1 accelerated the migration and invasion of U87 cells by decreasing miR-200s or activating ZEB1 (Figure 5E). Figure 5F and 5G show the quantification of migrated and invaded cells. Therefore, ZEB1-AS1 inhibited the cell proliferation, migration, and invasion of U87 cells by inhibiting miR-200s and ZEB1 activation. Finally, the cell cycle arrest and apoptosis induced by ZEB1-AS1 silencing in U87 cells was verified by flow cytometry. The growth-suppressing effect of ZEB1-AS1 silencing and miR-200s introduction on glioma cells was reduced by inhibiting the G1/S-phase transition arrest (Figure 6A and 6B). Furthermore, the transfection of U87 cells with siZEB1-AS1 or miR-200s increased cell apoptosis (Figure 6C and 6D). As shown in Figure 6, ZEB1 overexpression also neutralized the siZEB1-AS1- or miR-200s-induced cell cycle arrest and apoptosis. Therefore, ZEB1-AS1 inhibited the G1 arrest and apoptosis in U87 cells through the inhibition of miR-200s and the activation of ZEB1.
Figure 5.
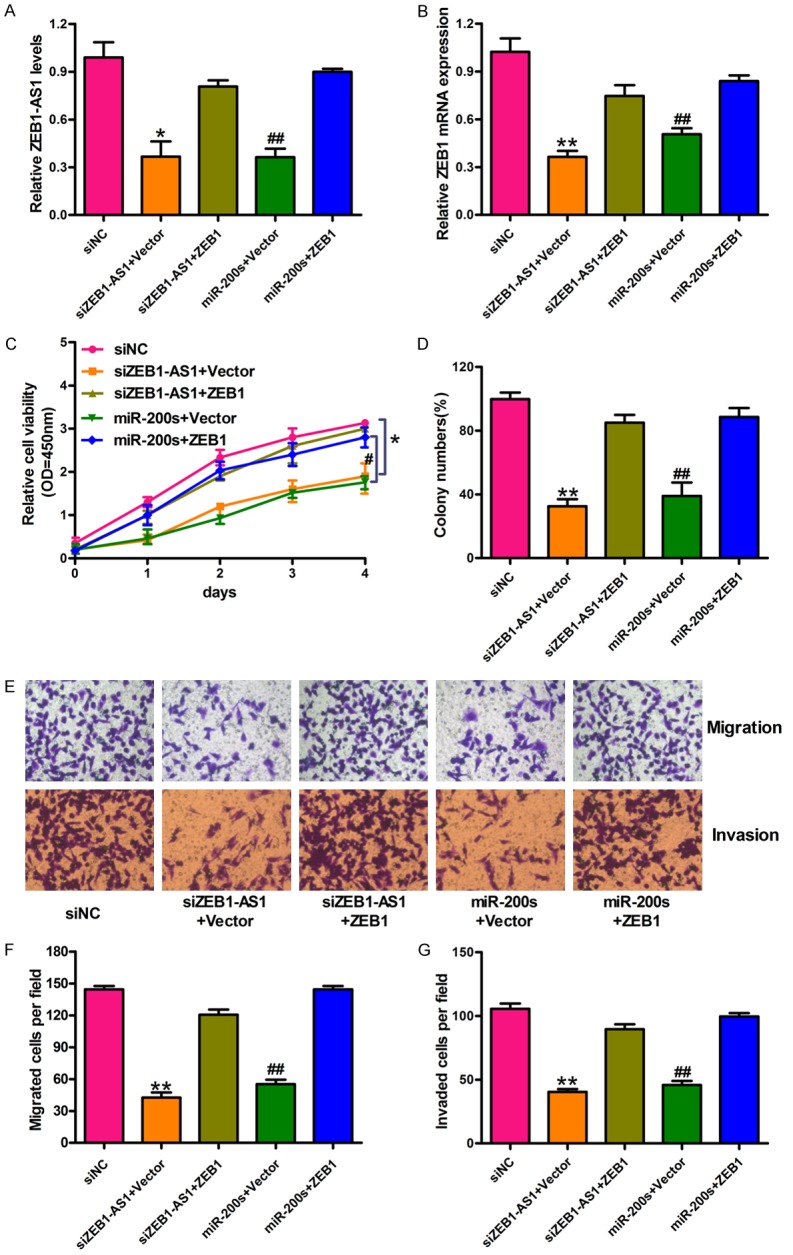
ZEB1-AS1 promotes glioma cancer cells proliferation, migration and invasion through inhibition of miRNA-200c/141 and activation of ZEB1. U87 cells were transfected with siNC, siZEB1-AS1 and vector, siZEB1-AS1 and ZEB1, miR-200s and vector, and miR-200s and ZEB1, respectively. qRT-PCR assay was performed to detect the expression of ZEB1-AS1 (A) and ZEB1 mRNA (B) in treated U87 cells. (C and D) Cell proliferation was detected by CCK-8 assay and colony formation assay in treated U87 cells. (E) Representative images of transwell assays in treated U87 cells. Total magnification of all images is 200 ×. (F and G) Quantification of migrated and invaded cells were presented. *P < 0.05, **P < 0.01, siZEB1-AS1 + ZEB1 versus siZEB1-AS1 + Vector; #P < 0.05, ##P < 0.01, miR-200s + ZEB1 versus miR-200s + Vector. siNC: siRNA negative control; siZEB1-AS1 + Vector: ZEB1-AS1 stably deleted cells transfected with vector; siZEB1-AS1 + ZEB1: ZEB1-AS1 stably deleted cells transfected with ZEB1; miR-200s + Vector: U87 cells transfected with the mixtures of miR-200c and miR-141; miR-200s + ZEB1: U87 cells transfected with miR-200s and ZEB1. Data are shown as mean ± SD based on at least three independent experiments.
Figure 6.
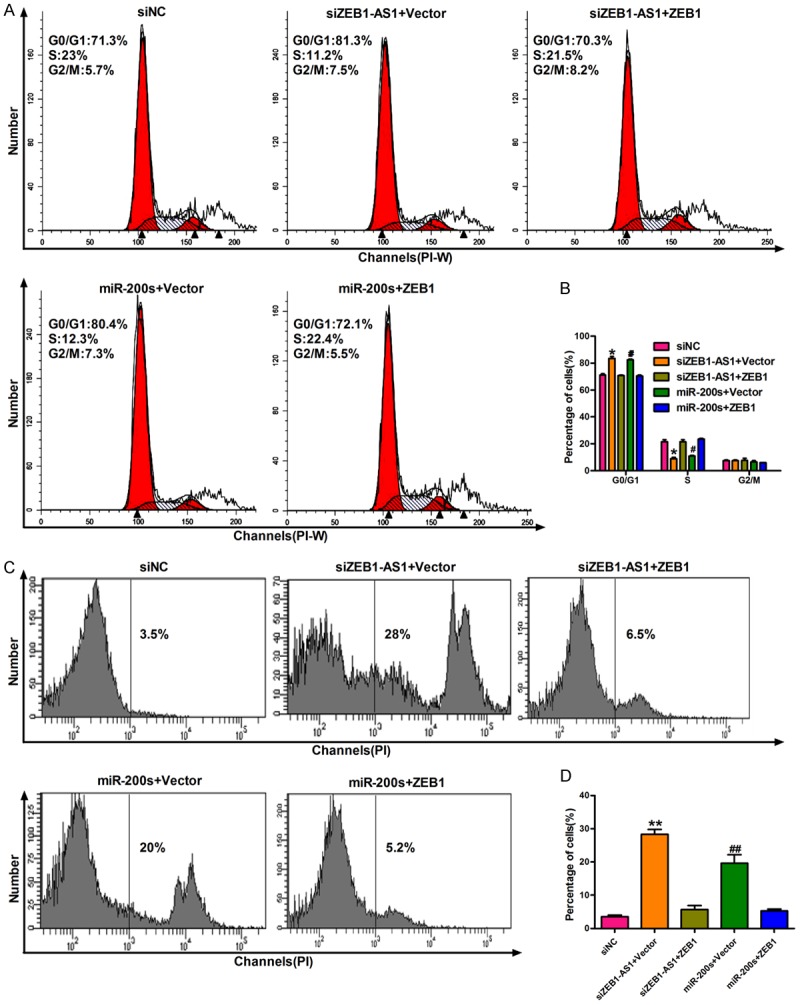
ZEB1-AS1 inhibits cell cycle arrest and apoptosis in glioma cancer cells through inhibition of miR-200c/141 and activation of ZEB1. A. Flow cytometry was used to detect cell cycle distribution in treated U87 cells. B. The percentage of treated U87 cells in G0/G1, S and G2/M phase were quantified, respectively. C. Apoptosis was detected by PI staining in treated U87 cells. D. The percentage of apoptosis cells in each group was quantified. *P < 0.05, **P < 0.01, siZEB1-AS1 + ZEB1 versus siZEB1-AS1 + Vector; #P < 0.05, ##P < 0.01, miR-200s + ZEB1 versus miR-200s + Vector. siNC: siRNA negative control; siZEB1-AS1 + Vector: ZEB1-AS1 stably deleted cells transfected with vector; siZEB1-AS1 + ZEB1: ZEB1-AS1 stably deleted cells transfected with ZEB1; miR-200s + Vector: U87 cells transfected with the mixtures of miR-200c and miR-141; miR-200s + ZEB1: U87 cells transfected with miR-200s and ZEB1.
Possible signaling pathways of ZEBA-AS1/miR-200c/141-ZEB1 involved in glioma cancer progression
As ZEB1 is an important transcription factor in regulating epithelial to mesenchymal transition (EMT), we speculated that ZEB1-AS1 might promote glioma cancer progression by regulating the ZEB1-EMT axis. We performed Western blot assays to detect related molecules protein expressions, such as E-cadherin, Vimentin, matrix metalloproteinase 2 (MMP2), Snail, and ZEB1. The protein expressions of Vimentin, Snail, and ZEB1 in U87 cells transfected with siZEB1-AS1 and miR-200s were expressed less than those in cells transfected with siNC. The inhibition effect can be restored by ZEB1 overexpression. In contrast, E-cadherin was significantly elevated in the same treatment of U87 (Figure 7A). Moreover, the protein levels of the key regulators of G1/S transition (Cyclin D1, Rb, and CDK2) and the apoptosis-relative proteins (Bax and Bcl-2) were also detected. The Cyclin D1, CDK2, and Bcl-2 protein expressions were significantly downregulated by siZEB1-AS1 and miR-200s in U87 cells and were restored by ZEB1 overexpression, whereas the expressions of Rb and Bax were elevated significantly. The original whole film of western blotting in Figure 7A was presented in Supplementary Figure 1. Figure 7B shows a potential regulation model of the ZEB1-AS1/miR-200c/141-ZEB1 axis in glioma cancer development. Therefore, ZEB1-AS1 promotes the translation of ZEB1 through the inhibition of the miR-200c/141 expression and promotes glioma cancer-cell growth, migration, and invasion. Conversely, miR-200c/141 down-regulates the ZEB1 expression by directly binding to its 3’-UTR in the absence of ZEB1-AS1 and suppresses glioma cancer cell proliferation, cell cycle, motility, and apoptosisresistance.
Figure 7.
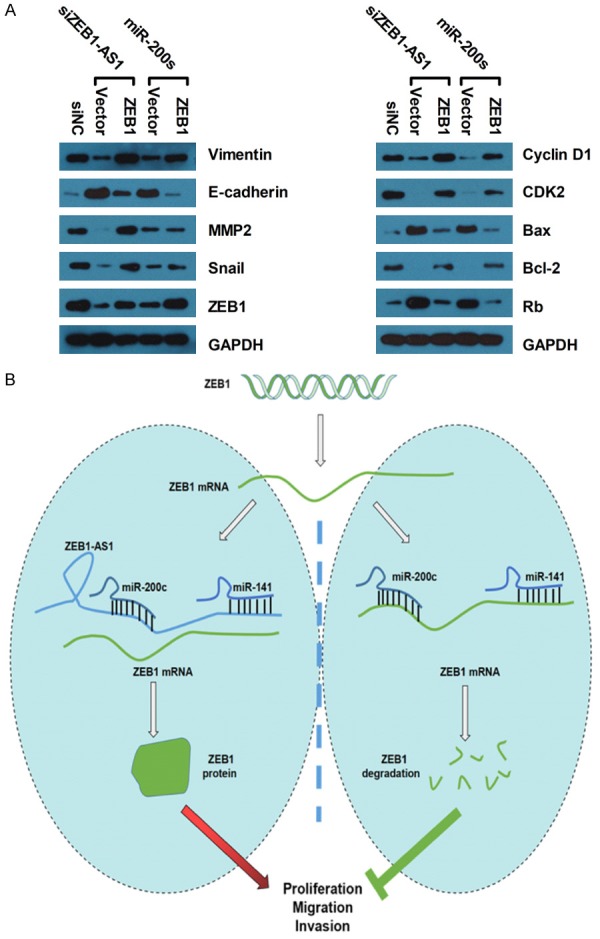
Possible signaling pathways of ZEBA-AS1/miR-200c/141-ZEB1 involved in glioma cancer progression. (A) Representative western blot results of EMT (E-cadherin, Vimentin, MMP2, Snail and ZEB1), cell cycle (Cyclin D1, Rb and CDK2) and apoptosis (Bax and Bcl-2) markers. GAPDH was used as the normal control. The original whole film of western blotting in (A) was presented in Supplementary Figure 1. (B) A diagram exhibiting the role and regulatory mechanisms of the ZEB1-AS1/miR-200c/141-ZEB1 axis in glioma cancer. In the ZEB1-AS1 depletion cells, upregulated miR-200c/141 inhibits glioma cancer cell proliferation, cell cycle, motility, and apoptosis-resistance by targeting ZEB1. On the contrary, ZEB1-AS1 overexpression suppresses miR-200c/141 expression, which then activates ZEB1 and promotes glioma cancer cell proliferation, cell cycle, motility, and apoptosis-resistance.
Identification of a positive correlation between the low expression of ZEB1-AS1 and poor prognosis of GBM through bioinformatics analysis
We first used GEPIA to detect the expressions of ZEB1 and ZEB1-AS1 between GBM and healthy people, and Figure 8A and 8B indicated that compared to healthy people, the expression levels of ZEB1 and ZEB1-AS1 were significantly increased in cancer patients. We then performed a pairwise gene correlation analysis between ZEB1 and ZEB1-AS1. As shown in Figure 8C, ZEB1 was positively correlated with ZEB1-AS1 (P = 0, R = 0.62). Finally, we performed survival analysis based on the gene expression levels. Figure 8D and 8E showed that the patients with low levels of ZEB1-AS1 had higher overall survival rates than those with high expressions of ZEB1-AS1, whereas the overall survival analysis showed no significantly difference between patients with high and low expressions of ZEB1.
Figure 8.
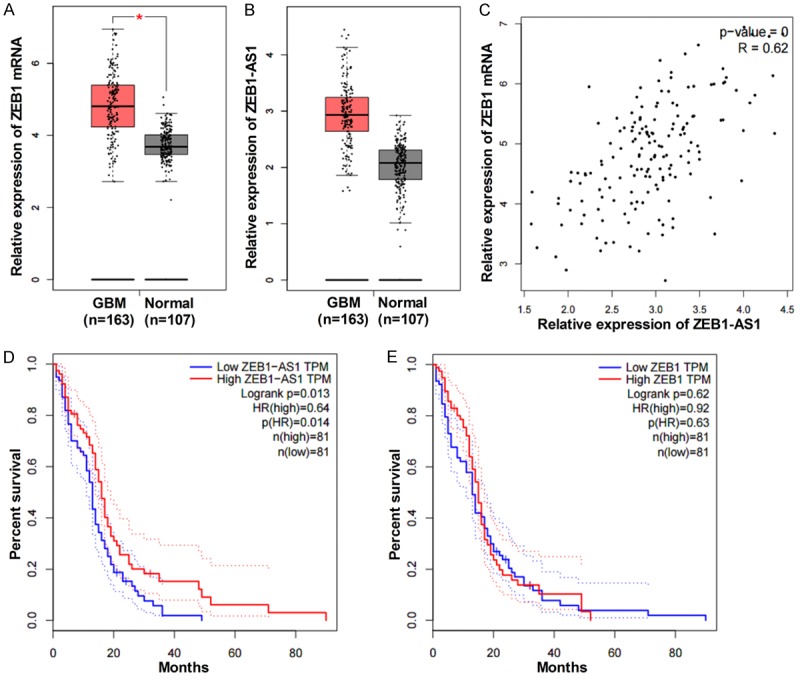
Identification of the correlation between ZEB1-AS1 level and prognosis of GBM through bioinformatics analysis. A and B. The expression levels of ZEB1 and ZEB1-AS1 in GBM patients (n = 163) and normal tissues (n = 107) using the GEPIA. C. The correlation between ZEB1 and ZEB1-AS1 has been showed based on the gene expression levels (P = 0, R = 0.62). D. The overall survival analysis with high (n = 81) and low (n = 81) levels of ZEB1-AS1 (P = 0.013). E. The overall survival analysis with high (n = 81) and low (n = 81) levels of ZEB1 (P = 0.63).
Discussion
In this study, we demonstrated that ZEB1-AS1 promotes the tumorigenesis of glioma cancer cells through modulating the miR-200c/141-ZEB1 axis. First, ZEB1-AS1 was up-regulated in glioma cancer tissues and cells. The high expression of ZEB1-AS1 was positively associated with tumor size and advanced tumor stages, but inversely correlated with the prognosis for glioma cancer patients. Second, ZEB1-AS1 silencing inhibited the growth and motility of glioma cancer cells in vitro and of tumorigenesis in vivo. Finally, we confirmed that ZEB1-AS1 promotes glioma cancer-cell growth and motility and inhibits cell cycle arrest and apoptosis through the inhibition of miR-200c/141 and the activation of ZEB1. Collectively, ZEB1-AS1 is a positive regulator in the progression of glioma cancer. Furthermore, targeting the ZEB1-AS1/miR-200c/141-ZEB1 axis may be an effective strategy for glioma cancer therapy.
Glioma is one of the most frequent malignancies with high morbidity and mortality globally [1,2]. Thus, identifying new molecular abnormalities concerning glioma progression is necessary to establish a specific target for individual therapeutic strategies of glioma treatment. Several reviews have focused on the functional roles of lncRNAs in various human cancers [9,10,12-15]. ZEB1-AS1 has been found to play clinically significant roles in several cancers [16-20,22,23]. Li et al. demonstrated that increased ZEB1-AS1 is markedly correlated with microvascular invasion, short overall survival, and high recurrence rates. It acts as an independent prognostic marker in HCC [17]. ZEB1-AS1 overexpression is a poor prognostic biomarker of ESCC [18]. The high expression of ZEB1-AS1 indicates poor prognoses of prostatic and colorectal cancers [19,20]. Furthermore, the amplification of ZEB1-AS1 is involved in the poor prognosis of gastric cancers [21]. Additionally, ZEB1-AS1 up-regulation is correlated with the poor overall survival of glioma and osteosarcoma cancer patients [22,23]. Consistently, in the present study, ZEB1-AS1 expression was markedly elevated in glioma cancer tissues. The up-regulation of ZEB1-AS1 was positively associated with tumor size and advanced histological grades, but negatively correlated with the outcomes of glioma cancer patients. This suggests the clinical significance of ZEB1-AS1 in glioma cancer.
Numerous studies have reported that ZEB1-AS1 contributes to cancer progression. For instance, ZEB1-AS1 was identified as an oncogenic regulator in diverse malignancies [16]. Increased ZEB1-AS1 promotes HCC cell growth and motility in vitro and facilitates tumor growth and metastasis in vivo [17]. Reportedly, ZEB1-AS1 knockdown inhibits the proliferation and migration of prostate cancer cells [19]. In CRC, the high expression of lncRNA ZEB1-AS1 promotes the colorectal cancer cell proliferation partially by suppressing the p15 expression [20]. Moreover, Lv et al. corroborated that silencing ZEB1-AS1 dramatically suppresses the proliferation, migration, and invasion of glioma cells and promotes cellular apoptosis [22]. In accordance with these results, we found that ZEB1-AS1 expression was much higher in glioma cancer cell lines than in the normal human astrocyte cells (NHA). Silencing ZEB1-AS1 using siRNA technology resulted in the significant decrease in proliferation, migration, invasion, and apoptosis resistance of U87 cells in vitro. Additionally, ZEB1-AS1 depletion dramatically suppressed the tumor growth and metastasis in vivo. These results indicated that ZEB1-AS1 promotes the aggressive behaviors of glioma cancer.
The miR-200 family has been revealed to have critical roles in many cancers [24]. Previous studies revealed that the aberrant expression of miR-200s is associated with the pathogenesis of glioma [28-30]. As important members of the miR-200 family, miR-200c and miR-141 show a potential suppressive effect on glioma cell proliferation, migration, and invasion by inhibiting ZEB1 [28]. Other researchers indicated that miR-141 inhibits glioma cell proliferation and metastasis by targeting TGF-β2 [30]. In line with these results, the introduction of miR-200c/141 inhibited U87 cells proliferation, migration, invasion, and apoptosis resistance. Given the widespread presence of miRNA-binding sites on mammalian mRNAs, competing endogenous RNAs (ceRNA) has been considered as a new mechanism of post-transcriptional regulation through the competitive binding of miRNA [21,23,31-33]. Different RNA species can compete for the same pool of available miRNAs and indirectly regulate each other [31]. Evidence has confirmed the different roles of ceRNA crosstalk on essential cellular processes and its imbalance in response to various diseases [33]. The downregulation of miR-335-5p by ZEB1-AS1 in gastric cancer promotes tumor proliferation and invasion [21]. MiR-200s overexpression partially abolishes the inhibitory effects of ZEB1-AS1 on osteosarcoma cell proliferation and metastasis [23]. MiR-141 regulates osteoblastic cell proliferation by modulating the target gene of lncRNA H19 and lncRNA H19-derived miR-675 [32]. In our follow-up study on ZEB1-AS1, the inhibition of miR-200c/141 restored the inhibition effects on cell proliferation, migration, invasion, and apoptosis resistance induced by ZEB1-AS1 silencing. We also found a negative reciprocal regulation between ZEB1-AS1 and miR-200c/141, implying that ZEB1-AS1 can bind to the miR-200c/141 site directly. The inhibitory effect of miR-200c/141 on glioma cancer cell carcinogenesis can be reversed by ZEB1-AS1.
ZEB1 is a transcriptional factor that plays a crucial role in tumorigenesis and cellular processes [36]. As a famous epithelialmesenchymal transition (EMT) promoter, ZEB1 not only takes part in biological tumor behaviors, such as EMT, cell apoptosis-resistance, chemo- and radio-resistance, and cell cycle, but also stem-cell property induction [37,38]. Accumulating evidence suggests that ZEB1 is a direct target of miR-200c/141 in glioma cancer [28]. Similarly, in this study, ZEB1 was directly targeted by tumor suppressor miR-200c/141 in glioma cancer. Moreover, ZEB1 overexpression also neutralized the inhibitory effects on cell proliferation and invasion and the induction effects on cell cycle arrest and apoptosis caused by siZEB1-AS1 or miR-200s in glioma cancer cells [22]. These results support the inter-regulation between ZEB1-AS1 and the miR-200c/141-ZEB1 axis. Moreover, the loss of balance between ZEB1-AS1, ZEB1, and miR-200c/141 in glioma cancer may contribute to glioma cancer progression. Additionally, Lv et al. discovered that siZEB1-AS1 increases the expressions of E-cadherin and Bax and decreases the ZEB1, N-cadherin, Integrin-β1, MMP2, MMP9, Bcl-2, cyclin D1, and CDK2 levels in glioma cancer cells. We found similar results in ZEB1-AS1 silencing and miR-200c/141 introduction, whereas ZEB1 overexpression can restore these changes. In line with these results, ZEB1-AS1 regulated the proliferation and progression of glioma cells by activating ZEB1.
In summary, we demonstrated that ZEB1-AS1 was a positive regulator in the tumorigenesis of glioma cancer. Clinically, ZEB1-AS1 was significantly associated with tumor size, advanced tumor stages, and poor prognosis for glioma cancer patients. In vitro and in vivo experiments showed that silencing the ZEB1-AS1 expression inhibits glioma cancer progression. ZEB1-AS1 regulated the processes of glioma tumorigenesis through the miR-200c/141-ZEB1 axis. Overall, these findings provided an insight into an effective strategy for glioma cancer therapy by targeting the ZEB1-AS1/miR-200c/141-ZEB1 axis.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro Oncol. 2017;19:v1–v88. doi: 10.1093/neuonc/nox158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sulman EP, Ismaila N, Armstrong TS, Tsien C, Batchelor TT, Cloughesy T, Galanis E, Gilbert M, Gondi V, Lovely M, Mehta M, Mumber MP, Sloan A, Chang SM. Radiation therapy for glioblastoma: american society of clinical oncology clinical practice guideline endorsement of the American society for radiation oncology guideline. J. Clin. Oncol. 2017;35:361–369. doi: 10.1200/JCO.2016.70.7562. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ Jr, Mehta MP. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perry JR, Laperriere N, O’Callaghan CJ, Brandes AA, Menten J, Phillips C, Fay M, Nishikawa R, Cairncross JG, Roa W, Osoba D, Rossiter JP, Sahgal A, Hirte H, Laigle-Donadey F, Franceschi E, Chinot O, Golfinopoulos V, Fariselli L, Wick A, Feuvret L, Back M, Tills M, Winch C, Baumert BG, Wick W, Ding K, Mason WP. Short-course radiation plus temozolomide in elderly patients with glioblastoma. N Engl J Med. 2017;376:1027–1037. doi: 10.1056/NEJMoa1611977. [DOI] [PubMed] [Google Scholar]
- 6.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 10.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 12.Sakurai K, Reon BJ, Anaya J, Dutta A. The lncRNA DRAIC/PCAT29 locus constitutes a tumor-suppressive nexus. Mol Cancer Res. 2015;13:828–838. doi: 10.1158/1541-7786.MCR-15-0016-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik R, Patel L, Prensner JR, Shi Y, Iyer MK, Subramaniyan S, Carley A, Niknafs YS, Sahu A, Han S, Ma T, Liu M, Asangani IA, Jing X, Cao X, Dhanasekaran SM, Robinson DR, Feng FY, Chinnaiyan AM. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res. 2014;12:1081–1087. doi: 10.1158/1541-7786.MCR-14-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Li Z, Leng K, Xu Y, Ji D, Huang L, Cui Y, Jiang X. ZEB1-AS1: a crucial cancer-related long non-coding RNA. Cell Prolif. 2018:51. doi: 10.1111/cpr.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 18.Wang YL, Bai Y, Yao WJ, Guo L, Wang ZM. Expression of long non-coding RNA ZEB1-AS1 in esophageal squamous cell carcinoma and its correlation with tumor progression and patient survival. Int J Clin Exp Pathol. 2015;8:11871–11876. [PMC free article] [PubMed] [Google Scholar]
- 19.Su W, Xu M, Chen X, Chen N, Gong J, Nie L, Li L, Li X, Zhang M, Zhou Q. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. 2017;16:142. doi: 10.1186/s12943-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong H, Wen H, Zhu X, Lian Y, Yang X, Qian Z, Zhu J. High expression of long noncoding RNA ZEB1-AS1 promotes colorectal cancer cell proliferation partially by suppressing p15 expression. Tumour Biol. 2017;39:1010428317705336. doi: 10.1177/1010428317705336. [DOI] [PubMed] [Google Scholar]
- 21.Zhang LL, Zhang LF, Guo XH, Zhang DZ, Yang F, Fan YY. Downregulation of miR-335-5p by long noncoding RNA ZEB1-AS1 in gastric cancer promotes tumor proliferation and invasion. DNA Cell Biol. 2018;37:46–52. doi: 10.1089/dna.2017.3926. [DOI] [PubMed] [Google Scholar]
- 22.Lv QL, Hu L, Chen SH, Sun B, Fu ML, Qin CZ, Qu Q, Wang GH, He CJ, Zhou HH. A long noncoding RNA ZEB1-AS1 promotes tumorigenesis and predicts poor prognosis in glioma. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17091431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Pan C, Cai Y, Wang H. Interplay between long noncoding RNA ZEB1-AS1 and miR-200s regulates osteosarcoma cell proliferation and migration. J Cell Biochem. 2017;118:2250–2260. doi: 10.1002/jcb.25879. [DOI] [PubMed] [Google Scholar]
- 24.Feng X, Wang Z, Fillmore R, Xi Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014;344:166–173. doi: 10.1016/j.canlet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P, Sastre-Garau X, Mechta-Grigoriou F. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–1635. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- 27.Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472–6498. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo E, Wang Z, Wang S. MiR-200c and miR-141 inhibit ZEB1 synergistically and suppress glioma cell growth and migration. Eur Rev Med Pharmacol Sci. 2016;20:3385–3391. [PubMed] [Google Scholar]
- 29.Lee JS, Ahn YH, Won HS, Sun S, Kim YH, Ko YH. Prognostic role of the MicroRNA-200 family in various carcinomas: a systematic review and meta-analysis. Biomed Res Int. 2017;2017:1928021. doi: 10.1155/2017/1928021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng T, Zhang S, Li W, Fu S, Luan Y, Zuo L. MicroRNA-141 inhibits glioma cells growth and metastasis by targeting TGF-beta2. Am J Transl Res. 2016;8:3513–3521. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He P, Zhang Z, Huang G, Wang H, Xu D, Liao W, Kang Y. miR-141 modulates osteoblastic cell proliferation by regulating the target gene of lncRNA H19 and lncRNA H19-derived miR-675. Am J Transl Res. 2016;8:1780–1788. [PMC free article] [PubMed] [Google Scholar]
- 33.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caramel J, Ligier M, Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.