Abstract
Background: Remote ischemic postconditioning (RIPostC) is an effective strategy for preventing key organs from becoming impaired due to an ischemia/reperfusion injury. In the current study, we investigated how remote exosome transfer of microRNAs (miRs) may contribute to the treatment effect of RIPostC on the central nerve system (CNS). Methods: Human umbilical vein endothelial cells (HUVECs) were subjected to hypoxia/reoxygenation (H/R) and their miR expression profiles were investigated using the microarray method. The pathways associated with dysregulated miRs were analyzed by gene ontology (GO) annotation of the target genes and a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The role played by the most significantly down-regulated miR (miR-21-3p) in the protective effect of HUVEC-derived exosomes on H/R-treated neural cells was further investigated. The pathway mediating the effect of miR-21-3p was then explored by focusing on activity of autophagy-related 12 (ATG12) protein. Results: The miR expression profile of HUVECs significantly changed after H/R administration, with 104 miRs becoming upregulated and 249 miRs becoming downregulated. Based on the GO and KEGG analyses, the target genes of 8 selected miRs were involved in multiple biological pathways, including the hippo signaling pathway and longevity regulating pathway. Further studies showed that inhibition of miR-21-3p by HUVEC-derived exosomes or a specific inhibitor could the block apoptotic process in H/R-treated neural cells. Molecular level studies showed that the effect of miR-21-3p inhibition depended on the restored function of ATG12, which resulted in the activation of autophagy and suppression of apoptosis. Conclusion: Taken together, these results suggest that H/R caused significant changes of miR expression in exosomes derived from H/R-treated HUVECs, and the exosomes protect neurons against H/R-induced injuries by suppressing miR-21-3p.
Keywords: ATG12, autophagy, exosome, hypoxia/reoxygenation, microarray, miR-21-3p
Introduction
Cerebral injuries due to ischemia always result in the death of massive numbers of neurons in the central nerve system (CNS). These injuries are responsible for the most common type of acute cerebrovascular disease, and the damaged tissue recovers very little of it original function [1]. Ischemia-induced cerebral injuries rank third among the leading causes of mortality and morbidity worldwide [1]. A host of intrinsic and extrinsic factors are thought to regulate the survival of neurons during ischemia of the cerebrum, and these factors provide targets for new drugs and treatments that alleviate cerebral ischemia [2,3]. However, most of the current interventions involve the transient stimulation of injured CNS tissues by exogenous means, making the beneficial effect of those interventions very tentative. Therefore, it is imperative to develop novel methods that have few side effects for treating cerebral ischemia. In recent years, scientists have proposed a theory that ischemic CNS tissue can be mobilized to develop endogenous protective mechanisms that combat injuries and promote the repair of damaged tissue, and this theory has been validated in other organs such as the heart [4]. Based on this theory, remote ischemic conditioning, that is universally used for treating heart ischemic injuries, has been introduced for use in the treatment of cerebral ischemia injuries [5,6]. In 1986, Murry et al. [7] proposed the definition of “ischemic preconditioning” (IPC) [7], and inferred that IPC can attenuate heart injuries induced by ischemia [7]. However, due to the unpredictability of an ischemia attack occurring in the clinic, the application of IPC has been gradually replaced by “ischemic postconditioning” (IPostC) [8]. To reduce its invasiveness and expand the application of this technique, IPostC was first performed with less important organs, such as limbs, to determine whether it could produce the effects needed for treating vital organs [9]. This topic was investigated in many experiments as well as clinical trials, and numerous outcomes were reported [10].
As remote IPostC (RIPostC) became more extensively used in the clinic, the mechanism responsible for its beneficial effect in cerebral injuries was comprehensively studied [11,12]. Nevertheless, most studies only reported on pathways in the brain that were modulated by RIPostC, while the mechanism by which RIPostC remotely transferred its affects remained unknown. A study by Yamaguchi et al. [4] inferred that repeated RIPostC attenuated left ventricular remodeling via exosome-mediated intracellular transfer of miR-29a [4]. Those results suggested that the effect of RIPostC on cerebral injuries might also rely on the exosome-mediated transfer of microRNAs (miRs).
Exosomes are small-membrane vesicles with a size of 30-120 nm in diameter [13]. They can be generated by several types of cells that contain not only surface membrane proteins but also mRNA and miR molecules [14]. Many cells in the CNS release exosomes in the form of extracellular membrane vesicles, indicating that exosomes participate in the function, development, and pathologies of the nervous system [15]. Yang et al. [16], recently found that the exosome-mediated delivery of miR-124 could promote neurogenesis after ischemia. That finding partially supports our hypothesis that RIPostC might attenuate ischemia-induced cerebral injuries by facilitating the exosome-mediated remote transfer of miRs.
To validate that hypothesis, vascular endothelial cells (VECs) subjected to hypoxia/reoxygenation (H/R) were used as an in vitro model of remote cells receiving RIPostC. A profile of miR expression in the VECs was created by using data gathered from microarray assays. H/R-treated neural cells were incubated with exosomes from VECs and an inhibitor of miR-21-3p to determine the mechanism by which RIPostC exerts its anti-ischemia effect in the CNS.
Materials and methods
Chemicals and agents
Antibodies against ATG12 (#ab155589), LC3-B (#ab48394), Beclin-1 (#ab62557), and Bcl-2 (#ab32124) were purchased from Abcam (Cambridge, MA, USA). The GAPDH antibody (#KC-5G5) was purchased from Kangcheng (Shanghai, China), and goat anti-rabbit antibody (#BA1054) was purchased from Boster Biological Technology Co. Ltd (Wuhan, China). Lipofectamine™ 2000 (#52887) was obtained from Invitrogen (Carlsbad, CA, USA). Trizol was obtained from Takara Shuzo Co., Ltd. (Kyoto, Japan). Reverse transcription kits and real time PCR reagents were purchased from DBI® Bioscience (#DBI-2220, Shanghai, China). Cell lysate (#P0013) was purchased from Beyotime Biotechnology (Shanghai, China). Protein Concentration Determination kits (#23227) were purchased from Thermo Fisher (Waltham, MA, USA). Hoechst staining kits (#C1011) were purchased from Beyotime (Shanghai, China). Specific inhibitors targeting miR-21-3p (5’-GUUGUGGUCAGCUACCCG-3’) and ATG12 (5’-GAAGGACUUACGGAUGUCU-3’), and non-targeting versions of negative controls were purchased from GenePharma (Suzhou, China). MiR-21-3p mimics (5’-CGGGUAGCUGACCACAAC-3’) were purchased from GenePharma (Suzhou, China). Dual Luciferase Assay Kits (#C8021) were purchased from Promega (Madison, WI, USA).
Cell lines
Human umbilical vein endothelial cells (HUVECs) (cat. no. PCS-100-010) were obtained from ATCC (Manassas, VA, USA) and cultured in DMEM supplemented with 10% FBS and 10% growth factors consisting of epidermal growth factor, fibroblast growth factor-2, cAMP, heparin, hydrocortisone, penicillin, streptomycin and amphotericin-B. Human SH-SY5Y (CRL-2266, ATCC) nerve cells were cultured in Ham’s F12 medium + DMEM supplemented with 10% FBS and a 1% v/v antibiotic mixture (penicillin and streptomycin) and incubated in a humidified atmosphere of 5% CO2 at 37°C. The HEK293T cells were cultured in 2 mL of Dulbecco’s modified Eagle’s medium that was supplemented with 100 U/mL penicillin, 100 U/mL streptomycin, and 10% FBS.
Establishment of an H/R model
H/R was performed by incubating SH-SY5Y cells or HUVECs under hypoxic conditions (1% O2, 5% CO2) at 37°C for 6 h, followed by 24 h of reoxygenation.
Exosome preparation
Exosomes were obtained from the cell culture medium of HUVECs. The medium was centrifuged at 3,000 g for 20 min at 4°C to obtain plasma, and then centrifuged again at 10,000 g for 20 min at 4°C to remove cells and platelets. After two more centrifugations at 100,000 g for 70 min at 4°C, 5 µg of exosomes were incubated with 1.25 µL of aldehyde/sulfate latex beads for 15 min. Next, the shape and size of the exosomes were imaged by electron microscopy (JEOL 1010) at × 49,000 magnification.
Total RNA extraction and miRNA microarray
The total mRNA in exosomes was extracted by using Trizol according to the manufacturer’s instructions, and the mRNA concentrations were quantified using a Nanodrop 2000 spectrometer (Thermo Scientific). Microarray hybridization and analysis were performed with the use of Illumina NextSeq 500, based on the miRDeep2 database. The data output was received in Excel spreadsheets containing the normalized micro-RNA expression profiles. Differentially expressed miRs in H/R HUVECs were filtered to exclude those that changed their expression by < 2.0-fold when compared with miRs in the parental HUVECs.
Real-time reverse transcription-PCR (RT2-PCR)
The expression levels of 8 miRs that showed the most significant changes in upregulation or downregulation were further analyzed by RT2-PCR. cDNA templates were obtained by reverse transcription of RNA performed using a reverse transcription kit according to the manufacturer’s instructions. The final 20 μL reaction mixture contained 10 μL of Bestar® SybrGreen qPCRmasterMix, 0.5 μL of each primer (Table 1), 1 μL of cDNA template, and 8 μL of Rnase-free H2O. The thermal cycling parameters used for amplification were as follows: denaturation at 94°C for 2 min, followed by 40 cycles of amplification at 94°C for 20 s, 58°C for 20 s, and 72°C for 20 s. A melting curve was generated and then analyzed in the region between 62°C and 95°C. The relative expression levels of the target genes were calculated using a Real-time PCR Detection System (Mx3000P, Agilent, Santa Clara, CA, USA) and the 2-ΔΔct method.
Table 1.
Primers used for RT2-PCR
| ID | Sequence (5’-3’) |
|---|---|
| U6 F | CTCGCTTCGGCAGCACA |
| U6 R | AACGCTTCACGAATTTGCGT |
| All R | CTCAACTGGTGTCGTGGA |
| hsa-miR-21-3p RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAGCCC |
| hsa-miR-21-3p F | ACACTCCAGCTGGGCAACACCAGTCGATGGGC |
| hsa-miR-769-5p RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACCCATG |
| hsa-miR-769-5p F | ACACTCCAGCTGGGTGAGACCTCTGGGTTCTG |
| hsa-miR-30a-5p RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTTCCAGT |
| hsa-miR-30a-5p F | ACACTCCAGCTGGGTGTAAACATCCTCGACTG |
| hsa-miR-221-5pRT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAATCTAC |
| hsa-miR-221-5p F | ACACTCCAGCTGGGACCTGGCATACAATGTAG |
| hsa-miR-362-5p RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACTCACACC |
| hsa-miR-362-5p F | ACACTCCAGCTGGGAATCCTTGGAACCTAGGT |
| hsa-miR-425-5p RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAACGGG |
| hsa-miR-425-5p F | ACACTCCAGCTGGGAATGACACGATCACTCCC |
| hsa-miR-29b-3p RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACACTGA |
| hsa-miR-29b-3p F | ACACTCCAGCTGGGTAGCACCATTTGAAATCA |
| hsa-miR-374a-3p RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAATTACAA |
| hsa-miR-374a-3p F | ACACTCCAGCTGGGCTTATCAGATTGTATTGT |
| hsa-miR-122-5p RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAAACACC |
| hsa-miR-122-5p F | ACACTCCAGCTGGGTGGAGTGTGACAATGGTG |
| hsa-miR-1246 RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCTGCTC |
| hsa-miR-1246 F | ACACTCCAGCTGGGAATGGATTTTTGGAGC |
| hsa-miR-6087 RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCTCGCC |
| hsa-miR-6087 F | ACACTCCAGCTGGGTGAGGCGGGGGGGCG |
| hsa-miR-novel-chr18_11121 RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAGACAG |
| hsa-miR-novel-chr18_11121 F | ACACTCCAGCTGGGCACGTGAAACCCTGT |
| hsa-miR-novel-chr2_20390 RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTACTACA |
| hsa-miR-novel-chr2_20390 F | ACACTCCAGCTGGGGAGTTCTGGGCTGTA |
| hsa-miR-novel-chr8_33797 RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAGTCATA |
| hsa-miR-novel-chr8_33797 F | ACACTCCAGCTGGGCCATCTGTGGGATTATGA |
| hsa-miR-novel-chr5_27530 RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACTCTTCA |
| hsa-miR-novel-chr5_27530 F | ACACTCCAGCTGGGAGTACTCAAGAGGCTGAA |
| hsa-miR-novel-chr16_9147 RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACTCTTCA |
| ATG12 F | TAGAGCGAACACGAACCATCC |
| ATG12 R | CACTGCCAAAACACTCATAGAGA |
| GAPDH F | TGTTCGTCATGGGTGTGAAC |
| GAPDH R | ATGGCATGGACTGTGGTCAT |
RT: primers used for reverse transcription; All R: PCR reverse primer used to detect miRNAs; R: reverse primer; F: forward primer. U6: U6 small nuclear RNA, as internal reference; GAPDH: glyceraldehyde-3-phosphate dehydrogenase, as internal reference.
Prediction of target genes
The potential target genes of the specifically selected dysregulated miRs were predicted using TargetScan7.1 and mirdbv5. Considering that the prediction programs often suffer from high false positive rates, only the target genes predicted by both tools were taken into account.
Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis and gene ontology (GO) annotation of the target genes
A KEGG pathway enrichment analysis and GO analysis were also performed to compare the specific miRs targets with the whole reference gene background. A count number > 2 and a Bonferroni P-value < 0.05 were chosen as cut-off criteria. Cytoscape 3.1.1 was utilized to construct the possible functional network of the selected miR.
Exosome administration and cell transfection
To investigate the underlying the mechanism by which RIPostC protects the CNS, SH-SY5Y cells were subjected to H/R and then exposed to exosomes or transfected with an miR-21-3p inhibitor by using LipofectamineTM 2000 according to the manufacturer’s instructions.
Immunofluorescent assay
SH-SY5Y cells were seeded in 14-well chambers. They were then fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 for 30 min. Next, the cells were incubated with primary rabbit polyclonal antibodies against ATG12 (1:500) for 1 h; after which, fluorescein isothiocyanate-labeled secondary antibodies (1:20000) were added and the cells were incubated for additional 1 h. After incubation, the cells were washed and stained with 4,6-diamino-2-phenyl indole (DAPI) for 5 min at room temperature. The distribution and expression of ATG12 in the SH-SY5Y cells were imaged with a fluorescence microscope (LSM 710, Germany) at × 120 magnification.
Hoechst 33258 staining
SH-SY5Y cells from different groups were cultured in 12-well plates (5 × 104 cells/well) for 24 h at 37°C. Next, 0.5 mL of Hoechst 33258 staining solution was added to each well and incubated for 5 min. Morphological changes in the cell nucleus were observed under a LSM microscope at × 120 magnification. Cells that displayed a bright illumination were identified as actively undergoing apoptosis.
Western blotting
Total proteins were extracted using a Total Protein Extraction Kit according to the manufacturer’s instructions, and western blotting assays were performed as follows: 40 μg of protein from each sample was subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the separated protein bands were transferred onto a polyvinylidene membrane, which was then blocked with skin milk. Next, the membrane was incubated with primary antibodies against ATG12 (1:2000), LC3-B (1:1000), Beclin-1 (1:1000), Bcl-2 (1:1000), and GAPDH (1:10000) (internal reference protein) for 1 h at room temperature, and then with secondary HRP-conjugated IgG antibodies (1:20000) for 40 min at 37°C. The blots were developed using Beyo ECL Plus reagent, and images were recorded in the Gel Imaging System.
Dual luciferase assay
Direct interactions between miR-21-3p and ATG12 were detected by using a dual luciferase assay kit according to the manufacturer’s instructions. The wild type and mutant sequences of ATG12 3’UTR were synthesized and each sequence was ligated to a psiCHECK-2 plasmid. Transfection for different research purposes was performed using LipofectamineTM 2000 according to the manufacturer’s instructions. Co-transfection of Renilla luciferase plasmid (psi-CHECK2) was used as an internal control for determining transfection efficiency. After 48 h of transfection with the different combinations of vectors and inhibitor, fluorescence intensity was detected using a GloMax Microplate Reader (Promega, Madison, WI, USA).
Statistical analysis
All data were analyzed using IBM SPSS Statistics for Windows, Version 19.0 (IBM Corp., Armonk, NY, USA) and results are expressed as the mean ± SD (n = 3). One-way ANOVA and post hoc multiple comparisons were analyzed using Student’s t-test, and a two-tailed P-value ≤ 0.05 was considered statistically significant.
Results
Profiling of miR expression in exosomes isolated from H/R HUVECs
Morphological features of the exosomes isolated from HUVECs and H/R HUVECs are shown in Figure 1A. No obvious differences were detected between exosomes from the two different sources. MiR expression data was collected from the miRDeep2 database. Following the normalization of raw sequencing data, 353 miRs were found to be differentially expressed in healthy HUVECs vs. H/R HUVECs; these included 104 miRs that were upregulated and 249 that were downregulated (Figure 1B and 1C). At chromosomal the level, autosome No. 2 contained the highest number of novel differentially expressed miRs and autosome No. 18 contained lowest number (Figure 1D). To assess the validity of microarray data, RT2-PCR was performed to compare the expression status of miRs in the two cell types. A total of 16 miRs, including the eight most significantly dysregulated miRs in either group that were upregulated (miR-122-5p, chr18_11121, chr5_27530, chr2_20390, chr8_33797, miR-1246, chr16_9147, and miR-6087) or downregulated (miR-211-5p, miR-29b-5p, miR-362-5p, miR-425-5p, miR-30a-5p, miR-21-3p, miR-374a-3p, miR-769-5p) were selected as possible candidates for further investigation. As shown in Figure 2, the RT2-PCR results were consistent with the microarray results. Among the downregulated miRs, miR-21-3p was the most significantly dysregulated (Figure 2A), and among the upregulated miRs, miR-122-5p was the most significantly dysregulated (Figure 2B). Based on results of the RT2-PCR validation study, miR-21-3p was selected for further exploration.
Figure 1.

The miR expression profile in HUVEC-derived exosomes. HUVECs were subjected to H/R and their exosomes were collected. The expression profile of miRs in the exosomes was analyzed by microarray. A. Electron microscopy identification of exosomes isolated from control HUVECs and H/R HUVECs; magnification, × 49,000. B. Heat map depicting the parental- and H/R HUVC-derived exosomal miRs that showed a statistically significant (P < 0.05) change. C. A scatter plot of microarray data comparing exosomal miRs detected in parental HUVECs (x axis) and H/R HUVECs (y axis). D. Distribution of novel miRs identified on different HUVEC chromosomes after H/R treatment.
Figure 2.
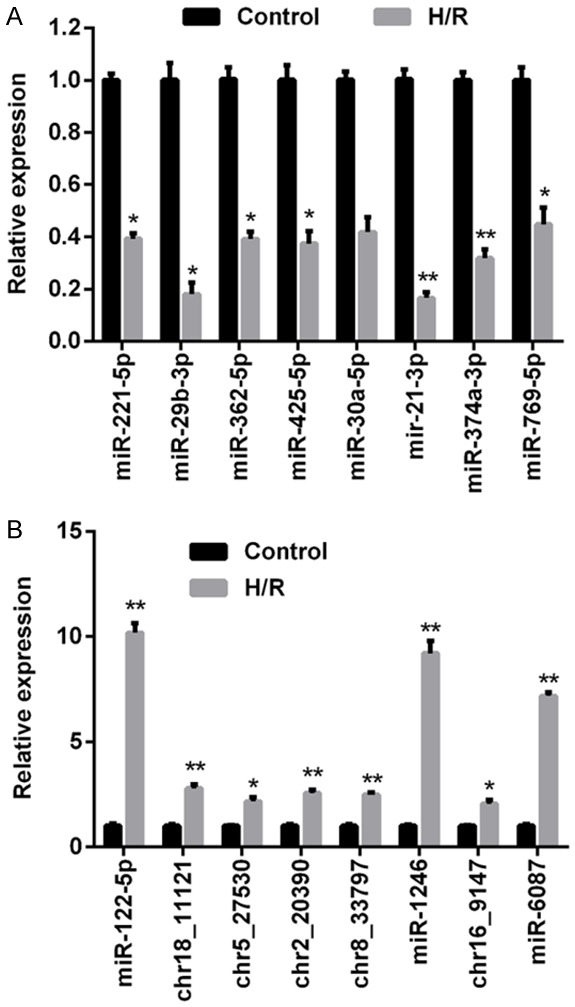
Verification of miR expression status based on microarray data by RT2-PCR. A. RT2-PCR was carried out to validate the expression status of selected miRs that were downregulated based on microarray data; these included miR-211-5p, miR-29b-5p, miR-362-5p, miR-425-5p, miR-30a-5p, miR-21-3p, miR-374a-3p, and miR-769-5p. B. RT2-PCR was carried out to validate the expression status of selected miRs that were upregulated based on microarray data; these included miR-122-5p, chr18_11121, chr5_27530, chr2_20390, chr8_33797, miR-1246, chr16_9147, and miR-6087. “*”, P < 0.05 vs. control group. “**”, P < 0.01 vs. control group.
Target gene prediction and GO and KEGG analysis
The potential targets of eight differentially expressed miRs (up-regulated: miR-122-5p, miR-1246, miR-224-5p, and miR-60-87; down-regulated: miR-21-3p, miR-29b-3p, miR-374a-3p, and miR-425-5p) were predicted using Target-Scan7.1 and miRBase Targets Release Version v5, and the results are shown in Figures S1 and S2. The GO and KEGG analyses were performed based on target prediction data, and the results showed that a broad range of biological processes were associated with the target genes (Tables 2 and 3; Figure 3). The downregulated genes were primarily involved in pathways related to cancers, PI3K/Akt signaling, amyotrophic lateral sclerosis, focal adhesion, hippo signaling, MAPK signaling, FoxO signaling, and phospholipase D signaling (Figure 3A), while the upregulated genes were mainly involved in the hippo signaling pathway, longevity regulating pathway, synaptic vesicle cycle, AMPK signaling pathway, adrenergic signaling in cardiomyocytes, melanogenesis, vibrio cholera infection, axon guidance, and the cAMP signaling pathway (Figure 3B).
Table 2.
GO analysis of target genes for downregulated miRs
| GO Term | GO Name | No. of Gene | FDR |
|---|---|---|---|
| Biological process | |||
| GO: 0007275 | Multicellular organism development | 1887 | 5.99E-15 |
| GO: 0060255 | Regulation of macromolecule metabolism | 2229 | 9.16E-15 |
| GO: 0048731 | System development | 1675 | 1.10E-14 |
| GO: 0007399 | Nerve system development | 899 | 1.01E-13 |
| GO: 0019222 | Regulation of metabolic process | 2340 | 1.01E-13 |
| GO: 0048522 | Positive regulation of cellular process | 1814 | 1.49E-13 |
| GO: 0048518 | Positive regulation of biological process | 1999 | 1.49E-13 |
| GO: 0048856 | Anatomical structure development | 2074 | 1.93E-13 |
| GO: 0080090 | Regulation of primary metabolic process | 2202 | 8.96E-13 |
| GO: 0035556 | Intracellular signal transduction | 1074 | 1.52E-12 |
| Cellular Component | |||
| GO: 0005622 | Intracellular | 5050 | 3.40E-19 |
| GO: 0044424 | Intracellular part | 4927 | 2.44E-17 |
| GO: 0005737 | Cytoplasm | 3851 | 4.70E-10 |
| GO: 0043227 | Membrane-bounded organelle | 4326 | 5.79E-10 |
| GO: 0042331 | Intracellular membrane-bounded organelle | 3930 | 6.73E-10 |
| GO: 0045202 | Synapse | 334 | 6.73E-10 |
| GO: 0043229 | Intracellular organelle | 4275 | 1.95E-09 |
| GO: 0030054 | Cell junction | 580 | 2.95E-09 |
| GO: 0043226 | Organelle | 4623 | 3.07E-09 |
| GO: 0012505 | Endomembrane system | 1465 | 3.56E-08 |
| Molecular function | |||
| GO: 0007071 | Nucleic acid binding transcription factor | 537 | 5.32E-10 |
| GO: 0003700 | Transcription factor activity sequence | 536 | 5.32E-10 |
| GO: 0000987 | RNA polymerase II transcription factor | 309 | 1.48E-09 |
| GO: 0005488 | Binding | 4759 | 4.03E-09 |
| GO: 0043565 | Sequence-specific | 470 | 4.31E-09 |
| GO: 0000976 | Transcription regulatory region sequence | 310 | 7.93E-08 |
| GO: 0046872 | Metal ion binding | 1587 | 8.90E-08 |
| GO: 0001067 | Regulatory region nucleic acid binding | 373 | 9.88E-08 |
| GO: 0044212 | Transcription regulatory region DNA binding | 371 | 1.04E-07 |
| GO: 0000975 | Regulatory region DNA binding | 372 | 1.04E-07 |
Table 3.
GO analysis of target genes for upregulated miRs
| GO Term | GO Name | No. of Gene | FDR |
|---|---|---|---|
| Biological process | |||
| GO: 0006355 | Regulation of transcription, DNA-templated | 602 | 5.77E-07 |
| GO: 1903506 | Regulation of nucleic acid-templated | 604 | 5.77E-07 |
| GO: 2001141 | Regulation of RNA biosynthetic process | 605 | 6.74E-07 |
| GO: 0051252 | Regulation of RNA metabolic process | 619 | 1.15E-06 |
| GO: 2000112 | Regulation of cellular macromolecules | 652 | 1.15E-06 |
| GO: 0031323 | Regulation of cellular metabolic process | 914 | 1.15E-06 |
| GO: 0009889 | Regulation of biosynthetic process | 701 | 1.26E-06 |
| GO: 0006351 | Transcription, DNA-templated | 617 | 1.48E-06 |
| GO: 0031326 | Regulation of cellular biosynthetic process | 692 | 1.51E-06 |
| GO: 0010556 | Regulation of macromolecule biosynthetic process | 665 | 1.72E-06 |
| Cellular Component | |||
| GO:00043231 | Intracellular membrane-bounded organelle | 1601 | 5.58E-10 |
| GO: 0043227 | Membrane-bounded organelle | 1740 | 9.18E-09 |
| GO: 0005622 | Intracellular | 1977 | 1.44E-07 |
| GO: 0043229 | Intracellular organelle | 1704 | 1.56E-06 |
| GO: 0044424 | Intracellular part | 1926 | 1.69E-06 |
| GO: 0005634 | Nucleus | 1043 | 3.56E-06 |
| GO: 0043226 | Organelle | 1827 | 1.78E-05 |
| GO: 0030054 | Cell junction | 237 | 0.00053388 |
| GO: 0044440 | Endosomal part | 88 | 0.00193758 |
| GO: 0010008 | Endosome membrane | 82 | 0.00253877 |
| Molecular function | |||
| GO: 0003700 | Transcription factor activity, sequence | 267 | 6.04E-14 |
| GO: 0001071 | Nucleic acid binding transcription factor | 267 | 6.04E-14 |
| GO: 0003677 | DNA binding | 448 | 1.20E-08 |
| GO: 0000987 | RNA polymerase II transcription factor | 146 | 8.48E-08 |
| GO: 0001012 | RNA polymerase II regulatory region DNA | 132 | 7.90E-07 |
| GO: 0000977 | RNA polymerase II regulatory region sequence | 131 | 1.11E-06 |
| GO: 0043565 | Sequence-specific DNA binding | 209 | 1.77E-06 |
| GO: 0001228 | Transcriptional activator activity, RNA | 79 | 1.01E-05 |
| GO: 0000982 | Transcription factor activity, RNA polymerase | 82 | 1.01E-05 |
| GO: 0001067 | Regulatory region nucleic acid binding | 167 | 1.35E-05 |
Figure 3.
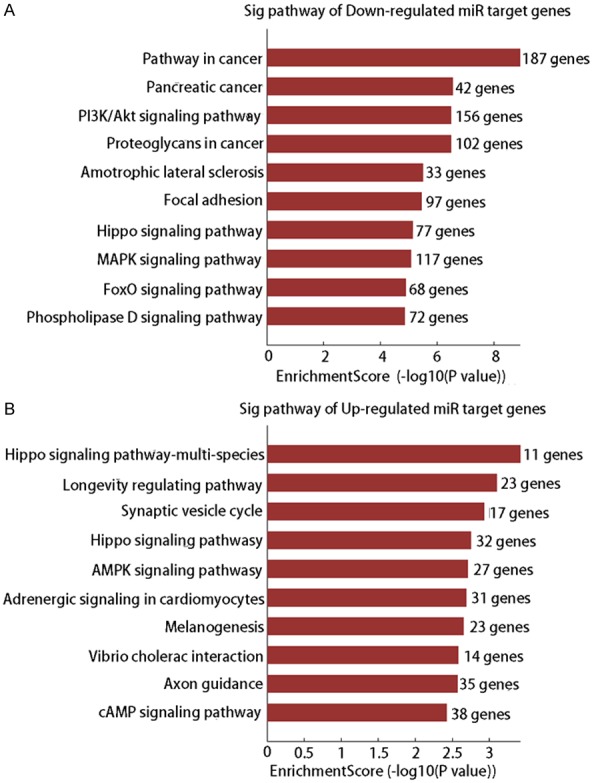
An analysis of KEGG pathways associated with the target genes for differently expressed miRs. A. KEGG pathway-associated target genes for selected downregulated miRs, including miR-21-3p, miR-29b-3p, miR-374a-3p, and miR-425-5p. B. KEGG pathway-associated target genes for selected upregulated miRs, including miR-122-5p, miR-1246, miR-224-5p, and miR-6087.
MiR-21-3p directly targeted ATG12
The RT2-PCR data indicated that miR-21-3p was the most significantly downregulated miR member. Results obtained from additional bioinformatics analyses (http://www.targetscan.org/cgi-bin/targetscan/vert_71/targetscan.cgi?species = Human&gid = &mir_sc = &mir_c = &mir_nc = &mir_vnc = &mirg = miR-21-3p) suggested the ATG12 gene, which is essential for cell survival, as a target for miR-21-3p. A dual luciferase assay was performed to assess interaction between miR-21-3p and the ATG12 gene. The relative activity of luciferase was lowest in cells co-transfected with miR-21-3p mimics and the plasmid which carried ATG12 3’UTR sequences, while the luciferase activity in cells transfected with miR-21-3p mimics and the mutant ATG12 3’UTR sequence remained unchanged (Figure 4). These results suggested a specific binding of miR-21-3p to the 3’UTR sequence of ATG12. The regulatory effect of miR-21-3p on ATG12 expression was also verified in SHSY5Y cells, where the H/R process induced miR-21-3p expression, which was associated with reduced levels of ATG12 expression (Figure S3). This result confirmed that miR-21-3p contributes to H/R-induced neural cell damage.
Figure 4.

Detection of direct interaction between miR-21-3p and ATG12 by the dual luciferase assay. HEK293T cells were transfected with different combinations of NC mimics, miR-21-3p mimics, and psiCHECK-2 plasmid carrying the wild type ATG12 3’UTR sequence and a psiCHECK-2 plasmid carrying the mutant type ATG12 3’UTR sequence, and then subjected to the dual luciferase assay. “**”, P < 0.01 vs. NC.
RIPostC attenuated H/R-induced apoptosis in SH-SY5Y cells by activating the miR-21-3p-mediated ATG12 pathway
To further investigate the role played by miR-21-3p in RIPostC in the CNS, H/R-treated SH-SY5Y cells were incubated with an miR-21-3p inhibitor and (or) exosomes collected from H/R-treated HUVECs. As shown in Figure 5A, although the inhibitory effect of exosome treatment was weaker than that of the specific inhibitor, the addition of HUVEC-derived exosomes alone also suppressed miR-21-3p expression in H/R-treated SH-SY5Y cells when compared with expression in the control group (P < 0.05). The inhibition of miR-21-3p in H/R-treated SH-SY5Y cells was synchronized with the up-regulation of ATG12 expression (Figure 5B and 5C). Based on RT2-PCR and immunofluorescent detection results, the levels of mRNA for ATG12 protein and the levels of ATG12 gene expression were both upregulated by the miR-21-3p inhibitor and (or) HUVEC-derived exosomes (Figure 5B). The production and distribution of ATG12 protein was also increased by both treatments (Figure 5C), which supported results of the dual luciferase assay. Given the fact that ATG12 was critical for the survival of cells, the expression of downstream factors associated with autophagy (LC3-B and Beclin-1) and apoptosis (Bcl-2) was also investigated. Results of western blot studies showed that the levels of molecules involved in autophagy activation and apoptosis suppression were all upregulated by treatment with the miR-21-3p inhibitor and (or) HUVCE-derived exosomes (Figure 6A). Moreover, Hoechst 33258 staining results showed that treatment with either the miR-21-3p inhibitor or HUVCE-derived exosomes inhibited apoptosis in H/R-treated SHSY5Y cells (Figure 6B). Although the apoptosis inhibiting effect of exosomes was weaker than that of the miR-21-3p inhibitor, it still helped to alleviate H/R-induced injures in SH-SY5Y cells.
Figure 5.
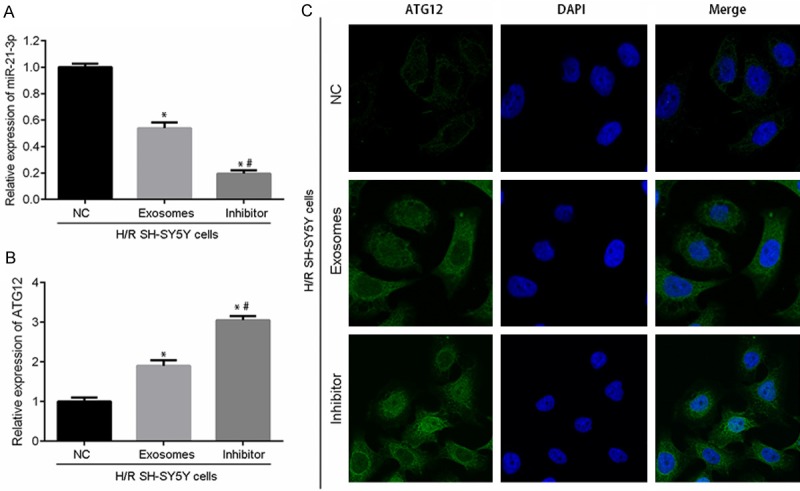
Inhibition of miR-21-3p increased ATG12 expression in H/R SH-SY5Y cells. H/R SH-SY5Y cells were post-treated with an NC inhibitor, HUVEC-derived exosomes, or an miR-21-3p inhibitor. A. RT2-PCR detection of miR-21-3p. B. RT2-PCR detection of ATG12. C. Immunofluorescent detection of ATG12 distribution and production in SH-SY5Y cells treated with the NC inhibitor, HUVEC-derived exosomes, or miR-21-3p inhibitor. Magnification, × 120. “*”, P < 0.01 vs. SH-SY5Y cells + H/R + NC. “#”, P < 0.05 vs. SH-SY5Y cells + H/R + exosomes.
Figure 6.
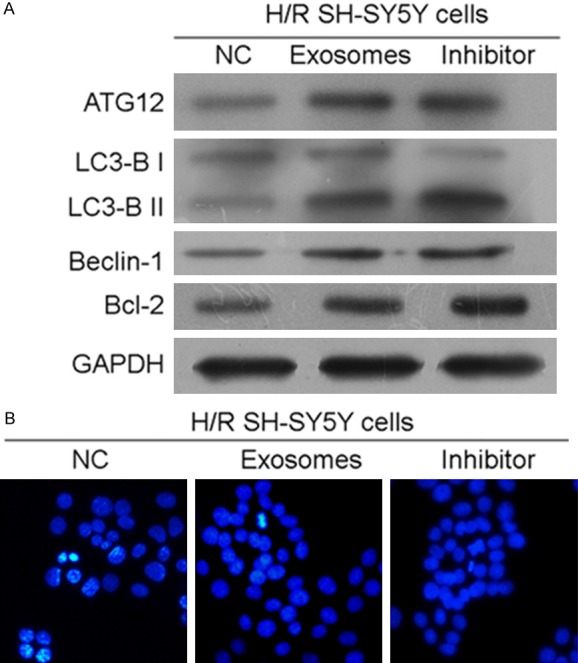
ATG12 upregulation induced autophagy and inhibited apoptosis in H/R SH-SY5Y cells. A. Western blot detection of ATG12, LC3-B, Beclin-1, and Bcl-2 expression in H/R SH-SY5Y cells treated with an NC inhibitor, HUVEC-derived exosomes, or miR-21-3p inhibitor. B. Hoechst 33258 staining detection of apoptosis in SH-SY5Y cells. Cells actively undergoing apoptosis are brightly illuminated. Magnification, × 120.
Discussion
RIPostC is a well-known strategy for protecting vital organs against ischemia/reperfusion (I/R) injuries. The technique was first reported by Kerendi et al. [16] as a method for treating coronary heart disease [17]. Following that report, several other studies showed the beneficial effects of RIPostC on I/R-induced brain injuries [11,18]. The attractive potential of using RIPostC for treating I/R-induced brain injuries has promoted further exploration of the mechanism associated with that effect. Several molecular pathways are known to be involved in the protective effect of RIPostC on cerebral tissues [11,12]. However, most previous studies only focused on signaling changes in the brain and ignored the mechanism driving the remote transfer involved in the treatment effect of RIPostC. Yamaguchi et al. [4] showed that repeated RIPostC attenuated left ventricular remodeling via exosome-mediated remote transfer of miR-29a [4]. Numerous cells in the CNS can release exosomes in the form of extracellular membrane vesicles [15], and Yang et al. [18] recently showed that exosome-mediated delivery of miR-124 could promote neurogenesis after ischemia [19]. Based on the above results, we subjected HUVECs to H/R for the purpose of imitating the effect of RIPostC stimulation on remote organs. We found that H/R treatment substantially changed the expression profiles of miRs, which was consistent with previous reports [6]. A total of 353 differentially expressed miRs were detected based on a comparison between HUVECs and H/R-treated HUVECs. Subsequent GO and KEGG analyses revealed that the target genes of the most significantly dysregulated miRs were involved in multiple key biological processes, including the hippo signaling pathway, longevity regulating pathway, pathways related to cancers, and the PI3K/Akt signaling pathway. Moreover, based on RT2-PCR validation data, miR-21-3p was selected as an candidate for exploring the treatment mechanism of RIPostC.
MiR-21-3p can attenuate brain injuries via multiple mechanisms [20,21]. However in the current study, the administration of H/R downregulated miR-21-3p expression in HUVECs, suggesting that miR-21-3P might harm the structure and function of the CNS. To verify this hypothesis, H/R-treated neural cells were incubated with HUVEC-derived exosomes and a miR-21-3p inhibitor to interrupt the miR-21-3p balance in SH-SY5Y cells. It was found that both treatments suppressed miR-21-3p expression and inhibited apoptosis in H/R-treated SH-SY5Y cells. Expression of the miR-21-3p downstream effector, ATG12, was also detected in the current study due to its involvement in regulating cell survival via autophagy pathways. Dual luciferase assay results showed that miR-21-3p directly binds to the 3’UTR sequence of the ATG12 gene and suppresses ATG12 signaling. ATG12 forms a conjugate with ATG5 and thereby plays a key role in the formation of autophagosomes. The role of ATG12 in regulating apoptosis is complex. Generally, inhibition of ATG12 suppresses autophagy and induces apoptosis [22,23]. Nevertheless, ATG12 also directly regulates the apoptotic pathway by inactivating pro-survival Bcl-2 family members [24]. Our current results are hard to explain based on the above findings. We found that inhibition of miR-21-3p induced the expression of ATG12 and indicators of autophagy, and also increased the levels of Bcl-2 expression. Although these results support the apoptosis inhibition via autophagy activation theory, it appears that the regulatory effect of ATG12 on Bcl-2 is more complicated than previously reported, and may differ depending on the type of organ tissue involved. Our study confirmed the exosome-mediated regulation of miR expression in RIPostC treated cells and also identified some miR candidates for future studies; however, the mechanism underlying the interruption of miR-21-3p balance in H/R-treated SH-SY5Y cells was only partially revealed. Although our data suggest that miR-21-3p suppression by RIPostC inhibited apoptosis in neural cells, the factors that contribute to miR-21-3p suppression in H/R-treated HUVECs, and whether those factors were encompassed in exosomes remains unknown.
In summary, we performed H/R treatments in vitro to imitate the RIPostC process, and found that exosomes secreted by cells in remote organs or tissues can influence cell damage in the CNS. Our results showed that miR-21-3p could promote H/R-induced apoptosis in neural cells. Furthermore, we found that exosomes secreted by H/R-treated HUVECs induced the suppression of the miR-21-3p, which inhibited apoptosis by activating ATG12-mediated autophagy and increasing Bcl-12 expression in SH-SY5Y cells. A shortcoming of the current study is that we failed to identify the miR-21-3p inhibiting factors in HUVEC-derived exosomes. Further comprehensive studies based on the current results will be performed to further reveal the treatment mechanism of RIPostC.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81660420), the Construction Plan of the Superior Science and Technology Innovation Team of Jiangxi Province (No. 20161BCB24009), the Foreign Science and Technology Cooperation Plan of Jiangxi Province (No. 20151BDH80009), and the Key Projects of Natural Science Foundation of Jiangxi Province (No. 20171ACB20035).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Zong Y, Jiang L, Zhang M, Zhou F, Qi W, Li S, Yang H, Zou Y, Xia Q, Zhou X. Limb remote ischemic postconditioning protects cerebral ischemia from injury associated with expression of HIF-1α in rats. BMC Neurosci. 2015;16:1–8. doi: 10.1186/s12868-015-0235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitch MT, Silver J. CNS injury, glial scars, and inflammation: inhibitory extracellular matrices and regeneration failure. Exp Neurol. 2008;209:294–301. doi: 10.1016/j.expneurol.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun F, He Z. Neuronal intrinsic barriers for axon regeneration in the adult CNS. Curr Opin Neurobiol. 2010;20:510–518. doi: 10.1016/j.conb.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaguchi T, Izumi Y, Nakamura Y, Yamazaki T, Shiota M, Sano S, Tanaka M, Osada-Oka M, Shimada K, Miura K. Repeated remote ischemic conditioning attenuates left ventricular remodeling via exosome-mediated intercellular communication on chronic heart failure after myocardial infarction. Int J Cardiol. 2015;178:239–246. doi: 10.1016/j.ijcard.2014.10.144. [DOI] [PubMed] [Google Scholar]
- 5.Ren C, Gao M, Dornbos D, Ding Y, Zeng X, Luo Y, Ji X. Remote ischemic post-conditioning reduced brain damage in experimental ischemia/reperfusion injury. Neurol Res. 2011;33:514–519. doi: 10.1179/016164111X13007856084241. [DOI] [PubMed] [Google Scholar]
- 6.Slagsvold KH, Rognmo O, Høydal M, Wisløff U, Wahba A. Remote ischemic preconditioning preserves mitochondrial function and influences myocardial microRNA expression in atrial myocardium during coronary bypass surgery. Circ Res. 2014;114:851–859. doi: 10.1161/CIRCRESAHA.114.302751. [DOI] [PubMed] [Google Scholar]
- 7.Murry CE, Jennings RB, Reimer KA. Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation. 1986;74:1124–1136. doi: 10.1161/01.cir.74.5.1124. [DOI] [PubMed] [Google Scholar]
- 8.Zhao ZQ, Corvera JS, Halkos ME, Kerendi F, Wang NP, Guyton RA, Vintenjohansen J. Inhibition of myocardial injury by ischemic postconditioning during reperfusion: comparison with ischemic preconditioning. Am J Physiol Heart Circ Physiol. 2003;285:579–588. doi: 10.1152/ajpheart.01064.2002. [DOI] [PubMed] [Google Scholar]
- 9.Pan J, Li X, Peng Y. Remote ischemic conditioning for acute ischemic stroke: dawn in the darkness. Rev Neurosci. 2016;27:501–510. doi: 10.1515/revneuro-2015-0043. [DOI] [PubMed] [Google Scholar]
- 10.Przyklenk K, Bauer B, Ovize M, Kloner RA, Whittaker P. Regional ischemic ‘preconditioning’ protects remote virgin myocardium from subsequent sustained coronary occlusion. Circulation. 1993;87:893–899. doi: 10.1161/01.cir.87.3.893. [DOI] [PubMed] [Google Scholar]
- 11.Ren C, Yan Z, Wei D, Gao X, Chen X, Zhao H. Limb remote ischemic postconditioning protects against focal ischemia in rats. Brain Res. 2009;1288:88–94. doi: 10.1016/j.brainres.2009.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng B, Guo QL, He ZJ, Ye Z, Yuan YJ, Wang N, Zhou J. Remote ischemic postconditioning protects the brain from global cerebral ischemia/reperfusion injury by up-regulating endothelial nitric oxide synthase through the PI3K/Akt pathway. Brain Res. 2012;1445:92–102. doi: 10.1016/j.brainres.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 13.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Baskin YK, Dietrich WD, Green EJ. Two effective behavioral tasks for evaluating sensorimotor dysfunction following traumatic brain injury in mice. J Neurosci Methods. 2003;129:87–93. doi: 10.1016/s0165-0270(03)00212-7. [DOI] [PubMed] [Google Scholar]
- 15.Kalani A, Tyagi A, Tyagi N. Exosomes: mediators of neurodegeneration, neuroprotection and therapeutics. Mol Neurobiol. 2014;49:590–600. doi: 10.1007/s12035-013-8544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Zhang X, Chen X, Wang L, Yang G. Exosome mediated delivery of miR-124 promotes neurogenesis after Ischemia. Mol Ther Nucleic Acids. 2017;7:278–287. doi: 10.1016/j.omtn.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerendi F, Kin H, Halkos ME, Jiang R, Zatta AJ, Zhao ZQ, Guyton RA, Vintenjohansen J. Remote postconditioning. Brief renal ischemia and reperfusion applied before coronary arter reperfusion reduces myocardial infarct size via endogenous activation of adenosine receptors. Basic Res Cardiol. 2005;100:404–421. doi: 10.1007/s00395-005-0539-2. [DOI] [PubMed] [Google Scholar]
- 18.Hess DC, Hoda MN, Bhatia K. Remote limb perconditioning [corrected] and postconditioning: will it translate into a promising treatment for acute stroke? Stroke. 2013;44:1191–1197. doi: 10.1161/STROKEAHA.112.678482. [DOI] [PubMed] [Google Scholar]
- 19.Barteneva NS, Maltsev N, Vorobjev IA. Microvesicles and intercellular communication in the context of parasitism. Front Cell Infect Microbiol. 2013;3:1–11. doi: 10.3389/fcimb.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge XT, Lei P, Wang HC, Zhang AL, Han ZL, Chen X, Li SH, Jiang RC, Kang CS, Zhang JN. miR-21 improves the neurological outcome after traumatic brain injury in rats. Sci Rep. 2014;4:1–11. doi: 10.1038/srep06718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge X, Han Z, Chen F, Wang H, Zhang B, Jiang R, Lei P, Zhang J. MiR-21 alleviates secondary blood-brain barrier damage after traumatic brain injury in rats. Brain Res. 2015;1603:150–157. doi: 10.1016/j.brainres.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D, Souquere S, Yoshimori T, Pierron G, Codogno P, Kroemer G. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


