Abstract
This study aims to identify the pivotal microRNAs (miRNAs) and genes, and their potential regulatory mechanisms in pancreatic ductal adenocarcinoma (PDAC) through bioinformatics analysis and experimental verification. We comprehensively analyzed two miRNA microarray datasets (GSE32678 and GSE43796) and three gene microarray datasets (GSE28735, GSE41368 and GSE71989), which were downloaded from the Gene Expression Omnibus (GEO) database, and identified the total of 8 differentially expressed miRNAs (DEMs) and 257 differentially expressed genes (DEGs) in common. Next, a new miRNA-mRNA regulatory network was constructed by bioinformatics methods, including 7 miRNAs, 58 putative target genes and 80 interaction pairs of miRNA-mRNA. Scrutinized by OncoLnc and GEPIA, it was found that 3 of 7 miRNAs (miR-21, miR-196b and miR-203) and 20 of 58 genes (MXRA5, EPYC, ECT2, COL12A1, SLC6A14, SLC7A2, BTG2, PDK4, CTNND2, NRP2, PXDN, CD109, TGFBI, LRRN1, ITGA2, DKK1, GREM1, EFNB2, SEMA3C and NT5E) were notably associated with prognosis in patients with PDAC. Furthermore, EFNB2 was significantly upregulated in PDAC compared with normal controls from different public databases. Cellular function experiments demonstrated that EFNB2 knockdown inhibited cell proliferation, migration and invasion in SW1990 cells. Western blot and luciferase reporter assays revealed that miR-557 negatively regulated the expression of EFNB2 by directly binging its 3’ UTR. In conclusion, we performed integrated analysis for multiple expression profiles, and provided novel candidate miRNAs and genes to be exploited for functional studies. In addition, our findings suggested that EFNB2 contributes to PDAC progression by acting as the target gene of miR-557. It is useful for uncovering miRNA-based treatments in PDAC.
Keywords: EFNB2, miR-557, miRNA-mRNA network, bioinformatics analysis, pancreatic ductal adenocarcinoma
Introduction
MicroRNAs (miRNAs) are endogenous, single stranded, noncoding RNA molecules consisting of 19-24 nucleotides. It was known that miRNAs negatively regulated gene expression by binding to complementary sequences in the 3’ untranslated region (UTR) of target mRNAs, which can result in degradation of the mRNA transcript or suppression of the protein translation process [1]. Accumulating evidence suggested that miRNAs play acritical regulatory role in most aspects ofhomeostasis and cell differentiation [2]. More than that, the dysregulation of miRNAs were involved in various pathologies, including carcinogenesis, metastasis, and chemoresistance of tumor cells, with loss of normal function giving rise to aberrant expression of key oncogenes or tumor suppressor genes [3,4].
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-related death in the United States, with more than 53,000 new cases predicted in 2017, and this number has been firmly increasing over the past few decades [5]. Due to no specific symptomsand lack of effective biomarkers, majority of patients have local invasion and even distant metastasis at the initial diagnosis, making them inoperable [6]. Only less than 20% of PDAC patients diagnosed at early stage can be considered as candidates for surgical excision [7].Despite receiving curative resection, numerous patients still die of invisible metastases and relapses at last. Hence, its 5-year survival rate less than 10% as before in the context of multimodality treatments [8]. However, its pathogenesis at the molecular level remains fragmentary and incomplete at present. Consequently, it is imperative to implement a more integrated analysis of PDAC to exploit the effective biomarkers for cancer diagnosis and treatment.
Recently, high-throughput technologies provide the new and significant insights into diagnosis, prognosis and individualized medication of PDAC in an unprecedented manner [9]. Online analysis tools and publicly available databases were designed for favoring researchers to conduct data mining and exploration, which greatly accelerated our understanding of miRNAs and genes in the initiation and development of PDAC.
In this study, our research team synthetically analyzed multiple datasets to establish a novel regulatory network of miRNA-mRNA. We analyzed functional, pathway and prognostic value among the differentially expressed miRNAs (DEMs) and differentially expressed genes (DEGs). More importantly, our data revealed that the expression level of EFNB2 was significantly upregulated in PDAC tissues and cell lines, and EFNB2 knockdown inhibited the cell proliferation, migration and invasion.Furthermore, we found that the overexpression of miR-557 decreased EFNB2 level by directly binding the 3’ UTR of EFNB2 for the first time.
Materials and methods
Collection of microarray datasets and data processing
Two miRNA microarray datasets (GSE32678 and GSE43796) contained 30 PDAC samples and 13 normal samples. Three gene microarray datasets (GSE28735, GSE41368 and GSE71989) contained 64 PDAC samples and 59 normal samples. A total of five microarray datasetswere obtained from GeneExpression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) [10]. The main features of these five datasets were shown in Table 1. The GEO2R online analysis tool based on the GEOquery and limma R packages was utilized to identify the DEMs and DEGs between PDAC samples and normal samples [11]. We used P value <0.01 and |logFC| > 1 as cutoff criteria for screening DEMs, and P value <0.05 and |logFC| > 1 as cutoff criteria for screening DEGs. Venn diagrams were drawn by Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/).
Table 1.
The main features of five selected datasets
| GEO accession | Datasets type | Platform | Samples in total (PDACs/ANTs) | Country | PMID |
|---|---|---|---|---|---|
| GSE32678 | miRNA | GPL7723 | 32 (25/7) | USA | 22261810 |
| GSE43796 | miRNA | GPL15159 | 11 (5/6) | South Korea | 24072181 |
| GSE28735 | Gene | GPL6244 | 90 (45/45) | USA | 23918603 |
| GSE41368 | Gene | GPL6244 | 12 (6/6) | Italy | 24120476 |
| GSE71989 | Gene | GPL570 | 21 (13/8) | USA | - |
Abbreviations: GEO, Gene Expression Omnibus; ANTs, adjacent normal tissues; PMID: PubMed unique identifier.
Construction of miRNA-mRNA interaction network
The miRNAs targets were predicted by miRecords, which is an integrative tool based on eleven established miRNA target prediction programs (DIANA-microT, MicroInspector, miRanda, MirTarget2, miTarget, NBmiRTar, PicTar, PITA, RNA22, RNAhybrid, and TargetScan/TargertScanS) [12]. We only considered miRNA target genes that were predicted by at least four of eleven programs. In addition, the miRNA-mRNA regulatory network was visualized by Cytoscape 3.6.0. [13].
Functional enrichment analysis and protein-protein interaction (PPI) network
For functional enrichment analysis of the 58 shared DEGs, an online program, Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) [14], was applied to perform Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis [15,16]. P<0.05 was set as the cutoff criterion. To assess the interactive associations among DEGs, the 58 common DEGs were mapped via using The Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org), which is an useful online tool designed to explore and analyze the protein-protein interaction (PPI) information [17]. Hide disconnected nodes in the network and confidence score ≥ 0.4 were selected.
Survival analysis
The overall survival curves of overlaping DEMs were performed by OncoLnc (http://www.oncolnc.org) [18], allowing investigators to quickly explore survival correlations for up to 21 cancers based on The Cancer Genome Atlas (TCGA). We divided into low and high expression groups according to the median value, namely 50% vs 50%. Thus, there were 87 patients with PDAC in each group. The overall survival and disease free survival curves of screening DEGs were generated by GEPIA (http://gepia.cancer-pku.cn/) [19], which is a newly developed interactive web server for analyzing the gene expression data and prognostic value based on TCGA and the GTEx by using a standard processing pipeline. We selected median as the group cutoff for survival plots.
Data mining from public databases
The function of EFNB2 and related networks with other genes were predicted by geneMANIA (http://genemania.org/), which is a flexible and user-friendly web interface for generating hypotheses about gene function [20]. The basic expression level of EFNB2 in different human organs or tissues were downloaded from The National Center for Biotechnology Information (NCBI) Gene database (https://www.ncbi.nlm.nih.gov/gene/) [21]. The clinical information of patients with PDAC in regard to the expression level of EFNB2 were provided by Oncomine (http://www.oncomine.org), which is a web-based data mining platform aimed at promoting discovery from genome-wide expression analyses [22]. The immunofluorescent and immunohistochemistry staining of EFNB2 were obtained from the Human Protein Atlas database (https://www.proteinatlas.org/) [23].
Cell culture and transfection
Human pancreatic cancer cell lines (MiaPaCa-2, PANC-1, BxPC-3 and SW1990) and an immortalized pancreatic ductal epithelial cells (HPDE6) were cultured in DMEM medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin in a humidified 5% CO2 atmosphere at 37°C. The small interfering RNA of EFNB2 (si-EFNB2), scramble siRNA of EFNB2 (negative control, NC), miR-557 mimics and negative control oligonucleotides (miR-NC) were constructed by GenePharma (Shanghai, China). Transfection was performed using the Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. The efficiencywas tested at 48 hours after transfection.
Real-time PCR analysis
Total RNA was extracted from pancreatic cancer cells by using Trizol solution (Invitrogen, USA) according to the manufacturer’s protocols, and then was converted into cDNA by using the Reverse Transcription System (Promega, USA). The expression of EFNB2 and miR-557 were detected by quantitative real-time PCR (qRT-PCR) with SYBR Green PCR Kit (TaKaRa, Japan) following the manufacturer’s instructions, and GAPDH as an internal control. For the purpose of reduction of bias, each sample were performed in triplicate. Fold changes in expression were calculated relative to the control group. The primer sequences were as follows: EFNB2, 5’-ACCAGTCCTTGTCCAGGTAGAA-3’ (forward) and 5’-TCCGTGTGGAAGTACTGCTG-3’ (reverse); miR-557, 5’-GTTTGCACGGGTGGGC-3’ (forward) and 5’-GAACATGTCTGCGTATCTC-3’ (reverse); GAPDH, 5’-TGCCATCAATGACCCCTTC-3’ (forward) and 5’-CATCGCCCCACTTGATTTTG-3’ (reverse).
Western blot analysis
Western blot was performed as previously published [24], the primary antibodies specific for EFNB2 or GAPDH purchased from Abcam (Cambridge, MA, USA).
Cell proliferation assays
The proliferation ability of cells was evaluatedby the Cell Counting Kit-8 (CCK-8) (Dojindo, Japan) according to the manufacturer’s instructions. Briefly, the transfected cells were seeded into 96-well plates and the absorbance was monitored at 450 nm every 24 h on a microplate reader to assess the number of viable cells.
Migration and invasion assays
The wound healing assays were utilized to measure the migration ability of tumor cells. Cells were seeded on the 6 well plate untilformed a single cell layer, and the wounds were gently scratched using a 200 μl sterile pipette tip across cell monolayer. Afterwards, exfoliated cells and debris were removed by washing with PBS, and were replaced with serum-free DMEM medium. The migration of the cells into the wound was photographed at five randomly chosen areas every 24 h. The invasion ability of cells was measured by using transwell assay in line with our previous study [25]. The proportion of migration and the number of invasive cells were calculated with Image J software.
Luciferase reporter assay
SW1990 cells were cultured in a 96 well plate and were co-transfected with 100 ng luciferase reporter vector containing the wild type or mutant 3’ UTR of EFNB2 with 50 nM miR-557 mimics or miR-NCs. Cells were harvested at 48 h after transfection, luciferase activity was measured by using adual-luciferase reporter assay system (Promega, USA) according to the kit instructions. Relative luciferase activity were normalized to respective Renilla luciferase.
Statistical analysis
Each experiment was performed at least three times. Statistical graphs and analyzes were performed by GraphPad Prism 6.0 (Lajolla, CA, USA) and all data were expressed as mean ± standard deviation (SD). Comparisons of two groups was conducted using the two-tailed student’s t-test. Overall survival were analyzed by the Kaplan-Meier method and differences were analyzed by log-rank test. Statistical significance was defined as P<0.05.
Results
Identification of DEMs and DEGs, and construction of the miRNA-mRNA regulation network in PDAC
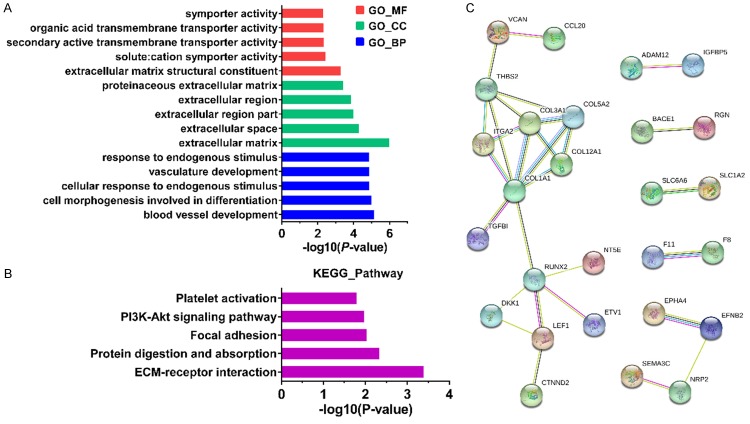
We selected two miRNA microarray datasets (GSE32678 and GSE43796) and three gene microarray datasets (GSE28735, GSE41368 and GSE71989) from GEO database for identification of DEMs and DEGs, respectively. The main features of five datasets were summarized in Table 1. A total of 8 shared DEMs (miR-21, miR-135b, miR-196b, miR-203, miR-210, miR-222, miR-224 and miR-557) and 257 common DEGs were obtained and showed by using Venn diagrams (Figure 1A and 1B). Of the eight miRNAs, only miR-557 was downregulated, and the remaining seven miRNAs were upregulated in PDAC. Next, we predicted 408, 811, 323, 2364, 41, 373, 433 and 1014 target genes for miR-21, miR-135b, miR-196b, miR-203, miR-210, miR-222, miR-224 and miR-557, respectively, by means of miRecords. In order to find potential miRNA-mRNA interactions, the predicted target genes datasets of 8 miRNAs were intersected with 257 DEGs to identify the overlapping genes, respectively (Figure 1C). Among them, the predicted target gene dataset of miR-210 and DEGs had no intersections. Finally, a total of 58 related genes and 80 putative miRNA-mRNA pairs were obtained. The seven miRNAs and corresponding target genes were shown in Table 2, and the regulatory network between them was visualized by the Cytoscape software (Figure 1D).
Figure 1.
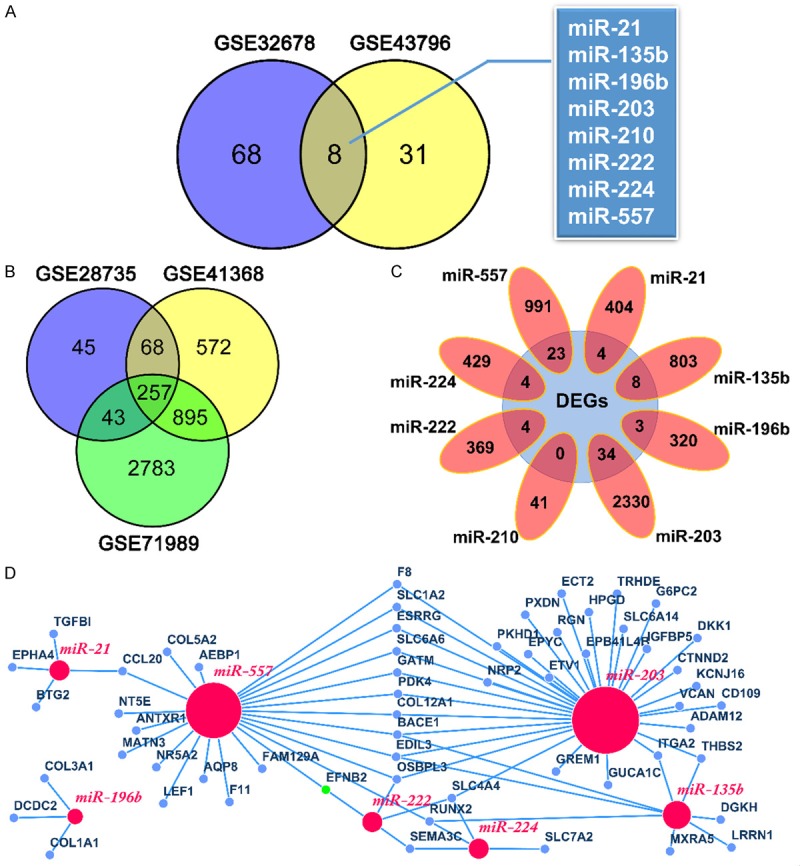
Identification of DEMs and DEGs, and construction of the miRNA-mRNA regulation network in PDAC. A. A total of 8 common DEMs were screened were obtained from GSE32678 and GSE43796. B. A total of 257 shared DEGs were identified from GSE28735, GSE41368 and GSE71989. C. Eight target gene datasets intersected with 257 DEGs, respectively. D. The regulatory network between miRNAs and target genes was visualized by the Cytoscape. Red nodes represent miRNAs and blue nodes represent genes. DEMs, differentially expressed miRNAs.DEGs, differentially expressed genes.
Table 2.
The seven screened DEMs and predicted target genes
| miRNA | Expression | Number of targets | Target genes |
|---|---|---|---|
| miR-21 | Up | 4 | BTG2, EPHA4, TGFBI, CCL20 |
| miR-135b | Up | 8 | BACE1, LRRN1, RUNX2, MXRA5, DGKH, ITGA2, EDIL3, THBS2 |
| miR-196b | Up | 3 | DCDC2, COL1A1, COL3A1 |
| miR-203 | Up | 34 | GUCA1C, TRHDE, KCNJ16, GATM, PKHD1, SLC4A4, BACE1, SLC1A2, RGN, ESRRG, G6PC2, EPB41L4B, F8, PDK4, CTNND2, DKK1, SLC6A6, NRP2, PXDN, CD109, EPYC, ETV1, OSBPL3, HPGD, ECT2, ADAM12, ITGA2, IGFBP5, GREM1, EDIL3, COL12A1, THBS2, SLC6A14, VCAN |
| miR-222 | Up | 4 | SLC4A4, EFNB2, OSBPL3, SEMA3C |
| miR-224 | Up | 4 | SLC4A4, SLC7A2, RUNX2, SEMA3C |
| miR-557 | Down | 23 | F11, GATM, NR5A2, AQP8, BACE1, SLC1A2, ESRRG, FAM129A, F8, PDK4, RUNX2, MATN3, SLC6A6, AEBP1, EFNB2, COL5A2, OSBPL3, NT5E, CCL20, EDIL3, ANTXR1, COL12A1, LEF1 |
Abbreviation: DEMs, differentially expressed microRNAs.
Functional enrichment analysis and PPI network
To gain a deeper understanding with regard to the function of shared miRNAs based on 58 target genes, we performed GO analysis and KEGG pathway analysis by using the DAVID. The GO analysis found that molecular function (MF) group was principally enriched in extracellular matrix structural constituent, symporter activity and transmembrane transporter activity.Biological process (BP) terms werenotably involved in blood vessel development, cell differentiation and cellular response to endogenous stimulus. Cellular component (CC) terms were mainly implicated in extracellular matrix and extracellular region (Figure 2A). In addition, the KEGG pathway analysis indicated that 58 target genes were observably enriched in ECM-receptor interaction, protein digestion and absorption, focal adhesion and PI3K-Akt signaling pathway (Figure 2B). For purpose of mining of 58 target gene-related proteins, we performed the PPI network by using the STRING online database (Figure 2C).
Figure 2.
Functional enrichment analysis and PPI network. A, B. Gene ontology analysis and KEGG pathway enrichment analysis on 58 target genes of selected miRNAs. BP, biological process; CC, cell component; MF, molecular function. C. PPI network on target genes of selected miRNAs.
Screening of potential indicators associated with prognosis in PDAC
OncoLnc was used to evaluate the prognostic value of the screened eight miRNAs in patients with PDAC. We found that high expression of miR-21, miR-196b and miR-203 were dramatically correlated with worse overall survival (OS) (P=0.0234, 0.0002 and 0.0010, respectively) (Figure 3A-C). There was no significant association between the expression of remaining five miRNAs and OS (data not shown).
Figure 3.
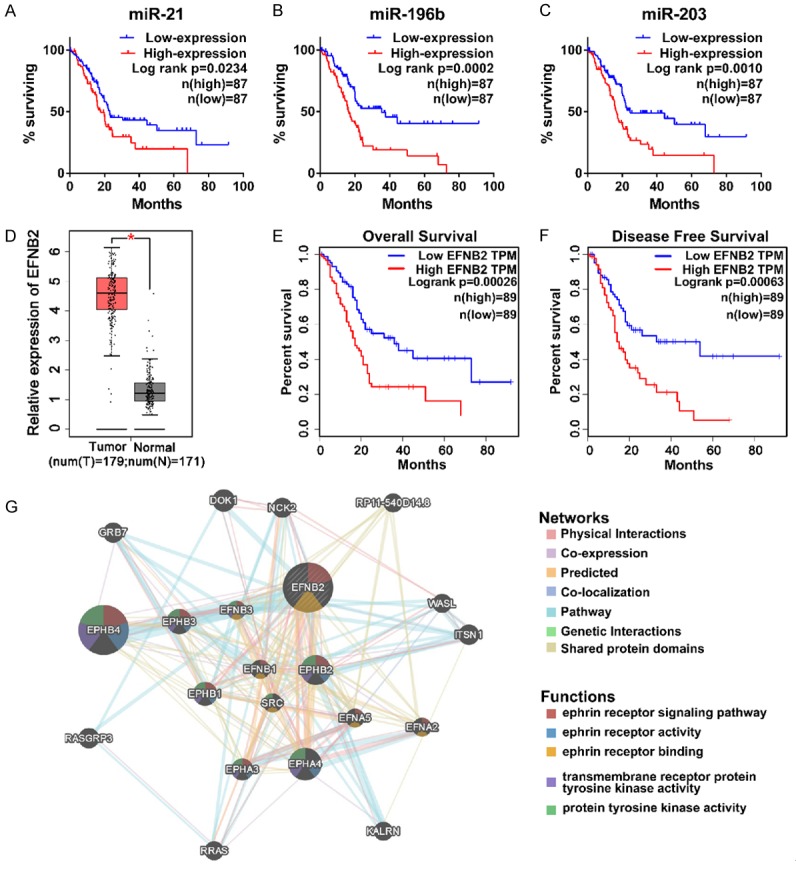
Screening of potential indicators associated with prognosis in PDAC. A-C. The overall survival curves of miR-21 (P=0.0234), miR-196b (P=0.0002) and miR-203 (P=0.0010). Patients were divided into low- and high-expression group according to median value. Both low- and high-expression groups contain 87 patients with PDAC. D. The expression level of EFNB2 between PDAC samples (n=179) and normal pancreatic samples (n=171). P<0.05. E, F. The overall survival and disease free survival curves of EFNB2 (P=0.00026 and P=0.00063, respectively). Both low- and high-expression groups contain 89 patients with PDAC. TPM, transcripts per million. G. The function of EFNB2 and relevant networks with other genes were predicted by geneMANIA.
GEPIA, a web-based tool for data mining based on TCGA and GTEx data, was applied to investigate prognostic value of 58 genes in patients with PDAC. As shown in Table 3, a total of 20 genes were associated with prognosis in patients with PDAC. Among them, either MXRA5, EPYC, ECT2, COL12A1, SLC6A14 high expression groups or SLC7A2 low expression group had observably poorer OS, while their expression levels might not affect the patients’ disease-free survival (DFS). Either downregulation of BTG2, PDK4 and CTNND2 or upregulation of NRP2, PXDN and CD109 were remarkably associated with shorter DFS time and earlier relapsed, while had no significant correlation with OS. The remaining eight genes (TGFBI, LRRN1, ITGA2, DKK1, GREM1, EFNB2, SEMA3C and NT5E) were upregulated in PDAC patients, and were significantly associated with OS and DFS.
Table 3.
Expression levels and prognosis-related P values of 20 genes in patients with PDAC
| Genes | Expression | P value | |
|---|---|---|---|
|
| |||
| Overall survival | Disease-free survival | ||
| MXRA5 | Upregulated | 0.039* | 0.14 |
| EPYC | Upregulated | 0.0029** | 0.87 |
| ECT2 | Upregulated | 0.005** | 0.12 |
| COL12A1 | Upregulated | 0.0055** | 0.057 |
| SLC6A14 | Upregulated | 0.0081** | 0.36 |
| SLC7A2 | Downregulated | 0.027* | 0.48 |
| BTG2 | Downregulated | 0.83 | 0.0084** |
| PDK4 | Downregulated | 0.34 | 0.0093** |
| CTNND2 | Downregulated | 0.15 | 0.029* |
| NRP2 | Upregulated | 0.07 | 0.014* |
| PXDN | Upregulated | 0.38 | 0.026* |
| CD109 | Upregulated | 0.11 | 0.039* |
| TGFBI | Upregulated | 0.047* | 0.033* |
| LRRN1 | Upregulated | 0.032* | 0.021* |
| ITGA2 | Upregulated | 0.0015** | 0.0039** |
| DKK1 | Upregulated | 0.0044** | 0.0014** |
| GREM1 | Upregulated | 0.026* | 0.029* |
| EFNB2 | Upregulated | 0.00026*** | 0.00063*** |
| SEMA3C | Upregulated | 0.024* | 0.0052** |
| NT5E | Upregulated | 0.023* | 0.00046*** |
P<0.05;
P<0.01;
P<0.001.
More importantly, Kaplan-Meier survival analysis demonstrated that EFNB2 had the extremely low log-rank P values in both OS and DFS analysis (P=0.00026 and P=0.00063, respectively) (Figure 3D-F), which caught the attention of our research team. We used geneMANIA to predict the function of EFNB2 and relevant networks with other genes (Figure 3G). We found that EFNB2 was related to ephrin receptor signaling pathway, and physical interacted and coexpressed with ephrin family (EPHB4, EPHA4, EPHB3 and EPHB2), which were implicated in the initiation and progression of cancer [26-28].
Data mining of EFNB2 from public databases
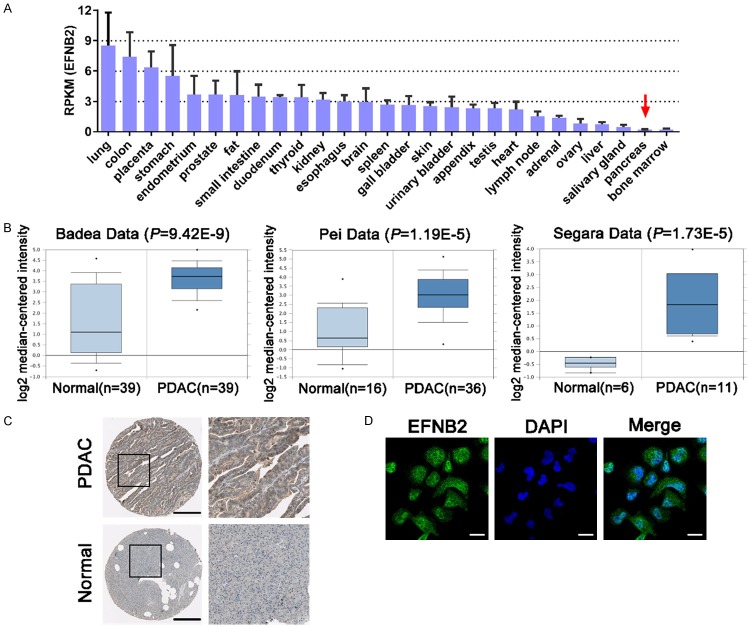
The data from NCBI Gene database revealed that the basic expression level of EFNB2 in normal pancreas was much lower than that in other organs or tissues (Figure 4A). Badea, Pei and Segara’s data indicated that the expression level of EFNB2 was elevated in PDAC compared with normal pancreatic tissues through employing the Oncomine (Figure 4B). Consistently, immunohistochemistry staining demonstrated that EFNB2 was positively expressed in PDAC tissues and negatively expressed in normal pancreatic tissues (Figure 4C). In addition, immunofluorescent analysis manifested that EFNB2 distributed in the nucleoplasm and cytosol by utilizing the Human Protein Atlas (Figure 4D). Taken together, the above results suggested that EFNB2 might play a crucial role in PDAC progression.
Figure 4.
Data mining of EFNB2 from public databases. A. The basic expression level of EFNB2 in different organs or tissues. RPKM, reads per kilobase per million reads. B. The expression level of EFNB2 between PDAC tissues and normal pancreatic tissues from the Oncomine. C. Immunohistochemistry staining of EFNB2 between PDAC samples and normal pancreatic samples. Black bar, 200 μm. D. Localization of EFNB2 in cells by immunofluorescent analysis. White bar, 20 μm.
Inhibition of EFNB2 impairs cell proliferation, migration and invasion
To demonstrate the expression of EFNB2 in PDAC patients, GSE28735 was used as an example, indicating that the expression of EFNB2 was significantly elevated in PDAC tissues compared with paracancerous tissues (Figure 5A). Furthermore, it was found that the expression of EFNB2 was markedly enhanced in the PDAC cell lines (PANC-1, BxPC-3 and SW1990) compared with the HPDE6 (Figure 5B). Due to the expression level of EFNB2 in SW1990 was the highest, we chose it for the next experiment. In order to ascertain the effects of EFNB2 on PDAC cells, we transfected SW1990 cells with specific siRNA to reduce its expression (Figure 5C). CCK-8 assays revealed that the proliferation rate of cells was notably impaired after downregulation of EFNB2 (Figure 5D). The wound healing assays showed that migration rate of cells was observably attenuated in si-EFNB2 group compared with negative control group (Figure 5E). Transwell invasion assays turned out that depletion of EFNB2 suppressed the invasive ability of cells (Figure 5F).
Figure 5.
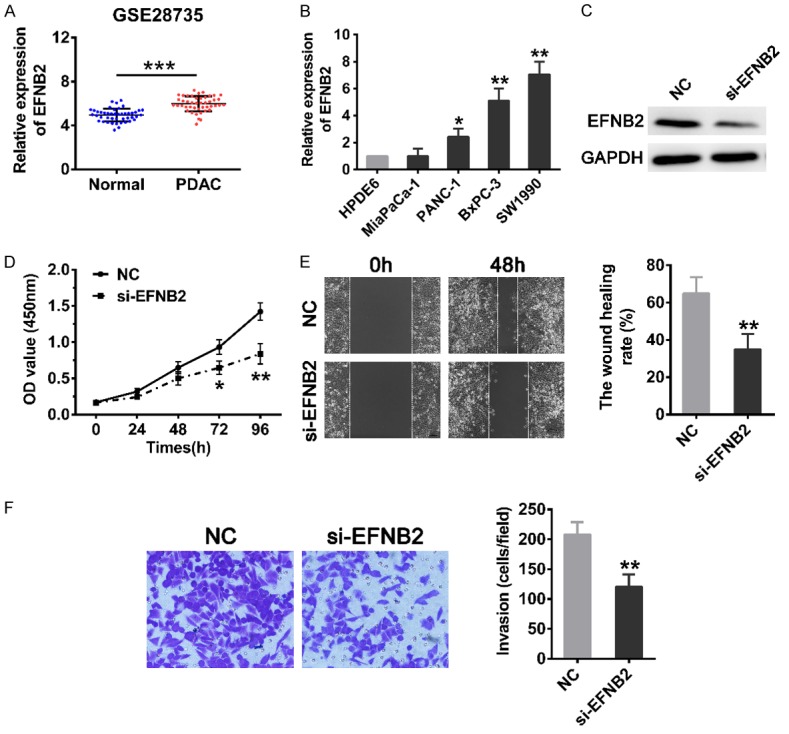
Inhibition of EFNB2 impairs cell proliferation, migration and invasion. A. The expression of EFNB2 between PDAC tissues (n=45) and matched normal tissues (n=45) in GSE28735. B. The expression level of EFNB2 in human pancreatic ductal epithelial cells (HPDE6) and PDAC cells (MiaPaCa-2, PANC-1, BxPC-3 and SW1990) were assessed by qRT-PCR. C. The protein level of EFNB2 in SW1990 cells after transfection of siRNA was examined by Western blot analysis. D. CCK-8 assays were carried out to compare the proliferation ability of SW1990 cells transfected with NC or siRNA. E. Wound healing assays were utilized to measure the migration ability of SW1990 cells transfected with NC or siRNA (magnification: 100×). Cells were photographed at 0 and 48 h after wounding. F. Transwell invasion assays were used to detected the invasion ability of cells. The invasive cells were counted from five random areas at 200× magnification. Mean ± SD, *P<0.05, **P<0.01, ***P<0.001.
MiR-557 inhibited EFNB2 expression by directly binding to its 3’ UTR
From the interaction network in Figure 1D, we found that EFNB2 had two putative miRNAs that could regulate it, namely miR-222 and miR-557. Because miR-222 was markedly increased in PDAC and had no negative relationship with the expression of EFNB2, we chose the miR-557 whose expression was significantly decreased in PDAC tissues to explore (Figure 6A and 6B). Meanwhile, the miR-557 expressionwas dramatically downregulated in all four PDAC cell lines compared to that of HPDE6 (Figure 6C). We transfected SW1990 cells with miR-557 mimics to augment its expression (Figure 6D). The 3’ UTR of EFNB2 mRNA contained putative binding site for miR-557 which located at 464-470 nt was predicted by TargetScan (Figure 6E). The SW1990 cells were co-transfected with the miR-557 mimics or miR-NCs and wild-type (WT) or mutant-type (MUT) 3’ UTR of EFNB2 luciferase plasmid. The luciferase reporter assay showed that luciferase activity of EFNB2 mRNA with WT 3’ UTR was prominently inhibited after transfecting with miR-557 mimics, but this effect was not detected in EFNB2 with the MUT 3’ UTR (Figure 6F). Furthermore, western blotting analysis discovered that overexpression of miR-557 significantly mitigated the protein level of EFNB2 in PDAC cells (Figure 6G). These data suggested that miR-557 can directly target EFNB2 through its 3’ UTR.
Figure 6.
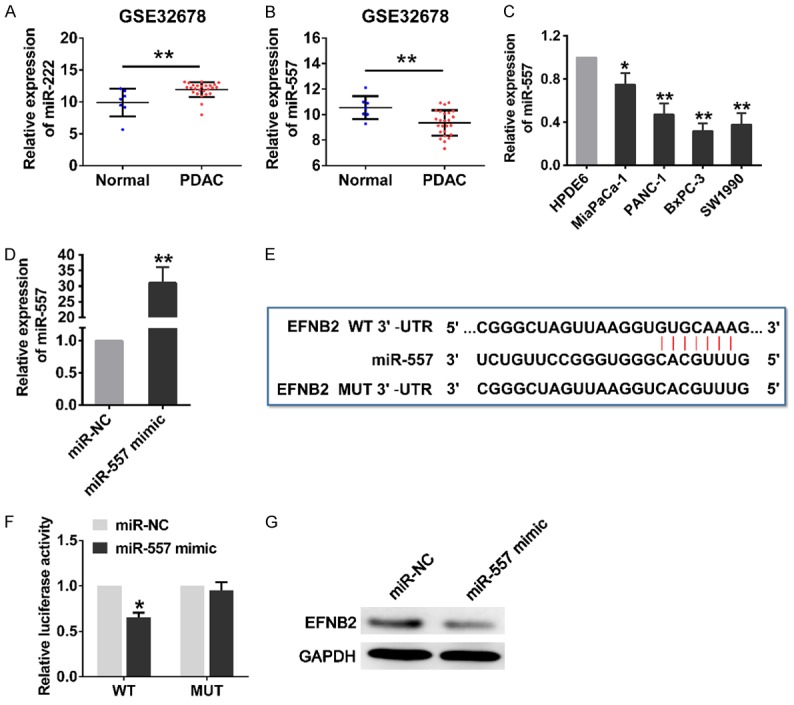
MiR-557 inhibited EFNB2 expression by directly binding to its 3’ UTR. A, B. The expression level of miR-222 and miR-557 between PDAC tissues (n=25) and normal pancreatic tissues (n=7) from GSE32678. C. The relative expression level of miR-557 in HPDE6 and PDAC cell lines (MiaPaCa-2, PANC-1, BxPC-3 and SW1990). D. The expression level of miR-557 was detected for 48 h in SW1990 cells transfected with miR-557 mimics or miR-NCs by qRT-PCR. E. EFNB2 mRNA contains a putative miR-557 binding region, located at 464-470 nt of 3’ UTR, and the mutant type with 7 altered nucleotides. F. Luciferase activity assays with the wild-type 3’ UTR or mutated 3’ UTR of EFNB2 co-transfected with miR-NCs or miR-557 mimics. G. The endogenous expression level of EFNB2 were determined by western blotting after the overexpression of miR-557. WT, wild type; MUT, mutant type; UTR, untranslated region. Mean ± SD, *P<0.05, **P<0.01.
Discussion
In the present study, a novel of miRNA-mRNA regulatory network including 7 significant miRNAs, 58 target genes and 80 miRNA-mRNA interactions was constructed after integrating and screening multiple expression profiles of miRNAs and genes. KEGG pathway analysis revealed that the significant miRNAs involved in the classical signaling pathway based on the putative target genes, such as ECM-receptor interaction, focal adhesion, PI3K-Akt signaling pathway. ECM acts as a key regulator factor in terms of cellular differentiation, proliferationand migration, its alterations can accelerate cancer development and progression [29].Focal adhesion signaling hubsare constitutive of multifarious molecules including integrins and growth factor receptors, regulating cellbehavior and affecting cancer cell survival [30].The PI3K-Akt pathway is one of major signaling pathway participated in the oncogenesis of many kinds of cancers, including PDAC [31]. It was indicated that screened DEMs might play an important role in PDAC.
To corroborate the results of bioinformatics analysis, we analyzed the prognostic value of screened miRNA and genes in patients with PDAC by OncoLnc and GEPIA. Consequently, 3 miRNAs (miR-21, miR-196b and miR-203) and 20 genes (MXRA5, EPYC, ECT2, COL12A1, SLC6A14, SLC7A2, BTG2, PDK4, CTNND2, NRP2, PXDN, CD109, TGFBI, LRRN1, ITGA2, DKK1, GREM1, EFNB2, SEMA3C and NT5E) were screened out. Earlier studies indicated that the expression of miR-21, miR-196b and miR-203 are enhanced in pancreatic cancer, promoting cell proliferation, migration and invasion of pancreatic cancer, and significantly associated with the overall survival of patients with PDAC [32-34]. Furthermore, many of the above-listed genes are involved in the initiation and progression of pancreatic cancer. For example, DKK1 acted as a negative regulator of Wnt signaling and might serve as a useful diagnostic predictor for pancreatic cancer [35]. The high expression of SEMA3C could facilitate tumorigenesis and metastasis by activating extracellular signal-regulated kinase1/2 signaling pathway [36]. Taken together, these findings are coincide with our results analyzed by bioinformatics methods, augmenting the validity and believability of our results in a way.
EFNB2 is a membrane-anchored ligand and belongs to the largest family of receptor tyrosine kinase. In neoplasms, EFNB2 was participated in neovascularization by promoting vascular endothelial precursor cell adhered to thespecific site of tumor, and lymphangiogenesis by inducing vascular endothelial growth factor receptor [37,38]. Besides, several studies have indicated that EFNB2 can promote cell migration, invasion and angiogenesis in glioma and melanoma [39,40], and is a poor prognostic indicator in ovarian cancer and esophageal squamous cell carcinoma [41,42]. Alam and colleagues found that EFNB2 knockdown suppressed colorectal cancer cell growth, reversedmalignant phenotype and impaired drug resistance [43]. Sasabe and colleagues showed that the upregulated EFNB2 could activate the ephrin-B2 reverse signaling pathway in microenvironment, and promote cancer progression by interaction with surrounding cells and reinforcement of malignant potential inoral squamous cell carcinoma [44]. However, the research work focused on the role of EFNB2 in PDAC was very limited, and remained to be illustrated. Our findings demonstrated that the basic expression of EFNB2 was at an extremely low level in normal pancreas relative to other organs or tissues. However, when normal pancreatic epithelial tissues transformed into cancerous tissues, the expression level of EFNB2 was remarkably enhanced in PDAC tissues, which confirmed by different public databases. Moreover, deficiency of EFNB2 dramatically dampened cell proliferation, migration and invasion in PDAC cells through cellular function assays. It was indicated that EFNB2 functioned as an oncogene and consistent with the roles of EFNB2 in other cancers.
To vertify the miRNA-mRNA regulatory network, we explored the regulatory relationship between miR-557 and EFNB2. Razaviyan and workmates found that the expression of miR-557 was downregulated and might exert its function by targeting the S6K1 gene in triple-negative breast cancer [45]. Qiu and partners discovered that miR-557 acted as a tumor suppressor via negatively adjusting the expression of LEF1 in lung cancer [46]. In this study, we found that the expression of miR-557 was also notably downregulated in PDAC cells. More than that, luciferase reporter assays confirmed that the overexpression of miR-557 decreased the protein level of EFNB2 by directly targeting the 3’ UTR of EFNB2 in PDAC cells.
In conclusion, we integrated multiple expression profiles of miRNAs and genes from GEO database with functional, pathway enrichment, and survival analysis to identify novel biomarkers among screened DEMs and DEGs in PDAC compared with those in normal controls. Moreover, we constructed a novel miRNA-mRNA regulatory network including 7 miRNAs and 58 significant target genes by comprehensive analysis. This study also indicated that EFNB2 promoted PDAC progression through enhancement of malignant behavior including cell proliferation, migration and invasion, and miR-557 might play a tumor suppressor role in PADC by targeting EFNB2. It is worth to further investigate their clinical significance and molecular mechanism for the pathophysiology and carcinogenesis of PDAC, and may be the promising targets for diagnosis and treatment of PDAC.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China [No. 81572388].
Disclosure of conflict of interest
None.
Abbreviations
- EFNB2
ephrin B2
- PDAC
pancreatic ductal adenocarcinoma
- DEGs
differentially expressed genes
- DEMs
differentially expressed miRNAs
- UTR
untranslated region
- BP
biological process; CC, cell component
- MF
molecular function
- PPI
protein-protein interactions
- HPDE6
human pancreatic ductal epithelial cells
- NC
negative control
- OS
overall survival
- FS
disease-free survival
- ECM
extracellular matrix
- RPKM
reads per kilobase per million reads
- TPM
transcripts per million
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 3.Yonemori K, Kurahara H, Maemura K, Natsugoe S. MicroRNA in pancreatic cancer. J Hum Genet. 2017;62:33–40. doi: 10.1038/jhg.2016.59. [DOI] [PubMed] [Google Scholar]
- 4.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 6.Chrystoja CC, Diamandis EP, Brand R, Ruckert F, Haun R, Molina R. Pancreatic cancer. Clin Chem. 2013;59:41–46. doi: 10.1373/clinchem.2012.196642. [DOI] [PubMed] [Google Scholar]
- 7.Wagner M, Redaelli C, Lietz M, Seiler CA, Friess H, Buchler MW. Curative resection is the single most important factor determining outcome in patients with pancreatic adenocarcinoma. Br J Surg. 2004;91:586–594. doi: 10.1002/bjs.4484. [DOI] [PubMed] [Google Scholar]
- 8.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 9.Li MH, Fu SB, Xiao HS. Genome-wide analysis of microRNA and mRNA expression signatures in cancer. Acta Pharmacol Sin. 2015;36:1200–1211. doi: 10.1038/aps.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 12.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao X, Sherman BT, Huang da W, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVIDWS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science. 2016;2:e67. [Google Scholar]
- 19.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlen M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnstrom H, Glimelius B, Sjoblom T, Edqvist PH, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang R, Luo G, Ai K. Long noncoding RNA SNHG1 promotes cell proliferation through PI3K/AKT signaling pathway in pancreatic ductal adenocarcinoma. J Cancer. 2018;9:2713–2722. doi: 10.7150/jca.26207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma C, Nong K, Wu B, Dong B, Bai Y, Zhu H, Wang W, Huang X, Yuan Z, Ai K. miR-212 promotes pancreatic cancer cell growth and invasion by targeting the hedgehog signaling pathway receptor patched-1. J Exp Clin Cancer Res. 2014;33:54. doi: 10.1186/1756-9966-33-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noren NK, Foos G, Hauser CA, Pasquale EB. The EphB4 receptor suppresses breast cancer cell tumorigenicity through an Abl-Crk pathway. Nat Cell Biol. 2006;8:815–825. doi: 10.1038/ncb1438. [DOI] [PubMed] [Google Scholar]
- 27.Saintigny P, Peng S, Zhang L, Sen B, Wistuba II, Lippman SM, Girard L, Minna JD, Heymach JV, Johnson FM. Global evaluation of Eph receptors and ephrins in lung adenocarcinomas identifies EphA4 as an inhibitor of cell migration and invasion. Mol Cancer Ther. 2012;11:2021–2032. doi: 10.1158/1535-7163.MCT-12-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu ST, Chang KJ, Ting CH, Shen HC, Li H, Hsieh FJ. Over-expression of EphB3 enhances cell-cell contacts and suppresses tumor growth in HT-29 human colon cancer cells. Carcinogenesis. 2009;30:1475–1486. doi: 10.1093/carcin/bgp133. [DOI] [PubMed] [Google Scholar]
- 29.Venning FA, Wullkopf L, Erler JT. Targeting ECM disrupts cancer progression. Front Oncol. 2015;5:224. doi: 10.3389/fonc.2015.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Furukawa T. Molecular targeting therapy for pancreatic cancer: current knowledge and perspectives from bench to bedside. J Gastroenterol. 2008;43:905–911. doi: 10.1007/s00535-008-2226-1. [DOI] [PubMed] [Google Scholar]
- 32.Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, Takahata S, Toma H, Nagai E, Tanaka M. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8:1067–1074. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 33.Kanno S, Nosho K, Ishigami K, Yamamoto I, Koide H, Kurihara H, Mitsuhashi K, Shitani M, Motoya M, Sasaki S, Tanuma T, Maguchi H, Hasegawa T, Kimura Y, Takemasa I, Shinomura Y, Nakase H. MicroRNA-196b is an independent prognostic biomarker in patients with pancreatic cancer. Carcinogenesis. 2017;38:425–431. doi: 10.1093/carcin/bgx013. [DOI] [PubMed] [Google Scholar]
- 34.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 35.Han SX, Zhou X, Sui X, He CC, Cai MJ, Ma JL, Zhang YY, Zhou CY, Ma CX, Varela-Ramirez A, Zhu Q. Serum dickkopf-1 is a novel serological biomarker for the diagnosis and prognosis of pancreatic cancer. Oncotarget. 2015;6:19907–19917. doi: 10.18632/oncotarget.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, Zhao Z, Guo S, Li J, Liu S, You Y, Ni B, Wang H, Bie P. Increased semaphorin 3c expression promotes tumor growth and metastasis in pancreatic ductal adenocarcinoma by activating the ERK1/2 signaling pathway. Cancer Lett. 2017;397:12–22. doi: 10.1016/j.canlet.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 37.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, Adams S, Davy A, Deutsch U, Luthi U, Barberis A, Benjamin LE, Makinen T, Nobes CD, Adams RH. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 39.Nakada M, Anderson EM, Demuth T, Nakada S, Reavie LB, Drake KL, Hoelzinger DB, Berens ME. The phosphorylation of ephrin-B2 ligand promotes glioma cell migration and invasion. Int J Cancer. 2010;126:1155–1165. doi: 10.1002/ijc.24849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer S, Hafner C, Guba M, Flegel S, Geissler EK, Becker B, Koehl GE, Orso E, Landthaler M, Vogt T. Ephrin-B2 overexpression enhances integrin-mediated ECM-attachment and migration of B16 melanoma cells. Int J Oncol. 2005;27:1197–1206. [PubMed] [Google Scholar]
- 41.Castellvi J, Garcia A, de la Torre J, Hernandez J, Gil A, Xercavins J, Ramón y Cajal S. Ephrin B expression in epithelial ovarian neoplasms correlates with tumor differentiation and angiogenesis. Hum Pathol. 2006;37:883–889. doi: 10.1016/j.humpath.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 42.Tachibana M, Tonomoto Y, Hyakudomi R, Hyakudomi M, Hattori S, Ueda S, Kinugasa S, Yoshimura H. Expression and prognostic significance of EFNB2 and EphB4 genes in patients with oesophageal squamous cell carcinoma. Dig Liver Dis. 2007;39:725–732. doi: 10.1016/j.dld.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 43.Alam SK, Yadav VK, Bajaj S, Datta A, Dutta SK, Bhattacharyya M, Bhattacharya S, Debnath S, Roy S, Boardman LA, Smyrk TC, Molina JR, Chakrabarti S, Chowdhury S, Mukhopadhyay D, Roychoudhury S. DNA damage-induced ephrin-B2 reverse signaling promotes chemoresistance and drives EMT in colorectal carcinoma harboring mutant p53. Cell Death Differ. 2016;23:707–722. doi: 10.1038/cdd.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasabe E, Tomomura A, Tomita R, Sento S, Kitamura N, Yamamoto T. Ephrin-B2 reverse signaling regulates progression and lymph node metastasis of oral squamous cell carcinoma. PLoS One. 2017;12:e0188965. doi: 10.1371/journal.pone.0188965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Razaviyan J, Hadavi R, Tavakoli R, Kamani F, Paknejad M, Mohammadi-Yeganeh S. Expression of miRNAs targeting mTOR and S6K1 genes of mTOR signaling pathway including miR-96, miR-557, and miR-3182 in triple-negative breast cancer. Appl Biochem Biotechnol. 2018;186:1074–1089. doi: 10.1007/s12010-018-2773-8. [DOI] [PubMed] [Google Scholar]
- 46.Qiu J, Hao Y, Huang S, Ma Y, Li X, Li D, Mao Y. MiR-557 works as a tumor suppressor in human lung cancers by negatively regulating LEF1 expression. Tumour Biol. 2017;39:1010428317709467. doi: 10.1177/1010428317709467. [DOI] [PubMed] [Google Scholar]