Abstract
Cross-Linking Immunoprecipitation associated to high-throughput sequencing (CLIP-seq) is a technique used to identify RNA directly bound to RNA-binding proteins across the entire transcriptome in cell or tissue samples. Recent technological and computational advances permit the analysis of many CLIP-seq samples simultaneously, allowing us to reveal the comprehensive network of RNA–protein interaction and to integrate it to other genome-wide analyses. Therefore, the design and quality management of the CLIP-seq analyses are of critical importance to extract clean and biological meaningful information from CLIP-seq experiments. The application of CLIP-seq technique to Argonaute 2 (Ago2) protein, the main component of the microRNA (miRNA)-induced silencing complex, reveals the direct binding sites of miRNAs, thus providing insightful information about the role played by miRNA(s). In this review, we summarize and discuss the most recent computational methods for CLIP-seq analysis, and discuss their impact on Ago2/miRNA-binding site identification and prediction with a regard toward human pathologies.
Keywords: RNA immunoprecipitation; bioinformatics workflow, computational guideline, large-scale analysis, quality management, microRNAs, human pathologies
Introduction
RNA-binding proteins (RBPs) bind RNAs to regulate their fate, function, localization or secondary structure [1] to ultimately modulate many biological processes including cell apoptosis, growth, fate and differentiation [2–4]. RBPs possess modular structure composed by at least one domain to directly bind either single- or double-stranded RNA, such as the RNA recognition motif, the zinc-finger domain, the KH domain and the double-stranded RNA-binding domain [5]. RNA-binding domains recognize primarily the RNA sequence, the RNA shape or both [6]. Because of the high versatility of the RNA-binding domains and the presence of some of them in each RBP [7], the full understanding of the RBP mode of binding is a challenging quest. To comprehensively uncover the RNA–protein interactions network in a genome-wide manner, Cross-Linking ImmunoPrecipitation associated to high-throughput sequencing (CLIP-seq) was recently developed [8]. Nowadays, CLIP-seq analysis has become one of the mainstream method to study RNA metabolism and has led to important discoveries in different fields of molecular and cellular biology [9–14]. Of particular interest for the community of RNA biologists and beyond is the application of CLIP-seq methods to Argonaute 2 (Ago2) protein, which together with microRNAs (miRNAs) form the miRNA-induced silencing complex (miRISC) [15]. miRISC targets mRNAs through partial sequence complementarity between the miRNA and the target sequence, promoting degradation or translation inhibition of the target mRNA [16]. miRNAs regulate many biological processes, including cell proliferation, differentiation and death in both physiological and pathological events [16]. Because CLIP-seq analysis of Ago2 is meant to identify the precise binding sites of miRNAs [9, 12, 17] but not of the protein itself, the downstream and validation steps of the analysis are different from those performed for other RBPs: binding features of miRNAs follow different rules to that of RBPs. On the other hand, if we consider each miRNA loaded into Ago2 as a different RBP [18], Ago2 CLIP-seq may be taken as universal example of the mode of binding to cognate RNAs.
CLIP-seq protocol has many steps involving sample preparation, sequencing and bioinformatics analysis. Briefly, RNA of ultraviolet cross-linked cells or tissue lysates is partially digested by enzymatic reaction into small fragments of about 50–100 nucleotides (nts), the RBP of interest is immunoprecipitated and the RBP-RNA complex is isolated by sodium dodecyl sulfate polyacrylamide gel electrophoresis migration. Afterward, RNA from the RBP-RNA complex is recovered by acid phenol/chloroform extraction, RNA adapters are ligated, RNA template is reverse transcribed and finally high-throughput sequenced. Several protocol variants for sample preparation have been proposed, which mainly include the most popular HIgh-Throughput Sequencing of RNA isolated by CLIP (HITS-CLIP) [9, 10], the PhotoActivable-Ribonucleoside-enhanced-CLIP (PAR-CLIP) [11–13] and the individual-nucleotide resolution CLIP (iCLIP) [19]. Detailed differences among these three variants and other more recent protocols have been previously discussed by others [20, 21]. An additional variant of the CLIP-seq, named CLASH, has been developed by Helwak et al. [22], and it is particularly appropriate to Ago2 CLIP-seq. Briefly, CLASH allows high-throughput mapping of small RNA::RNA-binding interaction by adding an intramolecular RNA ligation step during the sample preparation. CLASH approach and analysis have been previously reviewed by Broughton and Pasquinelli [23].
After high-throughput sequencing, the bioinformatics analysis workflow starts by the preprocessing to filter out the low quality and duplicate reads, and to map them onto the genome or the transcriptome of reference. Afterward, to assess real signal over the noise background, the reads are processed by peak-calling programs. Called peaks are further analyzed for functional, structural and biochemical characterizations of the RNA–protein interaction, including motif discovery, expression profile and gene ontology.
Importantly, because recent advances in sequencing technologies and bioinformatics analyses enable us to handle many CLIP-seq samples simultaneously, it is important to optimize a bioinformatic pipeline that can facilitate the work of researchers in obtaining unbiased and high-quality data. Despite great efforts from researchers to streamline CLIP-seq analysis, much remains to be improved on both experimental (e.g. the quality of the antibody used for the immunoprecipitation) and computational procedures. In fact, because of the complexity of the large data set coming from high-throughput technologies such as CLIP-seq, a correct interpretation of the data is facilitated by a proper and an accurate data analysis with refined and optimized computational tools. In this review, we describe the computational protocol for CLIP-seq analysis, discuss the latest bioinformatics developments for data processing and mining and provide advices for data analysis interpretation. We discuss the validity and the limitations of emerging programs for each step of the CLIP-seq analysis and the quality measurements currently available for specific tasks, by providing concrete examples on a case study: an in-house Ago2 HITS-CLIP data set generated from stem cells [24] (GEO accession: GSE85219). For simplicity and limited space, we focus the scope of this review in providing a valuable computational guideline for the bioinformatics analysis of the three main variants of CLIP-seq analysis, namely, HITS-CLIP, PAR-CLIP and iCLIP. To help the readers, we provide in Supplementary Table S1 the Web links to download all the programs cited in the present review. Finally, we discuss how Ago2 CLIP-seq analyses have improved the miRNA-binding site prediction and the understanding of miRNA function in human pathologies.
Bioinformatics workflow for CLIP-seq analysis
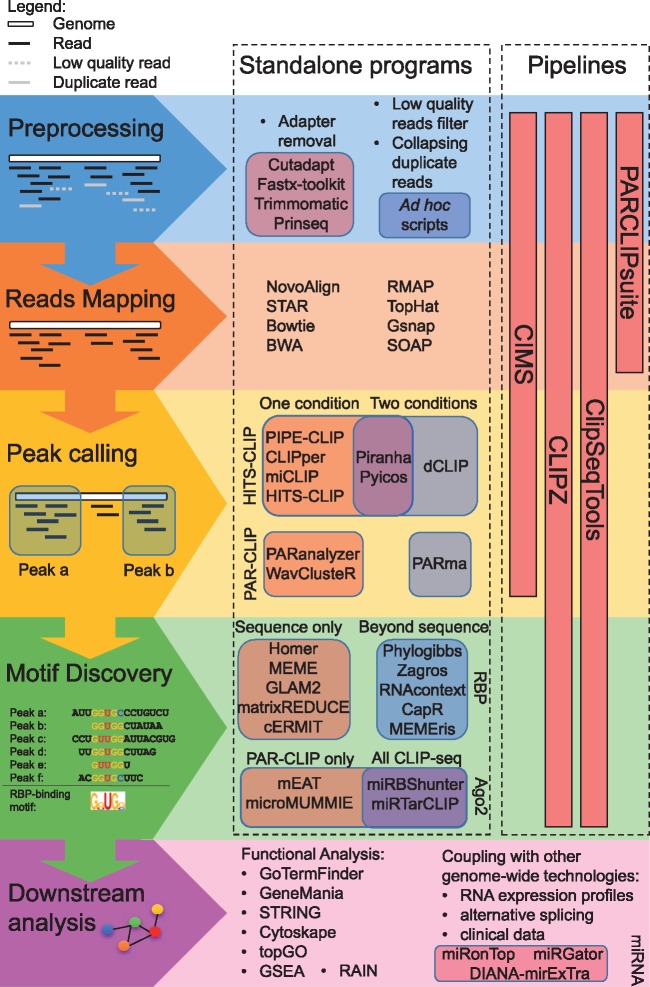
In Figure 1, we have summarized the main computational steps for CLIP-seq analysis. Recently, few computational pipelines have been developed such as CIMS [25], CLIPSeqTools [26], CLIPZ [27] and PARCLIPsuite [11], which provide useful resources to deal with preprocessing steps and some of the main steps of the analysis, including peak-calling procedure. In the following sections, we describe step-by-step the bioinformatics workflow of the CLIP-seq analysis giving a quality overview of existing software and providing practical examples on an in-house data set for Ago2 HITS-CLIP experiments on P19 stem cells [24].
Figure 1.
Main steps of the bioinformatics workflow to analyze CLIP-seq data with the main software or pipeline to use.
Preprocessing and read mapping onto the reference genome
The first step of the analysis is the preprocessing that involves adapter removal, filtering raw data according to read quality scores and collapsing reads with the exact sequence. While for the adapter removal, specific programs have been developed such as cutadapt [28] or Trimmomatic [29], for the quality filtering, usually bioinformaticians develop ad hoc scripts. However, lately few programs have been developed, such as FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and PRINSEQ [30]. To quantify the differences of the strategies used by different preprocessing programs to filter reads based on quality scores and collapse duplicated reads, we applied FASTX-Toolkit, PRINSEQ and the CIMS pipelines to a published in-house Ago2 CLIP-seq data set from mouse P19 stem cells [24] (GEO accession: GSE85219). This analysis was run with the default tuning or the recommended parameters (see Supplemental Information). While some programs have tunable parameters, we forgo parameter optimization, which might have improved the results for some data sets, as this task may be beyond the ken of most users. The highest number of reads that survive the preprocessing step for the three replicates was obtained using PRINSEQ (Table 1). To inspect the quality of the reads after the preprocessing, we used the program FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ developed by S. Andrews). As shown in Supplementary Figure S1, although FASTX-Toolkit yielded reads with best quality score per sequence and per base, this program filtered out much more reads compared with the other two programs (Table 1). These data indicate that FASTX-Toolkit is stringent to select high-quality reads. Altogether, PRINSEQ showed the best balance between quality and amount of reads, suggesting that this program uses a strategy that fits better for CLIP-seq data preprocessing than the other two programs. However, the balance of stringency and sensitivity can be tuned by changing the parameters of the programs to meet the needs of the researcher, such as the minimal quality of reads or their minimal length to be selected. Finally, the three methods achieved similar results regarding the collapse of duplicates, to remove redundant fractions of the data. A high redundancy of the data may be a consequence of low library complexity, which often occurs when samples are prepared from small amount of starting material or by performing too many polymerase chain reaction cycles for library construction.
Table 1.
Number of reads obtained using different preprocessing and mapping tools on the in-house Ago2 HITS-CLIP data set from P19 stem cells
| # of reads before preprocessing | Preprocessing programs | # of reads after preprocessing | Mapping tools | # of reads after mapping |
||
|---|---|---|---|---|---|---|
| Unique mapping (%) | Multiple mapping (%) | No mapping (%) | ||||
| Replicate 1 | ||||||
| 38 865 698 | PRINSEQ | 3 783 764 | Novoalign | 2 120 447 (56.0) | 1 300 950 (34.4) | 34 565 (0.9) |
| STAR | 1 142 926 (30.21) | 1 579 235 (41.74) | 1 061 603 (28.05) | |||
| FASTX-Toolkit | 933 985 | Novoalign | 596 192 (63.8) | 315 845 (33.8) | 7 004 (0.7) | |
| STAR | 372 062 (39.84) | 472 457 (50.59) | 89 466 (9.58) | |||
| CIMS | 2 467 666 | Novoalign | 1 355 710 (54.9) | 784 179 (31.8) | 25 516 (1.0) | |
| STAR | 787 349 (31.91) | 1 297 371 (52.57) | 382 946 (15.51) | |||
| Replicate 2 | ||||||
| 34.094.384 | PRINSEQ | 3 924 075 | Novoalign | 2 196 430 (56.0) | 1 321 658 (33.7) | 34 607 (0.9) |
| STAR | 1 311 574 (33.42) | 2 036 793 (51.91) | 575 708 (14.67) | |||
| fastXtoolkit | 957 572 | Novoalign | 634 824 (65.1) | 316 995% (32.1) | 6 529 (0.7) | |
| STAR | 423 300 (43.39) | 461 865 (47.34) | 72 407 (7.42) | |||
| CIMS | 2 584 470 | Novoalign | 1 425 553 (55.2) | 801 740 (31.0) | 25 557 (1.0) | |
| STAR | 871 764 (33.73) | 1 314 533 (50.86) | 398 173 (15.41) | |||
| Replicate 3 | ||||||
| 32.904.107 | PRINSEQ | 3 668 439 | Novoalign | 2 001 844 (54.6) | 1 264 702 (34.5) | 35 261 (1.0) |
| STAR | 1 152 648 (31.42) | 1 956 392 (53.33) | 559 399 (15.25) | |||
| fastXtoolkit | 888 186 | Novoalign | 562 070 (63.3) | 300 647 (33.8) | 6 308 (0.7) | |
| STAR | 358 485 (40.36) | 442 132 (49.78) | 87 569 (9.86) | |||
| CIMS | 2 359 084 | Novoalign | 1 264 546 (53.6) | 750 678 (31.8) | 25 402 (1.1) | |
| STAR | 740 554 (31.39) | 1 242 835 (52.68) | 375 695 (15.92) | |||
Reads that survive the preprocessing steps are mapped onto the reference sequences that can be the complete genome, the transcriptome or sequences belonging to specific categories, such as 3ʹ untranslated regions (UTRs), long noncoding RNAs, small RNAs, etc. The most common algorithms used to perform this task are Novoalign (http://www.novocraft.com/products/novoalign/), STAR [31], Bowtie [32], RMAP [33], TopHat [34], Gsnap [35], SOAP [36] and BWA [37]. A few precautions have to be considered while setting the parameters of these programs to perform a gapped alignment onto the genome. In fact, these parameters permit to map reads that include deletions, mutations or insertions caused by enzymatic errors occurring during the sample preparation. Depending on the sample, it might be important to use adequate parameters to map reads on the exon junctions. In addition, an important issue concerns the consideration of multiple mapped reads (reads that map in many loci). Although allowing for multiple read mapping would increase the number of usable reads and the sensitivity of peak detection, this may also cause an increase of false-positive peaks, as it was also suggested for ChIP-seq analysis [38].
To provide guidelines about the program to use, we ran two popular programs that achieved the best performances on RNA-seq data [39], namely, Novoalign and STAR, on the in-house Ago2 CLIP-seq data set from P19 stem cells after preprocessing with the three aforementioned programs. For this analysis, we set the recommended parameters for Novoalign and used similar ones for STAR (Supplemental Information). As shown in Table 1, regardless the preprocessing program used, Novoalign mapped uniquely between 20 and 30% more reads than STAR. To test whether there is a qualitative difference on the genomic location of the reads mapped by Novoalign and STAR, we divided the genome in bins of 100 nucleotides and counted the number of bins in which reads map by the two programs. As shown in Supplementary Table S2, we found a relative small number of bins in common between the two programs. These data indicate that Novoalign is able to map reads in about 10% more of the genomic locations than STAR. Therefore, we concluded that differences in both quantitative and qualitative mapping performances exist between Novoalign and STAR. Giving that higher numbers of correctly and uniquely mapped reads would have a beneficial effect on the peak-calling step to discriminate real peaks over the background, we would recommend to use Novoalign. However, the full version of Novoalign is not freely downloadable, and the computational analysis can take much longer with the uncomplete free version compared with STAR, which is freely downloadable. An alternative strategy is to map the reads with one program and afterward run the unmapped reads with a second program. Because this strategy was not benchmarked yet, it is unclear whether it is really advantageous. Finally, the mappability of multiple mapped reads can be dealt by tuning the minimal length of reads or using paired-end sequencing [38].
Peak calling
Assessing peaks is a central step of the analysis to determine specific signal over the noise background for the identification of real binding sites. The number of identified peaks increases with the sequencing depth because weaker sites become statistically significant with a greater number of reads [40]. However, the optimal sequencing depth can only be experimentally evaluated, as it depends on the noise background of the antibody [38].
Diverse methods of peak calling have been used by different programs. The most common strategy is to analyze distribution profiles to find clusters of reads that belong to the same peak. This strategy is used by different programs, including PIPE-CLIP [41], Pyicoclip [42], Piranha [43] and CLIPper [44], for all CLIP-seq protocol variants, and WavClusteR [45] and PARanalyzer [46] for only PAR-CLIP data. PIPE-CLIP and Pyicoclip group the reads based on positional overlap, while Piranha, MiCLIP [47] and HITS-CLIP data analyses (http://qbrc.swmed.edu/softwares.html) bin on genomic portions by fixed size. On the other hand, CIMS focuses on the identification of read clusters containing mutated cross-linked nucleotides [25]. To discriminate enriched read clusters over the background, the peak-calling programs use different statistical models. For instance, PIPE-CLIP and Piranha use the zero-truncated negative binomial likelihoods, including also additional covariates to refine the peak detection, such as identification of cross-linked-dependent mutations or transcript abundance. On the other hand, Pyicoclip performs a background estimation implementing the modified false discovery rate procedure to determine which clusters of reads are significantly enriched in a list of genomic regions, by randomly placing the same number of reads within the region and iterating the process many times. MiCLIP [47] and HITS-CLIP data analyses use hidden Markow models (HMMs) to model the spatial dependency of the reads that map in the cluster. Finally, CIMS assesses statistical significance using a permutation-based model. Specific statistical models are used for PAR-CLIP analysis. For instance, PARanalyzer uses a nonparametric kernel-density estimate classifier to identify RNA-protein interaction sites using T to C conversion rate and read density, while wavClusteR uses a two-step algorithm consisting of a nonparametric two-component mixture model and a wavelet-based procedure.
Importantly, different methods can give different results; thus, to help researchers in the choice of the right program for their peak-calling analysis, we recently performed a comprehensive, quantitative and qualitative comparative evaluation of four different publicly available programs for HITS-CLIP peak-calling step, including CIMS, PIPE-CLIP, Piranha and Pyicoclip, on four published Ago2 HITS-CLIP data sets [9, 48, 49] and one in-house Ago2 HITS-CLIP data set generated from P19 stem cells [24]. By tuning the programs in default parameters, we found that Pyicoclip outperformed the other programs in terms of sensitivity, positional accuracy, agreement with TargetScan miRNA-binding site prediction program, specificity and for consistency in finding the same results on different data sets from the same tissue [24]. Nevertheless, depending on the biological question and sample conditions, scientists may need to tune the parameters of the different peak-calling programs to find the best set to perform this task such as the P value and the minimal number of reads to select significant peaks. Alternatively, we suggested to rank the detected peaks according to different statistics, such as number of reads, fold of enrichment over the background or P-value, and apply arbitrarily thresholds according to the desired stringency [24].
Finally, although not always possible, ideally the addition of control conditions, such as knockout or knockdown, or stimulation versus nonstimulation, would significantly improve the accuracy and the quality of the peak-calling results [50, 51]. Accordingly, a few number of programs have been developed to analyze differential CLIP-seq experiments. These programs include the HMM-based model dCLIP [52], Piranha that uses different statistics to model reads distribution allowing the comparison of two different conditions through the addition of covariates, Pyicoenrich [42] that uses a strategy based on the MA plot as for expression profile analysis and PARma [53] that was specifically designed for PAR-CLIP data and uses a probabilistic model for the identification of differential peaks.
Motif discovery and other features
Following the peak calling, the analysis mainly focuses on the characterization of the RBP-RNA interactions, especially looking for possible binding sequence signature(s), using a candidate screening or a de novo motifs identification. For the candidate screening approach, programs like FIMO [54] can be used to screen peak sequences for the identification of known RNA-binding motifs, such as those from the database of Ray et al. [55]. If the user is looking for unknown RNA-binding motifs, a de novo motif identification could be performed. For this task, two main parameters should be calibrated before launching the analysis. The first parameter is the nucleotide length of the motif. Mitchell and Parker [56] recently showed that different RNA-binding domains bind in average a precise number of nucleotides. Thus, the length of the motif can be set according to the domain composition of the protein. The second parameter to take into account is the so-called ‘background sequences’ that can be used as negative template in which it is not expected to contain the enriched motif sequence(s). Two main strategies can be applied to select the appropriate background sequences: (i) to randomly scramble the CLIP-seq peak sequences; (ii) to define a set of sequences not bound by the protein of interest [57]. In the first strategy, a few constraints can be imposed to the background sequences, such as the dinucleotides frequency to avoid underestimation of false-positive rates in RNA prediction. As for the second strategy, instead, it is recommended that the background sequences possess the same size, dinucleotide frequency and the GC content of the target sequences used to perform the analysis. Besides these general options, each program allows to set its own parameters that may depend on the statistical/mathematical model used by the algorithm, including the distribution to model motif sites (i.e. the number of motif expected per sequence) and the threshold/score associated to the statistical model (i.e. P-value, enrichment score and probability). If successful, the final output of the de novo motif discovery analysis would be a list of sequences enriched in the CLIP-seq data set. On motif clusterization [58], these lists of enriched sequences can be represented as position weight matrices or position-specific affinity matrices that visualize the different affinity of the RBP for each sequence. Among the most popular programs for de novo motif discovery, we can cite MEME [59] and Homer [60]. Other programs that may be used are MatrixREDUCE [61], GLAM2 [62] and cERMIT [63]. In addition to the sequence motifs, some programs take into account other sequence parameters, including secondary structure prediction, discovering therefore a structural motif that combines sequence composition with secondary structure. For such analyses, one may consider to use Zagros [64], RNAcontext [65], MEMEris [66], PhyloGibbs [67] and CapR [68]. The identification of the RBP-binding motif(s) permits the prediction of the RBP binding in different transcriptome analyses.
Because Ago2 binds to miRNAs that determine the sequence specificity of the binding to target RNAs, researchers often analyze Ago2 CLIP-seq peaks with prediction programs for miRNA-binding sites, including Targetscan, PITA and Miranda. However, such an approach can mislead to high rates of false-positive and false-negative targets [69, 70]. Moreover, these programs only predict canonical miRNA-binding sites, which are defined by a perfect complementarity match between miRNA seed sequence (between second and eighth nts of miRNA sequence) and the 3ʹ UTR [16], or seed-like motifs allowing one mismatch or 1-nt bulge in the miRNA seed sequence [22, 71]. Lately, few programs have been developed to search for binding sites of highly expressed miRNAs from Ago2-CLIP-seq peaks. These programs mainly look for canonical or seed-like binding sites, such as miRTarClip for all CLIP-seq techniques, which are limited to 3ʹ UTR [72], or microMUMMIE [73] and mEAT [46] that are limited to PAR-CLIP data sets. A similar approach was also adopted by Clark et al. [74]; however, only the precomputed results for miRNAs expressed in 34 CLIP-seq data sets are available, but not the program. Finally, we have recently developed a novel method, called miRBShunter, that uses de novo motif search for an unbiased identification of miRNA-binding sites from Ago2 CLIP-seq data sets [24]. miRBShunter identifies any potential miRNA::RNA heteroduplexes for both canonical and noncanonical miRNA-binding sites, which involves portions of the miRNA sequence outside the seed or with seed-like binding, by searching for de novo motifs. Potential miRNA::RNA heteroduplexes are then ranked according to a heteroduplex score, which takes into account the following parameters of the heteroduplex: (i) free energy, (ii) the number of paired nucleotides, (iii) the number of paired nucleotides in the motif found, (iv) the number of paired nucleotides in the seed region and (v) the number of bulge nucleotides in the seed.
Downstream analysis
The last step of CLIP-seq analysis involves functional characterization of the target RNAs identified to provide clues about the molecular function of the RBP(s) or the miRNA(s) of interest. Many programs/databases address this task, including GoTermFinder [75] and topGO [76] to perform GO Term enrichment; GeneMania [77] to predict the function of a set of genes; STRING [78] and Cytoscape [79] to predict and visualize the protein interaction networks; GSEA [80] to determine whether a set of genes show similar expression differences between two biological conditions; and RAIN that integrates noncoding RNAs and protein–protein interaction networks [81]. Furthermore, the results from CLIP-seq analysis can be coupled with data sets from other genome-wide technologies, including RNA expression profile or alternative splicing.
Although not always routinely updated, many resources have been developed for the functional analysis of RBPs and miRNAs. For instance, miRonTop is an online Java Web tool that integrates DNA microarrays or high-throughput sequencing data to identify the potential miRNA target mRNAs by complementary between the seed and the 3ʹ UTR sequences. The list of potential miRNA targets can be used to assess specific biological functions of miRNAs by performing Gene Ontology enrichment [82]. DIANA-mirExTra performs a combined differential expression analysis of mRNAs and miRNAs to uncover miRNAs and transcription factors that play regulatory roles between two conditions [83]. Finally, miRGator is a portal collecting high-throughput sequencing miRNA data integrated with target expression profiles [84]. This portal includes 73 deep-sequencing data sets on human samples from Gene Expression Omnibus [85], Short Read Archive (SRA) (SRA:http://www.ncbi.nlm.nih.gov/sra/), The Cancer Genome Atlas archives (http://cancergenome.nih.gov/) and several supporting programs. Among those programs, we mention miR-seq browser that provides short-read alignment with the predicted secondary structure of transcripts, read count and different features to study iso-miRs and miRNA posttranscriptional modifications.
Validation of CLIP-seq analysis
CLIP-seq experiments can be validated using different technical approaches by either candidate or genome-wide approaches [86]. For practical reasons, the candidate approach is feasible only to a limited number of targets, usually top-scored targets from statistical significance tests or identified by machine learning algorithms, or to a subset of targets with a particular biological relevance. The candidate approach can be performed to validate the direct interaction between the protein of interest and the target RNA(s) or the function played by the protein (or miRNA) on the target RNA(s). The interaction can be validated by a plethora of wet laboratory techniques, such as the in vitro electrophoretic mobility shift assay or RNA immuno precipitation followed by either northern bloting or reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) from cell or tissue extracts. Functional validation may include knockdown/knockout or overexpression experiments on cells or tissues of the protein (or miRNA) of interest followed by RT-qPCR or northern bloting to check on the expression levels of the target RNA(s), or minigene assay to check on alternative splicing events.
The same functional validation can be also performed at the genome-wide scale assays, which may include RNA-seq or microarray experiments followed by an appropriate data analysis that depends on the function investigated. While the latter approach is more accurate and comprehensive, the cost can be higher.
Implication for miRNA-binding prediction and human pathogenesis
miRNAs are small noncoding RNAs of about 22 nts that associate to Ago2 to bind to RNA for degradation and/or translation block [16]. About 1000 miRNAs have been experimentally validated in human [87], which regulate many biological processes during physiopathological events and development [3, 4]. In this part of the review, we discuss the latest developments in the field of miRNA target prediction and mode of action in human pathologies made by the use of Ago2 CLIP-seq analysis.
Ago2 CLIP-seq data ameliorate miRNA target prediction
To date, the main miRNA target prediction programs take into account the following miRNA::RNA interaction features: (i) occurrence of perfect complementarity match between miRNA seed sequence and target mRNAs, (ii) sequence conservation of the target sequence across species, (iii) the free energy of the miRNA::RNA heteroduplex and (iv) the target site accessibility [88–91]. Even considering all these features, the rate of false positives and negatives is still high [70], indicating that more is needed to better predict miRNA target sequences. The presence of >15 Ago2 CLIP-seq analyses performed in several cells or tissues and deposited in the Starbase database [92, 93] provides an important resource for a genome-wide investigation of the miRNA targeting features. Based on these studies, a second generation of miRNA target prediction programs has been developed. Although the second-generation programs seem to perform better than the first one, a major limitation for both of them is the lack of an accurate list of bona fide miRNA-binding sites to calculate true- and false-positive rates.
Here, we briefly describe the recent improvements of the prediction programs for miRNA-binding sites, based on CLIP-seq data (Table 2). Recently developed second-generation programs propose new models/parameters for the implementation of new algorithms. For instance, TargetSpy uses for the first time Ago2-CLIP-seq data to train a machine learning algorithm [100]. MIRZA develops a biophysical model through the parametrization of miRNA::mRNA target alignments and the free energy of the binding optimized using CLIP-seq data [101]. STarMir implements a logistic prediction models based on thermodynamic parameters of the miRNA::RNA heteroduplexes and the secondary structure features of the target mRNAs from CLIP-seq analyses [103]. MiRTar2GO is a rule-based machine learning approach to predict cell type-specific miRNA target mRNAs, which are ranked using validated binding sites from luciferase assay or Ago CLASH data sets [105]. Lu and Leslie [106] developed the program chimiRic that uses a discriminative machine learning approach on Ago2 CLIP-seq and CLASH data to train a novel miRNA target prediction model. On the other hand, some of the first-generation miRNA prediction programs have been refined and updated thanks to Ago2 CLIP-seq analyses. For example, DIANA-micro-T-CDS [102] is an extension of the first-generation algorithm DIANA-micro-T [107] that uses a machine learning approach to identify the most relevant features of miRNA targeting from CLIP-seq data sets. Finally, the latest version of miRDB contains miRNA target prediction based on an updated version of the MirTarget computational model by including CLIP ligation (cross-linking and immunoprecipitation followed by RNA ligation) data in the training data set [104].
Table 2.
Main characteristics of first- and second-generation algorithms to predict miRNA-binding sites
| Binding site position on mRNA |
Type of
binding site |
Features |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Program name | Type of resource | 5ʹ UTR | CDS | 3ʹ UTR | Seed | NCSL | Conservation | Free energy | Accessibility | CLIP data | Reference | Web site |
| First generation | ||||||||||||
| Targetscan | WS | No | No | Yes | Yes | No | Yes | No | No | No | [93] | http://www.targetscan.org/vert_71/ |
| PicTar | WS | No | No | Yes | Yes | SL | No | Yes | No | No | [94] | http://www.pictar.org/ |
| PITA | WS/SA | No | No | Yes | Yes | No | Yes | Yes | Yes | No | [95] | https://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html |
| miRanda | D/SA | No | No | Yes | Yes | No | Yes | Yes | No | No | [96] | http://www.microrna.org/microrna/home.do |
| RNAhybrid | WS/SA | No | No | Yes | Yes | Yes | No | Yes | No | No | [97] | https://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid?id=rnahybrid_view_download |
| RNA22 | WS/SA | Yes | Yes | Yes | Yes | SL | No | Yes | No | No | [98] | https://cm.jefferson.edu/rna22/ |
| Second generation | ||||||||||||
| TargetSpy | WS/D | No | No | Yes | Yes | Yes | No | Yes | Yes | Yes | [99] | http://webclu.bio.wzw.tum.de/targetspy/index.php?search=true |
| MIRZA | SA/WS | No | No | Yes | Yes | SL | No | Yes | No | Yes | [100] | http://www.clipz.unibas.ch/downloads/mirza/ |
| DIANA-micro-T-CDS | WS/SA | No | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | [101] | http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index |
| STarMir | WS | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | [102] | http://sfold.wadsworth.org/cgi-bin/starmir.pl |
| miRDB | D/WS | Yes | Yes | Yes | Yes | No | Yes | Yes | No | Yes | [103] | http://www.mirdb.org/miRDB/ |
| miRTar2GO | D/WS | No | No | Yes | Yes | No | Yes | Yes | No | Yes | [104] | http://www.mirtar2go.org/ |
| chimiRic | SA | No | No | Yes | Yes | Yes | No | Yes | No | Yes | [105] | https://bitbucket.org/leslielab/chimiric |
WS, Web server; D, database; SA, stand-alone software; CDS, Protein-coding sequence; NC, Noncanonical; SL, seed-like.
In addition to the features within the miRNA::RNA heteroduplex identified by CLIP-seq analyses, few reports showed that the binding activity of miRNAs is also modulated by RBPs that sit on the sequence surrounding miRNA-binding sites [108–110]. This has led to the concept of a sequence microenvironment surrounding miRNA-binding sites that can play an important role in regulating miRNA activity [89]. However, much remains to be explored about the use of RBP-binding motifs to improve the prediction of miRNA target sequences. A first step toward this direction was made by Incarnato et al. [111]. Briefly, the authors used Pumilio-binding motif to predict miRNA-binding sites within a distance of 100 nts. Validation of this analysis was carried out by RNA expression profile. We foresee that the full incorporation of RBP-binding motifs may pave the way to a third generation of miRNA target prediction programs.
Application of Ago2 CLIP-seq in human pathologies
Several publications reported miRNA targetome in different cells and tissues [12, 48–50, 110, 112–116]. Ago2 CLIP-seq experiments significantly contributed to our knowledge about the role of miRNAs in human pathogenesis. For instance, recently several reports use Ago2 CLIP-seq analysis to study the role of either host or viral miRNAs during viral infection [117]. Noteworthy, Kim et al. [118] combined Ago2 CLIP-seq and bioinformatics to identify miRNA targetomes of the human cytomegalovirus miRNAs during infection. This study reveals that viral miRNAs can regulate multiple pathways and cooperatively function with the host human miRNAs to promote viral replication [118].
Surprisingly for some human pathologies, such as cardiovascular diseases, despite the vast literature and the well-established roles of small RNAs in their pathogenesis [2, 119–121], genome-wide studies of miRNA regulatory networks are poorly developed. Indeed, only two Ago2 CLIP-seq studies have been performed in cardiovascular diseases, including one in the heart of transgenic mice overexpressing miR-133a and miR-499 [122], and the other one in the left ventricular cardiac tissue from six men with cardiomyopathy [123]. Interestingly, in ventricular tissue of patients with cardiomyopathy, about 4000 Ago2-binding sites that contain seed sequence complementarity for the most highly expressed cardiac miRNAs have been identified. The authors deeply characterized the targetome of miR-133, known to be enriched in many pathological conditions of the heart and characterized new roles for miR-29 [123]. In particular, they found that miR-29 targets several mRNAs, including Ryr2, Serca2 and Junctin that are key regulators of sarcoplasmic reticulum Ca2+ in cardiomyocytes, PIK3R1 (p85-alpha) and Med13 that are involved in cardiomyocyte growth and metabolic signaling and Lama2 that plays a more general role in extracellular matrix composition of muscle cells. The authors speculate that these target genes suggest new roles for miR-29 in cardiomyocyte growth and calcium handling, which may have significant clinical relevance to cardiac hypertrophy and contractile dysfunction in cardiomyopathy patients. The novel aspect of these findings is also based on the fact that prior studies have focused on miR29 function in cardiac fibroblasts ignoring the cardiomyocytes [124]. Further preclinical and clinical investigations may strengthen these findings and eventually propose miR-29-based therapeutic approach to cure or prevent cardiopathies.
We foresee that Ago2 CLIP-seq experiments from biopsies of cardiovascular case-control studies and animal models will unravel the global role of miRNAs in the pathogenesis of cardiovascular disease and other human pathologies. A major limitation in the application of this approach is the limited amount of primary tissues derived from biopsies. Future technological advances to increase the depth of single-cell sequencing would overcome this limitation [125].
Concluding remarks
In this review, we discuss the recent computational developments of CLIP-seq analysis and highlight key points associated with each step, providing useful guideline to nonexpert users. We stress that a quality check of the data at each step is important to properly perform the analysis. While our manuscript was in revision, Uhl et al. [126] published a review on the CLIP-seq data analysis, which mainly focuses on the peak-calling step, whereas our review provides a more general point of view of the computational workflow. In addition, by addressing recent applications of CLIP-seq analysis for Ago2/miRNA target identification and prediction in physiological conditions or in human pathologies, our review provides practical examples of direct applications of this technique in biomedical fields. We believe that this review benefits scientists in RNA biology, gene expression fields and beyond.
CLIP-seq analysis provides a huge amount of data that often are not fully exploited by the researchers because of the lack of time or tools dedicated to perform/integrate different analyses. A further effort should be done to make these data available in a user-friendly format and resource databases to collect them. Some databases, such as StarBase [92], CLIPdb [127] and CLIPZ [27], collect raw data of CLIP-seq studies and provide peak sequences and/or coordinates. However, they are not always updated and use standard pipelines that may not be suitable for every RBPs. Therefore, the development of integrated platform of CLIP-seq data analysis and databases could be a direction to be taken in the near future. In particular, these platforms should combine multiple software, such as pipelines covering multiple steps of data analysis or multiple databases including other high-throughput techniques as RIP-seq, ChIP-seq, RNA-seq and quantitative proteomics data, making the analysis faster and more comprehensive. On the other hand, improvement of the experimental protocol or conditions, such as quantification of the benefits of using replicates and/or control experiments, is needed to improve the reproducibility of the data and increase the efficiency of the data mining.
Overall, we have shown that CLIP-seq experiments associated to sophisticated bioinformatics analysis have become nowadays an essential instrument to gain insight into the direct regulatory network(s) of RBP-RNA interactions to address central questions of RNA biology and gene expression control in normal and pathological events of physiology or development. We foresee that progression in software development will take the stage in the near future to render the CLIP-seq analysis more integrated to other genome-wide approaches and more accessible to nonexpert users.
Key Points
Recent developments in sequencing technologies and bioinformatics analyses enable us to handle many CLIP-seq samples simultaneously; thus, it is important to optimize bioinformatics pipelines that can facilitate the work of researchers to obtain unbiased and high-quality data.
Despite great effort from researchers to streamline this CLIP-seq analysis, much remains to be improved on computational procedures.
In this review, we discuss the validity and the limitations of emerging programs for CLIP-seq analysis and the quality measurements currently available for specific tasks by providing concrete examples on an in-house Ago2 HITS-CLIP data set generated in stem cells.
We have focused the scope of this review in providing a valuable computational guideline for the bioinformatics analysis of the three main variants of CLIP-seq analysis, namely, HITS-CLIP, PAR-CLIP and iCLIP.
We discuss how Ago2 CLIP-seq analyses have improved the miRNA-binding site prediction and the understanding of miRNA function in human pathologies.
Supplementary Material
Acknowledgements
The authors would like to thank the UCA Genomix platform.
Funding
The FRM (grant #DEQ20140329551) and ANR through the ‘Investments for the Future’ # ANR-11-LABX-0028-01 (LABEX SIGNALIFE) (to M.T.). The FRM (ING20140129224) (to E.R.).
Biographies
Silvia Bottini is a research associate at Inserm. Her research interests involve computational approaches to study the RNA–protein interaction network both in vitro and in vivo systems.
David Pratella is a graduate student at the Université Côte d’Azur. His research interests involve data analysis/mining and program development for high-throughput sequencing data.
Valerie Grandjean is a senior research associate at Inserm. Her research interests involve epigenetic inheritance mediated by small RNAs.
Emanuela Repetto is a research associate at Inserm. Her research interests involve the expression control and function of small RNAs in cardiovascular diseases.
Michele Trabucchi is a principal investigator at Inserm. His research interests involve the molecular mechanisms regulating the fate and the function of noncoding RNAs.
References
- 1. Cech TR, Steitz JA.. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157:77–94. [DOI] [PubMed] [Google Scholar]
- 2. Hizir Z, Bottini S, Grandjean V, et al. RNY (YRNA)-derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death Dis 2017;8:e2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Repetto E, Briata P, Kuziner N, et al. Let-7b/c enhance the stability of a tissue-specific mRNA during mammalian organogenesis as part of a feedback loop involving KSRP. PLoS Genet 2012;8:e1002823.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siddeek B, Lakhdari N, Inoubli L, et al. Developmental epigenetic programming of adult germ cell death disease: polycomb protein EZH2-miR-101 pathway. Epigenomics 2016;8: 1459–79. [DOI] [PubMed] [Google Scholar]
- 5. Lunde BM, Moore C, Varani G.. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 2007;8:479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castello A, Fischer B, Frese CK, et al. Comprehensive identification of RNA-binding domains in human cells. Mol Cell 2016;63:696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clery A, Boudet J, Allain FH.. Single-stranded nucleic acid recognition: is there a code after all? Structure 2013;21:4–6. [DOI] [PubMed] [Google Scholar]
- 8. Licatalosi DD, Mele A, Fak JJ, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008;456:464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chi SW, Zang JB, Mele A, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009;460:479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA 2010;1: 266–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garzia A, Meyer C, Morozov P, et al. Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods 2017;118–119:24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010;141:129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spitzer J, Hafner M, Landthaler M, et al. PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation): a step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzymol 2014;539:113–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang C, Darnell RB.. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol 2011;29:607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hauptmann J, Meister G.. Argonaute regulation: two roads to the same destination. Dev Cell 2013;25:553–4. [DOI] [PubMed] [Google Scholar]
- 16. Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012;13:271–82. [DOI] [PubMed] [Google Scholar]
- 17. Zisoulis DG, Lovci MT, Wilbert ML, et al. Comprehensive discovery of endogenous Argonaute binding sites in caenorhabditis elegans. Nat Struct Mol Biol 2010;17:173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The Long Tail of mRNA Regulation. Cell 2017;168:335–8. [DOI] [PubMed] [Google Scholar]
- 19. Konig J, Zarnack K, Rot G, et al. iCLIP–transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp 2011;50:e2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang T, Xiao G, Chu Y, et al. Design and bioinformatics analysis of genome-wide CLIP experiments. Nucleic Acids Res 2015;43:5263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y, Xie S, Xu H, et al. CLIP: viewing the RNA world from an RNA-protein interactome perspective. Sci China Life Sci 2015;58:75–88. [DOI] [PubMed] [Google Scholar]
- 22. Helwak A, Kudla G, Dudnakova T, et al. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013;153:654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Broughton JP, Pasquinelli AE.. A tale of two sequences: microRNA-target chimeric reads. Genet Sel Evol 2016;48:31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bottini S, Hamouda-Tekaya N, Tanasa B, et al. From benchmarking HITS-CLIP peak detection programs to a new method for identification of miRNA-binding sites from Ago2-CLIP data. Nucleic Acids Res 2017, doi: 10.1093/nar/gkx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moore MJ, Zhang C, Gantman EC, et al. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc 2014;9:263–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maragkakis M, Alexiou P, Nakaya T, et al. CLIPSeqTools-a novel bioinformatics CLIP-seq analysis suite. RNA 2015;22:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khorshid M, Rodak C, Zavolan M.. CLIPZ: a database and analysis environment for experimentally determined binding sites of RNA-binding proteins. Nucleic Acids Res 2011;39: D245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen C, Khaleel SS, Huang H, et al. Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol Med 2014;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmieder R, Edwards R.. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011;27:863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2012;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith AD, Chung WY, Hodges E, et al. Updates to the RMAP short-read mapping software. Bioinformatics 2009;25:2841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trapnell C, Pachter L, Salzberg SL.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu TD, Nacu S.. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010;26: 873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li R, Li Y, Kristiansen K, et al. SOAP: short oligonucleotide alignment program. Bioinformatics 2008;24:713–14. [DOI] [PubMed] [Google Scholar]
- 37. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakato R, Shirahige K.. Recent advances in ChIP-seq analysis: from quality management to whole-genome annotation. Brief Bioinform 2016;18:279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baruzzo G, Hayer KE, Kim EJ, et al. Simulation-based comprehensive benchmarking of RNA-seq aligners. Nat Methods 2017;14:135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sims D, Sudbery I, Ilott NE, et al. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 2014;15:121–32. [DOI] [PubMed] [Google Scholar]
- 41. Chen B, Yun J, Kim MS, et al. PIPE-CLIP: a comprehensive online tool for CLIP-seq data analysis. Genome Biol 2014;15:R18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Althammer S, Gonzalez-Vallinas J, Ballare C, et al. Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics 2011;27:3333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uren PJ, Bahrami-Samani E, Burns SC, et al. Site identification in high-throughput RNA-protein interaction data. Bioinformatics 2012;28:3013–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lovci MT, Ghanem D, Marr H, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol 2013;20:1434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Comoglio F, Sievers C, Paro R.. Sensitive and highly resolved identification of RNA-protein interaction sites in PAR-CLIP data. BMC Bioinformatics 2015;16:32.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Corcoran DL, Georgiev S, Mukherjee N, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol 2011;12:R79.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang T, Chen B, Kim M, et al. A model-based approach to identify binding sites in CLIP-Seq data. PLoS One 2014;9: e93248.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karginov FV, Hannon GJ.. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev 2013;27:1624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leung AK, Young AG, Bhutkar A, et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol 2011;18:237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loeb GB, Khan AA, Canner D, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell 2012;48:760–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xue Y, Zhou Y, Wu T, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 2009;36:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang T, Xie Y, Xiao G.. dCLIP: a computational approach for comparative CLIP-seq analyses. Genome Biol 2014;15:R11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Erhard F, Dolken L, Jaskiewicz L, et al. PARma: identification of microRNA target sites in AGO-PAR-CLIP data. Genome Biol 2013;14:R79.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grant CE, Bailey TL, Noble WS.. FIMO: scanning for occurrences of a given motif. Bioinformatics 2011;27:1017–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ray D, Kazan H, Cook KB, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 2013;499:172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mitchell SF, Parker R.. Principles and properties of eukaryotic mRNPs. Mol Cell 2014;54:547–58. [DOI] [PubMed] [Google Scholar]
- 57. Simcha D, Price ND, Geman D.. The limits of de novo DNA motif discovery. PLoS One 2012;7:e47836.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Heeringen SJ, Veenstra GJ.. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics 2011;27:270–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009;37: W202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Foat BC, Morozov AV, Bussemaker HJ.. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics 2006;22:e141–9. [DOI] [PubMed] [Google Scholar]
- 62. Frith MC, Saunders NF, Kobe B, et al. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput Biol 2008;4:e1000071.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Georgiev S, Boyle AP, Jayasurya K, et al. Evidence-ranked motif identification. Genome Biol 2010;11:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bahrami-Samani E, Penalva LO, Smith AD, et al. Leveraging cross-link modification events in CLIP-seq for motif discovery. Nucleic Acids Res 2015;43:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kazan H, Ray D, Chan ET, et al. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS Comput Biol 2010;6:e1000832.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bailey TL, Boden M, Whitington T, et al. The value of position-specific priors in motif discovery using MEME. BMC Bioinformatics 2010;11:179.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Siddharthan R. PhyloGibbs-MP: module prediction and discriminative motif-finding by Gibbs sampling. PLoS Comput Biol 2008;4:e1000156.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fukunaga T, Ozaki H, Terai G, et al. CapR: revealing structural specificities of RNA-binding protein target recognition using CLIP-seq data. Genome Biol 2014;15:R16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yue D, Liu H, Huang Y.. Survey of computational algorithms for MicroRNA target prediction. Curr Genomics 2009;10: 478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chi SW, Hannon GJ, Darnell RB.. An alternative mode of microRNA target recognition. Nat Struct Mol Biol 2012;19: 321–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chou CH, Lin FM, Chou MT, et al. A computational approach for identifying microRNA-target interactions using high-throughput CLIP and PAR-CLIP sequencing. BMC Genomics 2013;14(Suppl 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Majoros WH, Lekprasert P, Mukherjee N, et al. MicroRNA target site identification by integrating sequence and binding information. Nat Methods 2013;10:630–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Clark PM, Loher P, Quann K, et al. Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci Rep 2014;4:5947.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Boyle EI, Weng S, Gollub J, et al. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004;20:3710–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. R package version 2.26.0.
- 77. Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lotia S, Montojo J, Dong Y, et al. Cytoscape app store. Bioinformatics 2013;29:1350–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Junge A, Refsgaard JC, Garde C, et al. RAIN: RNA-protein association and interaction networks. Database 2017, doi: 10.1093/database/baw167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Le Brigand K, Robbe-Sermesant K, Mari B, et al. MiRonTop: mining microRNAs targets across large scale gene expression studies. Bioinformatics 2010;26:3131–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vlachos IS, Vergoulis T, Paraskevopoulou MD, et al. DIANA-mirExTra v2.0: uncovering microRNAs and transcription factors with crucial roles in NGS expression data. Nucleic Acids Res 2016;44:W128–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cho S, Jang I, Jun Y, et al. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res 2012;41:D252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res 2011;39:D1005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thomson DW, Bracken CP, Goodall GJ.. Experimental strategies for microRNA target identification. Nucleic Acids Res 2011;39:6845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Friedlander MR, Lizano E, Houben AJ, et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol 2014;15:R57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Akhtar MM, Micolucci L, Islam MS, et al. Bioinformatic tools for microRNA dissection. Nucleic Acids Res 2015;44:24–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jens M, Rajewsky N.. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet 2015;16:113–26. [DOI] [PubMed] [Google Scholar]
- 90. Maziere P, Enright AJ.. Prediction of microRNA targets. Drug Discov Today 2007;12:452–8. [DOI] [PubMed] [Google Scholar]
- 91. Peterson SM, Thompson JA, Ufkin ML, et al. Common features of microRNA target prediction tools. Front Genet 2014;5:23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yang JH, Li JH, Jiang S, et al. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res 2012;41:D177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet 2005;37:495–500. [DOI] [PubMed] [Google Scholar]
- 96. Kertesz M, Iovino N, Unnerstall U, et al. The role of site accessibility in microRNA target recognition. Nat Genet 2007;39:1278–84. [DOI] [PubMed] [Google Scholar]
- 97. Betel D, Koppal A, Agius P, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010;11:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rehmsmeier M, Steffen P, Hochsmann M, et al. Fast and effective prediction of microRNA/target duplexes. RNA 2004;10:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006;126:1203–17. [DOI] [PubMed] [Google Scholar]
- 100. Sturm M, Hackenberg M, Langenberger D, et al. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinformatics 2010;11:292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Khorshid M, Hausser J, Zavolan M, et al. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat Methods 2013;10:253–5. [DOI] [PubMed] [Google Scholar]
- 102. Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013;41:W169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rennie W, Liu C, Carmack CS, et al. STarMir: a web server for prediction of microRNA binding sites. Nucleic Acids Res 2014;42:W114–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics 2016;32:1316–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ahadi A, Sablok G, Hutvagner G.. miRTar2GO: a novel rule-based model learning method for cell line specific microRNA target prediction that integrates Ago2 CLIP-Seq and validated microRNA-target interaction data. Nucleic Acids Res 2017;45:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lu Y, Leslie CS.. Learning to predict miRNA-mRNA interactions from AGO CLIP sequencing and CLASH data. PLoS Comput Biol 2016;12:e1005026.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Maragkakis M, Reczko M, Simossis VA, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 2009;37:W273–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kedde M, Strasser MJ, Boldajipour B, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 2007;131:1273–86. [DOI] [PubMed] [Google Scholar]
- 109. Srikantan S, Tominaga K, Gorospe M.. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci 2012;13:372–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Xue Y, Ouyang K, Huang J, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013;152:82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Incarnato D, Neri F, Diamanti D, et al. MREdictor: a two-step dynamic interaction model that accounts for mRNA accessibility and Pumilio binding accurately predicts microRNA targets. Nucleic Acids Res 2013;41:8421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kishore S, Jaskiewicz L, Burger L, et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods 2011;8:559–64. [DOI] [PubMed] [Google Scholar]
- 113. Lipchina I, Elkabetz Y, Hafner M, et al. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev 2011;25:2173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–8. [DOI] [PubMed] [Google Scholar]
- 115. Cardinali B, Cappella M, Provenzano C, et al. MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis 2016;7:e2086.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Conte I, Merella S, Garcia-Manteiga JM, et al. The combination of transcriptomics and informatics identifies pathways targeted by miR-204 during neurogenesis and axon guidance. Nucleic Acids Res 2014;42:7793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Guo YE, Steitz JA.. Virus meets host microRNA: the destroyer, the booster, the hijacker. Mol Cell Biol 2014;34:3780–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kim S, Seo D, Kim D, et al. Temporal landscape of MicroRNA-Mediated Host-Virus crosstalk during productive human cytomegalovirus infection. Cell Host Microbe 2015;17:838–51. [DOI] [PubMed] [Google Scholar]
- 119. Boettger T, Braun T.. A new level of complexity: the role of microRNAs in cardiovascular development. Circ Res 2012;110:1000–13. [DOI] [PubMed] [Google Scholar]
- 120. Olson EN. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci Transl Med 2014;6:239ps233.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Repetto E, Lichtenstein L, Hizir Z, et al. RNY-derived small RNAs as a signature of coronary artery disease. BMC Med 2015;13:259.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Matkovich SJ, Van Booven DJ, Eschenbacher WH, et al. RISC RNA sequencing for context-specific identification of in vivo microRNA targets. Circ Res 2011;108:18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Spengler RM, Zhang X, Cheng C, et al. Elucidation of transcriptome-wide microRNA binding sites in human cardiac tissues by Ago2 HITS-CLIP. Nucleic Acids Res 2016;44:7120–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 2008;105:13027–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Romanov RA, Zeisel A, Bakker J, et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci 2017;20:176–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Uhl M, Houwaart T, Corrado G, et al. Computational analysis of CLIP-seq data. Methods 2017;118–119:60–72. [DOI] [PubMed] [Google Scholar]
- 127. Yang YC, Di C, Hu B, et al. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genomics 2015;16:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.