Abstract

DNA nanotechnology provides a versatile toolbox for creating custom and accurate shapes that can serve as versatile templates for nanopatterning. These DNA templates can be used as molecular-scale precision tools in, for example, biosensing, nanometrology, and super-resolution imaging, and biocompatible scaffolds for arranging other nano-objects, for example, for drug delivery applications and molecular electronics. Recently, increasing attention has been paid to their potent use in nanophotonics since these modular templates allow a wide range of plasmonic and photonic ensembles ranging from DNA-directed nanoparticle and fluorophore arrays to entirely metallic nanostructures. This Feature Article focuses on the DNA-origami-based nanophotonics and plasmonics—especially on the methods that take advantage of various substrates and interfaces for the foreseen applications.
Introduction
The rapid evolution of structural DNA nanotechnology1−5 along with advanced and automated techniques, computer-aided design software, powerful simulation tools, and reduced cost of synthesis6−10 have enabled the effortless fabrication of custom DNA nanoarchitectures for many applications in materials science and bio-oriented research.3,11,12
Examples of using versatile DNA shapes—especially DNA origami11,13−16—have been reported, for example, in molecular electronics,17−22 super-resolution imaging,23−26 and drug delivery.27−31 These customized DNA objects can also find use as molecular-scale diagnostic tools,32 dynamic devices,33−37 and templates for controlling chemical reactions,38−40 and importantly, these structures are inherently biocompatible and readily modifiable for application-specific purposes.28,30,41−45 These examples are enabled by virtue of the sub-nanometer structural accuracy of DNA origami and nanometer-scale patterning resolution of multiple molecular components.11,13−16 In general, the DNA origami technique provides a straightforward route from the target shape to a functional product via a self-assembly process. In a one-pot assembly, one can create a very large number of customized and well-defined DNA origami nanostructures ranging from nano- to micrometer size15 and from mega- to gigadalton scale16 for various user-defined tasks and implementations.
In addition to these examples, DNA nanostructures can be used as precise templates for arranging metal nanoparticles and creating plasmonic assemblies and nanophotonic devices.46−49 Like in all of the other above-mentioned examples, building nanophotonic structures using DNA is also based on taking advantage of the programmability, modularity, and high addressability of the DNA nano-objects. Arguably, there are several benefits of integrating the plasmonic DNA nanostructures with interfaces; for example, substrate-based methods are readily compatible with conventional top-down lithography methods, and they also enable characterization of individual nanodevices.50−53 Moreover, DNA lattice-based approaches can be improved by taking advantage of DNA structure diffusion on substrates.54,55 Importantly, anchoring of the nanostructures to interfaces facilitates coupling of nanodevices with outer circuitry,18 diagnostic and sensing tools,32 and fabrication of bioinspired substrates and metasurfaces.56
In this Feature Article, we summarize the key aspects of DNA origami plasmonics and provide an overview of placement and arrangement methods for self-assembled DNA nanostructures. Finally, we combine the methodology of these two sections and discuss the recent progress of DNA-origami-based nanophotonics at interfaces.
DNA-Nanostructure-Based Nanophotonics and Plasmonics: An Overview
The highly addressable DNA origami provides an excellent platform to organize nanophotonic components into systems with emerging optical properties because of its nanometer-scale precision control. The most common photonic objects arranged with DNA origami are metallic nanoparticles (NPs) and fluorophores. When NPs interact with light, their conduction-band electrons can enter a collective oscillation mode, that is, a so-called localized surface plasmon resonance (LSPR). The plasmon oscillation is highly sensitive to the polarizability of the particle; in other words, the composition, geometry, and surrounding media of the NP all have a significant influence on the resonance. In addition, when two NPs are in close proximity, their plasmon oscillations can couple with each other, resulting in intriguing optical properties and remarkable electromagnetic field enhancement. This coupling is strongly dependent on the spatial arrangement of NPs. For the tailoring of the optical response of the NP systems, techniques to position specific NPs with accurate spatial configuration are of vital importance. To date, DNA origami is one of the most promising candidates to tackle this issue with its sub-nanometer- to nanometer-scale positioning precision for molecules and nanoparticles.57,58 In addition to this extreme positioning resolution, the accessibility and absolute incorporation efficiency of the single strands in DNA origami can reach 95%.59
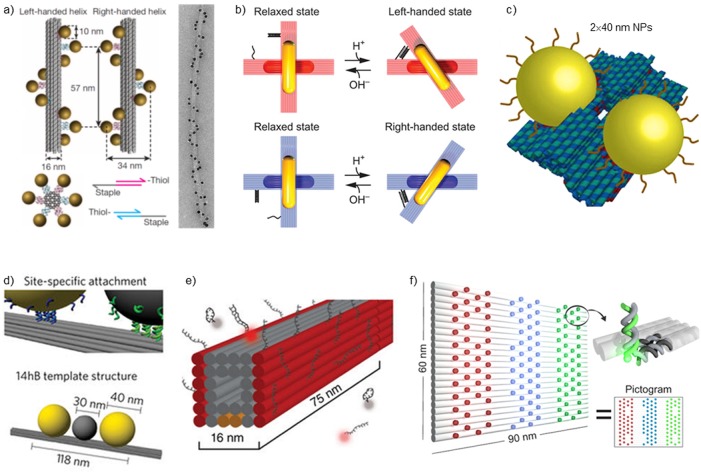
In 2010, the first DNA-origami-guided arrangement of spherical gold NPs (AuNPs) was demonstrated.60 The desired NP arrangement was achieved via coating AuNPs with oligonucleotides and further via their hybridization with complementary strands extended from the specific positions of origami. Later on, a plethora of NP assemblies with customized optical properties have been developed.46,48,49 A representative example of such a rationally designed plasmonic DNA device is a chiral plasmonic structure assembled from a rodlike DNA origami and nine AuNPs that go around the DNA rod in a helical fashion (Figure 1a).61 These chiral plasmonic nanostructures exhibited significant circular dichroism (CD) in the visible range, and their CD responses could be tuned by altering the NP size and the helical handedness. Spherical AuNPs can also be arranged into a ring conformation to obtain tunable electric and magnetic plasmon resonances at the visible frequencies.62 In addition to AuNPs, spherical silver NPs (AgNPs) have also been organized by DNA origami.63−65
Figure 1.
(a) Left: DNA-origami-based left- and right-handed chiral plasmonic nanoassemblies that show strong circular dichroism.61 Right: Transmission electron micrograph of stacked assembly. (b) Dynamic chiral metamolecule that is sensitive to the pH change of solution.36 (c) AuNP dimer with defined distance assembled on a DNA origami platform.78 (d) Heterotrimer assembly with a AgNP between two AuNPs on a DNA origami beam.81 (e) Bar-shaped DNA origami with docker strands on sides to capture transient imager strands for super-resolution imaging.25 (f) DNA origami with multiple programmable fluorescent dye binding sites acts as a metafluorophore.84 Part a is reprinted with permission from ref (61). Copyright 2012 Springer Nature. Part b is reprinted with permission from ref (36). Part c is reprinted with permission from ref (78). Part d is reprinted with permission from ref (81). Copyright 2017 Springer Nature. Part e is reprinted with permission from ref (25). Copyright 2014 Springer Nature. Part f is reprinted with permission from ref (84).
Since the LSPR is sensitive to the geometry of NPs, metallic NPs with shapes other than a sphere could offer more diverse optical properties and therefore further facilitate the design of nanophotonic devices. DNA origami conjugates with gold nanorods (AuNRs),34−36,66,67 gold nanotriangles (AuNTs),68 or hybrid assemblies of particles with different geometries69,70 have also been explored.
In addition to static nanophotonics, dynamic and reconfigurable plasmonic DNA devices have been developed. They are considered dynamic since the geometric configuration of such devices could be changed and switched after assembly. For example, by attaching two AuNRs to DNA origami beams connected via a single Holliday junction, a metamolecule with different rotational states—and therefore different CD signal states—can be formed. The reconfiguration of the states can be induced by the introduction of DNA displacement strands,34 by light,35 or by changing solution pH (see Figure 1b).36 In addition to these, Turek et al. have demonstrated how thermoresponsive polymer-equipped DNA tweezers could be used as an actuator for surface-enhanced fluorescence.71 Instead of reconfiguring the whole metamolecule, the AuNR itself can also “walk” between different sites on a DNA origami by fueling the system with displacement strands67 using a similar strategy as originally introduced by Yurke et al.72
Significant effort has been put into developing techniques to position NPs in extremely close proximity to form hot-spots for the field enhancement, which is essential for surface-enhanced Raman spectroscopy (SERS) and fluorescence enhancement (FE). To achieve the nanometer-scale distance, different strategies have been employed. For example, “seed-NPs” that are attached to DNA origami could be grown larger by chemical methods to shorten their distance.19,73,74 Upon conjugation of NP dimers on different sides of DNA origami, the DNA helices between NPs can act as binding sites and as a spacer of a few nanometer thickness.75−77 Rationally designed DNA origami structures could also be used to dock NP dimers with a gap of only a few nanometers, as shown in the work of Thacker et al. (see Figure 1c)78 and Roller et al.79 In these works, the local field was enhanced by several orders of magnitude and demonstrated by SERS or FE characterization.
To fully take advantage of the addressability and versatility of DNA origami, plasmonic systems with different metal compositions have also been assembled. These heteroparticle assemblies80,81 have shown optical modes that are challenging to obtain with any other method. For example, a nondissipative and ultrafast plasmon passage has been observed in a heterotrimer system consisting of both a AuNP and AgNP arranged on top of DNA origami (see Figure 1d).81
Fluorophores form another group of nanophotonic components that are widely combined with DNA nanostructures. DNA origami can work as a nanoadapter to enable manipulation of individual or a few fluorescent molecules. In addition to the well-known Förster resonance energy transfer (FRET) studies,49,82 DNA origami structures equipped with dye-labeled oligonucleotides have been used in DNA-point accumulation for imaging in nanoscale topography (DNA-PAINT) in super-resolution microscopy (see Figure 1e),24,25,83 and as a nanoruler for spatial calibration.23 Upon combination of various different fluorescence dyes on a single rectangular DNA origami, it is possible to assemble a so-called metafluorophore (see Figure 1f).84
DNA Origami Placement at Interfaces
As already explained, there are a number of advantages in anchoring the DNA-based devices to the substrates or incorporating them into larger systems for nanophotonics. This section deals with the approaches suitable for assembling DNA structures at the interfaces.
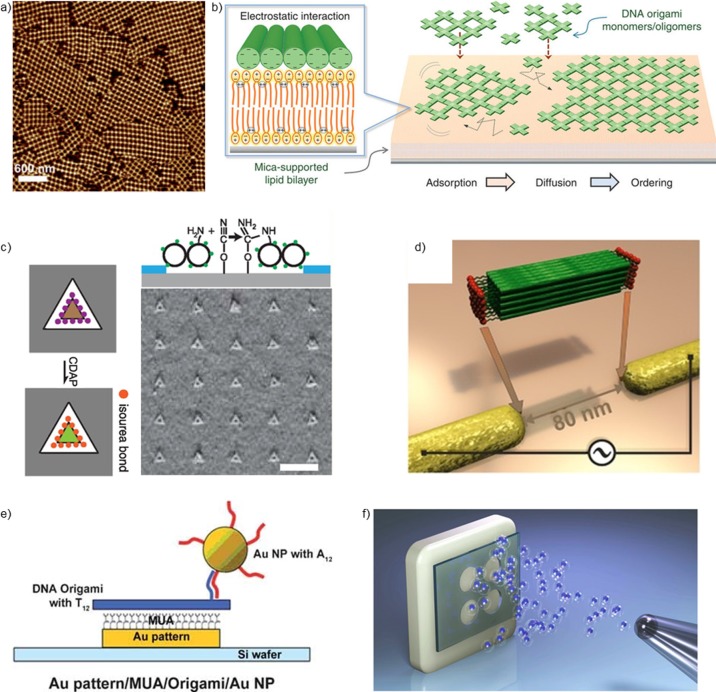
DNA nanostructures can be immobilized onto substrates merely by electrostatic interactions, and one of the most common methods is adhesion to mica with the presence of magnesium ions.85 Upon adjustment of the amount of cations of the deposition buffer, DNA nanostructures can be placed, for example, on silicon and silicon oxide.50,51,53 Interestingly, monovalent cations can be used for surface-assisted assembly of higher-order structures.54,55 Aghebat Rafat et al. used cross-tile structures (Figure 2a), and created 2D lattices by controlling the diffusion of the structures on a mica substrate with monovalent cations.54 The cross-tiles were similar to ones designed by Liu et al.,86 except that the tiles were “twist-corrected”, (i.e., the undesired twist caused by the square-lattice design87 was removed), and they formed the close-packed crystalline structures via blunt-end stacking interactions88,89 instead of sticky-end cohesion. A similar approach of lattice organization on the substrate has also been demonstrated for a rectangular origami55 and an origami triangle13 that has a void in the middle of the structure. The triangles form a close-packed lattice that could be further used as a removable mask for protein patterning of the substrate through the openings of the triangle layer.90 Moreover, blunt-end stacking of DNA origami objects could be applied to arranging DNA origami in 3D lattices. Recently, Zhang et al.91 used three-dimensional and rigid origami tensegrity triangles—similar to simpler tensegrity triangles in the very first 3D DNA crystal by Zheng et al.92—to form a 3D lattice with voids via blunt-end stacking. They also demonstrated the addressability of the origami lattice by attaching gold particles to the lattice voids, thus creating a hybrid DNA–gold nanoparticle superlattice.
Figure 2.
(a) AFM image of DNA origami cross-tiles assembled into a lattice configuration by cation-induced diffusion.54 (b) Lipid-bilayer-facilitated 2D-lattice formation from DNA origami cross-tiles.93 (c) Triangular DNA origami is covalently immobilized to binding sites patterned on a silicon substrate by lithography.50 (d) Dielectrophoretic trapping of 3D DNA origami between lithographically fabricated gold nanoelectrodes.18 (e) DNA origami equipped with a AuNP is selectively attached to a gold pattern on a silicon substrate.101 (f) Large-scale spray-deposition of DNA origami nanostructures to the selected substrate through a mask.103 Part a is reprinted with permission from ref (54). Copyright 2014 John Wiley and Sons. Part b is reprinted with permission from ref (93). Part c is reprinted with permission from ref (50). Copyright 2014 American Chemical Society. Part d is reprinted with permission from ref (18). Copyright 2015 John Wiley and Sons. Part e is reprinted with permission from ref (101). Copyright 2009 John Wiley and Sons. Part f is reprinted with permission from ref (103).
In addition to mica substrates, lipid bilayers can also be employed in the organization of DNA origami lattices and other higher-order assemblies93−95 as seen in Figure 2b. The lattice formation process can be controlled using cholesterol-modified DNA origami and connector staples94 or different lipid membrane phases (liquid-disordered or solid-ordered).95 Moreover, the growth of arrays can be triggered by adjusting monovalent cation concentration as well as the concentration of deposited origami structures.95
One approach to pattern substrates with the resolution beyond conventional lithography techniques is to combine top-down approaches with the high addressability of DNA origami. Kershner et al.96 fabricated arrays of DNA origami triangles by attaching origami to lithographically revealed precise triangular areas of silicon oxide in a hexamethyldisilazane (HDMS) film. To demonstrate the potential of the method in high-resolution nanoscale patterning, Hung et al.97 attached AuNPs to the corners of the origami triangle and organized them to the lithographically confined wells.
Later on, Gopinath & Rothemund50 optimized the assembly of DNA origami triangle nanoarrays (Figure 2c) on silicon and silicon oxide substrates. They reported how DNA origami triangles could be covalently coupled to the lithographically patterned wells with high yields using the isourea bond (as shown in Figure 2c) or amide bond for linking. Attaching DNA origami to silicon or silicon oxide usually requires deposition buffers with high magnesium content, but by employing covalent linking instead of simple electrostatic bonds between DNA origami and the substrate, a much wider magnesium concentration range could be used. Recently, Brassat et al.98 further studied DNA origami adsorption onto a silicon oxide surface in nanohole arrays by varying Mg and DNA origami concentration, buffer strength, adsorption time, and nanohole size. They observed that buffer strength plays a critical role in origami deposition.
In addition to fabricating origami-shaped wells in the films, one can also use other lithographic features for directing DNA origami to predefined locations with targeted geometries. One example is to take advantage of the (directional) polarizability of DNA origami and guide the structure by electromagnetic fields. Kuzyk et al.99 and Shen et al.18 have demonstrated how dielectrophoresis (DEP) can be used to trap various 2D and 3D DNA origami shapes between gold nanoelectrodes. By applying ac voltage to the nanoelectrodes, one can create a highly localized electric field that traps DNA origami in the electrode gap (see Figure 2d). The proper immobilization (and coupling to outer electrical circuitry) can be ensured through the covalent gold–sulfur bond by incorporating thiolated strands into DNA origami. Nevertheless, DEP could also allow transfer of the achieved trapping geometries to the chosen electrodeless substrate by the imprint technique.100
Moreover, Gerdon et al.101 used lithographically patterned gold patches functionalized with 11-mercaptoundecanoic acid (MUA) to selectively immobilize DNA origami (see Figure 2e). The carboxylic acid groups of the functionalized layer chelate magnesium ions in the deposition buffer, and thus, a salt bridge is formed between a negatively charged DNA origami and the positively charged Mg ions. However, compared to other lithographically achieved patterning methods, this technique lacks the control over spatial orientation of the delivered DNA origami. To achieve directionality and to align the structures in a controllable way, Ding et al.102 showed that small gold islands—patterned by e-beam lithography in different geometries—could be connected by thiol-modified DNA origami nanotubes. The nanotubes had exactly matching length with the distance of the neighboring gold islands.
To achieve large-scale deposition, Linko et al.103 developed a spray-coating technique (Figure 2f), which allows straightforward and efficient delivery of DNA origami to the substrate without a need of chemical treatments or washing steps. However, the majority of salt ions has to be removed from the origami solution45 before deposition. They demonstrated the feasibility of the method using various 2D and 3D origami structures, removed the residual salt by spin-filtering,45 and spray-coated large areas of glass and silicon substrates. Although one can control the coating procedure at large scales using mechanical masks, the structures are randomly oriented on the deposited areas. However, this technique might become compatible with other methods described in this section.
DNA Origami Plasmonics and Nanophotonics at Interfaces
By combining the DNA-based nanophotonic devices and different techniques to arrange DNA structures on interfaces described in the previous section, one can achieve plasmonic substrates with tailored optical properties. One goal in this direction would be to create metasurfaces using ordered assemblies, but nevertheless, the individual DNA-based objects and their properties should first be characterized.
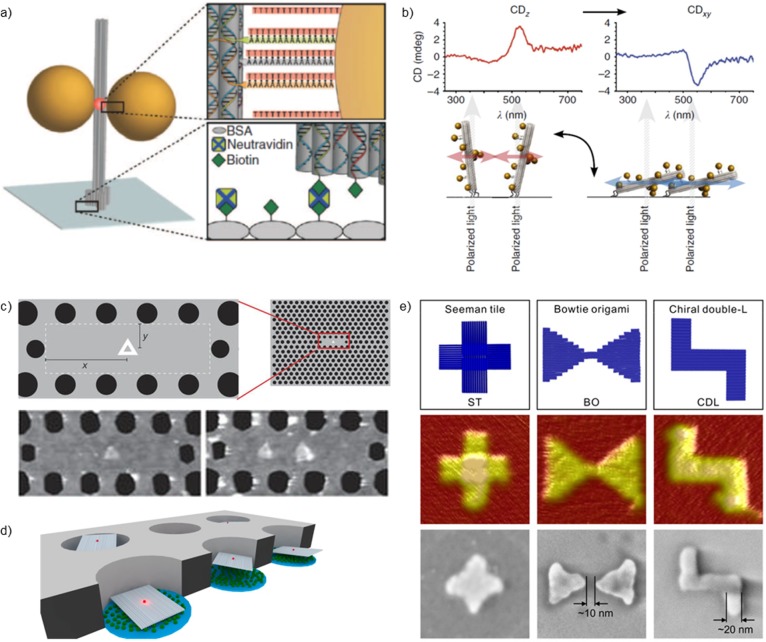
To probe single plasmonic devices, they need to be deposited onto a substrate after their self-assembly in buffer solution. In many reported experimental setups,76−78 plasmonic structures were immobilized via electrostatic interactions on a charged substrate (see the previous section). Another commonly used scheme is to take advantage of the strong interaction between biotin and avidin. As an example, Acuna et al.75 immobilized a rodlike DNA origami equipped with a AuNP dimer and a single fluorescent molecule in the gap to the substrate by biotin–neutravidin linking (see Figure 3a). The binding and unbinding events of short DNA strands, as well as the conformational dynamics of a Holliday junction in the hot-spot, were visualized in a form of enhanced fluorescence signals in real-time. Recently, a similar system has also been used to characterize single peridinin–chlorophyll a–protein complex.65
Figure 3.
(a) Gold nanoparticles and a rodlike DNA origami form a nanoantenna that is attached to the substrate via biotin–neutravidin interaction.75 (b) Chiral plasmonic nanostructures (similar to Figure 1a) arranged onto a substrate to achieve a switchable and directional circular dichroism (CD) effect.104 (c) DNA origami triangles equipped with fluorophores can be precisely placed into an optical cavity.51 The number of triangles and their position can be controlled and thus the fluorescence of the “pixel” (inset) in a large array of cavities. (d) DNA origami is used as a nanoadapter to place individual fluorescent molecules in lithographically fabricated metallic zero-mode waveguide.107 (e) DNA-assisted lithography (DALI).53 DNA origami (top panel) is deposited on a silicon chip (middle panel), and its shape is transferred into a metallic structure on a transparent substrate (bottom panel). Part a is reprinted with permission from ref (75). Copyright 2012 American Association for the Advancement of Science. Part b is reprinted with permission from ref (104). Part c is reprinted with permission from ref (51). Copyright 2016 Springer Nature. Part d is reprinted with permission from ref (107). Copyright 2014 American Chemical Society. Part e is reprinted with permission from ref (53).
The chiral plasmonic structures similar to the ones designed by Kuzyk et al.61 can also be immobilized by means of biotin–avidin interaction.104 Interestingly, upon anchoring of the structures on the substrate, their CD responses became switchable. When the sample was immersed in buffer, most of the structures were in an upstanding position, but when the buffer was dried, the structures lay horizontally on the surface. Their different relative orientations with respect to the exciting circularly polarized light could induce a change in the CD signal (see Figure 3b).
Another benefit for depositing DNA plasmonic structures on a surface is that they can be combined with materials with which it would be otherwise challenging to assemble in the solution phase. For example, an origami–AuNP dimer–graphene hybrid structure was formed by exfoliating a single layer of graphene on top of an immobilized AuNP nanoantenna.105 Such a hybrid system has demonstrated superior SERS performance compared to individual components.
Along the lines of using DNA origami as nanoadapters to position nanophotonic components beyond the accuracy of conventional lithography,50,96,97,106 Gopinath et al. recently demonstrated a reliable and controllable coupling of molecular emitters to photonic crystal cavities (PCCs) (see Figure 3c).51 Because of the high addressability of DNA origami, the location of the dye molecule was sufficiently precise to enable visualization of the local density of states within PCCs with a resolution of about 1/10 of a wavelength. Moreover, the intensity of the cavity emission could be digitally varied by changing the number of binding sites within a single cavity. Taking the high modularity of DNA origami into consideration, a great number of hybrid nanophotonic applications could be realized with this system.
In addition to relying just on the pattern with the same outline as DNA origami, circular openings in a metallic film can also host DNA origami adapters with size-selectivity. In the work of Pibiri et al.,107 individual dye molecules were placed inside a so-called zero-mode waveguide (ZMW) (see Figure 3d). In this way, the ZMW usage was optimized, and the photophysical properties of dyes were improved compared to stochastically immobilized molecules.
DNA objects can also be used as templates or stencils for producing accurate objects from different materials. One such example is creating custom-shaped features from inorganic oxides using DNA origami as a template.108 In this approach, Surwade et al.108 deposited DNA origami structures on a silicon oxide substrate, and by employing a chemical vapor deposition procedure, they created either oxide layers with DNA-origami-shaped openings or oxide shapes that have inherited the original origami shape (positive- and negative-tone patterns). To further employ this high-resolution and parallel substrate-based technique, Shen et al.52 showed that these origami-shaped openings in the grown oxide layer can be used as a mask for further lithography steps followed by metal evaporation. They demonstrated the versatility of the approach by fabricating gold, silver, and copper nanostructures having original DNA origami shapes (crosses and rectangles) on the silicon substrate. Later on, Shen et al.53 generalized the method to other substrates by fabricating, for example, gold bow-tie antennas and chiral gold shapes onto silicon nitride and sapphire (Figure 3e). The authors demonstrated that, by their DNA-assisted lithography (DALI) method, it is possible to produce transparent substrates with CD properties and a SERS capability for molecular diagnostics. In this work, individual metallic cross-shapes and antennas were also characterized, and these structures showed the plasmonic resonances at the visible wavelength range. However, the main drawback of this technique is that the metallic structures are randomly oriented on the substrate. Nevertheless, this method is compatible with the approaches presented by Gopinath et al.,50 and therefore parallel and large-scale patterning of plasmonic substrates with high resolution may become possible.
DNA origami has also been used in optical nanocavity fabrication and characterization. Chikkaraddy et al.109 constructed a nanocavity with <5 nm gap between a Au film and a AuNP. In such a device, a DNA origami plate between the two components has been employed not only to attach the AuNP to a substrate but also to precisely position Cy5 molecules with a nanometer-level lateral resolution. This enabled the precise mapping of the local density of optical states (LDOS) inside the nanocavity.
Conclusions
Although there are a plethora of examples of plasmonic structures that are assembled using DNA,46,48,49 so far only a few examples of DNA-based plasmonic interfaces or prototypes and approaches aiming toward DNA-assisted metasurfaces have been reported. However, despite all the challenges ahead, the future seems promising. As the cost of a single gram of mass-produced DNA origami has been reduced to 200 dollars,10 this means that covering a square meter of the substrate would cost only approximately 20 cents.106 This kind of vast area patterning may become possible by employing the presented methods to organize single DNA structures at interfaces or in 3D.91 Nevertheless, another route to achieve higher-order systems also exists, since approaches to create larger and larger DNA origami have recently been reported.15,16 This progress may lead to mass production of plasmonic DNA nanoantennas for sensing and intriguing optical metasurfaces consisting of miniature light scatterers.110,111
In general, using DNA origami as a bridge between bottom-up and top-down fabrication methods may help to go beyond the resolution limit of conventional lithography and solve the challenges of nanofabrication. This way it would be possible to achieve a whole class of novel hybrid materials with tunable optical and electronic properties.107,111 In addition to providing alternative approaches for solid-state nanofabrication, wet chemistry and structural DNA nanotechnology meet at the interface where DNA structures can be used in the fabrication of inorganic nanoparticles.22,53,112−114 Integrating these components into larger devices remains a challenge, but by merging the subfields with the help of DNA nanotechnology, we believe that the current achievements present just the beginning of a flourishing era of DNA-origami-based nanophotonics.
Acknowledgments
The financial support through the Academy of Finland (286845, 308578), Jane and Aatos Erkko Foundation, and Sigrid Jusélius Foundation is gratefully acknowledged. This work was carried out under the Academy of Finland Centers of Excellence Programme (2014–2019).
Biographies

Boxuan Shen received his M.Sc. (2012) and Ph.D. (2018) degrees in Physics from the University of Jyväskylä, Finland. Currently he works as a postdoctoral researcher in Biohybrid Materials research group at the Aalto University School of Chemical Engineering, Finland, under supervision of Dr. Veikko Linko. His research interests include structural DNA nanotechnology, nanolithography, self-assembled materials, plasmonics, and molecular electronics.

Mauri A. Kostiainen obtained his M.Sc. in organic chemistry from the University of Helsinki, Finland (2005), and the subsequent Ph.D. in engineering physics from the Helsinki University of Technology, Finland (2008). After receiving his doctoral degree, Kostiainen spent 2.5 years as a postdoctoral fellow at the Institute for Molecules and Materials (Radboud University Nijmegen, The Netherlands) developing new approaches for chemical and physical virology. He returned to Aalto University in 2011 as an Academy of Finland postdoctoral fellow and joined the faculty of School of Chemical Technology in 2013. Currently, he is an Associate Professor in the School of Chemical Engineering at Aalto University. His research interests focus on the integration of biological and synthetic building blocks in a designed manner to create biohybrid materials.

Veikko Linko received his M.Sc. (2007) and Ph.D. (2011) degrees in Physics from the University of Jyväskylä, Finland, and carried out a 2 year postdoctoral research period at the Technische Universität München, Germany, in Prof. Hendrik Dietz’s group (2011–2013). Since 2013, he has worked as a postdoctoral researcher in Biohybrid Materials group at the Aalto University School of Chemical Engineering under supervision of Prof. Mauri Kostiainen. In 2015 he was granted a Title of Docent at the University of Jyväskylä in the field of Molecular Nanotechnology (Physics) and in 2018 at the Aalto University in Bionanotechnology (Chemical Engineering). In 2017 he was a visiting scientist at the Universität Paderborn, Germany, in Dr. Adrian Keller’s Nanobiomaterials group. His research interests include structural DNA nanotechnology, self-assembled and biohybrid materials, drug-delivery applications, nanolithography, molecular electronics, and plasmonics.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
References
- Linko V.; Dietz H. The enabled state of DNA nanotechnology. Curr. Opin. Biotechnol. 2013, 24, 555–561. 10.1016/j.copbio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Jones M. R.; Seeman N. C.; Mirkin C. A. Programmable materials and the nature of the DNA bond. Science 2015, 347, 1260901. 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- Hong F.; Zhang F.; Liu Y.; Yan H. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 2017, 117, 12584–12640. 10.1021/acs.chemrev.6b00825. [DOI] [PubMed] [Google Scholar]
- Seeman N. C.; Sleiman H. F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 17068. 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- Nummelin S.; Kommeri J.; Kostiainen M. A.; Linko V. Evolution of structural DNA nanotechnology. Adv. Mater. 2018, 30, 1703721. 10.1002/adma.201703721. [DOI] [PubMed] [Google Scholar]
- Castro C. E.; Kilchherr F.; Kim D.-N.; Shiao E. L.; Wauer T.; Wortmann P.; Bathe M.; Dietz H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229. 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- Benson E.; Mohammed A.; Gardell J.; Masich S.; Czeizler E.; Orponen P.; Högberg B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- Veneziano R.; Ratanalert S.; Zhang K.; Zhang F.; Yan H.; Chiu W.; Bathe M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linko V.; Kostiainen M. A. Automated design of DNA origami. Nat. Biotechnol. 2016, 34, 826–827. 10.1038/nbt.3647. [DOI] [PubMed] [Google Scholar]
- Praetorius F.; Kick B.; Behler K. L.; Honemann M. N.; Weuster-Botz D.; Dietz H. Biotechnological mass production of DNA origami. Nature 2017, 552, 84–87. 10.1038/nature24650. [DOI] [PubMed] [Google Scholar]
- Wang P.; Meyer T. A.; Pan V.; Dutta P. K.; Ke Y. The beauty and utility of DNA origami. Chem. 2017, 2, 359–382. 10.1016/j.chempr.2017.02.009. [DOI] [Google Scholar]
- Bathe M.; Rothemund P. W. K. DNA nanotechnology: a foundation for programmable nanoscale materials. MRS Bull. 2017, 42, 882–888. 10.1557/mrs.2017.279. [DOI] [Google Scholar]
- Rothemund P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Dietz H.; Liedl T.; Högberg B.; Graf F.; Shih W. M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhomirov G.; Petersen P.; Qian L. Fractal assembly of micrometer-scale DNA origami arrays with arbitrary patterns. Nature 2017, 552, 67–71. 10.1038/nature24655. [DOI] [PubMed] [Google Scholar]
- Wagenbauer K. F.; Sigl C.; Dietz H. Gigadalton-scale shape-programmable DNA assemblies. Nature 2017, 552, 78–83. 10.1038/nature24651. [DOI] [PubMed] [Google Scholar]
- Maune H. T.; Han S.-P.; Barish R. D.; Bockrath M.; Goddard W. A. III; Rothemund P. W. K.; Winfree E. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat. Nanotechnol. 2010, 5, 61–66. 10.1038/nnano.2009.311. [DOI] [PubMed] [Google Scholar]
- Shen B.; Linko V.; Dietz H.; Toppari J. J. Dielectrophoretic trapping of multilayer DNA origami nanostructures and DNA origami-induced local destruction of silicon dioxide. Electrophoresis 2015, 36, 255–262. 10.1002/elps.201400323. [DOI] [PubMed] [Google Scholar]
- Tapio K.; Leppiniemi J.; Shen B.; Hytönen V. P.; Fritzsche W.; Toppari J. J. Toward single electron nanoelectronics using self-assembled DNA structures. Nano Lett. 2016, 16, 6780–6786. 10.1021/acs.nanolett.6b02378. [DOI] [PubMed] [Google Scholar]
- Uprety B.; Westover T.; Stoddard M.; Brinkerhoff K.; Jensen J.; Davis R. C.; Woolley A. T.; Harb J. N. Anisotropic electroless deposition on DNA origami templates to form small diameter conductive nanowires. Langmuir 2017, 33, 726–735. 10.1021/acs.langmuir.6b04097. [DOI] [PubMed] [Google Scholar]
- Uprety B.; Jensen J.; Aryal B. R.; Davis R. C.; Woolley A. T.; Harb J. N. Directional growth of DNA-functionalized nanorods to enable continuous, site-specific metallization of DNA origami templates. Langmuir 2017, 33, 10143–10152. 10.1021/acs.langmuir.7b01659. [DOI] [PubMed] [Google Scholar]
- Bayrak T.; Helmi S.; Ye J.; Kauert D.; Kelling J.; Schönherr T.; Weichelt R.; Erbe A.; Seidel R. DNA-mold templated assembly of conductive gold nanowires. Nano Lett. 2018, 18, 2116–2123. 10.1021/acs.nanolett.8b00344. [DOI] [PubMed] [Google Scholar]
- Steinhauer C.; Jungmann R.; Sobey T. L.; Simmel F. C.; Tinnefeld P. DNA origami as a nanoscopic ruler for super-resolution microscopy. Angew. Chem., Int. Ed. 2009, 48, 8870–8873. 10.1002/anie.200903308. [DOI] [PubMed] [Google Scholar]
- Jungmann R.; Steinhauer C.; Scheible M.; Kuzyk A.; Tinnefeld P.; Simmel F. C. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 2010, 10, 4756–4761. 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- Jungmann R.; Avendano M. S.; Woehrstein J. B.; Dai M.; Shih W. M.; Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11, 313–318. 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graugnard E.; Hughes W. L.; Jungmann R.; Kostiainen M. A.; Linko V. Nanometrology and super-resolution imaging with DNA. MRS Bull. 2017, 42, 951–959. 10.1557/mrs.2017.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S. M.; Bachelet I.; Church G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- Perrault S. D.; Shih W. M. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 2014, 8, 5132–5140. 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linko V.; Ora A.; Kostiainen M. A. DNA nanostructures as smart drug-delivery vehicles and molecular devices. Trends Biotechnol. 2015, 33, 586–594. 10.1016/j.tibtech.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Surana S.; Shenoy A. R.; Krishnan Y. Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat. Nanotechnol. 2015, 10, 741–747. 10.1038/nnano.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ora A.; Järvihaavisto E.; Zhang H.; Auvinen H.; Santos H. A.; Kostiainen M. A.; Linko V. Cellular delivery of enzyme-loaded DNA origami. Chem. Commun. 2016, 52, 14161–14164. 10.1039/C6CC08197E. [DOI] [PubMed] [Google Scholar]
- Castro C. E.; Dietz H.; Högberg B. DNA origami devices for molecular-scale precision measurements. MRS Bull. 2017, 42, 925–929. 10.1557/mrs.2017.273. [DOI] [Google Scholar]
- Marras A. E.; Zhou L.; Su H.-J.; Castro C. E. Programmable motion of DNA origami mechanisms. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 713–718. 10.1073/pnas.1408869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyk A.; Schreiber R.; Zhang H.; Govorov A. O.; Liedl T.; Liu N. Reconfigurable 3D plasmonic metamolecules. Nat. Mater. 2014, 13, 862–866. 10.1038/nmat4031. [DOI] [PubMed] [Google Scholar]
- Kuzyk A.; Yang Y.; Duan X.; Stoll S.; Govorov A. O.; Sugiyama H.; Endo M.; Liu N. A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat. Commun. 2016, 7, 10591. 10.1038/ncomms10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyk A.; Urban M. J.; Idili A.; Ricci F.; Liu N. Selective control of reconfigurable chiral plasmonic metamolecules. Sci. Adv. 2017, 3, e1602803. 10.1126/sciadv.1602803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijäs H.; Nummelin S.; Shen B.; Kostiainen M. A.; Linko V. Dynamic DNA origami devices: from strand-displacement reactions to external-stimuli responsive systems. Int. J. Mol. Sci. 2018, 19, 2114. 10.3390/ijms19072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linko V.; Nummelin S.; Aarnos L.; Tapio K.; Toppari J. J.; Kostiainen M. A. DNA-based enzyme reactors and systems. Nanomaterials 2016, 6, 139. 10.3390/nano6080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothelf K. V. Chemical modifications and reactions in DNA nanostructures. MRS Bull. 2017, 42, 897–903. 10.1557/mrs.2017.276. [DOI] [Google Scholar]
- Grossi G.; Jaekel A.; Andersen E. S.; Saccà B. Enzyme-functionalized DNA nanostructures as tools for organizing and controlling enzymatic reactions. MRS Bull. 2017, 42, 920–924. 10.1557/mrs.2017.269. [DOI] [Google Scholar]
- Kiviaho J. K.; Linko V.; Ora A.; Tiainen T.; Järvihaavisto E.; Mikkilä J.; Tenhu H.; Nonappa; Kostiainen M. A. Cationic polymers for DNA origami coating – examining their binding efficiency and tuning the enzymatic reaction rates. Nanoscale 2016, 8, 11674–11680. 10.1039/C5NR08355A. [DOI] [PubMed] [Google Scholar]
- Agarwal N. P.; Matthies M.; Gür F. N.; Osada K.; Schmidt T. L. Block copolymer micellization as a protection strategy for DNA origami. Angew. Chem., Int. Ed. 2017, 56, 5460–5464. 10.1002/anie.201608873. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy N.; Bastings M. M. C.; Nathwani B.; Ryu J. H.; Chou L. Y. T.; Vinther M.; Li W. A.; Anastassacos F. M.; Mooney D. J.; Shih W. M. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017, 8, 15654. 10.1038/ncomms15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvinen H.; Zhang H.; Nonappa; Kopilow A.; Niemelä E. H.; Nummelin S.; Correia A.; Santos H. A.; Linko V.; Kostiainen M. A. Protein coating of DNA nanostructures for enhanced stability and biocompatibility. Adv. Healthcare Mater. 2017, 6, 1700692. 10.1002/adhm.201700692. [DOI] [PubMed] [Google Scholar]
- Kielar C.; Xin Y.; Shen B.; Kostiainen M. A.; Grundmeier G.; Linko V.; Keller A. On the stability of DNA origami nanostructures in low-magnesium buffers. Angew. Chem., Int. Ed. 2018, 57, 9470–9474. 10.1002/anie.201802890. [DOI] [PubMed] [Google Scholar]
- Tan S. J.; Campolongo M. J.; Luo D.; Cheng W. Building plasmonic nanostructures with DNA. Nat. Nanotechnol. 2011, 6, 268–276. 10.1038/nnano.2011.49. [DOI] [PubMed] [Google Scholar]
- Julin S.; Nummelin S.; Kostiainen M. A.; Linko V. DNA nanostructure-directed assembly of metal nanoparticle superlattices. J. Nanopart. Res. 2018, 20, 119. 10.1007/s11051-018-4225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N.; Liedl T. DNA-assembled advanced plasmonic architectures. Chem. Rev. 2018, 118, 3032–3053. 10.1021/acs.chemrev.7b00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyk A.; Jungmann R.; Acuna G. P.; Liu N. DNA origami route for nanophotonics. ACS Photonics 2018, 5, 1151–1163. 10.1021/acsphotonics.7b01580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath A.; Rothemund P. W. K. Optimized assembly and covalent coupling of single-molecule DNA origami arrays. ACS Nano 2014, 8, 12030–12040. 10.1021/nn506014s. [DOI] [PubMed] [Google Scholar]
- Gopinath A.; Miyazono E.; Faraon A.; Rothemund P. W. K. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature 2016, 535, 401–405. 10.1038/nature18287. [DOI] [PubMed] [Google Scholar]
- Shen B.; Linko V.; Tapio K.; Kostiainen M. A.; Toppari J. J. Custom-shaped metal nanostructures based on DNA origami silhouettes. Nanoscale 2015, 7, 11267–11272. 10.1039/C5NR02300A. [DOI] [PubMed] [Google Scholar]
- Shen B.; Linko V.; Tapio K.; Pikker S.; Lemma T.; Gopinath A.; Gothelf K. V.; Kostiainen M. A.; Toppari J. J. Plasmonic nanostructures through DNA-assisted lithography. Sci. Adv. 2018, 4, eaap8978. 10.1126/sciadv.aap8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghebat Rafat A.; Pirzer T.; Scheible M. B.; Kostina A.; Simmel F. C. Surface-assisted large-scale ordering of DNA origami tiles. Angew. Chem., Int. Ed. 2014, 53, 7665–7668. 10.1002/anie.201403965. [DOI] [PubMed] [Google Scholar]
- Woo S.; Rothemund P. W. K. Self-assembly of two-dimensional DNA origami lattices using cation-controlled surface diffusion. Nat. Commun. 2014, 5, 4889. 10.1038/ncomms5889. [DOI] [PubMed] [Google Scholar]
- Arbabi A.; Horie Y.; Bagheri M.; Faraon A. Dielectric metasurfaces for complete control of phase and polarization with subwavelength spatial resolution and high transmission. Nat. Nanotechnol. 2015, 10, 937–943. 10.1038/nnano.2015.186. [DOI] [PubMed] [Google Scholar]
- Funke J. J.; Dietz H. Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotechnol. 2016, 11, 47–52. 10.1038/nnano.2015.240. [DOI] [PubMed] [Google Scholar]
- Hartl C.; Frank K.; Amenitsch H.; Fischer S.; Liedl T.; Nickel B. Position accuracy of gold nanoparticles on DNA origami structures studied with small-angle X-ray scattering. Nano Lett. 2018, 18, 2609–1615. 10.1021/acs.nanolett.8b00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M. T.; Schueder F.; Haas D.; Nickels P. C.; Jungmann R. Quantifying absolute addressability in DNA origami with molecular resolution. Nat. Commun. 2018, 9, 1600. 10.1038/s41467-018-04031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B.; Deng Z.; Yan H.; Cabrini S.; Zuckermann R. N.; Bokor J. Gold nanoparticle self-similar chain structure organized by DNA origami. J. Am. Chem. Soc. 2010, 132, 3248–3249. 10.1021/ja9101198. [DOI] [PubMed] [Google Scholar]
- Kuzyk A.; Schreiber R.; Fan Z.; Pardatscher G.; Roller E.-M.; Högele A.; Simmel F. C.; Govorov A. O.; Liedl T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 2012, 483, 311–314. 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- Roller E.-M.; Khorashad L. K.; Fedoruk M.; Schreiber R.; Govorov A. O.; Liedl T. DNA-assembled nanoparticle rings exhibit electric and magnetic resonances at visible frequencies. Nano Lett. 2015, 15, 1368–1373. 10.1021/nl5046473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S.; Deng Z.; Ding B.; Yan H.; Liu Y. DNA-origami-directed self-assembly of discrete silver-nanoparticle architectures. Angew. Chem., Int. Ed. 2010, 49, 2700–2704. 10.1002/anie.201000330. [DOI] [PubMed] [Google Scholar]
- Eskelinen A.-P.; Moerland R. J.; Kostiainen M. A.; Törmä P. Self-assembled silver nanoparticles in a bow-tie antenna configuration. Small 2014, 10, 1057–1062. 10.1002/smll.201302046. [DOI] [PubMed] [Google Scholar]
- Kaminska I.; Bohlen J.; Mackowski S.; Tinnefeld P.; Acuna G. P. Strong plasmonic enhancement of a single peridinin–chlorophyll a–protein complex on DNA origami-based optical antennas. ACS Nano 2018, 12, 1650–1655. 10.1021/acsnano.7b08233. [DOI] [PubMed] [Google Scholar]
- Lan X.; Su Z.; Zhou Y.; Meyer T.; Ke Y.; Wang Q.; Chiu W.; Liu N.; Zou S.; Yan H.; Liu Y. Programmable supra-assembly of a DNA surface adapter for tunable chiral directional self-assembly of gold nanorods. Angew. Chem., Int. Ed. 2017, 56, 14632–14636. 10.1002/anie.201709775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C.; Duan X.; Liu N. A plasmonic nanorod that walks on DNA origami. Nat. Commun. 2015, 6, 8102. 10.1038/ncomms9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan P.; Wen T.; Wang Z.; He Y.; Shi J.; Wang T.; Liu X.; Lu G.; Ding B. DNA origami directed assembly of gold bowtie nanoantennas for single-molecule surface-enhanced raman scattering. Angew. Chem., Int. Ed. 2018, 57, 2846–2850. 10.1002/anie.201712749. [DOI] [PubMed] [Google Scholar]
- Liu B.; Song C.; Zhu D.; Wang X.; Zhao M.; Yang Y.; Zhang Y.; Su S.; Shi J.; Chao J.; Liu H.; Zhao Y.; Fan C.; Wang L. DNA-origami-based assembly of anisotropic plasmonic gold nanostructures. Small 2017, 13, 1603991. 10.1002/smll.201603991. [DOI] [PubMed] [Google Scholar]
- Heck C.; Prinz J.; Dathe A.; Merk V.; Stranik O.; Fritzsche W.; Kneipp J.; Bald I. Gold nanolenses self-assembled by DNA origami. ACS Photonics 2017, 4, 1123–1130. 10.1021/acsphotonics.6b00946. [DOI] [Google Scholar]
- Turek V. A.; Chikkaraddy R.; Cormier S.; Stockham B.; Ding T.; Keyser U. F.; Baumberg J. J. Thermo-responsive actuation of a DNA origami flexor. Adv. Funct. Mater. 2018, 28, 1706410. 10.1002/adfm.201706410. [DOI] [Google Scholar]
- Yurke B.; Turberfield A. J.; Mills A. P. Jr.; Simmel F. C.; Neumann J. L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- Pilo-Pais M.; Goldberg S.; Samano E.; LaBean T. H.; Finkelstein G. Connecting the nanodots: programmable nanofabrication of fused metal shapes on DNA templates. Nano Lett. 2011, 11, 3489–3492. 10.1021/nl202066c. [DOI] [PubMed] [Google Scholar]
- Pilo-Pais M.; Watson A.; Demers S.; LaBean T. H.; Finkelstein G. Surface-enhanced Raman scattering plasmonic enhancement using DNA origami-based complex metallic nanostructures. Nano Lett. 2014, 14, 2099–2104. 10.1021/nl5003069. [DOI] [PubMed] [Google Scholar]
- Acuna G. P.; Möller F. M.; Holzmeister P.; Beater S.; Lalkens B.; Tinnefeld P. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science 2012, 338, 506–510. 10.1126/science.1228638. [DOI] [PubMed] [Google Scholar]
- Kühler P.; Roller E.-M.; Schreiber R.; Liedl T.; Lohmüller T.; Feldmann J. Plasmonic DNA-origami nanoantennas for surface-enhanced Raman spectroscopy. Nano Lett. 2014, 14, 2914–2919. 10.1021/nl5009635. [DOI] [PubMed] [Google Scholar]
- Simoncelli S.; Roller E.-M.; Urban P.; Schreiber R.; Turberfield A. J.; Liedl T.; Lohmüller T. Quantitative single-molecule surface-enhanced Raman scattering by optothermal tuning of DNA origami-assembled plasmonic nanoantennas. ACS Nano 2016, 10, 9809–9815. 10.1021/acsnano.6b05276. [DOI] [PubMed] [Google Scholar]
- Thacker V. V.; Herrmann L. O.; Sigle D. O.; Zhang T.; Liedl T.; Baumberg J. J.; Keyser U. F. DNA origami based assembly of gold nanoparticle dimers for surface-enhanced Raman scattering. Nat. Commun. 2014, 5, 3448. 10.1038/ncomms4448. [DOI] [PubMed] [Google Scholar]
- Roller E.-M.; Argyropoulos C.; Högele A.; Liedl T.; Pilo-Pais M. Plasmon-exciton coupling using DNA templates. Nano Lett. 2016, 16, 5962–5966. 10.1021/acs.nanolett.6b03015. [DOI] [PubMed] [Google Scholar]
- Weller L.; Thacker V. V.; Herrmann L. O.; Hemmig E. A.; Lombardi A.; Keyser U. F.; Baumberg J. J. Gap-dependent coupling of Ag-Au nanoparticle heterodimers using DNA origami-based self-assembly. ACS Photonics 2016, 3, 1589–1595. 10.1021/acsphotonics.6b00062. [DOI] [Google Scholar]
- Roller E.-M.; Besteiro L. V.; Pupp C.; Khorashad L. K.; Govorov A. O.; Liedl T. Hotspot-mediated non-dissipative and ultrafast plasmon passage. Nat. Phys. 2017, 13, 761–765. 10.1038/nphys4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli F.; Barth A.; Bae W.; Neukirchinger F.; Crevenna A. H.; Lamb D. C.; Liedl T. Directional photonic wire mediated by homo-Förster resonance energy transfer on a DNA origami platform. ACS Nano 2017, 11, 11264–11272. 10.1021/acsnano.7b05631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueder F.; Lara-Gutíerrez J.; Beliveau B. J.; Saka S. K.; Sasaki H. M.; Woehrstein J. B.; Strauss M. T.; Grabmayr H.; Yin P.; Jungmann R. Multiplexed 3D super-resolution imaging of whole cells using spinning disk confocal microscopy and DNA-PAINT. Nat. Commun. 2017, 8, 2090. 10.1038/s41467-017-02028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehrstein J. B.; Strauss M. T.; Ong L. L.; Wei B.; Zhang D. Y.; Jungmann R.; Yin P. Sub-100-nm metafluorophores with digitally tunable optical properties self-assembled from DNA. Sci. Adv. 2017, 3, e1602128. 10.1126/sciadv.1602128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfree E.; Liu F.; Wenzler L. A.; Seeman N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 1998, 394, 539–544. 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- Liu W.; Zhong H.; Wang R.; Seeman N. C. Crystalline two-dimensional DNA origami arrays. Angew. Chem., Int. Ed. 2011, 50, 264–267. 10.1002/anie.201005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y.; Douglas S. M.; Liu M. H.; Sharma J.; Cheng A.; Leung A.; Liu Y.; Shih W. M.; Yan H. Multilayer DNA origami packed on a square lattice. J. Am. Chem. Soc. 2009, 131, 15903–15908. 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerling T.; Wagenbauer K. F.; Neuner A. M.; Dietz H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 2015, 347, 1446–1452. 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- Kilchherr F.; Wachauf C.; Pelz B.; Rief M.; Zacharias M.; Dietz H. Single-molecule dissection of stacking forces in DNA. Science 2016, 353, aaf5508. 10.1126/science.aaf5508. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan S.; Subramaniam S.; Stewart A. F.; Grundmeier G.; Keller A. Regular nanoscale protein patterns via directed adsorption through self-assembled DNA origami masks. ACS Appl. Mater. Interfaces 2016, 8, 31239–31247. 10.1021/acsami.6b10535. [DOI] [PubMed] [Google Scholar]
- Zhang T.; Hartl C.; Fischer S.; Frank K.; Nickels P.; Heuer-Jungemann A.; Nickel B.; Liedl T. 3D DNA origami crystals. Adv. Mater. 2018, 30, 1800273. 10.1002/adma.201800273. [DOI] [PubMed] [Google Scholar]
- Zheng J.; Birktoft J. J.; Chen Y.; Wang T.; Sha R.; Constantinou P. E.; Ginell S. L.; Mao C.; Seeman N. C. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 2009, 461, 74–77. 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y.; Endo M.; Sugiyama H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nat. Commun. 2015, 6, 8052. 10.1038/ncomms9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabey S.; Kempter S.; List J.; Xing Y.; Bae W.; Schiffels D.; Shih W. M.; Simmel F. C.; Liedl T. Membrane-assisted growth of DNA origami nanostructure arrays. ACS Nano 2015, 9, 3530–3539. 10.1021/acsnano.5b00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y.; Endo M.; Morita M.; Takinoue M.; Sugiyama H.; Murata S.; Nomura S. M.; Suzuki Y. Environment-dependent self-assembly of DNA origami lattices on phase-separated lipid membranes. Adv. Mater. Interfaces 2018, 5, 1800437. 10.1002/admi.201800437. [DOI] [Google Scholar]
- Kershner R. J.; Bozano L. D.; Micheel C. M.; Hung A. M.; Fornof A. R.; Cha J. N.; Rettner C. T.; Bersani M.; Frommer J.; Rothemund P. W. K.; Wallraff G. M. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 2009, 4, 557–561. 10.1038/nnano.2009.220. [DOI] [PubMed] [Google Scholar]
- Hung A. M.; Micheel C. M.; Bozano L. D.; Osterbur L. W.; Wallraff G. M.; Cha J. N. Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat. Nanotechnol. 2010, 5, 121–126. 10.1038/nnano.2009.450. [DOI] [PubMed] [Google Scholar]
- Brassat K.; Ramakrishnan S.; Bürger J.; Hanke M.; Doostdar M.; Lindner J. K. N.; Grundmeier G.; Keller A.. On the adsorption of DNA origami nanostructures in nanohole arrays. Langmuir 2018, in press. 10.1021/acs.langmuir.8b00793. [DOI] [PubMed] [Google Scholar]
- Kuzyk A.; Yurke B.; Toppari J. J.; Linko V.; Törmä P. Dielectrophoretic trapping of DNA origami. Small 2008, 4, 447–450. 10.1002/smll.200701320. [DOI] [PubMed] [Google Scholar]
- Hakala T. K.; Linko V.; Eskelinen A.-P.; Toppari J. J.; Kuzyk A.; Törmä P. Field-enhanced nanolithography for high-throughput pattern transfer. Small 2009, 5, 2683–2686. 10.1002/smll.200901326. [DOI] [PubMed] [Google Scholar]
- Gerdon A. E.; Oh S. S.; Hsieh K.; Ke Y.; Yan H.; Soh H. T. Controlled delivery of DNA origami on patterned surfaces. Small 2009, 5, 1942–1946. 10.1002/smll.200900442. [DOI] [PubMed] [Google Scholar]
- Ding B.; Wu H.; Xu W.; Zhao Z.; Liu Y.; Yu H.; Yan H. Interconnecting gold islands with DNA origami nanotubes. Nano Lett. 2010, 10, 5065–5069. 10.1021/nl1033073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linko V.; Shen B.; Tapio K.; Toppari J. J.; Kostiainen M. A.; Tuukkanen S. One-step large-scale deposition of salt-free DNA origami nanostructures. Sci. Rep. 2015, 5, 15634. 10.1038/srep15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber R.; Luong N.; Fan Z.; Kuzyk A.; Nickels P. C.; Zhang T.; Smith D. M.; Yurke B.; Kuang W.; Govorov A. O.; Liedl T. Chiral plasmonic DNA nanostructures with switchable circular dichroism. Nat. Commun. 2013, 4, 2948. 10.1038/ncomms3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz J.; Matković A.; Pešić J.; Gajić R.; Bald I. Hybrid structures for surface-enhanced Raman scattering: DNA origami/gold nanoparticle dimer/graphene. Small 2016, 12, 5458–5467. 10.1002/smll.201601908. [DOI] [PubMed] [Google Scholar]
- Xu A.; Harb J. N.; Kostiainen M. A.; Hughes W. L.; Woolley A. T.; Liu H.; Gopinath A. DNA origami: the bridge from bottom to top. MRS Bull. 2017, 42, 943–950. 10.1557/mrs.2017.275. [DOI] [Google Scholar]
- Pibiri E.; Holzmeister P.; Lalkens B.; Acuna G. P.; Tinnefeld P. Single-molecule positioning in zeromode waveguides by DNA origami nanoadapters. Nano Lett. 2014, 14, 3499–3503. 10.1021/nl501064b. [DOI] [PubMed] [Google Scholar]
- Surwade S.; Zhou F.; Wei B.; Sun W.; Powell A.; O’Donnell C.; Yin P.; Liu H. Nanoscale growth and patterning of inorganic oxides using DNA nanostructure templates. J. Am. Chem. Soc. 2013, 135, 6778–6781. 10.1021/ja401785h. [DOI] [PubMed] [Google Scholar]
- Chikkaraddy R.; Turek V. A.; Kongsuwan N.; Benz F.; Carnegie C.; van de Goor T.; de Nijs B.; Demetriadou A.; Hess O.; Keyser U. F.; Baumberg J. J. Mapping nanoscale hotspots with single-molecule emitters assembled into plasmonic nanocavities using DNA origami. Nano Lett. 2018, 18, 405–411. 10.1021/acs.nanolett.7b04283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N.; Capasso F. Flat optics with designer metasurfaces. Nat. Mater. 2014, 13, 139–150. 10.1038/nmat3839. [DOI] [PubMed] [Google Scholar]
- Pilo-Pais M.; Acuna G. P.; Tinnefeld P.; Liedl T. Sculpting light by arranging optical components with DNA nanostructures. MRS Bull. 2017, 42, 936–942. 10.1557/mrs.2017.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B.; Tapio K.; Linko V.; Kostiainen M. A.; Toppari J. J. Metallic nanoshapes based on DNA nanostructures. Nanomaterials 2016, 6, 146. 10.3390/nano6080146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmi S.; Ziegler C.; Kauert D. J.; Seidel R. Shape-controlled synthesis of gold nanostructures using DNA origami molds. Nano Lett. 2014, 14, 6693–6698. 10.1021/nl503441v. [DOI] [PubMed] [Google Scholar]
- Sun W.; Boulais E.; Hakobyan Y.; Wang W. L.; Guan A.; Bathe M.; Yin P. Casting inorganic nanoparticles with DNA molds. Science 2014, 346, 1258361. 10.1126/science.1258361. [DOI] [PMC free article] [PubMed] [Google Scholar]