Abstract
Background
Auxin-induced genes regulate many aspects of plant growth and development. The Gretchen Hagen 3 (GH3) gene family, one of three major early auxin-responsive families, is ubiquitous in the plant kingdom and its members function as regulators in modulating hormonal homeostasis, and stress adaptations. Specific Auxin-amido synthetase activity of GH3 subfamily II genes is reported to reversibly inactivate or fully degrade excess auxin through the formation of amino acid conjugates. Despite these crucial roles, to date, genome-wide analysis of the GH3 gene family has not been reported in cotton.
Results
We identified a total of 10 GH3 subfamily II genes in G. arboreum, 10 in G. raimondii, and 20 in G. hirsutum, respectively. Bioinformatic analysis showed that cotton GH3 genes are conserved with the established GH3s in plants. Expression pattern analysis based on RNA-seq data and qRT-PCR revealed that 20 GhGH3 genes were differentially expressed in a temporally and spatially specific manner, indicating their diverse functions in growth and development. We further summarized the organization of promoter regulatory elements and monitored their responsiveness to treatment with IAA (indole-3-acetic acid), SA (salicylic acid), GA (gibberellic acid) and BL (brassinolide) by qRT-PCR in roots and stems. These hormones seemed to regulate the expression of GH3 genes in both a positive and a negative manner while certain members likely have higher sensitivity to all four hormones. Further, we tested the expression of GhGH3 genes in the BR-deficient mutant pag1 and the corresponding wild-type (WT) of CCRI24. The altered expression reflected the true responsiveness to BL and further suggested possible reasons, at least in part, responsible for the dramatic dwarf and shriveled phenotypes of pag1.
Conclusion
We comprehensively identified GH3 subfamily II genes in cotton. GhGH3s are differentially expressed in various tissues/organs/stages. Their response to IAA, SA, BL and GA and altered expression in pag1 suggest that some GhGH3 genes might be simultaneously involved in multiple hormone signaling pathways. Taken together, our results suggest that members of the GhGH3 gene family could be possible candidate genes for mechanistic study and applications in cotton fiber development in addition to the reconstruction of plant architecture.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1545-5) contains supplementary material, which is available to authorized users.
Keywords: Gossypium hirsutum, Gene family, GH3, Phytohormone, Expression patterns, cis-regulatory element
Background
As the first class of plant growth-promoting substances discovered more than 70 years ago, auxin is a critical plant hormone, playing pivotal roles in root and shoot architecture, vascular development, organ patterning, reproduction, hormonal crosstalk and responses to light, gravity and biotic and abiotic stimuli [1–4]. The effect of auxin on plant growth and development is achieved by regulating the expression of downstream target genes of auxin signaling. Among them, three major groups, auxin/indole-3-acetic acid (Aux/IAA), small auxin up RNA (SAUR) and Gretchen Hagen 3 (GH3) have been designated as early auxin-responsive gene families characterized by rapid expression changes on exposure to exogenous auxin [5–7]. Genome-wide isolation and characterization of early auxin-responsive family genes from many plant species has provided an opportunity for better understanding of the molecular mechanisms of auxin action [7–9].
The GH3 family of proteins were found to be ubiquitously present in numerous plant species and some vertebrates such as mouse, but not in the sequenced genomes of yeast, Caenorhabditis elegans or Drosophila [7, 10]. In 1984, the first GH3 gene was isolated from soybean by differential hybridization screening as an auxin-induced cDNA clone from etiolated hypocotyls [11]. GH3 family genes were first comprehensively identified in the model plant Arabidopsis [7]. To date, genome-wide analysis of the GH3 gene family has been completed in rice [8, 12], grape [13], rosids [14], tomato [15], legumes [10], Medicago truncatula [16], apple [17], and maize [4]. In general, the chromosomal distribution of GH3 genes is uneven and some members tend to be clustered [7, 16]. A majority of GH3 genes encode proteins of about 70 kDa and comprise 3–5 exons separated by introns of different lengths [6]. The promoter region of most GH3 genes contains a common cis-element (AuxREs, TGTCTC) recognized by auxin-response factors (ARFs). When treated with active auxin, the transcript level rapidly increases within 0.25–0.5 h and peaks a maximum of 2–4 h after treatment [11]. In addition to the specific response to auxin, GH3 genes are also transcriptionally regulated by stress-related hormones ABA (abscisic acid), JA (jasmonic acid) and SA (salicylic acid); growth-promoting hormones GA (gibberellins) and BRs (brassinosteroids); ripening/senescence-associated hormone ETH (ethylene); biotic stresses of pathogens; and abiotic stresses of light, salt, drought, cold, and so on [10, 16]. Moreover, GH3 genes are transcribed in a spatial- and temporal-specific manner, and the cytoplasmic-localized GH3 protein appears to be more abundant and stable than Aux/lAA and SAUR proteins [18]. Previous studies have shown that GH3 proteins maintain dynamic homeostasis or activity of IAA (indole-3-acetic acid), SA, and JA through the formation of hormone-amino acid conjugates [19, 20]. Notably, the fates of hormone conjugates seem to be diverse; formation of a JA-Ile conjugate can activate JA and enhance some jasmonate responses while several other forms of JA conjugates appear to be inactive [21]. The action of IAA-amido synthetases is more related to reducing the amount of excess active IAA by degradation (IAA-Asp/Glu) or temporary storage (IAA-Ala/Leu) [19]. Although some GH3 genes have been shown to form SA conjugates, their biochemical function remains unclear [20, 22–26].
Based on sequence similarity and substrate specificities, a total of 20 GH3 family genes in Arabidopsis have been classified into three subfamilies (I-III) [19]. In particular, 10 enzymes of subfamily III (AtGH3.7–3.8, AtGH3.12–3.16 and AtGH3.18–3.20) seem to be species specific and have only been identified in Arabidopsis to date, although some of these do not appear to be active in response to IAA or other plant hormones [20]. AtGH3.12/PBS3 is the only characterized member of subfamily III that is involved in SA metabolism, and is induced in response to avirulent or virulent Pseudomonas syringae pathogen infection [22, 23]. Subfamily I consists of AtGH3.10 and AtGH3.11/JAR1/FIN219, and the latter is the only enzyme functionally documented in subgroup I to activate JA by forming JA-amino acid conjugates [21]. The remaining members, AtGH3.1–3.6, AtGH3.9 and AtGH3.17, belong to subgroup II. All Arabidopsis subgroup II enzymes but AtGH3.1, which was not analyzed, were demonstrated to be IAA responsive [19]. It was noted that the IAA-amido synthetases are restricted to the eight enzymes of subfamily II in Arabidopsis, and the same may be true for GH3 enzymes of subfamily II from other plant species [19]. The subfamily II enzymes are involved in auxin homeostasis, and when the normal level of GH3 genes is disrupted, distinct but interrelated phenotypes can be observed [27]. The gain-of-function mutant ydk1-D, generated by an activation-tagged insertion proximal to the GH3.2 gene in Arabidopsis, shows a short primary root, reduced lateral root number, reduced apical dominance and short hypocotyl both in light and dark [28]. Wes1-D and gh3.5-1D, both resulting from activation of the GH3.5 gene by insertion of the 35S enhancer, exhibit auxin-deficient traits. The adult plant exhibits exaggerated dwarfism with small curly rosette leaves, short primary roots and a reduced number of lateral roots [24, 25]. With the exception of almost unchanged primary root length, a similar phenotype is found in the Arabidopsis dfl1-D mutant, in which the GH3.6 gene is over-expressed following a T-DNA insertion [29]. OsGH3.13/TLD1 is the rice ortholog of Arabidopsis GH3.17, and its activation results in alterations in plant architecture and tissue patterning, displaying a typical auxin-deficient phenotype of dwarfism, increased tillers, enlarged leaf angles, and improved drought tolerance [30].
Cotton (Gossypium spp.) is an important crop plant. The widely cultivated G. hirsutum (AD1-genome) is an allotetraploid originating from two diploid ancestor species, G. arboreum (A-genome) and G. raimondii (D-genome), through a natural hybridization and genome doubling process 1–2 MYA (Million Years Ago) [31, 32]. Cotton fiber, the most prevalent natural fiber used in the textile industry, is a single-celled seed hair derived from seed coat epidermal cells. The elongated fiber cells, composed of nearly pure cellulose, can reach lengths of nearly 6 cm [33]. With several commercially important products, cotton represents an important model plant and commercial crop for the study of polyploidy, cell elongation and cell wall synthesis [34]. Various studies on cotton fiber cell development have implicated auxin as a critical regulator of fiber development. IAA is an important component for fiber growth in in vitro-cultured cotton ovules [33]. Targeted expression of the IAA biosynthetic gene iaaM driven by the promoter of the FBP7 gene increased IAA levels in the epidermis of cotton ovules at the fiber initiation stage and substantially increased the number of lint fibers [35]. In addition, reconstruction of ideal plant architecture is imperative for mechanization in cotton production. Previous studies reported that altered IAA levels can change plant architecture in Arabidopsis and rice, which presented a new idea for regulating auxin homeostasis in vivo to rebuild cotton plant architecture [24, 28–30, 36]. The main function of GH3 family genes is to maintain dynamic homeostasis of active IAA. However, no genome-wide analysis of GH3 family genes in cotton has been published to date, especially of subfamily II GH3s with IAA-amino synthetase activity. The availability of genome sequences and the established GH3 gene family of other plant species provides opportunities for comprehensive analysis of the GH3 gene family in cotton.
In this study, we performed genome-wide identification and expression analysis of subfamily II GH3s in tetraploid G. hirsutum and two diploid species G. arboreum and G. raimondii. A phylogenetic tree was built to analyze the evolutionary relationships of GH3s between cotton and other plant species. In particular, the gene structure, conserved motif, expression pattern in root, stem, leaf, flower, fiber and ovule in different development stages, cis-regulatory elements of promoter sequences, and the responses to BL, GA, IAA and SA were examined systematically in G. hirsutum. The results of this study provide essential information on GH3 genes with potential application in studies of the mechanism of fiber initiation and development, fiber improvement, and plant architecture modification in cotton.
Results
Genome-wide identification and nomenclature of cotton GH3 subfamily II genes
The Hidden Markov Model (HMM) profile (PF03321.10) was employed to search against protein sequences of three cotton species (see below) [37–39]. In the first round of screening, using a threshold value of 10− 5, 49, 39, and 56 tentative genes of the GH3 family were obtained in G. arboreum (BGI, v2.0), G. raimondii (BGI, v1.0), and G. hirsutum (NAU, v1.1), respectively (Additional file 1: Table S1, sheet1–3). To identify the cotton orthologs of Arabidopsis GH3s, each set of tentative cotton GH3s was aligned with 20 AtGH3 proteins using ClustalX 1.83, and three neighbor-joining (NJ) phylogenetic trees were then constructed independently using PHYLIP 3.6 or MEGA7 based on the sequence alignment (Additional file 2: Figure S1 a, b and c). This strategy resulted in the identification of 17, 16, and 34 candidate genes of the GH3 family in G. arboreum, G. raimondii, and G. hirsutum, respectively (Additional file 1: Table S1, sheet1–3). Despite the presence of a putative GH3 domain, members clustered into AtGH3.20 in G. arboreum and G. raimondii were disregarded due to the incompleteness of AtGH3.20 and unexpected clustering compared with that in G. hirsutum. We focused on subfamily II GhGH3s because of their particular activity as IAA-amido synthetases. Members grouped with subfamily II Arabidopsis GH3 genes were considered theoretically as subfamily II GH3 genes in cotton. Conserved domain analysis using the online tool SMART showed the presence of a GH3 domain for all candidates. In total, 10, 10, and 20 candidate genes of subfamily II of the GH3 gene family were identified in G. arboreum, G. raimondii, and G. hirsutum, respectively (Table 1; Additional file 3: Table S2). Most of the GH3 genes identified here exhibited similar characteristics in ORF (Open Reading Frame) length, molecular weight, intron numbers and isoelectric point, except for GrGH3.5, which possessed a 3828 bp ORF according to the annotation of the G. raimondii genome (BGI, v1.0). We speculated that the abnormal length of GrGH3.5 was because of improper genome assembly. To confirm the observation, we used Gh_DGH3.5 from the Dt-subgenome of the G. hirsutum genome (NAU, v1.1) as a query to search against the G. raimondii genome (JGI, v2.0) and confirmed its homologous gene GrGH3.5 (locus_ID: Gorai.007G219500.1), which displayed, as expected, a normal ORF length of 1821 bp. We further tested 20 GhGH3 genes in the latest genome data of G. hirsutum (DOE-JGI, v3.1). Except for Gh_DGH3.4 in which two amino acids were deleted at the N-terminus, all GhGH3s shared the same peptide length, intron numbers, and chromosomal distribution (Additional file 4: Table S3). No frame shift mutation was found for any of the GhGH3 genes. In consideration of the highly similar characteristics of GH3 genes in the two genome assemblies of G. hirsutum (NAU, v1.1 and JGI, v3.1), we selected the widely used annotation of the G. hirsutum genome (NAU, v1.1) for subsequent analysis and presentation of subfamily II GH3s in cotton. All the identified proteins were named based on sequence identity with their orthologs in Arabidopsis and their chromosomal locations. The comprehensive information of GH3 genes in three cotton species, including gene name, locus_IDs, Arabidopsis orthologs, ORF length, intron numbers, chromosomal distribution as well as the length, molecular weight and isoelectric point of deduced polypeptides, are listed in Table 1 and Additional file 3: Table S2.
Table 1.
Characteristics of subfamily II GH3s in G. hirsutum (NAU-NBI, v1.1)
| Gene name | Locus_ID | Arabidopsis orthologs | ORF | Introns | Chr. | Position | Deduced polypeptide | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | Strand | Length (aa) | MW (Da) | pI | |||||||
| Gh_AGH3.1 | Gh_A03G1628 | AtGH3.1 | 1797 | 2 | A03 | 97,446,817 | 97,448,799 | – | 598 | 67,902.66 | 6.59 | |
| Gh_DGH3.1 | Gh_D02G2045 | AtGH3.1 | 1797 | 2 | D02 | 64,687,129 | 64,689,111 | – | 598 | 67,882.61 | 6.59 | |
| Gh_AGH3.2 | Gh_A12G0181 | AtGH3.1 | 1794 | 2 | A12 | 2,676,124 | 2,678,101 | – | 597 | 67,886.41 | 6.02 | |
| Gh_DGH3.2 | Gh_D12G0182 | AtGH3.1 | 1797 | 2 | D12 | 2,358,751 | 2,360,733 | – | 598 | 67,901.46 | 6.02 | |
| Gh_AGH3.3 | Gh_A13G0392 | AtGH3.1 | 1809 | 2 | A13 | 5,213,724 | 5,216,267 | + | 602 | 68,094.70 | 5.55 | |
| Gh_DGH3.3 | Gh_D13G0434 | AtGH3.1 | 1809 | 2 | D13 | 4,919,812 | 4,922,365 | + | 602 | 68,036.63 | 5.59 | |
| Gh_AGH3.4 | Gh_A11G0443 | AtGH3.1 | 1779 | 2 | A11 | 4,184,163 | 4,186,123 | + | 592 | 67,383.32 | 5.58 | |
| Gh_DGH3.4 | Gh_D11G0514 | AtGH3.1 | 1779 | 2 | D11 | 4,476,391 | 4,478,353 | + | 592 | 67,370.40 | 5.83 | |
| Gh_AGH3.5 | Gh_A11G1993 | AtGH3.1 | 1821 | 2 | A11 | 56,447,917 | 56,449,936 | – | 606 | 68,573.57 | 5.60 | |
| Gh_DGH3.5 | Gh_D11G1989 | AtGH3.1 | 1821 | 2 | D11 | 26,018,863 | 26,020,882 | + | 606 | 68,492.53 | 5.53 | |
| Gh_AGH3.6 | Gh_A01G0546 | AtGH3.5/3.6 | 1842 | 2 | A01 | 9,238,374 | 9,240,450 | + | 613 | 69,496.44 | 5.91 | |
| Gh_DGH3.6 | Gh_D01G0557 | AtGH3.5/3.6 | 1842 | 2 | D01 | 7,266,590 | 7,268,663 | + | 613 | 69,476.51 | 5.87 | |
| Gh_AGH3.7 | Gh_A01G0547 | AtGH3.5/3.6 | 1842 | 2 | A01 | 9,407,662 | 9,409,824 | + | 613 | 69,261.22 | 6.08 | |
| Gh_DGH3.7 | Gh_D01G0559 | AtGH3.5/3.6 | 1842 | 2 | D01 | 7,427,672 | 7,429,831 | + | 613 | 69,183.12 | 6.08 | |
| Gh_AGH3.8 | Gh_A03G1429 | AtGH3.5/3.6 | 1848 | 2 | A03 | 94,060,935 | 94,062,983 | + | 615 | 69,133.16 | 5.51 | |
| Gh_AGH3.9 | Gh_A11G1054 | AtGH3.9 | 1788 | 3 | A11 | 12,002,212 | 12,005,380 | + | 595 | 67,035.10 | 5.58 | |
| Gh_DGH3.9 | Gh_D11G1209 | AtGH3.9 | 1788 | 3 | D11 | 11,289,480 | 11,292,643 | + | 595 | 66,823.59 | 5.41 | |
| Gh_AGH3.17 | Gh_A03G1354 | AtGH3.17 | 1935 | 4 | A03 | 92,377,252 | 92,379,703 | – | 644 | 73,210.99 | 6.18 | |
| Gh_DGH3.17 | Gh_D02G1794 | AtGH3.17 | 1935 | 4 | D02 | 60,662,189 | 60,664,648 | – | 644 | 73,258.06 | 6.81 | |
| Gh_AGH3.18 | Gh_A04G0874 | AtGH3.17 | 1629 | 4 | A04 | 56,316,128 | 56,318,166 | – | 542 | 61,206.77 | 5.97 | |
Characteristics of GH3s in latest genomes of G. hirsutum (DOE-JGI, v3.1) is shown in Additional file 4 Table S3
Systematic bioinformatic analysis of cotton GH3 subfamily II genes
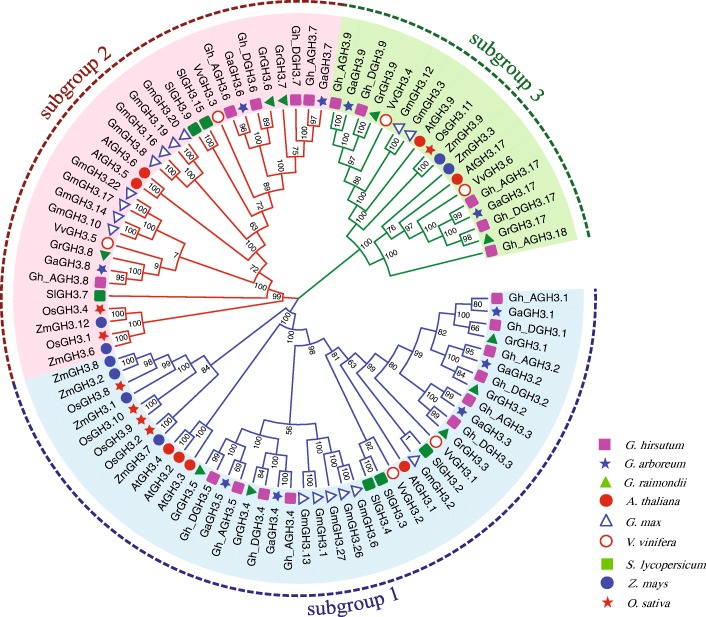
As described above, the GH3 gene family has been identified in numerous plant species. Various GH3 genes well-studied in model plants can help us to accurately search for and categorize them and predict their potential roles in cotton. Phylogenetic analysis was performed using neighbor-joining and a phylogenetic tree was constructed with subfamily II GH3s from different plant species, including 20 GhGH3s (G. hirsutum), 10 GrGH3s (G. raimondii), 10 GaGH3s (G. arboreum), 8 AtGH3s (A. thaliana), 6 SlGH3s (S. lycopersicum), 7 OsGH3s (O. sativa), 8 ZmGH3s (Z. mays), 16 GmGH3s (G. max) and 6 VvGH3s (V. vinifera). All sequences used in this study are supplied in Additional file 1: Table S1, sheet 4. The resulting tree exhibits clustering in three major groups (designated as subgroups 1–3 to differentiate them from the established subfamilies I-III in Arabidopsis). Gh_AGH3.1-Gh_DGH3.5, AtGH3.1-AtGH3.4 together with two VvGH3s, three SlGH3s, four ZmGH3s, four OsGH3s, and six GmGH3s fell into subgroup 1 while Gh_AGH3.9-Gh_AGH3.18, AtGH3.9, AtGH3.17, VvGH3.4/3.6, OsGH3.11, ZmGH3.3/3.9, and GmGH3.3/3.12 were allotted to subgroup 3 (Fig. 1). GH3 genes from nine kinds of plant species were present in almost all three subgroups with the exception that no members of SlGH3 genes were found in group 3. One out of 7 OSGH3 genes, OsGH3.11, was the only member existed in subgroup 3. Interestingly, VvGH3s, GmGH3s and cotton GH3 genes always clustered closely to each other, exhibiting closer relationships compared with other species, which is consistent with the close evolutionary relationships between these three plant species [40].
Fig. 1.
Phylogenetic relationships of subfamily II GH3s in plants. GH3s identified in three cotton species and established subfamily II GH3s from A .thaliana, S. lycopersicum, G. max, O. sativa, Z.mays and V. vinifera were included. Cotton GH3s identified here could be re-classified into subgroup 1–3. GH3s of other plants were unevenly distributed such that no SlGH3 and just one OsGH3 (OsGH3.11) clustered into subgroup 3. Members of VvGH3s, GmGH3s and cotton GH3s always clustered close to each other, exhibiting closer relationships. A neighbor-joining tree was built using MEGA 7.0 and visualized using online tool Evolview. The Gh_A and Gh_D in the upland cotton indicate the At- and Dt-subgenomes, respectively. GH3s belonging to one plant species are marked with the indicated leaf label decorations. Blocks of subgroup 1–3 are highlighted with light blue, pink and light green, respectively. Percentage bootstrap scores were calculated from 1000 iterations
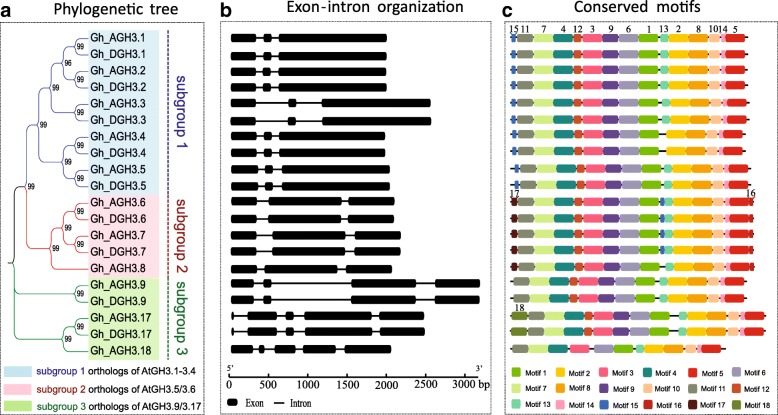
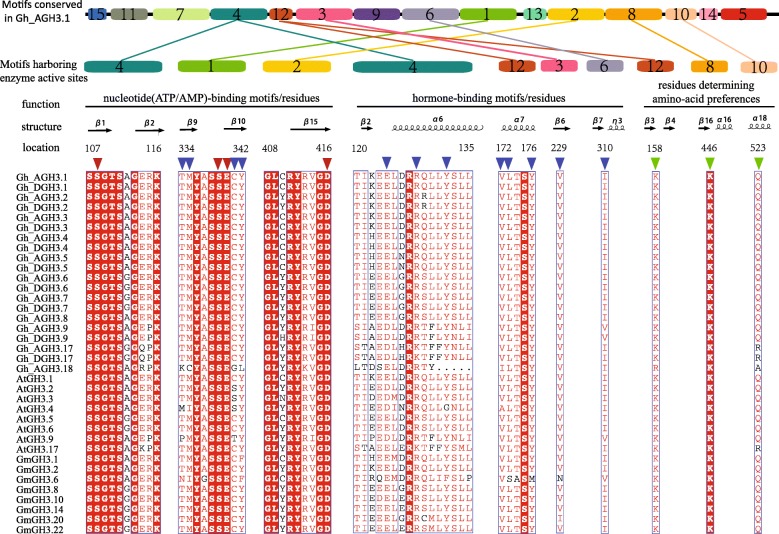
As described above, the 20 GhGH3 genes can be divided into three subgroups. The largest subgroup 1 contained Gh_AGH3.1-Gh_DGH3.5, showing close relationships to AtGH3.1. Gh_AGH3.6-Gh_AGH3.8 belonged to subgroup 2, which seem to be the orthologs of AtGH3.5/3.6 in cotton. The remaining members constituted subgroup 3 and displayed close relationships to AtGH3.9 (Fig. 2a). Gene structure analysis suggested that intron numbers were conserved among GhGH3 genes. Subgroup 1 and 2 possessed three exons while subgroup 3 had four or five. The length of the second exon in subgroup 2 GH3 genes was markedly longer than that of subgroup 1 (Fig. 2b). Conserved motif analysis showed that all 19 GhGH3 genes shared the same fourteen motifs except for GhGH3.18, among them eight conserved motifs containing known enzyme active sites, with some sites found in the same motifs. For example, motifs 1, 2 and 4 contained nucleotide (ATP/AMP)-binding motifs/residues while motifs 3, 4, 6 and 12 possessed hormone-binding motifs/residues. The residues contained in motifs 8, 10 and 12 determined amino-acid preferences (Fig. 2c; Fig. 3; Additional file 5: Figure S2 and Additional file 6: Figure S3).
Fig. 2.
Phylogenetic relationship, gene structure and motif analysis of 20 GhGH3s. a The small phylogenetic tree constructed with 20 GhGH3s, clearly exhibiting three subgroups. b Exon-intron organizations. Subgroup 1 and 2 have three exons while subgroup 3 genes have four or five. The length of the second exon in subgroup 2 is longer than that of subgroup 1. Black boxes and lines represent exons and introns, respectively. c Conserved motif analysis. Except for GhGH3.18, all of the remaining 19 GhGH3s share the same fourteen motifs ranging from 1 to 14. Necessary conserved motifs/residues for hormone-amido synthetase activity were analyzed, and motifs 1, 2 and 4 contain the nucleotide (ATP/AMP)-binding motifs/residues, motifs 3, 4, 6 and 12 possess the hormone-binding motifs/residues. The residues contained in motifs 8, 10 and 12 determine amino-acid preferences. The number above each motif represents its code as described below
Fig. 3.
Sequence alignment of conserved residues or motifs. Protein sequences of 20 GhGH3s were aligned using MAFFT with all members of subfamily II in Arabidopsis and some in Soybean. The nucleotide (ATP/AMP) and hormone-binding motifs/residues as well as residues determining amino-acid preferences are conserved, indicating their similar functions as the IAA-amido synthetase in cotton. The colored triangles represent active sites and secondary structures near the active sites are also shown. Numbering at the top indicates the location of residues or motifs corresponding to Gh_AGH3.1. Motifs are put above the corresponding alignment of conserved residues and conserved motifs in full-length Gh_AGH3.1 are also shown as an example to display the locations of all motifs. The same motifs are linked by lines with the same color. The number in each motif indicates its code as described in Fig. 2c. Note that the ends of the lines do not indicate the actual positions of enzyme active sites
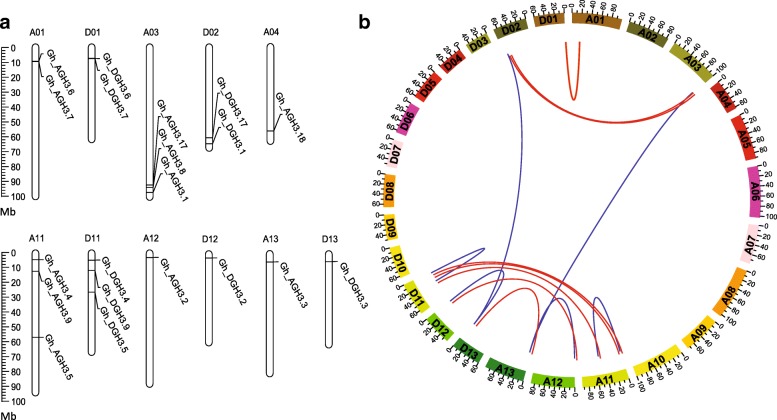
Furthermore, we analyzed the chromosomal distribution of GhGH3 genes. The 20 GhGH3 genes were unevenly distributed on 11 chromosomes; none were mapped on scaffold fragments (Fig. 4). Chromosomes A03, A11 and D11 each contained three GhGH3 genes while chromosomes A04, A12, A13, D12 and D13 only had one GhGH3 gene each. The putative GhGH3 genes of subfamily I were also mapped to the chromosomes. The largest gene clusters were found on chromosome D11, consisting of Gh_DGH3.4, Gh_DGH3.5, Gh_DGH3.9 as well as the unnamed locus_IDs of Gh_D11G1005 and Gh_D11G1006. Additionally, three genes were located on scaffold fragments (Additional file 7: Figure S4). Moreover, synteny analysis displayed the same chromosomal distribution, revealing that most GH3 loci were highly conserved between At- and Dt-subgenomes. Except for Gh_AGH3.8 and Gh_AGH3.18, all others from the A genome of G. arboreum and the D genome of G. raimondii were well matched to members of At- and Dt-subgenomes of G. hirsutum, forming orthologous pairs.
Fig. 4.
Mapping and synteny analysis of GhGH3 genes. GhGH3 genes were named based on sequence identity with their orthologs in Arabidopsis and their chromosomal locations. The capital letters and numbers indicate the At- or Dt-subgenome of G. hirsutum and chromosome numbers, respectively. a Illustrative diagram of chromosomal distribution of GhGH3 genes, showing uneven distribution on 11 out of 26 chromosomes. b CIRCOS figure of GhGH3 genes. The orthologous pairs from homologous chromosome pairs, At- and Dt-subgenome of G. hirsutum, are linked with red lines while the paralogous genes mapped on non-homologous chromosome pairs are linked with blue lines
Expression pattern analysis of cotton GH3 genes in vegetative and reproductive tissues/stages
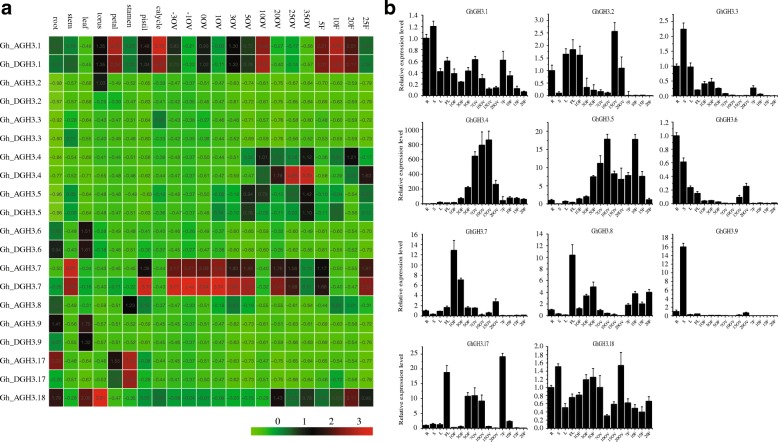
Gene expression is highly regulated, which is necessary for normal growth and development in plants. Precise expression patterns of candidate genes can give us important clues to their potential functions in growth and development. Trends of GhGH3 gene expression were shown using publicly available RNA-seq data, and the 20 GhGH3 genes displayed diverse expression patterns in 22 organs/tissues (Fig. 5a). We then verified these by quantitative RT-PCR (qRT-PCR) analysis, using custom-designed gene-specific primers (Additional file 8: Table S4). Allele pairs from the At- and Dt-subgenomes were designed as one and tested together due to the high sequence similarity of their transcription product. Thus, a total of 11 pairs of specific primers were used to check the expression profiles in roots (R), stems (S), leaves (L), flowers (FL), early developing ovules with attached fibers (1, 3, 5 OF, ovules plus fibers), and different stages of detached ovules and fibers (7, 10, 15, 20 O/F, ovules or fibers) (Fig. 5b). Based on the phylogenetic relationships and gene structure, we previously subdivided the GH3 genes into three subgroups with the hypothesis that members from the same subgroup should have similar expression patterns as well as functions (Fig. 2a; Fig. 6b). The results of RNA-seq and qRT-PCR analysis both revealed that GhGH3 genes were differentially expressed in various tissues/stages; unexpectedly, similar expression patterns were not observed for members of the same subgroup (Fig. 5). In our qRT-PCR experiment, GhGH3.1 and GhGH3.18 showed ubiquitous expression patterns in almost all tissues/stages. GhGH3.7 exhibited high accumulation during early stages of ovule and fiber development. GhGH3.8 is likely to be involved in the regulation of flower development and showed preferential expression in flowers. Expression levels of GhGH3.3 and GhGH3.6 were found to be high in vegetative organs (roots, stems and leaves). The transcripts of GhGH3.4 and GhGH3.5 were hardly detectable in vegetative organs but highly expressed during ovule development. Meanwhile, high transcript accumulation of GhGH3.5 throughout the whole process of fiber cell initiation and elongation was observed. The mRNA level of GhGH3.9 was high in a stem-specific manner, while GhGH3.17 was predominantly expressed in flowers, 5–10 DPA (days post anthesis) ovules and 7 DPA fibers. Results were similar for most cotton GH3 genes in RNA-seq data, and differences in expression pattern mainly existed in few organs/tissues. For example, GhGH3.1 and GhGH3.18 still showed ubiquitous expression patterns despite altered expression in different organs/tissues. The stem-preferential GhGH3.3 gene showed higher expression in calycle in RNA-seq data. GhGH3.4 had high transcript accumulation in developing fibers in addition to ovules. Although not being root-preferential and stem-specific genes, respectively, GhGH3.6 and GhGH3.9 also showed preferential expression in vegetative organs/tissue based on the RNA-seq data. In short, the results of analysis of RNA-seq data and qRT-PCR were basically identical, indicating the reliability and accuracy of RNA-seq data and qRT-PCR analysis, and further presenting the potential functions of GhGH3 genes in growth and development in corresponding organs/tissues. The original RNA-seq data for GhGH3 expression pattern analysis is supplied in Additional file 9: Table S5.
Fig. 5.
Expression profile of GhGH3s based on RNA-seq and qRT-PCR in various tissue/organs/stages. a Trends in GhGH3 gene expression based on publicly available RNA-seq data. Expression of individual genes are shown for 20 GhGH3s. Numbers on the x-axis indicate days post anthesis, with negative numbers implying days before anthesis. The prefix OV is for ovules and F for fiber. Green, black and red backgrounds represent low, intermediate and high expression levels, respectively. The original RPKM (reads per kilobase per million) values are normalized to 0–1 and shown in boxes. b Pattern of gene expression by qRT-PCR analysis. Expression of allele pairs or individual genes are shown. Values on the y-axis represent relative expression levels while the x-axis indicates days post anthesis. Materials from the field under normal conditions were tagged and sampled. The relative expression levels of GhGH3 genes were measured in roots (R), stems (S), leaves (L), flowers (FL), early stage developing ovules with attached fibers (1, 3, 5 DPA OF, ovules plus fibers), and different stages of detached ovules and fibers (7, 10, 15, 20 DPA O/F, ovules or fibers). GhHis 3 was used as an internal control to normalize the expression data. Error bars show standard deviation calculated from three replicates
Fig. 6.
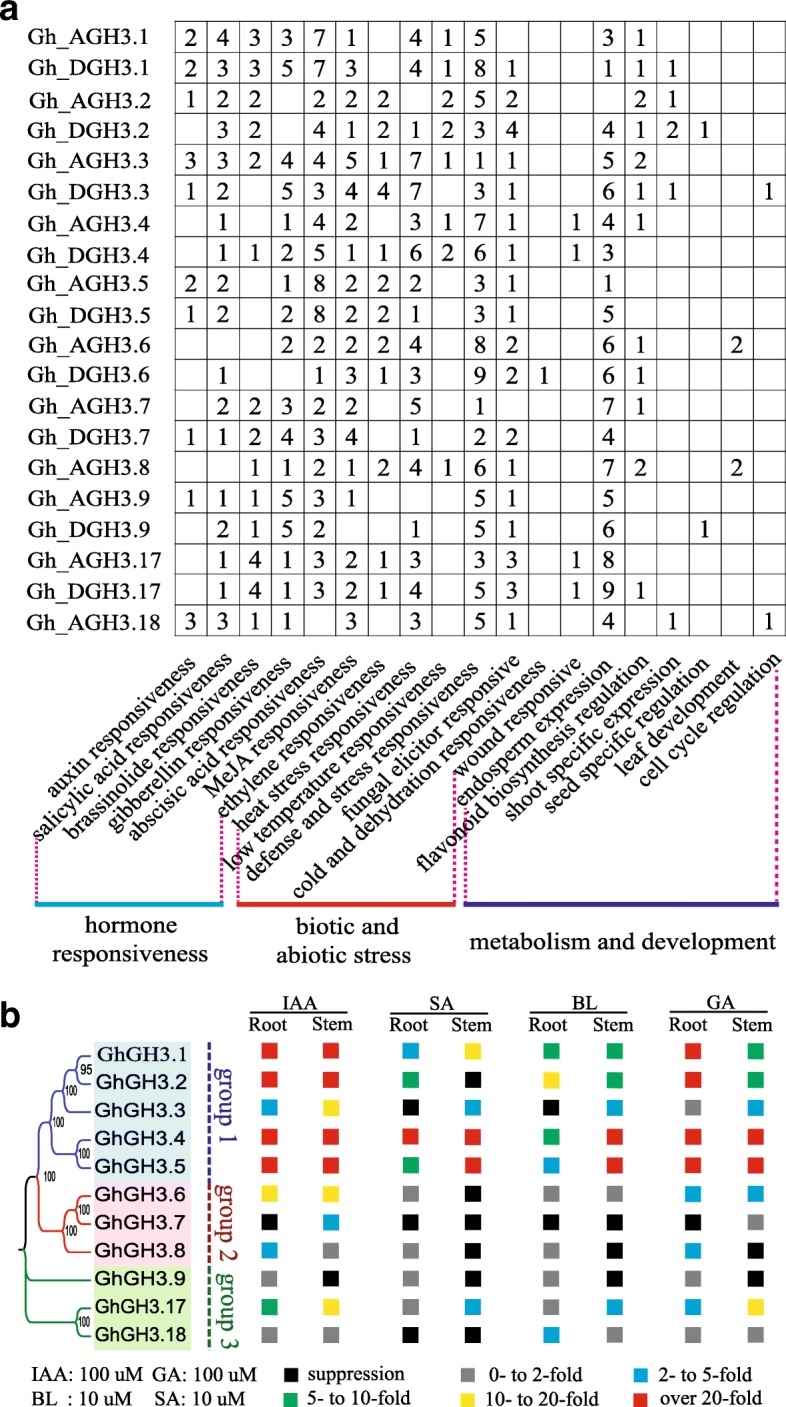
Organization of regulatory elements of GhGH3s and their expression in response to phytohormones. a The regulatory region of each GhGH3 gene was analyzed, the numbers indicate the sum of various cis-acting elements response to the same stimuli. b The summary of GhGH3 expression response to treatment with IAA, SA, BL and GA in roots (R) and stems (S). Each orthologous pair from At- and Dt-subgenome of G. hirsutum was designed as a single gene and tested together due to their highly identical sequences. In total, 11 analogue genes standing for 20 GhGH3s were generated and could be divided into 3 subgroups (subgroups 1–3). The expression pattern of 11 analogue genes was detected by qRT-PCR. The relative expression intensity, that is, the ratio of the highest expression level to that of the 0 h control, was allocated into six different grades ranging from suppression shown as black boxes to over 20-fold induction shown as red boxes
Cis-acting regulatory elements in GhGH3 promoters
Finely tuned gene expression is mainly regulated by corresponding promoters, and the action of promoters is further mediated by transcription factors through direct binding to cis-acting regulatory elements. Thus, analysis of upstream regulatory sequence will be helpful to understand the mode of target gene regulation and predict their potential functions [41]. To completely identify the putative cis-acting regulatory elements, about 3000 bp of non-coding sequence upstream of the predicted translation start site of each cotton GH3 gene was identified, extracted and scanned. Integrated utilization of the online software tools PLACE and PlantRegMap was performed to confirm the presence of abundant regulatory cores involved in hormone, stress and development-related responses (Fig. 6a; Additional file 9: Table S6).
Many hormone-related motifs were significantly enriched in most regulatory regions of tested cotton GH3 genes, including auxin (TGA-element, AuxRE-core), salicylic acid (TCA-element, SARE), brassinosteroid (E-box, BRRE-box), gibberellin (TATC-box, P-box and GARE-motif), abscisic acid (ABRE-box), methyl jasmonate (CGTCA-, TGACG-motif) and even ethylene (ERE-box). According to statistical analysis, two sets of stress-related motifs that are involved in abscisic acid and MeJA (methyl jasmonate) responsiveness were the most abundant cis-acting hormone responsive elements and were present in the promoters of all GhGH3 genes with the exception of Gh_AGH3.18 and Gh_DGH3.9, respectively. Gh_AGH3.6 and Gh_AGH3.8 lacked the abscisic acid responsive elements in their promoters. Both alleles of GhGH3.2 together with Gh_AGH3.6 showed an absence of typical gibberellin-related motifs in their regulatory regions. Interestingly, 14 out of 20 GhGH3 genes contained BR-response elements E-box or BRRE-box. The ethylene-responsive element ERE-box was also found in the promoters of 12 GhGH3 genes. As putative early auxin-responsive genes, intriguingly, half of the GhGH3 promoters identified here did not show the typical auxin responsive elements TGA-element or AuxRR-core.
In addition to hormone-related motifs, a large number of stress-related elements, as expected, were observed in the promoters of cotton GH3 genes. Specifically, defense and stress responsiveness TC-rich repeats and MBS sites can be found in all the promoters of GhGH3 genes. In the case of temperature responsiveness, 18 GhGH3 gene promoters contained HSE heat stress responsive elements, whereas eight possessed LTR low temperature responsive elements. Box-W1, a motif involved in fungal elicitor responsiveness, was another main component of stress-related motifs found in the promoters of all GhGH3s except Gh_AGH3.1 and Gh_AGH3.7. Allele pairs of GhGH3.4 and GhGH3.17 showed the presence of a WUN-motif wound responsive element. Gh_DGH3.6 was the only member containing the C-repeat/DRE core cold and dehydration responsive element. It is worth noting that some cis-elements involved in organ/tissue specific expression or metabolism, such as the Skn-1 motif, GCN4 motif for endosperm expression, as-2-box for shoot-specific expression, RY-element for seed specific regulation, MSA-like for cell cycle regulation, HD-Zip1/2 for leaf development, and MBS I/ II for flavonoid biosynthetic genes regulation, were also identified in some GH3 promoters. All the cis-acting regulatory elements predicted in the upstream 3 kb regions of GhGH3 family genes are summarized and listed in Fig. 6 and Additional file 9: Table S5, respectively. Promoter sequences of the 20 GhGH3 genes from two individual genome assemblies (NAU-NBI, v1.1 and DOE-JGI, v3.1), which share high sequence identity, are also supplied in Additional file 10: Table S6.
GhGH3 gene expression in response to IAA, SA, BL and GA treatments
GH3 genes exhibit a characteristic rapid response to active auxin within 0.25–0.5 h after treatment [11]. In addition to their typical feature of high sensitivity to auxin, these genes have also been reported to respond to SA, GA and BL (brassinolide) [10, 16]. Cotton GH3 genes were highly conserved both in protein-coding region and organization of cis-acting regulatory elements. The presence of plentiful hormone-related cis-acting regulatory elements in GhGH3s promoters further increased the possibility of response to hormones as stated above. Here, we monitored the expression changes of GhGH3 genes by qRT-PCR at five different time points (0, 0.5, 1, 3, 5 h) when exposed to 10 μM of BL and 100 μM of SA, IAA and GA. As described above, a total of 11 pairs of specific primers were employed to measure the gene expression profiles in response to hormone treatment both in roots and stems (Fig. 6b; Additional file 11: Figure S5). Under high concentration 100 μM IAA treatment, most GhGH3 genes were induced in the early stages of treatment both in roots and stems, especially GhGH3.1, GhGH3.2, GhGH3.4 and GhGH3.5, whose expressions were elevated hundreds- or even thousands-fold compared to that of the 0 h control (> 20-fold). GhGH3.7 and GhGH3.9 were found to be repressed in roots or stems, respectively. When exposed to SA, only GhGH3.4 exhibited up-regulation by tens- to hundreds-fold in both roots and stems, while similar up-regulation was also observed for GhGH3.5 in stems (> 20-fold). Transcript levels of some members, including GhGH3.2, GhGH3.3, GhGH3.6, GhGH3.8, GhGH3.9 and GhGH3.18, were slightly increased or even inhibited in roots or stems (< 2-fold or inhibition). Moreover, the transcript levels of GhGH3.7 and GhGH3.18 were down-regulated in tested organs/tissues.
BL treatment was also found to induce expression of some GhGH3 genes while suppressing others. The up-regulated expression of GhGH3.4 and GhGH3.5 was very significant in stems (> 20-fold) but moderate in roots (2- to 10-fold). Increased mRNA levels of GhGH3.1 and GhGH3.2 were also found at moderate levels both in roots and stems. Down-regulation of GhGH3.7 in both tested materials was observed after BL treatment. In the case of GA treatment, the expression of GhGH3.1, GhGH3.2, GhGH3.4 and GhGH3.5 showed significant up-regulation both in roots and stems, whereas most other GH3 genes exhibited only slight changes in expression level. All the results are summarized and displayed in Fig. 6 with different colored boxes. Interestingly, our qRT-PCR data showed that certain GhGH3 genes, including GhGH3.1, GhGH3.2, GhGH3.4, GhGH3.5 and GhGH3.17, exhibited concurrent sensitivity to all four hormone treatments with uneven expression levels.
Expression divergence in G. hirsutum BR-deficient mutant pag1
Pag1 is an artificial BR-deficient mutant derived from activation-tagged insertion in a BR metabolism-associated P450-like gene in the upland cotton CCRI24, which results in its over-expression and further elevated BR catabolism [42]. Compared to the wild type CCRI24, the mutant seedlings display short hypocotyls with small and wrinkled cotyledons. In the mature plant, short petioles, wrinkled leaves and shortened stems can be observed. Furthermore, the fiber length of the pag1 mutant is reduced compared to that of WT (Fig. 7a-e). These dramatic phenotypes in turn reveal the effect of BR on cotton plant growth and development, providing a rare opportunity, along with CCRI24, to study the functions of genes downstream of BR signaling and the interaction with other hormones in cotton.
Fig. 7.
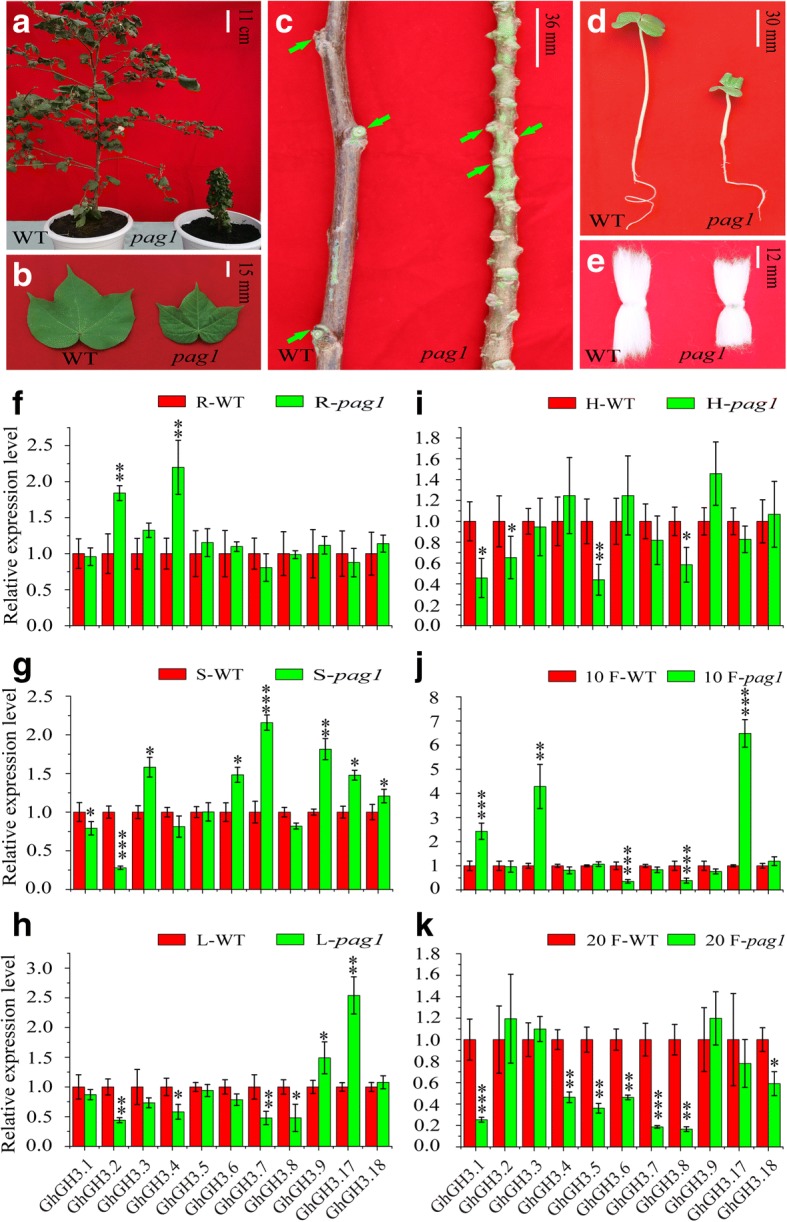
Comparison of phenotypes and expression differences in CCRI24 and pag1. a Phenotypes of WT (CCRI24) and pag1 plants at late development stages. b Mature leaves. c Phenotypes of stems in WT and pag1 plants. Green arrows indicate branch nodes. d Seedlings of WT and pag1 plants, showing short hypocotyl phenotype. e Adult fiber length of WT and pag1 plants. f-k the expression difference of GhGH3 genes in root (R), stem (S), leave (L), hypocotyl (H), 10 and 20 DPA fibers in WT and pag1 plants. The relative expression level of GhGH3 in WT organs/tissues was designated as 1, GhHis 3 was used as an internal control to normalize the expression data. Error bars show standard deviation calculated from three replications. Data points marked with asterisks (* P ≤ 0.05, ** P ≤ 0.01, and *** P ≤ 0.001) indicate statistically significant differences between control (GS) and other tissues
We previously analyzed BR-related cis-acting regulatory elements and the response of expression profiles to BL treatment. Here, we further tested expression differences of cotton GH3 subfamily II genes between pag1 and CCRI24. The expression pattern was monitored using qRT-PCR in roots, stems, leaves, hypocotyls, and 10 and 20 DPA fibers between the pair of materials on hand, and significant differences in expression of GhGH3 genes were observed (Fig. 7f-k). Opposite to the elevated expression in root after BL treatment, both GhGH3.2 and GhGH3.4 were induced in BR-deficient mutant pag1 compared to WT. GhGH3.1 and GhGH3.2 displayed down-regulated expression while GhGH3.7 and GhGH3.9 were up-regulated in pag1 stem, consistent with their nature of induction or inhibition in stem when exposed to exogenous BL. Inversely, some BL-induced GhGH3 genes in stems, including GhGH3.3, GhGH3.6, GhGH3.17 and GhGH3.18, were unexpectedly up-regulated in pag1 stem. The same was true for most GH3 genes in other organs/tissues, indicating the complex regulation or expression changes in response to reduced BR in pag1. As described above, BR-mediated gene expression was diverse depending on the organs and developmental stages of plants. Thus, it is difficult to speculate on its expression and evaluate the results without experimental evidence of the response of GH3 genes to BL in leaf, hypocotyl and fiber. In short, most GH3 genes having significant difference were inhibited in the pag1 mutant except for GhGH3.9 and GhGH3.17 in leaf, and GhGH3.1, GhGH3.3 and GhGH3.17 in 10 DPA fibers, respectively. The enhanced expression of GhGH3.1, GhGH3.3 and GhGH3.17 in 10 DPA fibers may be relevant to the short fiber in the pag1 mutant by reducing IAA levels in early fiber development. Additionally, whether most GH3 genes being inhibited in 20 DPA fibers had direct or indirect influence on fiber development in pag1 was not clear, but it may reflect the high expression of most GH3 genes in 20 DPA fiber of WT, which was consistent with the gradually reduced IAA level from 10 DPA. The true expression difference and the underlying molecular mechanisms need to be further investigated in the future.
Discussion
Conserved GH3 subfamily II genes in cotton
The Gretchen Hagen 3 (GH3) proteins, part of the acyl-adenylate/thioester-forming enzyme superfamily, maintain hormonal homeostasis of excess IAA, SA, and JA by forming conjugates with amino acids. GH3 proteins are a small multi-gene family widely found (not restricted to plants) and have been identified in numerous plant species ranging from mosses to angiosperms [10]. In the model plant Arabidopsis, 19 GH3 proteins together with an incomplete GH3.20 are subdivided into three subfamilies, and only the members of subfamily II show IAA-amido synthetase activity [19]. Similarly, it is speculated that subfamily II members of the soybean GH3 gene family corresponding to that of Arabidopsis have the capacity to conjugate excess auxin with amino acids based on the analysis of conserved hormone-binding residues [10]. In this study, we initially identified 17, 16, and 34 candidate genes of the GH3 gene family in G. arboreum, G. raimondii and G. hirsutum, respectively (Table 1; Additional file 3: Table S2). The number of GH3s in G. hirsutum was roughly equal to the sum of that in G. arboreum and G. raimondii. This was consistent with the fact that the allotetraploid (AD1 genome) species formed from hybridization of A- and D-genome ancestors around 1–2 million years ago [31, 32]. Because the IAA-amido synthetases are restricted to members of Group II in Arabidopsis and soybean, we paid special attention to systematic bioinformatic analysis of subfamily II GhGH3s. Members clustering with subfamily II of the Arabidopsis GH3 gene family were selected. Ultimately, 10, 10, and 20 candidate genes of subfamily II GH3s were identified in G. arboreum, G. raimondii and G. hirsutum, respectively. We analyzed various characteristics of GH3 genes in cotton. Similar to GH3s in other plant species, most GhGH3 genes encoded proteins with predicted molecular mass of 65–70 kDa, and isoelectric point of about 5.5–6.5 except for GrGH3.5 [7]. Most GhGH3 genes of subfamily II contained three exons and two introns. Members homologous to AtGH3.9 and AtGH3.17 seemed to have more exons, which was coherent with previous findings in maize. We also analyzed enzyme active sites according to the available crystal structures of GH3 proteins [10, 26, 43]. The residues involved in nucleotide (ATP/AMP)-binding, hormone-binding, and amino-acid preference were conserved in most GhGH3 genes, suggesting their similar functions as IAA-amido synthetases in cotton (Fig. 3; Additional file 6: Figure S3). The numbers of GH3 genes identified in different plants as well as in cotton are listed in Table 2. Members of subfamily III are species specific and have only been identified in Arabidopsis to date. Interestingly, the proportion of subfamily II GH3 genes to all GH3 members is relatively stable ranging from 0.4 to 0.67 in observed plants, indicating strict control of GH3 gene number in different genomes.
Table 2.
Proportion of subfamily II GH3s in various plant species
| species | I | II | III | total | ratio | Ref. |
|---|---|---|---|---|---|---|
| G. arboreum | 7 | 10 | – | 17 | 0.59 | – |
| G. raimondii | 6 | 10 | – | 16 | 0.63 | – |
| G. hirsutum | 14 | 20 | – | 34 | 0.59 | – |
| A. thaliana | 2 | 8 | 10 | 20 | 0.40 | [20] |
| S. lycopersicum | 9 | 6 | – | 15 | 0.40 | [15] |
| M. truncatula | 10 | 7 | – | 17 | 0.41 | [16] |
| G. max | 12 | 16 | – | 28 | 0.57 | [10] |
| O. sativa | 5 | 7 | – | 12 | 0.58 | [4, 12] |
| Z. mays | 5 | 8 | – | 13 | 0.62 | [4] |
| V. vinifera | 3 | 6 | – | 9 | 0.67 | [13] |
| M. domestica | 5 | 10 | – | 15 | 0.67 | [17] |
| S. bicolor | 7 | 9 | 16 | 0.56 | [3] |
Diverse expression pattern of GH3 genes in cotton
Determining spatial and temporal mRNA expression patterns of candidate genes can partly predict their biological functions in tissue/organ development. Numerous studies of GH3 gene expression patterns in various plants provided us with abundant reference data for analyzing GH3 expression in cotton [8, 10, 12–16]. In this study, the transcript level of GhGH3s was measured in 15 tissues/stages of vegetative organs, developing ovules and fibers. Similar to other plant species, cotton GH3 genes were differentially transcribed in a tissue-, stage-, and cell-specific manner. Some of the GhGH3 genes, including GhGH3.4, GhGH3.5, GhGH3.7, GhGH3.8, GhGH3.9 and GhGH3.17, showed low expression levels in leaves (Fig. 4b). This observation is similar to previous findings that all the ZmGH3s (Z. mays) and most of the MtGH3s (M. truncatula) display lower expression levels in leaves as compared with other organs [16]. Many SlGH3 (S. lycopersicum) genes also show lower expression levels in leaves [15]. Besides this, the accumulation of these GhGH3 mRNAs was low either in roots or stems, indicating their limited role in vegetative organ development. In Arabidopsis, ydk1-D and dfl1-D, two T-DNA insertion activation tagged mutants, show shortened primary roots and reduced lateral root number due to over-expression of AtGH3.1 and AtGH3.6, respectively [28, 29]. GhGH3.1 and GhGH3.6, the orthologs of AtGH3.1 and AtGH3.6, respectively, in cotton exhibited higher expression in roots, suggesting their possible role in root development.
In rice, eight out of 12 GH3 genes are predominately expressed in flowers [8]. GhGH3.8 displayed its highest expression level in flower, suggesting a role in flower development. In the process of fiber initiation and development, auxin undoubtedly is an important phytohormone that regulates fiber initiation, number and length [35]. Some GhGH3 genes like GhGH3.4, GhGH3.5 and GhGH3.7, exhibited high accumulation in ovules with attached fibers or in fibers. The enrichment of GH3 transcripts in certain cells may mediate free IAA content, thereby further controlling ovule and fiber development or acting as the signaling stimuli to regulate the transition from one stage to another. Cotton has a perennial and indeterminate growth habit and excessive vegetative growth can have a negative impact on cotton production. Cotton canopy is usually controlled by applications of excess plant growth regulator mepiquat chloride (1,1-dimethylpiperidinium chloride) [44]. In Arabidopsis, over-expression of most GH3 genes in subfamily II alters the homeostasis of active auxin, resulting in a dwarf phenotype. This gives us an important clue that cotton canopy could be controlled by modulating GH3 expression. Our qRT-PCR data show that GhGH3.1, GhGH3.3 and GhGH3.18 are predominantly expressed, and GhGH3.9 is specifically expressed in stem, indicating their possible roles in cotton growth and development.
Regulatory element organization implies GH3 family relevance to various stimuli
Gene expression or transcription is initiated through the action of an upstream regulatory promoter region, which can be considered as the combination of many cis-acting regulatory elements fused with a minimal basic start unit. Various combinations of regulatory cores confer the promoters with characteristics of strength, time-space specificity and response to stimuli. Therefore, analysis of regulatory elements of a target gene promoter can help us to predict its expression in response to various stimuli. Scanning of GhGH3 gene family promoters revealed the presence of numerous phytohormone-, stress- and development-related regulatory elements. AuxRE and TGA-element are two auxin-response regulatory elements. Previous studies have shown that the presence and absence of AuxRE can be correlated with inducibility or lack thereof for most GH3 genes [15, 27, 45]. The majority of GH3 gene promoters contain one or multiple AuxREs, suggesting their possible response to auxin [15, 16]. In cotton, five out of 20 GhGH3 genes contain AuxREs and half of them contain at least one AuxRE or TGA-element, implying their possible response to auxin. In addition to auxin responsive elements, the regulatory cores required for other growth-promoting hormone (BL and GA) responses were also widely distributed in the promoters of the GhGH3 gene family. E-box and BRRE are typical cis-elements that can be recognized by transcription factors downstream of the BR signaling pathway, BES1 or BZR1 [46]. In Arabidopsis, independent chromatin-immunoprecipitation microarray (ChIP-chip) experiments found that some members of GH3 gene family are the direct targets of BES1 or BZR1 [47, 48]. In sorghum and tomato, the expression of some GH3 genes in response to BRs has been confirmed [15]. These results suggest that BRs might directly or indirectly regulate the expression of some GhGH3 genes, as they harbor the cis-elements required for BR response. In the case of GA, the molecular mechanism by which some GH3 genes are regulated by the GA signaling pathway is not clear, but the real response of some GH3 genes to GA in tomato cannot be ignored [15]. We therefore present the possibility that GA response elements (TATC-, P-box and GARE-motif) enriched in the promoters of some GhGH3 genes may confer their response to GA. The ethylene response element ERE also can be seen in most GhGH3 gene family promoters, consistent with findings in pepper and tomato [15, 49]. The plentiful predicted hormone-related cis-acting regulatory elements that exist in most GhGH3 promoters indicate their important roles as core regulators in many hormone signaling pathways.
In addition to maintaining auxin homeostasis, GH3 gene family members are widely involved in disease resistance, biotic and abiotic stress responses, consistent with the enrichment of numerous stress-related cis-elements [3, 9, 24]. GhGH3 promoters contain many stress-related regulatory cores, including SA-responsive TCA-elements and SARE, ABA-responsive elements ABRE, MeJA-responsive elements CGTCA- and TGACG-motif, abnormal temperature-related elements HSE and LTR, defense and stress responsive elements TC-rich repeats and MBS, wound-responsive elements WUN-motif, and fungal elicitor responsive elements Box-W1. Among them, SARE and TCA-element, two typical SA-responsive cis-acting elements, were observed in 18 promoters out of 20 GhGH3 genes. Salicylic acid is a primary growth hormone that mediates plant disease resistance and abiotic stress responses [24, 50, 51]. Most established GH3 genes are more or less responsive to SA [4, 14–17]. Notably, some organ/tissue specific elements involved in shoot, seed, leaf or endosperm development were also observed. Except for Gh_AGH3.2, all members contained the endosperm expression elements Skn-1- and GCN4-motif. The promoters of Gh_DGH3.2 and Gh_DGH3.9 had the seed-specific regulation core RY-element, implying their potential functions in seed development. Some GhGH3 genes containing the shoot-specific expression element as-2-box are worth further study for their possible roles in shoot development.
Variable response of GH3 family to IAA, SA, BL and GA
Analysis of cis-acting regulatory elements showed large numbers of regulatory cores in the promoters of the GhGH3 gene family. To investigate their particular response to different hormones, qRT-PCR analysis was conducted to monitor the expression changes of GhGH3 genes in root and stem in response to IAA, SA, BL and GA. IAA treatment sharply up-regulated most GhGH3 genes, slightly inducing some members while repressing others in roots or stems. Similar expression trends have been observed in previous reports of the GH3 gene family in maize and sorghum [3]. In maize, the expression pattern of ZmGH3s responsive to auxin in root is different to that in stems. These differences include the induction intensity and the expression pattern of up-regulation or suppression. GH3s, especially the members of group II, are mainly involved in auxin homeostasis and stress response. The expression differences reveal subtle regulation of transcriptional levels of GhGH3 genes to maintain the free IAA content at an appropriate level for normal growth and development or in response to stress. Similar to IAA treatment, intricate expression differences can be seen on exposure to SA, BL and GA. In contrast, most GH3 genes are up-regulated in maize, M. truncatula (clover) and apple, whereas the majority of GH3 genes are down-regulated in tomato on exposure to SA treatment [3, 15–17]. Most of the tested GhGH3 genes were up-regulated in response to GA, however, these results are opposite to those in tomato [15]. The altered expression response to certain hormones may be due to the difference in plant species, suggesting novel functions in adaptation to changed circumstances in the process of evolution. In brief, all four tested hormones seemed to regulate the expression of GH3 genes in both a positive and negative manner in cotton. However, the response to hormones in roots and stems was different. Notably, we previously subdivided the genes of subfamily II into three subgroups based on their evolutionary relationships (Fig. 2a; Fig. 6b). Our qRT-PCR data show that the majority of subgroup 1 genes appeared to have higher sensitivity to stimuli. When treated with IAA, SA, GA or BL, the expression of most members of subgroup 1 was significantly up-regulated as compared to members of subgroup 2 and 3 (over 20-fold). These findings suggest strong functions for subgroup 1 members in growth and development, as well as in response to environmental stresses.
GH3s mediate fiber development, plant architecture, seed or somatic embryogenesis
GH3s, a small multi-gene family designated as the early auxin-responsive gene family, are widely distributed in higher plants. The particular function of subfamily II GH3 genes is to maintain auxin homeostasis by converting excess auxin to amino acid conjugates leading to inactivation or degradation [19]. Due to significant functional redundancy, loss-of-function mutations in one gene of subfamily II usually do not exhibit any visible phenotype or altered sensitivity to IAA. However, over-expression of one functional gene of subfamily II usually results in small or dwarf phenotypes across whole activation-tagged mutant or overexpression lines [25, 27–30, 36]. Previous findings imply that the dramatic effect on plant phenotype is caused by increased IAA-amido synthetase activity, which results in the reduction of free auxin content [30, 36, 52]. Notably, although the expression level of AtGH3.6 was enhanced by the activation-tagged insertion, free IAA content of dfl1-D was not significantly decreased [19]. In maize, most ZmGH3 genes were up-regulated in shoots in response to MeJA with no effect on free IAA content. These opposite findings may be attributed to small undetected or limited decline of free IAA in important tissues/organs. In addition, some other molecular mechanisms underlying this phenomenon may also exist and need further study. Taken together, GH3 gene expression changes usually alter free IAA content, with effects on plant growth and development as well as architecture.
Practical applications in the textile industry and as a model for studying single cell development make cotton a distinct crop plant. Numerous efforts have been made to discover the molecular mechanisms of fiber cell initiation and development in order to improve cotton fiber quality. Fiber cells are derived from ovule epidermal cells and their initiation and development seem to be more related to ovules themselves [53]. Ovules and fiber cells initiate accumulation of auxin at − 1 and 0 DPA, respectively, peak at + 2 DPA and gradually decrease to basal levels by + 10 DPA [35, 54]. This is consistent with the increasing expression of GhGH3.4 and GhGH3.5 with peaks also at 10 DPA, which may degrade and/or store excess IAA. In addition, enhanced expression of GH3 genes was not reported to have significant influence on seed morphology in Arabidopsis. Analogously, although dwarf plants and short siliques can be seen, transgenic Arabidopsis plants overexpressing GhGH3.5 gene do not display obvious changes in seed morphology (data not shown), and this may be the same case for cotton. The fact that GhGH3.4, GhGH3.5 and GhGH3.7 show high expression levels in developing ovules raises an interesting question about their biological function in ovule or fiber development. As accumulated IAA in fiber initials depends mainly on efflux transport from ovules rather than in situ synthesis, we speculate that some GhGH3 genes may play a role in fiber initiation and development by regulating the IAA content of ovules. Further, stable genetic transformation is necessary for functional characterization of these genes to explore their specific roles. Somatic embryogenesis is, to date, the most popular and stable transformation approach for cotton. However, this process is time-consuming, expensive, and labor-intensive, so that progress in increasing the regeneration rate has been limited [55]. The morphological and physiological characteristics of zygotic embryos are similar to those of somatic embryos, sharing sets of genes with similar expression patterns [56, 57]. Up-regulation of some GhGH3 genes during ovule development may imply their functions in seed or somatic embryo development, either directly or indirectly via altered free IAA content. Previous studies showed that over-expression of GH3 genes can alter plant architecture directly by local and/or systemic reduction of biologically active auxin in Arabidopsis and rice [24, 28–30, 36]. This is consistent with the most recent finding that low-level auxin zones in shoot apical meristem (SAM) organization center could break stem cell homeostasis and promote their proliferation, resulting in enlarged SAM and more lateral organs [58]. In this study, GhGH3.3 and GhGH3.9 were found to be expressed in a stem-preferential or -specific manner, thus we speculate that these GhGH3 genes together with others could be potential regulators for genetic modification of plant architecture in cotton. Finally, the presence of BR- and GA-related cis-acting regulatory elements and their particular response suggest that some GH3 genes might be involved in BR and GA signaling pathways [15]. We extrapolate that some dramatic phenotypes in BR- or GA-deficient mutants might be due to the altered expression of GH3 genes, which in turn reveal the applicability of GH3 genes to genetic modification of cotton plant architecture. In addition to the potential functions in architecture and development, as described above, cotton GH3 genes of subfamily II as well as I are also expected to be candidate genes for adversity-related studies and applications in cotton genetics and breeding.
Conclusion
In this study, a total of 10, 10, and 20 GH3 subfamily II genes were identified in G. arboreum, G. raimondii, and G. hirsutum, respectively. GhGH3 genes identified here clustered into subfamily II of the GH3 gene family and were conserved in intron numbers, protein motifs, active sites, and proportion in cotton GH3 genes. Expression pattern analysis showed that cotton GH3 genes were differentially expressed, suggesting their diverse roles in cotton plant growth and development. Promoter cis-element analysis and expression patterns under hormone treatments illustrated that cotton GH3 genes were also responsive to various stimuli. The variable expression of GhGH3 genes in BR-deficient mutant pag1 further confirms the conclusion stated above. Based on our experimental results, we conclude that some members of GhGH3 genes could be possible mediators for the study of cotton fiber development, fiber improvement, seed development, reconstruction of plant architecture, somatic embryogenesis, and stress tolerance. In short, our work provides comprehensive information on subfamily II of the cotton GH3 gene family to aid in future exploration of their functional divergence and genetic manipulation.
Materials and methods
Plant materials, growth conditions, and hormone treatments
G. hirsutum cultivar CCRI24, grown under experimental field conditions, was used to perform the expression of candidate genes. Flower buds were tagged or collected on the flowering day recorded as 0 DPA. The 7, 10, 15, and 20 DPA ovules and fibers were collected from the tagged buds. 1, 3, and 5 DPA fibers were sampled together with ovules because of the difficulty of separating them from each other. Young roots, stems and leaves were also collected from plants grown in the field after washing three times using distilled water. In order to compare GhGH3 expression differences between CCRI24 and pag1, 10 and 20 DPA fibers were collected as described above from the BR-deficient mutant pag1 that was grown simultaneously in the experimental field under normal conditions. The remaining materials were collected from sand-grown seedlings. In detail, hypocotyls of CCRI24 and pag1 were obtained from seedlings at the flattened-cotyledon stage. Materials of roots, stems and leaves were collected from CCRI24 and pag1 at the three-leaf stage. All samples were wrapped with DNase/RNase-free silver paper or tubes and frozen in liquid nitrogen immediately and stored at − 80 °C for subsequent use.
For phytohormone treatment, the CCRI24 seeds were planted in sand containers kept in a growth chamber at 28 °C with a photoperiod of 16 h light and 8 h dark. Flattened cotyledons were uprooted from the sealed container and washed with sterile water, then transferred into Hoagland’s solution with continuous air supply through an oxygen pump. Opaque containers were used and the nutrient solution was changed every 3 days until the third true-leaf stage. Seedlings were then immersed in new hydroponic culture solutions supplied with 100 μM IAA, SA, GA, or 10 μM BL. Root and stem samples at 0.5-h, 1-h, 3-h, and 5-h after applications of hormones were collected for qRT-PCR experiment. All samples were quick-frozen in liquid nitrogen and stored at − 80 °C for RNA isolation.
RNA extraction, cDNA synthesis, and qRT-PCR analysis
Total RNA was isolated using RNAprep Pure Plant kit (TIANGEN, Beijing, China). To avoid degradation all steps were carried out at low temperature. RNA quantity and quality was determined using both Nanodrop2000-C spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and gel electrophoresis. Approximately 1 μg of qualified total RNA of each sample was used to synthesize first strand complementary DNA according to the manufacturer’s instructions of the PrimeScript® RT Reagent kit after gDNA Eraser treatment (TaKaRa, Dalian, China).
Gene-specific primers for qRT-PCR were designed using the NCBI Primer-Blast Tool. Detailed information of all the primers used in this study is listed in Additional file 8: Table S4. qRT-PCR analysis was performed on a LightCycler TM 480 II System (Roche Applied Science, Basel, Switzerland) using LightCycler 480 SYBR green 1 Master mix (Roche). The relative amounts of candidate genes were calculated with the 2-ΔΔCT method. The cotton histone3 (GenBank: AF024716.1/locus_ID of NAU-NBI, v1.1: Gh_D03G0370) gene was used as the reference gene. All qRT-PCRs were performed in triplicate.
Genome-wide identification and nomenclature of GH3 subfamily genes in three cotton species
Available reference genome sequences, including G. hirsutum (NAU-NBI, v1.1; DOE-JGI, v3.1), G. arboreum (BGI, v2.0), and G. raimondii (BGI-CGP, v1.0; JGI, v2.0) were downloaded from COTTONGEN (http://www.cottongen.org/) or JGI (https://phytozome.jgi.doe.gov/pz/portal.html). Twenty protein sequences of the GH3 gene family of Arabidopsis were taken from the Arabidopsis Information Resource (TAIR, http://www.Arabidopsis.org/). Firstly, the Hidden Markov Model profile (PF03321.10, GH3 auxin-responsive promoter) of Arabidopsis GH3.1 (AtGH3.1) downloaded from pfam database (http://pfam.xfam.org/) was utilized as the query to search against the proteomes of the three cotton species using local HMM search program HMMER3.0. All genes with E-values below the threshold value of 10− 5 obtained from these three searches were presumed to be tentative genes for amino acid sequence retrieval by local blastdbcmd program (blast-2.2.31+). Further, the online Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de/) was employed to test the presence of a GH3 domain in all the candidate genes. Finally, in order to identify the orthologous genes of subfamily II of Arabidopsis GH3s in the three cotton species, each set of tentative cotton GH3s was aligned with the 20 GH3s of Arabidopsis using ClustalX 1.83 and three neighbor-joining (NJ) phylogenetic trees were constructed independently using MEGA7.0 (http://www.megasoftware.net/) or PHYLIP 3.6 (http://evolution.genetics.washington.edu/phylip.html). Bootstrap values were calculated from 1000 iterations. Only the genes that clustered with group II of the Arabidopsis GH3 gene family were considered theoretically as the group II genes of cotton and selected for further analysis.
Systematic bioinformatic analysis of cotton GH3 II subfamily
For phylogenetic analysis of cotton GH3 subfamily II with other plant species, proteomes of G. max (v9.0), V. vinifera (Genoscope.12X), S. lycopersicum (iTAGv2.40), Z. mays (v10), and O. sativa (v10) were downloaded from JGI (https://phytozome.jgi.doe.gov/pz/portal.html). All the GH3 protein sequences were obtained directly from the supplemental materials or retrieved from the proteomes based on the gene locus provided by previous studies. Similarly, each set of GH3s of the five plant species was aligned with the 20 GH3s of Arabidopsis; only the orthologous genes of subgroup II of the Arabidopsis GH3 family were selected. A neighbor-joining phylogenetic tree was constructed as mentioned above and visualized using Evolview (http://www.evolgenius.info/). For genetic structure analysis, the GFF (General Feature Format) files of 20 GhGH3 genes were retrieved from the genome database of G. hirsutum (NAU-NBI, v1.1). Intron-exon organization was then visualized using the online Gene Structure Display Server 2.0 (GSDS 2.0) (http://gsds.cbi.pku.edu.cn/). Conserved motifs were predicted by the online program MEME (http://meme-suite.org/). RNA-seq data were downloaded from the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA248163/) and analyzed as previously reported [59]. A heatmap was drawn using online software omicshare (http://www.omicshare.com/tools/Home/Soft/heatmap). The length, molecular weight, and isoelectric point of each GH3 protein was calculated using ExPasy (https://web.expasy.org/protparam/). Multiple sequence alignment of the enzyme active sites of selected GH3 protein sequences in Arabidopsis (8 AtGH3s), soybean (8 GmGH3s), and cotton (20 GhGH3s) was carried out using the local alignment tool of MAFFT7 (http://mafft.cbrc.jp/alignment/software/). The resulting output was then imported and visualized using the online tool ESPript (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Chromosomal distribution analysis was conducted using Mapchart (http://mapchart.software.informer.com/). CIRCOS software was employed to draw a collinearity map [59]. Analysis of cis-acting regulatory elements was performed using plantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PlantRegMap (http://plantregmap.cbi.pku.edu.cn/binding_site_prediction.php).
Additional files
Table S1. All protein sequences used in this study. (XLSX 71 kb)
Figure S1. Three NJ phylogenetic trees for identification of GH3 genes in cotton. GH3s in G. arboreum (a), G. raimondii (b) and G. hirsutum (c) are shown. All GH3s in Arabidopsis are marked by solid red circles, members of subfamily II GH3s in Arabidopsis are highlighted in lime. Gene name is located on the right side of the locus_ID. (PDF 1002 kb)
Table S2. Characteristics of subfamily II GH3s in G. arboreum and G.raimondii. Detailed characteristics of GH3s in two diploid cottons are shown. Locus_ID and sequence of GrGH3.5 in G. raimondii (BGI, v1.0) was replaced by that of its analogue in G. raimondii (JGI, v2.0) due to the abnormal length possibly caused by improper genomic assembly. (DOCX 24 kb)
Table S3. Comparison of characteristics of GH3s identified in two genome assemblies of G. hirsutum. (DOCX 19 kb)
Figure S2. Sequence logos of 18 motifs. (EPS 2278 kb)
Figure S3. Multiple sequence alignment of 20 GhGH3s. (PDF 35 kb)
Figure S4. Chromosomal distribution of subfamily II GhGH3 genes together with putative subfamily I GhGH3 genes. The orthologous pairs are linked with red dotted lines. Gray locus_IDs, the putative GhGH3 genes of subfamily I in G. hirsutum, are also mapped to the chromosomes. (EPS 1142 kb)
Table S4. Primers used in this study. (DOCX 23 kb)
Table S5. The original RNA-seq data for GhGH3 expression pattern analysis. (XLSX 20 kb)
Table S6. Organization of cis-acting regulatory elements in the promoters of subfamily II members of cotton GH3 gene family. The number represents the upstream distance to start codon. “+” and “-” represent the plus strand and minus strand, respectively. (XLSX 84 kb)
Figure S5. Expression of GhGH3 gene family in response to IAA, SA, BL and GA treatment. R, roots; S, stems. The expression pattern of each GhGH3 gene in response to IAA, SA, BL and GA treatment is also shown. (PDF 1630 kb)
Acknowledgments
We thank Venugopal Mendu and Ghulam Qanmber, two authors of this paper, for revising the manuscript. Thank Xiaona Li and Peng Huo (Zhengzhou Research Base, Institute of Cotton Research of CAAS, Zhengzhou) for technical assistance. We thank Bennet McComish, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript. We also appreciate the reviewers and editors for the patience to the work.
Funding
This work was supported by the Funds for Creative Research Groups of China (Grant No. 31621005) and Young Elite Scientist Sponsorship Program by CAST.
Availability of data and materials
The data sets of this article are included within the article and its additional file.
Abbreviations
- A. thaliana
Arabidopsis thaliana
- DPA
Days Post-Anthesis
- G. arboreum
Gossypium arboreum
- G. hirsutum
Gossypium hirsutum
- G. max
Glycine max
- G. raimondii
Gossypium raimondii
- GH3
Gretchen Hagen 3
- M. truncatula
Medicago truncatula
- O. sativa
Oryza sativa
- S. lycopersicum
Solanum lycopersicum
- V. vinifera
Vitis vinifera
- Z. mays
Zea mays
Authors’ contributions
FGL, ZRY and DQY conceived and designed the study; VM and QC contributed technical support in detailed experimental design, wrote part of the manuscript and made a large contribution to the manuscript revision with GQ; DQY, GQ and LLL carried out experiments; ZEY and LLW analyzed data; ZL, YL and JL prepared figures; DQY prepared the manuscript. All the authors have read, edited, and approved the current version of the manuscript.
Ethics approval and consent to participate
The plant materials (including seeds) were collected from Zhengzhou Research base, State key Laboratory of Cotton Biology and Institute of Cotton Research, CAAS. The experimental research on plants, including collection of plant material, was complied with the institutional, national, or international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Daoqian Yu, Email: yudaoqian88@163.com.
Ghulam Qanmber, Email: gqkhan12@gmail.com.
Lili Lu, Email: biolll@126.com.
Lingling Wang, Email: wll_198927@126.com.
Jie Li, Email: maggieli1106@126.com.
Zhaoen Yang, Email: yangzhaoen0925@126.com.
Zhao Liu, Email: liuzhaocaas@163.com.
Yi Li, Email: liyi20050901@163.com.
Quanjia Chen, Email: chqjia@126.com.
Venugopal Mendu, Email: venugopal.mendu@ttu.edu.
Fuguang Li, Email: aylifug@163.com.
Zuoren Yang, Phone: +86-377-55912760, Email: yangzuoren4012@163.com.
References
- 1.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot-London. 2005;95(5):707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126(3):467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Bai Y, Shen C, Wu Y, Zhang S, Jiang D, et al. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomic. 2010;10(4):533–546. doi: 10.1007/s10142-010-0174-3. [DOI] [PubMed] [Google Scholar]
- 4.Feng S, Yue R, Tao S, Yang Y, Zhang L, Xu M, et al. Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J Integr Plant Biol. 2015;57(9):783–795. doi: 10.1111/jipb.12327. [DOI] [PubMed] [Google Scholar]
- 5.Abel S, Theologis A. Early genes and auxin action. Plant Physiol 1996;111(1):9–17. [DOI] [PMC free article] [PubMed]
- 6.Guilfoyle TJ. Auxin-regulated genes and promoters. In: Hooykaas PJJ, Hall MA, Libbenga KR, editors. Biochemistry and molecular biology of plant hormones. Amsterdam: Elsevier; 1999. pp. 423–459. [Google Scholar]
- 7.Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49(3):373–385. [PubMed] [Google Scholar]
- 8.Jain M, Kaur N, Tyagi AK, Khurana JP. The auxin-responsive GH3 gene family in rice (Oryza sativa) Funct Integr Genomic. 2006;6(1):36. doi: 10.1007/s10142-005-0142-5. [DOI] [PubMed] [Google Scholar]
- 9.Jain M, Khurana JP. Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 2009;276(11):3148–3162. doi: 10.1111/j.1742-4658.2009.07033.x. [DOI] [PubMed] [Google Scholar]
- 10.Singh VK, Jain M, Garg R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front Plant Sci. 2015;5:789. [DOI] [PMC free article] [PubMed]
- 11.Hagen G, Kleinschmidt A, Guilfoyle T. Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta. 1984;162(2):147–153. doi: 10.1007/BF00410211. [DOI] [PubMed] [Google Scholar]
- 12.Terol J, Domingo C, Talón M. The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene. 2006;371(2):279–290. doi: 10.1016/j.gene.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Böttcher C, Boss PK, Davies C. Acyl substrate preferences of an IAA-amido synthetase account for variations in grape (Vitis vinifera L.) berry ripening caused by different auxinic compounds indicating the importance of auxin conjugation in plant development. J Exp Bot. 2011;62(12):4267–4280. doi: 10.1093/jxb/err134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okrent RA, Wildermuth MC. Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol Biol. 2011;76(6):489–505. doi: 10.1007/s11103-011-9776-y. [DOI] [PubMed] [Google Scholar]
- 15.Kumar R, Agarwal P, Tyagi AK, Sharma AK. Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum) Mol Gen Genomics. 2012;287(3):221–235. doi: 10.1007/s00438-011-0672-6. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Yue R, Sun T, Zhang L, Chen W, Zeng H, et al. Genome-wide identification, expression analysis of GH3 family genes in Medicago truncatula under stress-related hormones and Sinorhizobium meliloti infection. Appl Microbiol Biotechnol. 2015;99(2):841–854. doi: 10.1007/s00253-014-6311-5. [DOI] [PubMed] [Google Scholar]
- 17.Yuan H, Zhao K, Lei H, Shen X, Liu Y, Liao X, et al. Genome-wide analysis of the GH3 family in apple (Malus x domestica) BMC Genomics. 2013;14(1):297. doi: 10.1186/1471-2164-14-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright RM, Hagen G, Guilfoyle T. An auxin-induced polypeptide in dicotyledonous plants. Plant Mol Biol. 1987;9(6):625–634. doi: 10.1007/BF00020538. [DOI] [PubMed] [Google Scholar]
- 19.Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17(2):616. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staswick PE. Jasmonate response locus JAR1 and several related arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14(6):1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16(8):2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L, Wildermuth MC, et al. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007;144(2):1144. doi: 10.1104/pp.107.097691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okrent RA, Brooks MD, Wildermuth MC. Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J Biol Chem. 2009;284(15):9742–9754. doi: 10.1074/jbc.M806662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, et al. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem. 2007;282(13):10036–10046. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, et al. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007;145(2):450–464. doi: 10.1104/pp.107.106021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westfall CS, Zubieta C, Herrmann J, Kapp U, Nanao MH, Jez JM. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science. 2012;336(6089):1708. doi: 10.1126/science.1221863. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Tian CE, Duan J, Wu K. Research progresses on GH3s, one family of primary auxin-responsive genes. Plant Growth Regul. 2008;56(3):225–232. [Google Scholar]
- 28.Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, et al. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004;37(4):471–483. doi: 10.1046/j.1365-313x.2003.01973.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, et al. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001;25(2):213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang SW, Li CH, Cao J, Zhang YC, Zhang SQ, Xia YF, et al. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009;151(4):1889. doi: 10.1104/pp.109.146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wendel JF. New world tetraploid cottons contain old world cytoplasm. Proc Natl Acad Sci U S A. 1989;86(11):4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wendel JFCR. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. [Google Scholar]
- 33.Lee JJ, Woodward AW, Chen ZJ. Gene expression changes and early events in cotton fibre development. Ann Bot-London. 2007;100(7):1391–1401. doi: 10.1093/aob/mcm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492(7429):423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, et al. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol. 2011;29(5):453–458. doi: 10.1038/nbt.1843. [DOI] [PubMed] [Google Scholar]
- 36.Domingo C, Andrés F, Tharreau D, Iglesias DJ, Talón M. Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol Plant Microbe In. 2009;22(2):201–210. doi: 10.1094/MPMI-22-2-0201. [DOI] [PubMed] [Google Scholar]
- 37.Wang K, Wang Z, Li F, Ye W, Wang J, Song G, et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44:1098. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 38.Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 39.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 40.Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33(5):524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 41.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z, Zhang C, Yang X, Liu K, Wu Z, Zhang X, et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 2014;203(2):437–448. doi: 10.1111/nph.12824. [DOI] [PubMed] [Google Scholar]
- 43.Peat TS, Böttcher C, Newman J, Lucent D, Cowieson N, Davies C. Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell. 2012;24(11):4525. doi: 10.1105/tpc.112.102921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao L, Zhang L, Zhao X, Liu S, Werf WVD, Zhang S, et al. Crop growth, light utilization and yield of relay intercropped cotton as affected by plant density and a plant growth regulator. Field Crop Res. 2014;155:67–76. [Google Scholar]
- 45.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 46.Guo H, Li L, Aluru M, Aluru S, Yin Y. Mechanisms and networks for brassinosteroid regulated gene expression. Curr Opin Plant Biol. 2013;16(5):545–553. doi: 10.1016/j.pbi.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in arabidopsis. Dev Cell. 2010;19(5):765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65(4):634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 49.Liu K, Kang BC, Jiang H, Moore SL, Li H, Watkins CB, et al. A GH3-like gene, CcGH3, isolated from Capsicum chinense L. fruit is regulated by auxin and ethylene. Plant Mol Biol. 2005;58(4):447–464. doi: 10.1007/s11103-005-6505-4. [DOI] [PubMed] [Google Scholar]
- 50.Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy ASN, et al. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature. 2009;457:1154. doi: 10.1038/nature07612. [DOI] [PubMed] [Google Scholar]
- 51.Gao F, Dai R, Pike SM, Qiu W, Gassmann W. Functions of EDS1-like and PAD4 genes in grapevine defenses against powdery mildew. Plant Mol Biol. 2014;86(4):381–393. doi: 10.1007/s11103-014-0235-4. [DOI] [PubMed] [Google Scholar]
- 52.Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, et al. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 2008;20(1):228–240. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruan YL. Boosting seed development as a new strategy to increase cotton fiber yield and quality. J Integr Plant Biol. 2013;55(7):572–575. doi: 10.1111/jipb.12074. [DOI] [PubMed] [Google Scholar]
- 54.Zhang M, Zeng JY, Long H, Xiao YH, Yan XY, Pei Y. Auxin regulates cotton fiber initiation via GhPIN-mediated auxin transport. Plant Cell Physiol. 2016;58(2):385. doi: 10.1093/pcp/pcw203. [DOI] [PubMed] [Google Scholar]
- 55.Ge X, Zhang C, Wang Q, Yang Z, Wang Y, Zhang X, et al. iTRAQ protein profile differential analysis between somatic globular and cotyledonary embryos reveals stress, hormone, and respiration involved in increasing plantlet regeneration of Gossypium hirsutum L. J Proteome Res. 2015;14(1):268–278. doi: 10.1021/pr500688g. [DOI] [PubMed] [Google Scholar]
- 56.Dodeman VL, Ducreux G, Kreis M. Review article zygotic embryogenesis versus somatic embryogenesis. J Exp Bot. 1997;48(313):1493–1509. [Google Scholar]
- 57.Pullman GS, Johnson S, Peter G, Cairney J, Xu N. Improving loblolly pine somatic embryo maturation: comparison of somatic and zygotic embryo morphology, germination, and gene expression. Plant Cell Rep. 2003;21(8):747. doi: 10.1007/s00299-003-0586-9. [DOI] [PubMed] [Google Scholar]
- 58.Shi B, Guo X, Wang Y, Xiong Y, Wang J, Hayashi KI, et al. Feedback from lateral organs controls shoot apical meristem growth by modulating auxin transport. Dev Cell. 2018;44(11):204–216. doi: 10.1016/j.devcel.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 59.Yang Z, Gong Q, Qin W, Yang Z, Cheng Y, Lu L, et al. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017;17(1):113. doi: 10.1186/s12870-017-1065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. All protein sequences used in this study. (XLSX 71 kb)
Figure S1. Three NJ phylogenetic trees for identification of GH3 genes in cotton. GH3s in G. arboreum (a), G. raimondii (b) and G. hirsutum (c) are shown. All GH3s in Arabidopsis are marked by solid red circles, members of subfamily II GH3s in Arabidopsis are highlighted in lime. Gene name is located on the right side of the locus_ID. (PDF 1002 kb)
Table S2. Characteristics of subfamily II GH3s in G. arboreum and G.raimondii. Detailed characteristics of GH3s in two diploid cottons are shown. Locus_ID and sequence of GrGH3.5 in G. raimondii (BGI, v1.0) was replaced by that of its analogue in G. raimondii (JGI, v2.0) due to the abnormal length possibly caused by improper genomic assembly. (DOCX 24 kb)
Table S3. Comparison of characteristics of GH3s identified in two genome assemblies of G. hirsutum. (DOCX 19 kb)
Figure S2. Sequence logos of 18 motifs. (EPS 2278 kb)
Figure S3. Multiple sequence alignment of 20 GhGH3s. (PDF 35 kb)
Figure S4. Chromosomal distribution of subfamily II GhGH3 genes together with putative subfamily I GhGH3 genes. The orthologous pairs are linked with red dotted lines. Gray locus_IDs, the putative GhGH3 genes of subfamily I in G. hirsutum, are also mapped to the chromosomes. (EPS 1142 kb)
Table S4. Primers used in this study. (DOCX 23 kb)
Table S5. The original RNA-seq data for GhGH3 expression pattern analysis. (XLSX 20 kb)
Table S6. Organization of cis-acting regulatory elements in the promoters of subfamily II members of cotton GH3 gene family. The number represents the upstream distance to start codon. “+” and “-” represent the plus strand and minus strand, respectively. (XLSX 84 kb)
Figure S5. Expression of GhGH3 gene family in response to IAA, SA, BL and GA treatment. R, roots; S, stems. The expression pattern of each GhGH3 gene in response to IAA, SA, BL and GA treatment is also shown. (PDF 1630 kb)
Data Availability Statement
The data sets of this article are included within the article and its additional file.