Abstract
Background
MicroRNAs (miRNAs) play a crucial role in regulating diverse biological processes, including drug resistance. We investigated the potential roles of the miR-29 family in methotrexate (MTX) resistance in osteosarcoma.
Material/Methods
Two MTX-resistant osteosarcoma cell lines, MG-63/MTX and U2OS/MTX, were generated by continuous exposure to stepwise increasing concentrations of MTX. miR-29abc, COL3A1, and MCL1 mRNA expression levels were determined using quantitative real-time PCR (qRT-PCR). Protein expression levels of COL3A1 and MCL1 were detected by Western blot. Cell viability, IC50 value, and cell apoptosis were assessed by CCK-8 assay and flow cytometry, respectively. The target relationship between the miR-29 family and COL3A1 or MCL1 was confirmed by luciferase reporter assay.
Results
miR-29a, miR-29b, and miR-29c were significantly downregulated in MG-63/MTX and U2OS/MTX cells and in chemotherapy poor-response osteosarcoma tissues. Overexpression of the miR-29 family sensitized MG-63/MTX and U2OS/MTX cells to MTX and obviously promoted cell apoptosis compared with negative control. COL3A1 and MCL1 were identified to be target genes of the miR-29 family, and transfection with miR-29abc mimics in MG-63/MTX and U2OS/MTX cells decreased COL3A1 and MCL1 mRNA and protein expression. Meanwhile, overexpression of COL3A1 and MCL1 partly neutralized the effects of the miR-29 family on MTX resistance and cell apoptosis.
Conclusions
Taken together, our findings suggested a tumor-suppressor role of the miR-29 family in control of MTX resistance and cell apoptosis through regulating COL3A1 or MCL1. Targeting the miR-29 family might provide new strategies to overcome the high-dosage MTX-induced cytotoxicity in osteosarcoma treatment.
MeSH Keywords: Methotrexate, MicroRNAs, Osteosarcoma
Background
Osteosarcoma is the most common malignant bone tumor in children and adolescents [1,2]. In the last 20 years, unfortunately, little prognostic progress has been made [3]. The 5-year survival rate remains at 60–70% in osteosarcoma patients and approximately 40% of patients acquire drug resistance [4,5]. Therefore, seeking new therapeutic targets and understanding the molecular mechanism underlying the chemoresistance are of great importance for osteosarcoma treatment.
The current standard treatment for osteosarcoma is high-dose methotrexate (MTX) together with doxorubicin, cisplatin, and ifosfamide. MTX is a folate analog that inhibits dihydrofolate reductase. Despite its efficacy, high-dose MTX can cause many serious adverse effects, such as bone-marrow suppression, mucositis, and nephrotoxicity [6–8]. Control of chemoresistance and enhancing cell sensibility to MTX may allow a lower dosage and help to reduce the adverse effects of high MTX for osteosarcoma patients.
Previous studies have shown that the MTX resistance in osteosarcoma was associated decreased transport through the reduced folate carrier (RFC) and increased expression of dihydrofolate reductase (DHFR) [9,10]. Recently, several studies also indicated that the methylenetetetrahydrofolate reductase (MTHFR) 677C allele is associated with worse liver toxicity and that increased expression of high mobility group box 1 (HMGB1) and epidermal growth factor receptor (EGFR) contribute to MTX resistance in osteosarcoma through autophagy and survival signals, respectively [11–13]. These findings provided novel strategies to overcome MTX resistance for osteosarcoma treatment.
MicroRNAs (miRNAs) are small non-coding RNAs implicated in a growing number of human diseases, including osteosarcoma [14]. Several studies have shown that altered miRNAs can function as tumor suppressors or oncogenes through regulating many pathological and physiological processes, such as proliferation, migration, invasion, apoptosis, and multidrug resistance [15–17]. The present study aimed to investigate the potential involvement of the miR-29 family and MTX resistance in osteosarcoma. We examined the expression levels of miR-29a, miR-29b, and miR-29c in MTX-resistant osteosarcoma cell lines and tissues. Using gain- and loss-of-function approaches, we explored the underlying mechanisms of miR-29abc in control of MTX resistance. Our results will improve understanding of MTX resistance in osteosarcoma.
Material and Methods
Patients
Eighteen poor-response osteosarcoma tissues (<90% tumor necrosis after chemotherapy) and 18 stage-matched good-response osteosarcoma tissues (≥90% tumor necrosis after chemotherapy) were analyzed from the Second Affiliated Hospital of Soochow University. All patients were treated preoperatively with neoadjuvant chemotherapy. This study was approved by the Ethics Committee of the Second Affiliated Hospital of Soochow University. Written informed consent was obtained from each participant.
Cell culture and transfection
MG-63 and U2OS cells were purchased from ATCC. MTX-resistant cell lines MG-63/MTX and U2OS/MTX were generated by continuous exposure of drug-sensitive cells to stepwise increasing concentrations of MTX. Cells were incubated at a beginning concentration of 0.1 μg/ml MTX for 24 h and then we changed the medium. When the cells grew well, cells were exposed to 0.1 μg/ml MTX for 24 h and then we repeated the above steps 7 times. The concentrations of MTX were increased stepwise to 0.25, 0.5, 1, 5, and 10 μg/ml. To maintain the resistance, MG-63/MTX and U2OS/MTX were cultured with 1 μg/ml MTX supplement. Human COL3A1 and MCL1 cDNA were subcloned into the pcDNA 3.1 vector (Promega, Madison, WI). Cells were transiently transfected with miR-29abc mimics, miRNA negative control, pcDNA 3.1-COL3A1, pcDNA 3.1-MCL1, or pcDNA 3.1 using Lipofectamine 2000 (Life Technologies).
CCK-8 assay
Cell viability and cytotoxicity were evaluated using CCK-8 assay. Briefly, 1×104 cells/well were seeded into 96-well plates for 24-h incubation. Then, the cells were treated with various concentration of MTX. After 48 h, OD450 values were measured on a spectrophotometer (Bio-Rad, CA, USA) using the CCK-8 kit (Dojindo Molecular Technologies, Kumamoto, Japan) according to the manufacturer’s instructions. The IC50 value was calculated by the relative survival curve.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cultured cells or tissues using Trizol (Invitrogen). First-strand cDNA was synthesized with random primers using a RevertAid First-Strand cDNA Synthesis kit (Thermo Scientific, Waltham, MA, USA) or commercial miRNA reverse transcription PCR kit (RiboBio, Guangzhou, China). qRT-PCR analysis was performed using PowerUp™ SYBR Green Master Mix (Thermo-Fisher Scientific) on an Applied Biosystems 7500 Real-Time PCR System. miR-29a, miR-29b, miR-29c, and mRNA expression levels were normalized to U6 and β-actin. Each sample was assessed in triplicate. The relative expression of miRNA or mRNAs was calculated using the 2−ΔΔCt method. The primer sequences were as follows:
COL3A1 Forward: 5′-CGGCAATCCTGAACTTCCTG-3′,
Reverse: 5′-ATCAGCTTCAGGGCCTTCTT-3′;
MCL1 Forward: 5′-GCTGCATCGAACCATTAGCA-3′,
Reverse: 5′-ATGCCAAACCAGCTCCTACT-3′;
β-actin Forward: 5′-GGCATCCTCACCCTGAAGTA-3′,
Reverse: 5′-TAGCACAGCCTGGATAGCAA-3′.
Western blot
Total protein was extracted from cultured cells or tissues. A total of 30 mg of proteins was separated by SDS/PAGE (10% gel), transferred onto nitrocellulose membranes, and blocked by 5% non-fat milk. Then, the membranes were incubated with anti-COL3A1 (sc-514601, Santa Cruz) and anti-MCL1 (#94296, Cell Signaling Technology) overnight at 4°C and then incubated with horseradish peroxidase-labeled secondary antibodies for 2 h at room temperature. Bands were developed using chemiluminescence and β-actin (#4970, Cell Signaling Technology) was used as the control.
Apoptosis assay
After 24-h transfection, cells were treated with MTX for 48 h. Then, flow cytometry (Beckman Coulter, Brea, CA, USA) analysis was performed using an Annexin V-FITC apoptosis detection kit (Invitrogen) according to the manufacturer’s protocol. Briefly, cells were collected and dual-stained with 5 μl Annexin V and 5 μl propidium iodide for 30 min at room temperature. The stained cells were immediately analyzed by flow cytometry (BD, San Diego, CA, USA). Results are from 3 separate experiments.
Luciferase reporter assay
Luciferase reporters of wild-type (WT) COL3A1 3′ UTR and MCL1 3′ UTR containing predictive binding site of miRNA-29abc and mutant (Mut) COL3A1 3′ UTR and MCL1 3′ UTR were synthesized and cloned into pMIR-REPORT luciferase miRNA expression reporter vector (Ambion, TX, USA) between the SpeI and HindIII sites. Cells were co-transfected with luciferase reporters and miR-29abc mimics or miRNA negative control. After 48-h transfection, the luciferase activity was detected using a Luciferase Reporter Assay System kit (Promega).
Statistical analysis
One-way ANOVA was performed to analyze the differences between groups. The differences between 2 groups were compared using the independent-samples t test. All values are expressed as the mean ±SD. P value <0.05 was considered statistically significant. The statistical analyses were performed using SPSS 18.0 (SPSS, Inc., IL, USA).
Results
miR-29 family was down-regulated in MTX-resistant osteosarcoma cells and tissues
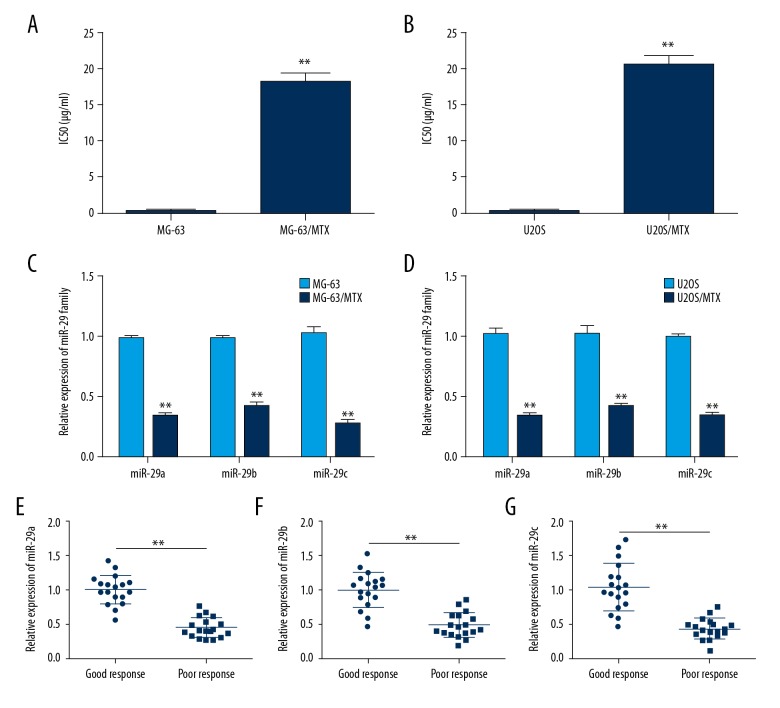
To explore the functional role of the miR-29 family in development of MTX resistance in osteosarcoma, 2 MTX-resistant osteosarcoma cell lines, MG-63/MTX and U2OS/MTX, were produced by continuous exposure of drug-sensitive cells to stepwise increasing concentrations of MTX. We first confirmed the MTX resistance of MG-63/MTX and U2OS/MTX cells treated with different concentration of MTX and found that their IC50 values were significantly enhanced by (112.42±42.93)-fold and (93.11±26.36)-fold compared with MG-63 and U2OS cells, respectively (Figure 1A, 1B). Then, we performed qRT-PCR assay to profile the expression levels of the miR-29 family. The results showed that miR-29abc expression level in MG-63/MTX cells was downregulated (2.97±0.30)-fold, (2.34±0.24)-fold and (3.81±0.76)-fold compared with that of MG-63 cells (Figure 1C). In U2OS/MTX cells, miR-29abc expression level was down-regulated by (3.19±0.50)-fold, (2.54±0.36)-fold, and (3.03±0.33)-fold compared with that of U2OS cells (Figure 1D). To further validate the expression level of miR-29abc, qRT-PCR assay was used in 18 poor-response osteosarcoma tissues and 18 stage-matched good-response osteosarcoma tissues. As shown in Figure 1E, miR-29abc expression level in poor-response osteosarcoma patients was also obviously decreased compared with that of good-response osteosarcoma patients. These findings suggested miR-29abc has a regulatory function in MTX resistance of osteosarcoma.
Figure 1.
Frequent downregulation of miR-29abc in MTX-resistant osteosarcoma cells and tissues. (A) IC50 value for MTX in MG-63 and MG-63/MTX cells determined using CCK-8 assay. (B) IC50 value for MTX in U2OS and U2OS/MTX cells determined using CCK-8 assay. (C) Relative expression levels of miR-29abc in MG-63 and MG-63/MTX cells assessed by qRT-PCR analysis. (D) Relative expression levels of miR-29abc in U2OS and U2OS/MTX cells assessed by qRT-PCR analysis. (E–G) Relative expression levels of miR-29abc in 18 poor-response osteosarcoma tissues (<90% tumor necrosis after chemotherapy) and 18 stage-matched good-response osteosarcoma tissues (≥90% tumor necrosis after chemotherapy) analyzed by qRT-PCR. ** P<0.01.
Overexpression of miR-29 family suppressed MTX resistance and promoted cell apoptosis
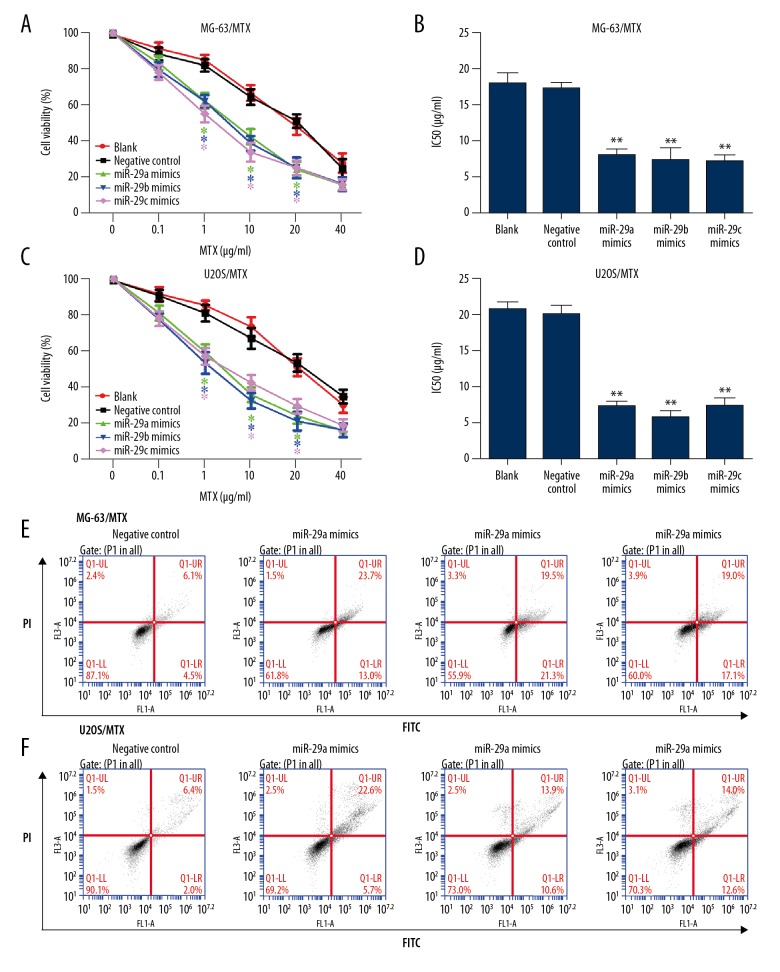
To confirm the relationship between miR-29 family and MTX resistance in osteosarcoma, miR-29abc mimics or miRNA negative control was transiently transfected into MG-63/MTX and U2OS/MTX cells. The transfection efficiency was first evaluated by qRT-PCR, as shown in Table 1. Then, we investigated the effect of miR-29abc on cell viability and MTX resistance using CCK-8 assay. As shown in Figure 2A and 2B, overexpression of miR-29abc significantly decreased cell viability and IC50 values in both MG-63/MTX cell lines. Consistent with MG-63/MTX cells, inhibitory effects of miR-29abc mimics on cell viability and MTX resistance were observed in U2OS/MTX cells (Figure 2C, 2D). Flow cytometry analysis showed dramatic apoptosis in MG-63/MTX and U2OS/MTX cells transfected with miR-29abc mimics compared with miRNA negative control (Figure 2E, 2F).
Table 1.
Transfection efficiency of miR-29abc mimics validated by qRT-PCR compared with miRNA negative control.
| miRNAs | Fold change | |
|---|---|---|
| MG63/MTX | U2OS/MTX | |
| miR-29a | 170.58±16.54 | 162.18±14.74 |
| miR-29b | 155.49±27.89 | 151.41±25.55 |
| miR-29c | 187.30±15.71 | 166.76±16.47 |
Figure 2.
Overexpression of miR-29 family suppressed MTX resistance and promoted cell apoptosis. (A) MG-63/MTX cells were transfected with miR-29abc or negative control and treated with different concentrations of MTX for 48 h. Cell viability was determined using CCK-8 assay. (B) IC50 value for MTX in MG-63/MTX cells transfected with miR-29abc or negative control and treated with different concentrations determined using CCK-8 assay. (C) U2OS/MTX cells were transfected with miR-29abc or negative control and treated with different concentrations of MTX for 48 h. Cell viability was determined using CCK-8 assay. (D) IC50 value for MTX in U2OS/MTX cells transfected with miR-29abc or negative control and treated with different concentrations determined using CCK-8 assay. (E) Flow cytometry analysis was performed to determine the effect of miR-29abc mimics on cell apoptosis induced by MTX in MG-63/MTX cells. (F) Flow cytometry analysis was performed to determine the effect of miR-29abc mimics on cell apoptosis induced by MTX in U2OS/MTX cells. * P<0.05, ** P<0.01 compared with negative control.
COL3A1 and MCL1 are target genes of miR-29 family
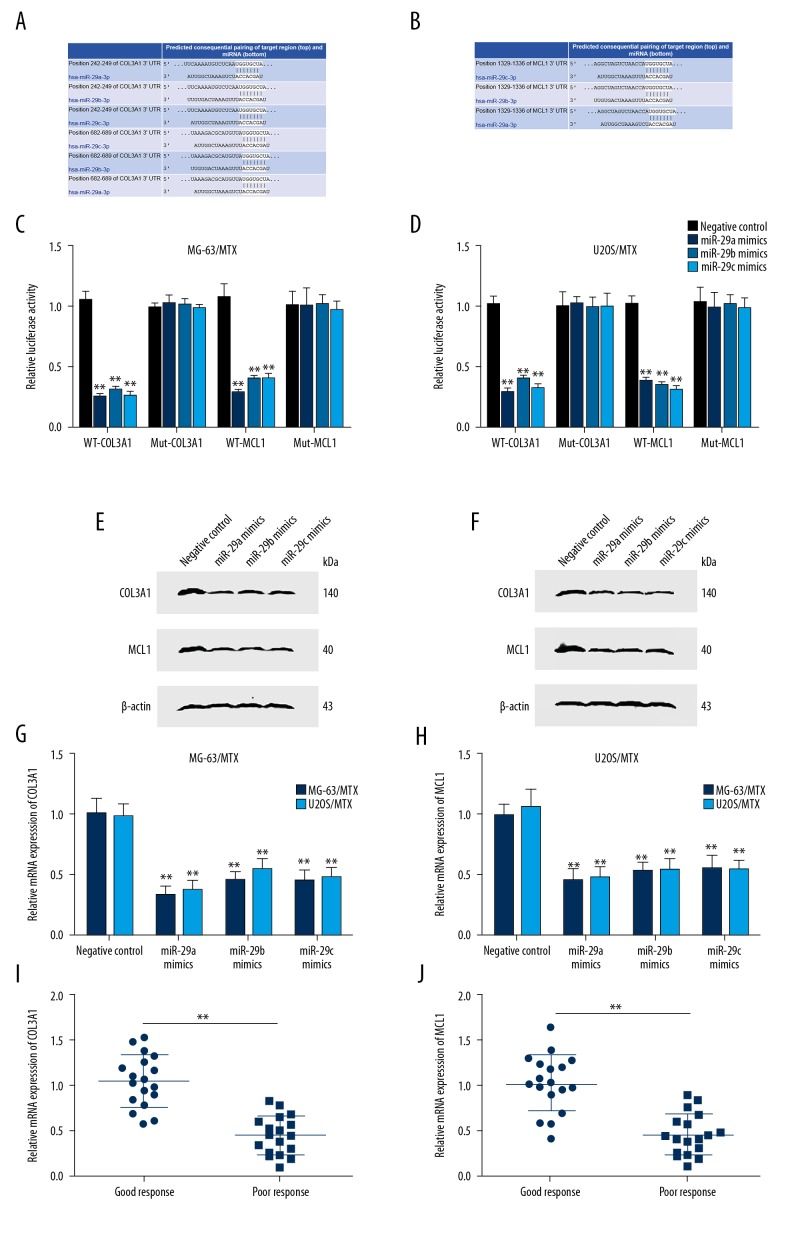
To uncover the potential mechanism in which miR-29 family inhibits MTX resistance and induces cell apoptosis in osteosarcoma, we searched for its target genes in TargetScan and found that the 3′UTR of COL3A1 and MCL1 contains binding sites of miR-29abc (Figures 3A, 3B). We constructed luciferase reporter vectors WT and Mut of COL3A1 or MCL1 3′UTR with predictive binding sites for miR-29abc. In MG-63/MTX and U2OS/MTX cells, miR-29abc mimics co-transfected with WT-COL3A1 or WT-MCL1 significantly reduced luciferase activity, but no obvious change was observed for Mut-COL3A1 or Mut-MCL1 (Figure 3C, 3D). qRT-PCR and Western blot analysis showed a significant decrease of COL3A1 and MCL1 mRNA and protein level in MG-63/MTX and U2OS/MTX cells transfected with miR-29abc mimics compared with miRNA negative control (Figure 3E–3H). These results reveal that miR-29abc directly regulates COL3A1 and MCL1 in osteosarcoma.
Figure 3.
miR-29 family directly targeted COL3A1 and MCL1. (A) Binding sites of the 3′UTR of COL3A1 for miR-29abc predicted by TargetScan. (B) Binding sites of the 3′UTR of MCL1 for miR-29abc predicted by TargetScan. (C) Luciferase reporter assay validated that miR-29abc directly targets the 3′UTR of COL3A1 and MCL1 in MG-63/MTX cells. (D) Luciferase reporter assay validated that miR-29abc directly targets the 3′UTR of COL3A1 and MCL1 in U2OS/MTX cells. (E) Overexpression of miR-29abc in MG-63/MTX cells reduced COL3A1 and MCL1 protein expression level determined by Western blot analysis compared with negative control. (F) Overexpression of miR-29abc in U2OS/MTX reduced COL3A1 and MCL1 protein expression level determined by Western blot analysis compared with negative control. (G) Transfection with miR-29abc mimics in MG-63/MTX and U2OS/MTX cells reduced COL3A1 mRNA expression level determined by qRT-PCR compared with negative control. (H) Transfection with miR-29abc mimics in MG-63/MTX and U2OS/MTX cells reduced MCL1 mRNA expression level determined by qRT-PCR compared with negative control. (I) Relative mRNA expression of COL3A1 in 18 poor-response osteosarcoma tissues (<90% tumor necrosis after chemotherapy) and 18 stage-matched good-response osteosarcoma tissues (≥90% tumor necrosis after chemotherapy) as analyzed by qRT-PCR. (J) Relative mRNA expression of MCL1 in 18 poor-response osteosarcoma tissues (<90% tumor necrosis after chemotherapy) and 18 stage-matched good-response osteosarcoma tissues (≥90% tumor necrosis after chemotherapy) as analyzed by qRT-PCR. ** P<0.01. (C, D, G, H) compared with negative control. (I, J) compared with good-response osteosarcoma tissues.
We also detected COL3A1 and MCL1 mRNA and protein level in 18 poor-response osteosarcoma tissues and 18 stage-matched good-response osteosarcoma tissues. A higher mRNA level of COL3A1 and MCL1 was observed in poor-response osteosarcoma patients compared with that of good-response osteosarcoma patients (Figure 3I, 3J). Person correlation analysis showed that miR-29abc was negatively correlated with COL3A1 and MCL1 mRNA levels in MTX-resistant osteosarcoma tissues (Table 2).
Table 2.
Person correlation analysis of miR-29abc with COL3A1 and MCL1 mRNA level in MTX-resistant osteosarcoma tissues.
| miRNAs | P value | R |
|---|---|---|
| miR-29a | 0.029 | −0.7569 |
| miR-29b | 0.0053 | −0.8311 |
| miR-29c | 0.015 | −0.5251 |
Overexpression of COL3A1 and MCL1 reversed miR-29abc-induced MTX resistance and cell apoptosis
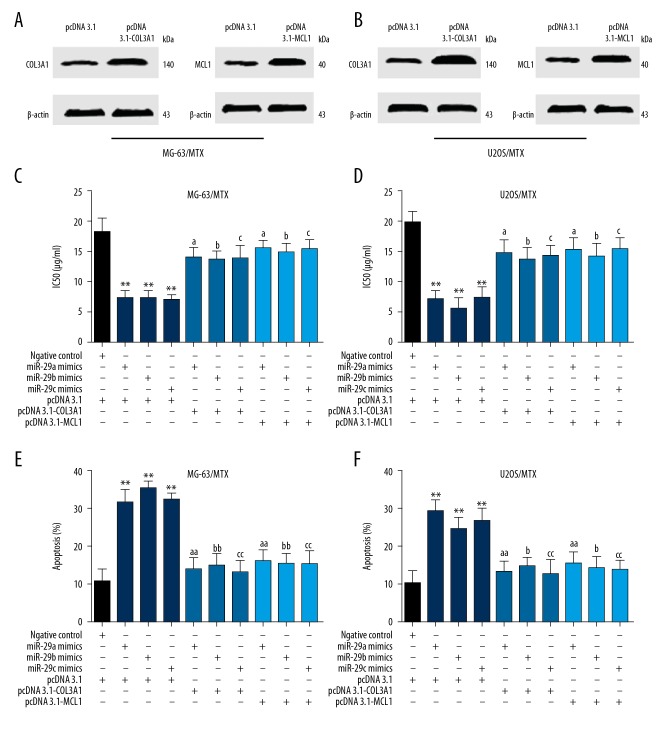
To further confirm the regulatory effect of miR-29abc on COL3A1 and MCL1 in the MTX resistance of osteosarcoma, we co-transfected miR-29abc or miRNA negative control with pcDNA3.1-COL3A1, pcDNA 3.1-MCL1 or vector negative control in MG-63/MTX and U2OS/MTX cells. We first transfected pcDNA3.1-COL3A1, pcDNA 3.1-MCL1, or pcDNA 3.1 vector into MG-63/MTX and U2OS/MTX cells, and Western blot results showed a significant increase protein level of COL3A1 and MCL1 after transfection with pcDNA 3.1-MCL1 and pcDNA 3.1, respectively (Figure 4A, 4B). As shown in Figure 4C and 4D, MTT assay results demonstrated that transfection with pcDNA 3.1-COL3A1 or pcDNA 3.1-MCL1 in MG-63/MTX and U2OS/MTX cells significantly increased cell viability and IC50 values compared with pcDNA 3.1 vector. However, lower cell viability and IC50 values were observed in MG-63/MTX and U2OS/MTX cells co-transfected with miR-29abc mimics and pcDNA3.1-COL3A1 or pcDNA 3.1-MCL1 compared with that of pcDNA 3.1 vector, suggesting COL3A1 and MCL1 are associated with miR-29abc-mediated MTX resistance. Flow cytometry analysis showed that overexpression of COL3A1 or MCL1 in MG-63/MTX and U2OS/MTX cells transfected with miR-29abc mimics significantly reduced the rate of cell apoptosis compared with that of pcDNA 3.1 and miR-29abc mimics (Figure 4C, 4D). These results further confirmed that the miR-29 family regulates MTX resistance and cell apoptosis through targeting COL3A1 and MCL1.
Figure 4.
Overexpression of COL3A1 and MCL1 reversed miR-29abc-induced MTX resistance and cell apoptosis. (A) Western blot analysis of COL3A1 and MCL1 protein level after transfection with pcDNA3.1-COL3A1 and pcDNA 3.1-MCL1 in MG-63/MTX cells. (B) Western blot analysis of COL3A1 and MCL1 protein level after transfection with pcDNA3.1-COL3A1 and pcDNA 3.1-MCL1 in U2OS/MTX cells. (C) MG-63/MTX cells were co-transfected miR-29abc mimics, miRNA negative control with pcDNA3.1-COL3A1, pcDNA 3.1-MCL1, or pcDNA 3.1 vector and treated with different concentrations of MTX for 48 h. IC50 values were then determined using CCK-8 assay. (D) U2OS/MTX cells were co-transfected miR-29abc mimics, miRNA negative control with pcDNA3.1-COL3A1, pcDNA 3.1-MCL1, or pcDNA 3.1 vector and treated with different concentrations of MTX for 48 h. IC50 values were then determined using CCK-8 assay. (E) Flow cytometry analysis was performed to determine the effect of COL3A1 or MCL1 overexpression on cell apoptosis induced by MTX in miR-29abc mimics transfected MG-63/MTX cells. (F) Flow cytometry analysis was performed to determine the effect of COL3A1 or MCL1 overexpression on cell apoptosis induced by MTX in miR-29abc mimics-transfected U2OS/MTX cells. ** P<0.01 compared with negative control. a P<0.05 and aa P<0.01 compared with miR-29a mimics. b P<0.05 and bb P<0.01 compared with miR-29b mimics. c P<0.05 and cc P<0.01 compared with miR-29c mimics.
Discussion
In this study, using osteosarcoma MTX-resistant cell lines and clinical samples, we first determined miR-29abc expression level by qRT-PCR and found that miR-29abc was frequently downregulated in MTX-resistant osteosarcoma cells and tissues. In addition, overexpression of miR-29abc in MG-63/MTX and U2OS/MTX cells significantly reduced cell viability, enhanced cell sensitivity to MTX, and promoted cell apoptosis induced by MTX. Through bioinformatics prediction, we found that COL3A1 and MCL1 might be functional targets of miR-29abc. Then, we confirmed this hypothesis by a series of molecular biology experiments. Finally, we found that overexpression of COL3A1 or MCL1 could neutralize, at least in part, the effect of inhibition of miRNA-29abc mimics on MTX resistance. These findings show the potential roles of miR-29abc in control of MTX resistance in osteosarcoma.
The miR-29 family includes 3 members: miR-29a, miR-29b (miR-29b-1 and miR-29b-2 are known as mature miRNAs and together are referred to as miR-29b), and miR-29c. miR-29a and miR-29b-1 are located on chromosome 1 while miR-29b-2 and miR-29c are located on chromosome 7. They are highly homologous and share a common seed sequence (ACCACGA). Many studies have shown that the miR-29 family can function as a tumor suppressor in gastric cancer, lung squamous cell carcinoma, and acute myeloid leukemia, but also acts as an oncogene in pancreatic cancer and breast cancer [18–22]. In addition, the miR-29 family also plays an important regulatory role in drug resistance, type 2 diabetes, and aging [23–25]. In osteosarcoma, the miR-29 family was implicated in cell proliferation, migration, and angiogenesis [26,27]. However, the relationship between the miR-29 family and MTX resistance in osteosarcoma was still unclear. The present study revealed the role of the miR-29 family in inhibiting MTX resistance and our results will broaden existing knowledge about the miR-29 family.
COL3A1 encodes the pro-alpha1 chains of type III collagen. Mutations in this gene are associated with Ehlers-Danlos syndrome type IV, intracranial aneurysms, and cervical artery aneurysms [28,29]. Recent studies have reported that upregulated COL3A1 is involved in the tumorigenesis and development of lung adenocarcinoma and renal cell carcinoma [30,31]. Here, we detected COL3A1 expression in MTX-resistant osteosarcoma tissues and observed a negative correlation between COL3A1 and miR-29 family, and found that overexpression of COL3A1 significantly enhanced IC50 for MTX compared with negative control.
MCL1 is a BCL2-related pro-survival protein originally isolated from human myeloid leukemia cells [32]. Frequent upregulation of MCL1 was observed across many solid and hematological malignancies [33]. There are several lines of evidence showing that MCL1 plays a critical role in multidrug resistance. For example, inhibition of MCL-1 in cisplatin-resistant non-small cell lung cancer cells increased the drug sensitivity [34]. In addition, miR-451 sensitizes lung cancer cells to cisplatin through targeting Mcl-1 [35]. Our research also suggests that MCL-1 is a direct target gene of the miR-29 family in regulating MTX-mediated cell apoptosis.
Conclusions
Our work demonstrates that the miR-29 family is dysregulated in MTX-resistant osteosarcoma cell lines and tissues and acts as a tumor suppressor through targeting COL3A1 or MCL1. Targeting the miR-29 family might provide new strategies to overcome high-dosage MTX-induced cytotoxicity in osteosarcoma treatment.
Footnotes
Source of support: This study was supported by the National Natural Science Foundation of China (No. 81501897) and the Shanghai Municipal Commission of Health and Family Planning of science and Research Fund (20154Y0070)
Conflicts of interest
None.
References
- 1.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–43. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Siegel HJ, Pressey JG. Current concepts on the surgical and medical management of osteosarcoma. Expert Rev Anticancer Ther. 2008;8:1257–69. doi: 10.1586/14737140.8.8.1257. [DOI] [PubMed] [Google Scholar]
- 4.Savage SA, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011 doi: 10.1155/2011/548151. 548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou AJ, Gorlick R. Chemotherapy resistance in osteosarcoma: Current challenges and future directions. Expert Rev Anticancer Ther. 2006;6:1075–85. doi: 10.1586/14737140.6.7.1075. [DOI] [PubMed] [Google Scholar]
- 6.Janeway KA, Grier HE. Sequelae of osteosarcoma medical therapy: A review of rare acute toxicities and late effects. Lancet Oncol. 2010;11:670–78. doi: 10.1016/S1470-2045(10)70062-0. [DOI] [PubMed] [Google Scholar]
- 7.Treon SP, Chabner BA. Concepts in use of high-dose methotrexate therapy. Clin Chem. 1996;42:1322–29. [PubMed] [Google Scholar]
- 8.Widemann BC, Balis FM, Kempf-Bielack B, et al. High-dose methotrexate-induced nephrotoxicity in patients with osteosarcoma. Cancer. 2004;100:2222–32. doi: 10.1002/cncr.20255. [DOI] [PubMed] [Google Scholar]
- 9.Guo W, Healey JH, Meyers PA, et al. Mechanisms of methotrexate resistance in osteosarcoma. Clin Cancer Res. 1999;5:621–27. [PubMed] [Google Scholar]
- 10.Serra M, Reverter-Branchat G, Maurici D, et al. Analysis of dihydrofolate reductase and reduced folate carrier gene status in relation to methotrexate resistance in osteosarcoma cells. Ann Oncol. 2004;15:151–60. doi: 10.1093/annonc/mdh004. [DOI] [PubMed] [Google Scholar]
- 11.Jabeen S, Holmboe L, Alnæs GI, et al. Impact of genetic variants of RFC1, DHFR and MTHFR in osteosarcoma patients treated with high-dose methotrexate. Pharmacogenomics J. 2015;15:385–90. doi: 10.1038/tpj.2015.11. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Ni J, Liu K, et al. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72:230–38. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- 13.Sevelda F, Mayr L, Kubista B, et al. EGFR is not a major driver for osteosarcoma cell growth in vitro but contributes to starvation and chemotherapy resistance. J Exp Clin Cancer Res. 2015;34:134. doi: 10.1186/s13046-015-0251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi E, Hornicek FJ, Duan Z. MicroRNA involvement in osteosarcoma. Sarcoma. 2012;2012 doi: 10.1155/2012/359739. 359739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G, Fang T, Huang Z, et al. MicroRNA-133a inhibits osteosarcoma cells proliferation and invasion via targeting IGF-1R. Cell Physiol Biochem. 2016;38:598–608. doi: 10.1159/000438653. [DOI] [PubMed] [Google Scholar]
- 16.Jiang R, Zhang C, Liu G, et al. MicroRNA-101 inhibits proliferation, migration and invasion in osteosarcoma cells by targeting ROCK1. Am J Cancer Res. 2017;7:88–97. [PMC free article] [PubMed] [Google Scholar]
- 17.Duan Z, Gao Y, Shen J, et al. miR-15b modulates multidrug resistance in human osteosarcoma in vitro and in vivo. Mol Oncol. 2017;11:151–66. doi: 10.1002/1878-0261.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong J, Li J, Wang Y, et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis. 2014;35:497–506. doi: 10.1093/carcin/bgt337. [DOI] [PubMed] [Google Scholar]
- 19.Mizuno K, Seki N, Mataki H, et al. Tumor-suppressive microRNA-29 family inhibits cancer cell migration and invasion directly targeting LOXL2 in lung squamous cell carcinoma. Int J Oncol. 2016;48:450–60. doi: 10.3892/ijo.2015.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong JN, Yu J, Lin HS, et al. The role, mechanism and potentially therapeutic application of microRNA-29 family in acute myeloid leukemia. Cell Death Differ. 2014;21:100–12. doi: 10.1038/cdd.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun XJ, Liu BY, Yan S, et al. MicroRNA-29a promotes pancreatic cancer growth by inhibiting tristetraprolin. Cell Physiol Biochem. 2015;37:707–18. doi: 10.1159/000430389. [DOI] [PubMed] [Google Scholar]
- 22.Wang C, Bian Z, Wei D, et al. MiR-29b regulates migration of human breast cancer cells. Mol Cell Biochem. 2011;352:197–207. doi: 10.1007/s11010-011-0755-z. [DOI] [PubMed] [Google Scholar]
- 23.Yu PN, Yan MD, Lai HC, et al. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells. Int J Cancer. 2014;134:542–51. doi: 10.1002/ijc.28399. [DOI] [PubMed] [Google Scholar]
- 24.Peng H, Zhong M, Zhao W, et al. Urinary miR-29 correlates with albuminuria and carotid intima-media thickness in type 2 diabetes patients. PLoS One. 2013;8:e82607. doi: 10.1371/journal.pone.0082607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ripa R, Dolfi L, Terrigno M, et al. MicroRNA miR-29 controls a compensatory response to limit neuronal iron accumulation during adult life and aging. BMC Biol. 2017;15:9. doi: 10.1186/s12915-017-0354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Zhang C, Liu L, et al. A key role of microRNA-29b in suppression of osteosarcoma cell proliferation and migration via modulation of VEGF. Int J Clin Exp Pathol. 2014;7:5701–8. [PMC free article] [PubMed] [Google Scholar]
- 27.Gao S, Cheng C, Chen H, et al. IGF1 3′UTR functions as a ceRNA in promoting angiogenesis by sponging miR-29 family in osteosarcoma. J Mol Histol. 2016;47:135–43. doi: 10.1007/s10735-016-9659-2. [DOI] [PubMed] [Google Scholar]
- 28.Superti-Furga A, Gugler E, Gitzelmann R, et al. Ehlers-Danlos syndrome type IV: A multi-exon deletion in one of the two COL3A1 alleles affecting structure, stability, and processing of type III procollagen. J Biol Chem. 1988;263:6226–32. [PubMed] [Google Scholar]
- 29.Kuivaniemi H, Prockop DJ, Wu Y, et al. Exclusion of mutations in the gene for type III collagen (COL3A1) as a common cause of intracranial aneurysms or cervical artery dissections: Results from sequence analysis of the coding sequences of type III collagen from 55 unrelated patients. Neurology. 1993;43:2652–58. doi: 10.1212/wnl.43.12.2652. [DOI] [PubMed] [Google Scholar]
- 30.Rothschild SI, Tschan MP, Federzoni EA, et al. MicroRNA-29b is involved in the Src-ID1 signaling pathway and is dysregulated in human lung adenocarcinoma. Oncogene. 2012;31:4221–32. doi: 10.1038/onc.2011.578. [DOI] [PubMed] [Google Scholar]
- 31.Su B, Zhao W, Shi B, et al. Let-7d suppresses growth, metastasis, and tumor macrophage infiltration in renal cell carcinoma by targeting COL3A1 and CCL7. Mol Cancer. 2014;13:206. doi: 10.1186/1476-4598-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozopas KM, Yang T, Buchan HL, et al. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci USA. 1993;90:3516–20. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma J, Zhao Z, Wu K, et al. MCL-1 is the key target of adjuvant chemotherapy to reverse the cisplatin-resistance in NSCLC. Gene. 2016;587:147–54. doi: 10.1016/j.gene.2016.04.054. [DOI] [PubMed] [Google Scholar]
- 35.Cheng D, Xu Y, Sun C, et al. MicroRNA-451 sensitizes lung cancer cells to cisplatin through regulation of Mcl-1. Mol Cell Biochem. 2016;423:85–91. doi: 10.1007/s11010-016-2827-6. [DOI] [PubMed] [Google Scholar]