Abstract
Background
Epithelial ovarian cancer (EOC) has a high mortality rate and is a common malignant tumor of women, seriously impairing health. Chemoresistance is one of the major causes of poor prognosis. Therefore, analyzing the molecular mechanism of paclitaxel resistance has great significance.
Material/Methods
We analyzed aberrantly expressed lncRNAs in chemoresistant EOC cells by microarray and confirmed LINC01118 expression by real-time PCR. The paclitaxel sensitivity alternation was analyzed by MTS, flow cytometry, and Transwell assay, while wound healing assays were performed to assess apoptosis, migration, and invasion in vitro. The interaction between LINC01118 and miR-134 was confirmed by luciferase assay.
Results
LINC01118 was highly expressed in EOC tissues and chemoresistant cells. Biological function experiments showed LINC01118 could facilitate paclitaxel resistance and promote migration and invasion while inhibiting apoptosis of EOC cells. Moreover, LINC01118 targets miR-134 and then affects ABCC1 expression.
Conclusions
LINC01118 acted as an oncogene and modulated EOC paclitaxel sensitivity by regulating miR-134/ABCC1.
MeSH Keywords: Drug Resistance; Ovarian Neoplasms; Paclitaxel; RNA, Long Noncoding
Background
Ovarian cancer (OC) is the seventh most common malignancy in women [1] yet its mortality rate is the highest among all gynecological cancers [2]. Although OC includes multiple histological types, such as epithelial ovarian cancer (EOC), sex cord-stromal tumor, and germ cell tumor, EOC is the deadliest gynecologic malignancy [3]. EOC is further divided into serous cystadenocarcinoma, mucinous cystadenocarcinoma, and endometrioid carcinoma, among which serous cystadenocarcinoma accounts for 75% of EOC. At present, the most widely accepted therapy of EOC is cytoreductive surgery supplemented with paclitaxel and platinum-based chemotherapy. Although patients initially respond well to the treatment, nearly all have multiple recurrences [4,5]. This is not only because the ovaries are located deep in the pelvic cavity, which lack of obvious clinical symptoms in the early stage, but also because there is no accurate and specific screening method for EOC [6,7]; the more important cause of recurrence is chemotherapy resistance. Accordingly, EOC still threatens women’s health, and its 5-year survival rate is less than 45% [8,9]. Therefore, it is urgent to elucidate the paclitaxel resistance mechanism, and it is crucial to improve the therapeutic effect and prognosis of OC patients.
Long noncoding RNA (lncRNA) is a kind of noncoding RNA. It is more than 200 nt in length and does not encode proteins [10]. Accumulating evidence reveals that numerous aberrantly expressed lncRNAs exist in chemoresistant tissues or cells in breast cancer, stomach cancer, lung cancer, and osteosarcoma [11–14]. Moreover, some of lncRNAs can modulate the sensitivity of tumors to chemotherapy, such as lnc-ATB [11], HANR [15], and MEG3 [16]. It was also reported that lncRNAs are involved in the development of OC and its platinum resistance [17,18]. However, the role of lncRNAs in EOC paclitaxel resistance remains uncertain, so it is essential to analyze the dysregulated expressed lncRNAs in paclitaxel resistance EOC and clarify its regulatory function in paclitaxel chemoresistance.
In this study we used microarray analysis to screen out a novel lncRNA-LINC01118, which is significantly overexpressed in chemoresistant EOC cells. We clarified its expression and functions in EOC tissues and cells. Moreover, we found that LINC01118 can affect paclitaxel resistance of EOC through regulating miR-134/ABCC1.
Material and Methods
Patients and tissues
Tissue samples were collected from patients in the Department of Gynecology of Shengjing Hospital of China Medical University from 2016 to 2017. Tissue samples were collected from 30 patients: 5 with normal ovarian tissues, 7 with serous cystadenoma, 5 with borderline cystadenoma, and 13 with serous cystadenocarcinoma. EOC patient ages were 39–79 years old (mean age, 48.46 years). According to the 2009 International Federation of Gynecology and Obstetrics (FIGO) criteria, 6 cases were stage I–II and 7 cases were stage III–IV. EOC patients had not received chemotherapy before surgery and were diagnosed by pathological examination. All patients signed informed consent and the study was approved by the Ethics Committee of Shengjing Hospital of China Medical University.
Cell culture
SKOV3-TR30 cells were provided by Women’s Hospital, School of Medicine, Zhejiang University, and other cells were obtained from the Tumor Cell Band Research Institute of the Chinese Academy of Medical Science. Human EOC cell lines SKOV3, A2780, and COC1 were cultured in McCoy’s 5A, DMED, and RPMI-1640 medium, respectively. Paclitaxel-resistant cell lines SKOV3-TR30 and A2780 were cultured in RPMI-1640 with Taxol 20 nM and 600 ng/ml separately. Platinum-resistant cell lines SKOV3-DDP and COC1/DDP were cultured in RPMI-1640 with cisplatin 1 μg/ml. 293T cells were cultured in DMEM. The entire medium contains 10% fetal bovine serum and 1% penicillin/streptomycin (100 U/ml). Cells cultured at 37°C, 5% CO2, and saturated humidity.
Real-time PCR
Total RNA was extracted from cells and tissues by RNAiso Plus (Takara Bio, China), and RNA reverse transcription for cDNA by PrimeScriptTMRT reagent kit with gDNA Eraser (Takara Bio, China). SYBR premix Ex TapTM II (Takara Bio, China) was used for quantitative PCR. Primers were synthesized by Sangon Biotech (Shanghai, China). All procedures were performed according to the manufacturer’s instructions. GAPDH and U6 were the internal references for lncRNA and microRNA. The relative expression level was normalized by 2−ΔΔCT method.
Cell transfection
LINC01118 siRNA was purchased from RiboBio (Guangzhou, China) and the overexpression plasmid Phb-H-h-LINC01118-1-gfp was from HanBio (Shanghai, China). miR-134 mimic and negative control were designed and compounded by GenePharma (Shanghai, China). Lipofectamine 2000 (Invitrogen, USA) was used for cell transfection in accordance with the product instructions.
Cell viability assay
The cells were seeded in 96-well plates, and different concentrations of paclitaxel were added after transfection. MTS assay was performed 48 h after incubation. We added 20 μl of MTS reagent (Promega, USA) to each 100 μl of medium, followed by incubation for 30 min at 37°C and 5% CO2, detected the absorbance at 490 nm, and calculated the cell paclitaxel inhibition rate.
Cell apoptosis assay
Adherent cells were digested with EDTA-free trypsin and washed twice with PBS. We resuspended the cells with binding buffer. The cells transfected with siRNA were double-stained with Annexin V FITC/PI (Key GEN BioTECH, Jiangsu, China). Flow cytometry was used to analyze apoptosis.
Wound healing and Transwell assay
To detect the wound healing condition, we incubated cells in 6-well plates at the appropriate density, and ensured that cells reached confluence overnight. We scratched the cell layers with a sterile pipette tip held as perpendicular as possible. We photographed the scratch healing condition and calculated the wound healing percentage.
To investigate the migration and invasion, the transfected cells were digested, resuspended in serum-free medium, added to the upper chamber of each insert (Corning, USA) of a Transwell device, and the complete medium containing 10% fetal bovine serum was added to the lower chamber. Migration experiments were performed in SKOV3 cells for 40 h and in SKOV3-TR30 for 30 h. For invasion experiments, we added 60 μl matrigel (1: 8 dilution). Then, we cultured SKOV3 cells for 48 h and SKOV3-TR30 cells for 40 h. After that, we fixed the cells with paraformaldehyde, stained them with 0.1% crystal violet, then photographed and counted them.
Luciferase assay
The pmirGLO Dual-Luciferase miRNA target expression vector (Promega, USA) was used for plasmid construction. We adjusted the number of 293T cells to approximately 4×105/well in 12-well plates; the cells were cultured until the confluence rate reached 60–80% and transfection began with addition of lipofectamine 2000 (Invitrogen, USA). The Dual-Luciferase Reporter Assay System (Promega, USA) was applied for detecting luciferase activity and the firefly luciferase level was normalized by Renilla luciferase activity.
Western blot
We preformed 10% SDS-PAGE gel electrophoresis with the same amount of total protein at 30 μg per well. The electrophoresis was performed at 80V, and then we transferred membranes by PVDF for 200 mA. Antibodies were incubated overnight at 4°C and secondary antibodies were incubated at room temperature. Results were photographed.
Statistical analysis
Data statistical analysis by statistical software IBM SPSS Statistics 21, the experimental results in the form of mean ±SD, 2 independent samples by t test; multiple data comparison was conducted by ANOVA analysis; P<0.05 was considered as statistically significant.
Results
Expression of LINC01118 in EOC cells and tissues
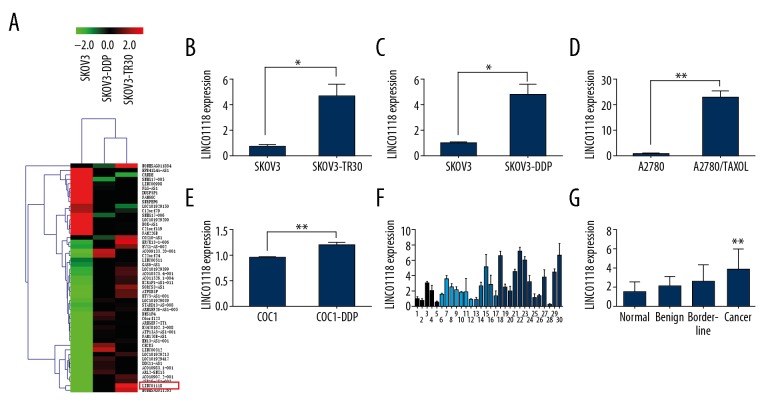
To inspect the chemoresistant lncRNAs in EOC, we performed microarray analysis comparing paclitaxel-resistant SKOV3-TR30, cisplatin-resistant SKOV3-DDP, and their parental cell line SKOV3. The outcome showed that there were 40830 aberrantly expressed lncRNAs between SKOV3-TR30 and SKOV3 (fold change ≥2 and P<0.05), among which LINC01118 was observed to be abnormally overexpressed in chemoresistant EOC cells (Figure 1A). To confirm this difference, we assessed the expression level of LINC01118 in 4 pairs of chemoresistant cell lines (paclitaxel resistance: SKOV3 and SKOV3-TR30, A2780 and A2780/TAXOL; cisplatin-resistance: SKOV3 and SKOV3-DDP, COC1 and COC1-DDP) by use of real-time PCR. The results showed that LINC01118 expression in paclitaxel- and cisplatin-resistant cells were higher than in the parental cells, which was consistent with the result of the microarray analysis (Figure 1B–1E). We also observed the expression of LINC01118 in 5 cases of normal ovaries, 7 cases of ovarian serous cystadenomas, 5 cases of borderline cystadenoma, and 13 cases of serous ovarian cancer tissues. The data demonstrated in Figure 1F and 1G reveal that the level of LINC01118 in ovarian cancer was significantly higher than that in other ovarian tissues, but the differences in expression levels among normal, benign, and borderline EOC tissues had no statistical significance. This suggests that LINC01118 is involved in regulation of paclitaxel resistance as well as the occurrence and development of EOC.
Figure 1.
LINC01118 expression in EOC cells and tissues. (A) Microarray analysis of SKOV3, SKOV3-TR30, and SKOV3-DDP. (B–E) LINC01118 was overexpressed in chemoresistant EOC cells of SKOV3-TR30, SKOV3-DDP, A2780/TAXOL, and COC1-DDP than in their parental cells. (F) Expression of LINC01118 in 5 cases of normal ovarian tissue (1–5), 7 cases of serous cystadenoma (6–12), 5 cases of borderline cystadenoma (13–17), and 13 cases of serous cystadenocarcinoma (18–30) by real-time PCR. (G) LINC01118 was overexpressed in ovarian cancer tissues compared to normal, benign, and borderline groups, ** P<0.01.
LINC01118 regulated paclitaxel sensitivity in EOC cells
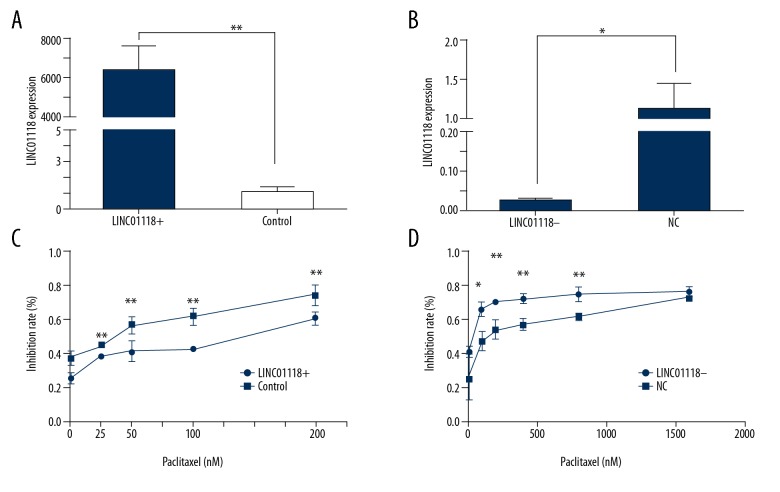
We overexpressed the expression of LINC01118 in SKOV3 by plasmids and suppressed it with siRNA, and the regulatory expression levels were confirmed by real-time PCR (Figure 2A, 2B). To clarify the impact of LINC01118 on paclitaxel sensitivity in EOC, we upregulated LINC01118 in SKOV3 and downregulated LINC01118 in SKOV3-TR30, thereby showing the inhibition rates of different concentrations of paclitaxel. We found that in the same concentration, the inhibition rate of downregulated cells was higher than that in the control group, which means the sensitivity to paclitaxel was increased, while chemoresistance was reversed. On the contrary, the LINC01118-overexpressed cells showed a decrease in paclitaxel inhibition rate, meaning that sensitive ovarian cancer became paclitaxel resistant (Figure 2C, 2D). The MTS assay showed that LINC01118 can affect paclitaxel sensitivity in EOC cells and it can be a target of chemoresistance reversion.
Figure 2.
LINC01118 regulated paclitaxel sensitivity in EOC cells. (A) The expression level of LINC01118 in SKOV3-TR30 was significantly upregulated by plasmid. (B) LINC01118 was knocked down by specific siRNA. (C) Overexpression of LINC01118 in SKOV3 decreased the paclitaxel inhibition rate. (D) Downregulation of LINC01118 in SKOV3-TR30 cells promoted the inhibitory effect of paclitaxel on cell proliferation. * P<0.05 and ** P<0.01.
LINC01118 regulated EOC apoptosis, migration, and invasion
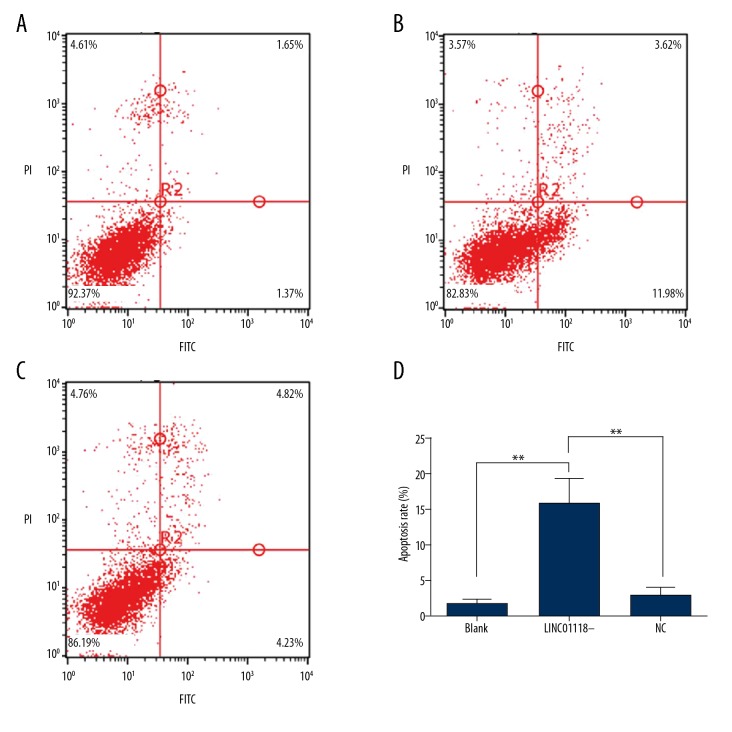
We confirmed the influence of LINC01118 on paclitaxel sensitivity of EOC; however, as a novel lncRNA, the biological functions were still unknown. Therefore, we concentrated on the apoptosis of EOC regulated by LINC01118 and analyzed the early apoptosis rate of the Q4 zone. Flow cytometry showed that downregulation of LINC01118 in SKOV3-TR30 cells facilitated apoptosis compared with its NC group (15.810±3.433 vs. 2.996±1.123, P<0.01, Figure 3).
Figure 3.
LINC01118 regulated apoptosis of EOC. (A) The apoptosis of SKOV3-TR30 cells without transfection. (B) Apoptosis of SKOV3-TR30 with si-LINC01118. (C) The negative control group of siRNA-LINC01118. (D) Inhibition of LINC01118 expression promoted ovarian cancer cell apoptosis, ** P<0.01.
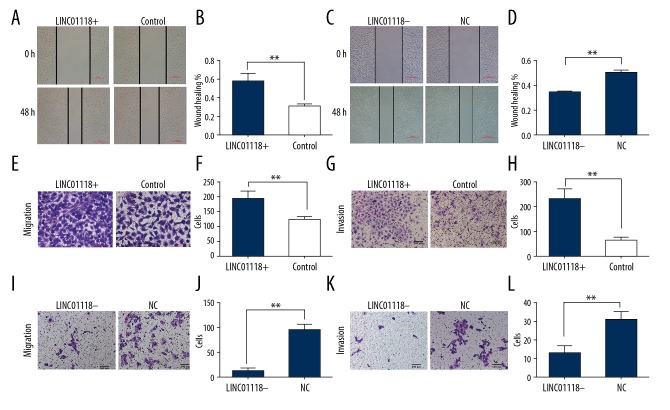
Wound healing and Transwell assays were carried out to estimate the EOC cell abilities of migration and invasion. The results showed that overexpression of LINC01118 in SKOV3 not only accelerated wound healing, but also promoted more cells to migrate from upper chamber to the lower one. Inhibition of LINC01118 expression resulted in the exact opposite of overexpression. Results of invasion and migration experiments were consistent and are shown in Figure 4.
Figure 4.
LINC01118 enhanced invasion and migration in EOC cells. (A) Wound healing assay in SKOV3 cells with LINC01118 overexpression plasmid compared with negative control group. (B) The wound healing percentage of plasmid group was significantly higher than in the control group. (C) Wound healing results in SKOV3-TR30 cells with siRNA-LINC01118 compared with NC. (D) Downregulation of LINC01118 significantly inhibited the wound healing. (E, F) The migration ability was significantly enhanced compared with the control group when LINC01118 was overexpressed in SKOV3. (G, H) The invasion was promoted compared with the control group when LINC01118 was overexpressed in SKOV3. (I, J) The migration was decreased compared with NC group in SKOV3-TR30. (K, L) The invasion ability was decreased compared with NC group in SKOV3-TR30. * P<0.05 and ** P<0.01.
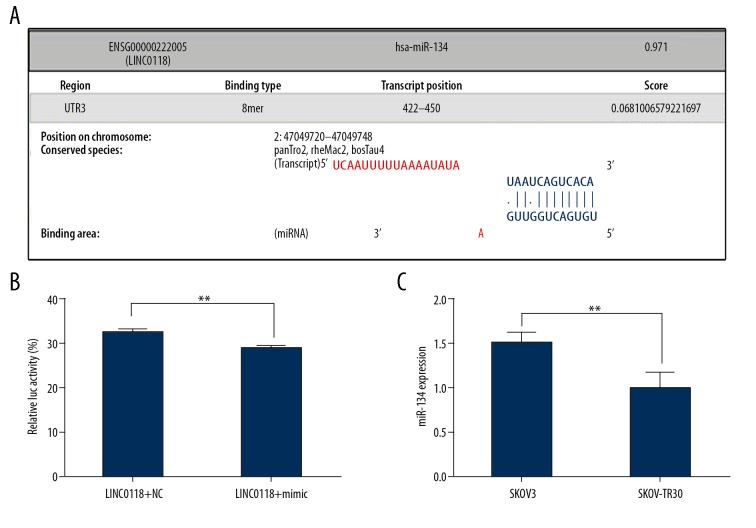
LINC01118 was a direct target for miR-134
The biological function study of LINC01118 revealed that it could regulate the sensitivity of EOC to paclitaxel, but the mechanism was unclear. So, we searched LncBase Predicted v.2 [19] for microRNAs that might interact with LINC01118, and found that miR-134 was a possible candidate (Figure 5A). The luciferase reporter gene assay confirmed that miR-134 targeted LINC01118, which was in accordance with the prediction (Figure 5B). Subsequently, we affirmed that the expression pattern of miR-134 was contrary to LINC01118 in SKOV3 and SKOV3-TR30 cells (Figure 5C), and it was overexpressed in paclitaxel sensitive SKOV3 cells (1.510±0.092 vs. 1.000±1.148, P<0.01). We found that LINC01118 can affect paclitaxel resistance by targeting miR-134.
Figure 5.
miR-134 was a direct target of LINC01118. (A) The prediction binding area between LINC01118 and miR-134. (B) miR-134 targeted the 3′UTR of LINC01118. (C) miR-134 was underexpressed in SKOV3-TR30 cells compared with SKOV3.
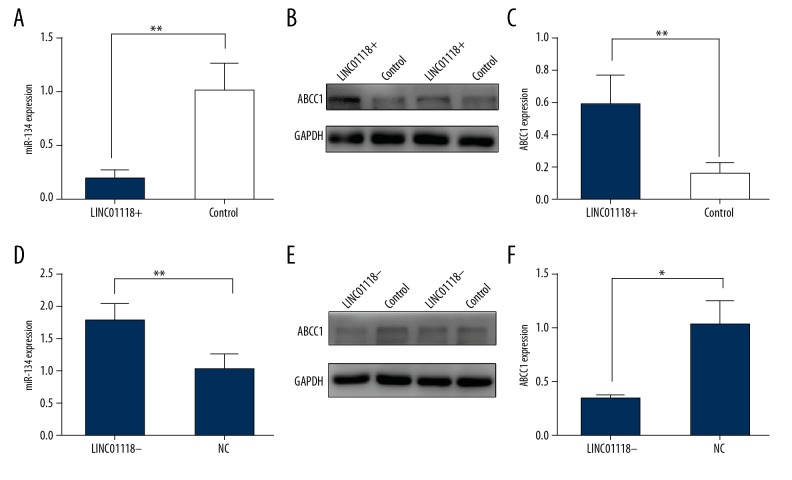
LINC01118 modulated miR-134 and ABCC1
It had been clarified that ABCC1 was directly regulated by miR-134, and our results illuminated the relationship between miR-134 and LINC01118. To elucidate the function of LINC01118 in EOC paclitaxel resistance, we assessed the expression of miR-134 and ABCC1 by Realtime PCR and western blot, respectively, under the condition of upregulation and downregulation of LINC01118. Surprisingly, we found that when LINC01118 was up-regulated, miR-134 was restrained and ABCC1 expression increased (Figure 6A–6C), and when LINC01118 was knocked down, there was the opposite change in expression of miR-134 and ABCC1 (Figure 6D–6F).
Figure 6.
LINC01118 regulated ABCC1 via miR-134. (A) miR-134 was downregulated when LINC01118 was overexpressed. (B, C) ABCC1 was upregulated by overexpression of LINC01118. (D) miR-134 was negatively controlled by LINC01118. (E, F) ABCC1 was less translated with the decrease of LINC01118. * P<0.05 and ** P<0.01.
Discussion
The mortality rate of EOC is the highest among gynecological malignant tumors, and acquiring chemotherapy resistance is considered to be the major cause of death [20]. Currently, the combination of platinum and paclitaxel usually has a good effect initially, but most patients will relapse, with a median progression-free survival of only 18 months [21], and this recurrence is usually incurable [22]. Therefore, the study of EOC chemoresistance, especially the molecular mechanism of paclitaxel resistance, is a key factor in improving the prognosis of ovarian cancer. In our research, we analyzed the molecular difference of lncRNAs in EOC chemoresistant cells by microarray assay. LINC01118, which was highly expressed in both cisplatin and paclitaxel resistant cells, attracted our attention.
Accumulating research shows that many lncRNAs are dysregulated in OC and they might play important roles. HOXD-AS1, an OC-related lncRNA, was overexpressed in both OC tissues and cells and it predicted poor prognosis [23, 24]. However, there are no related reports on LINC01118 in OC or even in any cancers, while only Bouckenheimer et al. found that LINC01118 cells were abundant in metaphase II oocytes [25]. As a novel lncRNA without reports on chemoresistance, we targeted it for further research. To verify LINC01118 results in microarray and to avoid being misled by false-positive results, we confirmed its expression in 4 pairs of paclitaxel- and cisplatin-resistant EOC cells, finding that LINC01118 was highly expressed in chemoresistant EOC cells. We also found that LINC01118 was highly overexpressed in OC clinical tissues. Consequently, LINC01118 might be closely associated with chemoresistance and it has a regulatory effect in development of EOC.
Previous studies have shown that lncRNA often has a strong function and the regulatory mechanism is complex in OC. For example, Lnc-OC1 has the function of promoting OC progression, and this oncogenic function might depend on suppressing miR-34a and miR-34c [26]; another lncRNA – RP11-552M11.4 – was not only associated with poor prognosis, but also could promote EOC cell proliferation and migration and inhibit apoptosis by targeting BRCA2 [27]. As a novel lncRNA, it insufficient to just clarify the expression model of LINC01118. Thus, we examined its biological function and found that its upregulation could promote cell viability, invasion, and migration, while inhibiting apoptosis and vice versa. LINC01118 might be an oncogene in EOC and its biological function might be the basis for regulating paclitaxel resistance.
The role of lncRNAs in modulating chemotherapy sensitivity had been confirmed in various cancers. For example, downregulation of the classical lncRNA NEAT1 improved the sensitivity to cisplatin in osteosarcoma [28] and MALAT1 enhanced docetaxel resistance of prostate cancer [29]. Similarly, our data revealed that LINC01118 could facilitate paclitaxel resistance, so we further explored the specific mechanism. Recent evidence suggests that the relationship between lncRNA and microRNA modulates lncRNAs. Cao et al. found that lncRNA ATB negatively regulated miR-494 and promoted proliferation and migration [30]. Another study reported that H19 can sponge miR-17 as a competing endogenous RNA and affect STAT3 expression, thereby promoting non-small-cell lung cancer development [31]. Thus, we searched for a candidate for LINC01118 and surprisingly found that miR-134 might have binding sites with it. In addition, miR-134 had been proved to be extremely relevant to paclitaxel resistance in OC [32] and the alteration of paclitaxel sensitivity in EOC was achieved by miR-134 modulating Pak2 [33]. Several transcription factors can also affect Taxol sensitivity by targeting miR-134 [34]. Fortunately, our luciferase reporter gene assay result was consistent with the prediction, and we found that miR-134 binds to the 3′UTR of LINC01118. Although the interaction was affirmed, it remained unclear how LINC01118 altered paclitaxel sensitivity through miR-134. Surprisingly, Wang et al. validated that lncRNA could regulate the classical resistance protein ABCB1 by targeting microRNA and made breast cancer cells paclitaxel-resistant [35]. Previous studies suggested that ATP Binding Cassette C1 (ABCC1), which is also referred to as multidrug resistance-associated protein 1 (MRP1), is a target gene of miR-134 [36]. Fortunately, Lu et al. not only confirmed that miR-134 could directly combine to ABCC1, but also found that it could negatively control the expression of ABCC1 [37]. It is well known that ABCC1 can invalidate chemotherapy by prompting the efflux of intracellular chemotherapeutic agents, which directly led to the occurrence of chemoresistance [38]. We found that when LINC01118 was upregulated, miR-134 was downregulated and ABCC1 was overexpressed; moreover, when LINC01118 was knocked down, the chain reaction was in the opposite direction. The present study shows that LINC01118 modulates paclitaxel resistance via miR-134/ABCC1.
Conclusions
LINC01118 appears to act as an oncogene in EOC, especially in serous cystadenocarcinoma, and it can regulate paclitaxel resistance by regulating miR-134/ABCC1. LINC01118 might be a key target in the treatment of ovarian cancer.
Footnotes
Source of support: This work was supported by the Outstanding Scientific Fund of Shengjing Hospital (grant numbers 201705)
Conflicts of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Kurosaki A, Hasegawa K, Kato T, et al. Serum folate receptor alpha as a biomarker for ovarian cancer: Implications for diagnosis, prognosis and predicting its local tumor expression. Int J Cancer. 2016;138:1994–2002. doi: 10.1002/ijc.29937. [DOI] [PubMed] [Google Scholar]
- 4.Holmes D. The problem with platinum. Nature. 2015;527:S218–19. doi: 10.1038/527S218a. [DOI] [PubMed] [Google Scholar]
- 5.Holmes D. Ovarian cancer: Beyond resistance. Nature. 2015;527:S217. doi: 10.1038/527S217a. [DOI] [PubMed] [Google Scholar]
- 6.Han C, Bellone S, Siegel ER, et al. A novel multiple biomarker panel for the early detection of high-grade serous ovarian carcinoma. Gynecol Oncol. 2018;149(3):585–91. doi: 10.1016/j.ygyno.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nie L, Li F, Huang X, et al. Folic acid targeting for efficient isolation and detection of ovarian cancer CTCs from human whole blood based on two-step binding strategy. ACS Appl Mater Interfaces. 2018;10(16):14055–62. doi: 10.1021/acsami.8b02583. [DOI] [PubMed] [Google Scholar]
- 8.Stewart SL, Harewood R, Matz M, et al. Disparities in ovarian cancer survival in the United States (2001–2009): Findings from the CONCORD-2 study. Cancer. 2017;123(Suppl 24):5138–59. doi: 10.1002/cncr.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timmermans M, Sonke GS, Van de Vijver KK, et al. No improvement in long-term survival for epithelial ovarian cancer patients: A population-based study between 1989 and 2014 in the Netherlands. Eur J Cancer. 2018;88:31–37. doi: 10.1016/j.ejca.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 10.Zhang XD, Huang GW, Xie YH, et al. The interaction of lncRNA EZR-AS1 with SMYD3 maintains overexpression of EZR in ESCC cells. Nucleic Acids Res. 2018;46:1793–809. doi: 10.1093/nar/gkx1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi SJ, Wang LJ, Yu B, et al. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–63. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Ben Q, Lu E, et al. Long noncoding RNA PANDAR blocks CDKN1A gene transcription by competitive interaction with p53 protein in gastric cancer. Cell Death Dis. 2018;9:168. doi: 10.1038/s41419-017-0246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Hu B, Wang Q, et al. Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR-216a. Cell Death Dis. 2018;9:85. doi: 10.1038/s41419-017-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kun-Peng Z, Xiao-Long M, Chun-Lin Z. LncRNA FENDRR sensitizes doxorubicin-resistance of osteosarcoma cells through down-regulating ABCB1 and ABCC1. Oncotarget. 2017;8:71881–93. doi: 10.18632/oncotarget.17985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao J, Lv Y, Jin F, et al. LncRNA HANR promotes tumorigenesis and increase of chemoresistance in hepatocellular carcinoma. Cell Physiol Biochem. 2017;43:1926–38. doi: 10.1159/000484116. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Wan L, Lu K, et al. The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS One. 2015;10:e114586. doi: 10.1371/journal.pone.0114586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao F, Tian X, Ruan S, et al. miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. FASEB J. 2018 doi: 10.1096/fj.201800495RR. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Song J, Zhang W, Wang S, et al. A panel of 7 prognosis-related long non-coding RNAs to improve platinum-based chemoresistance prediction in ovarian cancer. Int J Oncol. 2018;53:866–76. doi: 10.3892/ijo.2018.4403. [DOI] [PubMed] [Google Scholar]
- 19.Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–38. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Bao W, Liu Y, et al. miR-98-5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1. Cell Death Dis. 2018;9:447. doi: 10.1038/s41419-018-0390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barriga-Rivera A, Morley JW, Lovell NH, Suaning GJ. Cortical responses following simultaneous and sequential retinal neurostimulation with different return configurations. Conf Proc IEEE Eng Med Biol Soc. 2016;2016:5435–38. doi: 10.1109/EMBC.2016.7591956. [DOI] [PubMed] [Google Scholar]
- 22.Yang K, Hou Y, Li A, et al. Identification of a six-lncRNA signature associated with recurrence of ovarian cancer. Sci Rep. 2017;7:752. doi: 10.1038/s41598-017-00763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Zhang W, Wang Y, Wang S. HOXD-AS1 promotes cell proliferation, migration and invasion through miR-608/FZD4 axis in ovarian cancer. Am J Cancer Res. 2018;8:170–82. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Dun Y, Zhou S, Huang XH. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/beta-catenin signaling pathway. Biomed Pharmacother. 2017;96:1216–21. doi: 10.1016/j.biopha.2017.11.096. [DOI] [PubMed] [Google Scholar]
- 25.Bouckenheimer J, Fauque P, Lecellier CH, et al. Differential long non-coding RNA expression profiles in human oocytes and cumulus cells. Sci Rep. 2018;8:2202. doi: 10.1038/s41598-018-20727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao F, Tian X, Lu M, Zhang Z. A novel lncRNA, Lnc-OC1, promotes ovarian cancer cell proliferation and migration by sponging miR-34a and miR-34c. J Genet Genomics. 2018;45:137–45. doi: 10.1016/j.jgg.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Huang K, Geng J, Wang J. Long non-coding RNA RP11-552M11.4 promotes cells proliferation, migration and invasion by targeting BRCA2 in ovarian cancer. Cancer Sci. 2018;109(5):1428–46. doi: 10.1111/cas.13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Yang Q, Wang L, et al. Knockdown of the oncogene LncRNA NEAT1 restores the availability of miR-34c and improves the sensitivity to cisplatin in osteosarcoma. Biosci Rep. 2018;38(3) doi: 10.1042/BSR20180375. pii: BSR20180375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue D, Lu H, Xu HY, et al. Long noncoding RNA MALAT1 enhances the docetaxel resistance of prostate cancer cells via miR-145-5p-mediated regulation of AKAP12. J Cell Mol Med. 2018;22(6):3223–37. doi: 10.1111/jcmm.13604. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Cao Y, Luo X, Ding X, et al. LncRNA ATB promotes proliferation and metastasis in A549 cells by down-regulation of microRNA-494. J Cell Biochem. 2018;119(8):6935–42. doi: 10.1002/jcb.26894. [DOI] [PubMed] [Google Scholar]
- 31.Huang Z, Lei W, Hu HB, et al. H19 promotes non-small-cell lung cancer (NSCLC) development through STAT3 signaling via sponging miR-17. J Cell Physiol. 2018;233(10):6768–76. doi: 10.1002/jcp.26530. [DOI] [PubMed] [Google Scholar]
- 32.Zhu H, Yang SY, Wang J, et al. Evidence for miR-17–92 and miR-134 gene cluster regulation of ovarian cancer drug resistance. Eur Rev Med Pharmacol Sci. 2016;20:2526–31. [PubMed] [Google Scholar]
- 33.Shuang T, Wang M, Shi C, et al. Down-regulated expression of miR-134 contributes to paclitaxel resistance in human ovarian cancer cells. Febs Lett. 2015;589:3154–64. doi: 10.1016/j.febslet.2015.08.047. [DOI] [PubMed] [Google Scholar]
- 34.Shuang T, Wang M, Zhou Y, et al. NF-kappaB1, c-Rel, and ELK1 inhibit miR-134 expression leading to TAB1 upregulation in paclitaxel-resistant human ovarian cancer. Oncotarget. 2017;8:24853–68. doi: 10.18632/oncotarget.15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, Zhang T, Yang Z, et al. Long non-coding RNA FTH1P3 activates paclitaxel resistance in breast cancer through miR-206/ABCB1. J Cell Mol Med. 2018 doi: 10.1111/jcmm.13679. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuang T, Jianlei W, Yanli Z, et al. [Detection and analysis of differential expression of microRNA expression in drug-resistant and sensitive ovarian cancer tissues]. Chin J Prac Gynecol Obstetr. 2013:33–37. [in Chinese] [Google Scholar]
- 37.Lu L, Ju F, Zhao H, Ma X. MicroRNA-134 modulates resistance to doxorubicin in human breast cancer cells by downregulating ABCC1. Biotechnol Lett. 2015;37:2387–94. doi: 10.1007/s10529-015-1941-y. [DOI] [PubMed] [Google Scholar]
- 38.Li S, Yang J, Wang J, et al. Down-regulation of miR-210-3p encourages chemotherapy resistance of renal cell carcinoma via modulating ABCC1. Cell Biosci. 2018;8:9. doi: 10.1186/s13578-018-0209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]