Abstract
Background
A mosquito-based arbovirus surveillance system was set up at Barkedji, Senegal after the first outbreak of Rift valley fever in West Africa in 1988. This system was recently updated using more sampling methods and collecting in greater number of ponds and villages sites.
Methods
For the current study, mosquitoes were sampled biweekly between July and December 2012 and 2013 using CDC+CO2 light traps set at ground and canopy level, mosquito nets baited with goat, sheep, human or chicken, light traps baited with goat, sheep and chicken; bird-baited traps using pigeons or chickens placed either at the ground or canopy level. Collected mosquitoes were identified, pooled and screened for arboviruses.
Results
A total of 42,969 mosquitoes in 4,429 pools were processed for virus isolation. Ten virus species were identified among 103 virus isolates. West Nile virus (WNV; 31 isolates), Barkedji virus (BARV; 18), Sindbis virus (SINV; 13), Usutu virus (USUV; 12), Acado virus (ACAV; 8), Ndumu virus (NDUV; 9), Sanar virus (SANV; 7), Bagaza virus (BAGV; 3), Rift valley fever virus (RVFV; 1), and Yaounde virus (YAOV; 1) were isolated from 9 ponds (91 strains) and 7 villages (12 strains). Only 3 virus species (WNV, NDU and SINV) were isolated from villages. The largest numbers of isolates were collected in October (29.1% of total isolates) and November (50.5%). Viruses were isolated from 14 mosquito species including Cx. neavei (69.9% of the strains), Cx. antennatus (9.7%), and Ma. uniformis (4.8%). NDUV, ACAV, and SINV are herein reported for the first time in the Barkedji area. Isolation of ACAV and SANV from a pool of male Ma. uniformis and USUV and BARV from a pool of male Cx. neavei, are reported for the first time to our knowledge.
Conclusion
Our data indicate that the Barkedji area is characterized by a high diversity of viruses of medical, veterinary and unknown importance. Arboviruses were first detected in July at the beginning of the rainy season and peaked in abundance in October and November. The Barkedji area, an enzootic focus of several potentially emerging arboviruses, should be surveilled annually to be prepared to deal with future disease emergence events.
Keywords: Arbovirus, Surveillance, Mosquitoes, Aampling methods, Ponds, Villages
Background
The Barkedji area, located in the Sahelian biogeographic area of Senegal is known as a focus of enzootic transmission of several arthropod-borne viruses (arboviruses). Indeed, a surveillance program set up in this area after the first outbreak of Rift valley fever in Senegal in 1988 [1, 2] resulted in the isolation of several viruses including 2 alphaviruses (Semliki Forest and Babanki), 6 flaviviruses (Barkedji (BARKV), Bagaza (BAGV), Usutu (USUV), Yaounde (YAOV), West Nile (WNV), Koutango and Saboya), 2 bunyaviruses (Bunyamwera, and Ngari), 2 phleboviruses (Rift valley fever (RVFV), and Gabek Forest), 1 orbivirus (Sanar (SANV)), 1 rhabdovirus (Chandipura) and 1 unclassified virus (ArD95537). Some of these arboviruses (RVFV, USUV, and WNV) are of medical and/or veterinary importance, while the potential health impact of the others is still unknown [2–7]. Enzootic cycles of these viruses generally involve mosquito vectors and domestic and/or wild vertebrates as amplifying or reservoirs hosts [5, 8]. Human are known to be incidentally affected by several of these viruses but are considered as dead-end hosts that do not support ongoing transmission. The viruses listed above cause clinical syndromes of varying severity, ranging from acute benign fevers of short duration to life-threatening encephalitis and/or hemorrhagic fever [8].
Arboviruses surveillance program can monitor viruses in vertebrate hosts and/or arthropod vectors. The surveillance of virus in vertebrate hosts has several drawbacks relative to surveillance of arthropods, including the limited accessibility of hosts, small sample sizes of hosts, and variation of susceptibility of different vertebrate species to the same virus [9]. Surveillance of mosquitoes for circulating arboviruses can be used as an early warning system because it allows the detection of seasonal initiation or increase of virus circulation. Thus, the implementation of mosquito surveillance can give decision makers enough time to enact efficient measures for outbreak control [10]. Mosquito-based virus surveillance programs are also important tools in the study of the eco-epidemiology and transmission of viruses by enabling identification of potential vectors of a given virus as well as the spatio-temporal dynamics of the virus. Mosquito surveillance can also be useful for identifying new arbovirus species of medical and veterinary importance [9, 11, 12].
The mosquito surveillance program for detection of arboviruses that has been active in Barkedji since 1990 used to collect mosquitoes each month using human landing catch, CDC+CO2 light trap set at the ground level, and animal + light baited traps in a very few temporary ponds and villages [1, 13]. This program resulted in the identification of all the arbovirus species listed above, the vectors of RVFV and description of the RVFV enzootic cycle. Previous studies, in the Barkedji area, have shown a high diversity of the mosquito fauna in the study area with a dominance of Aedes vexans and Culex poicilipes and a switch over of dominance of these two species between years [1, 14, 15]. They also showed that Ae. vexans was always the most abundant species at the beginning of the rainy season, while Cx. poicilipes dominated the end of the rainy season. These species preferred host seeking in barren and temporary ponds rather than villages and other land cover Classes were rare [15, 16]. These vectors are temporary ponds breeders [1, 14, 16, 17]. The maximum flight distances, from these ponds, of the main vectors were estimated, to be around 650 m for Ae. vexans and 550 m for Cx. poicilipes [16]. Diallo and others [15], predicted that Ae. vexans, Cx. poicilipes and Cx. neavei mosquitoes would not disperse up to 1,500 m to the nearest ponds. The seasonal dynamics of these vectors were positively correlated with rainfall for Ae. vexans, after a lag time of one month for the Culex species. All the vectors had their highest abundances and parity rates between September and November [15].
However, identification and description of arboviruses circulating in the area are still incomplete. To date, data are still lacking on spatial extent of arbovirus circulation and the complete suite of potential vectors for each virus. In 2012 and 2013, the program was modified to expand the number of trap types and baits used, increase the frequency of trapping from monthly to biweekly, and increase the number of ponds and villages sampled. This paper reports the result of this modified surveillance program in 2012 and 2013. Our specific objectives were to detect arboviruses circulating in the Barkedji area, their associated vectors and their spatio-temporal dynamics.
Methods
Study Area
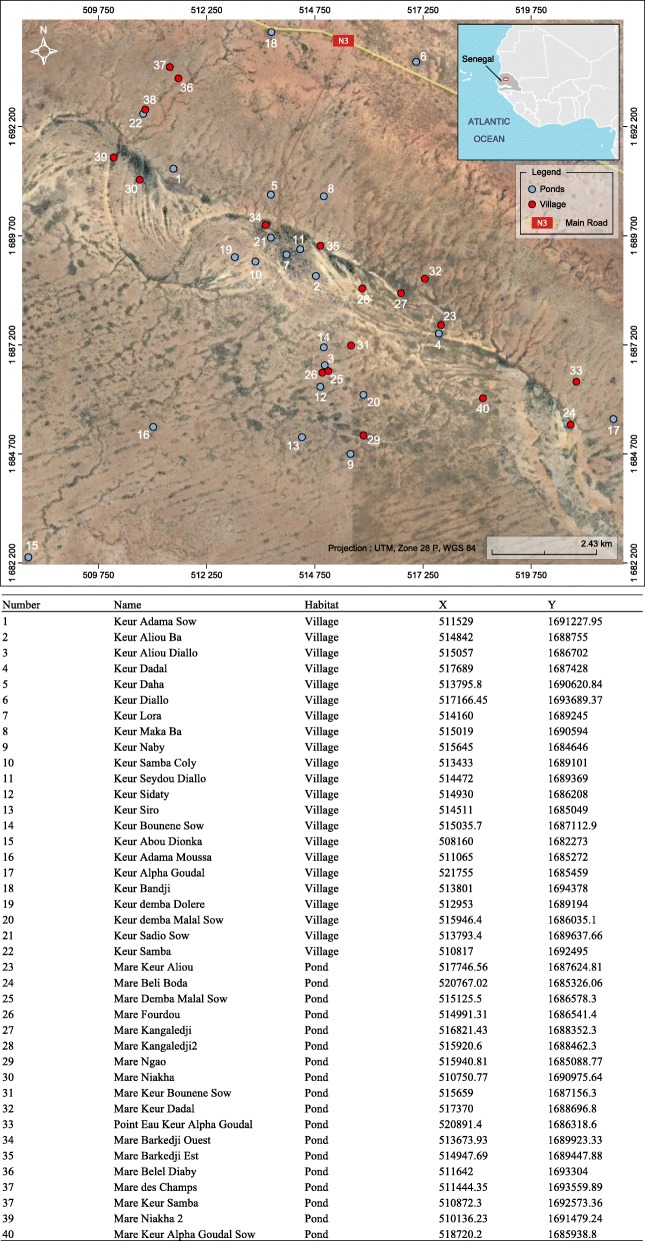
The study was conducted in the Ferlo region of Senegal in an area located around the Barkedji village (14° 53' 0" N, 15° 55' 0" W) from late July to late December in 2012 and 2013. This area (Fig. 1) is semi-desert, characterized mainly as shrubby savanna, with a hot dry climate with a long and severe dry season and short rainy season that generally occurs between July and September. Annual rainfall is approximately 300 mm. A network of temporary ponds of different sizes is flooded by the first rains. Small ponds are flooded and drained after each rain. Large ponds are flooded at the beginning of the rainy season, remain inundated for a long period and are covered by hydrophytes. These ponds are the main source of water for herders and their livestock in this period. These ponds are also the natural habitats of many vertebrate species (birds, reptiles and rodents) and mosquito vectors of arboviruses. Horses and donkeys serve for transportation and animal traction. They congregate around ponds in the afternoon and at night. The area is covered with a rich carpet grass used by local and nomadic populations during the rainy season. The main activities of people are millet cultivation and sheep, goats, and cattle grazing. Family groups and their herds may become seasonal nomads, temporarily relocating for a few months during which they use grass huts for shelter. Barkedji is the only substantial and populated village in the area, while the other villages are located near ponds and composed of just a few huts.
Fig. 1.
Study area with the names of villages and ponds sampled and their geographic coordinates in UTM (Modified from Google Earth)
Mosquito collection
In 2012, mosquitoes were collected using animal+light-baited traps [18], mosquito net-baited traps [17] and CDC+CO2 light traps [19] set on the shore of 18 ponds, and using CDC+CO2 light traps in 22 villages around Barkedji. Mosquito were collected monthly in villages and biweekly in ponds. In 2013, ponds were sampled biweekly with the traps listed above as well as bird-baited traps [20]. In November 2013, 3 ponds and 1 village were sampled once in Thiargny (50 km from Barkedji) following suspicion of an arbovirus outbreak. The sampling methods used were CDC+CO2 light traps set at ground and canopy level; mosquito-net baited with goat, sheep, human or chicken; light trap baited with goat, sheep and chicken; bird baited trap using pigeons or chickens at the ground and canopy level. Each fortnight, traps were set from dusk to dawn. In the field laboratory, mosquitoes were sorted, identified and pooled (maximum of 89 and average of 9.8 mosquitoes per pool) by species, sex and collection site on a chill table using several morphological keys [21–26]. Mosquito pools were then stored in liquid nitrogen and shipped to the Institut Pasteur in Dakar for virus testing.
Virus isolation and identification
Mosquito pools were homogenized in 3 ml of L-15 medium (Gibco BRL, GrandIsland, NY, USA), using chilled tissue-grinders, supplemented with 20% fetal bovine serum (Gibco) and clarified by centrifugation at 1500 g, 4°C for 10 min. Following centrifugation, the supernatant was filtered using a 1 ml syringe (Artsana, Como, Italy) and sterilized with 0.20 μm filters (Sartorius, Göttingen, Germany).
Following procedures described previously in Digoutte et al. [27], the clarified and filtered suspensions were inoculated into Aedes pseudoscutellaris (AP61) and Vero cell lines and incubuated for 7-8 days. The presence of virus was detected by indirect immunofluorescence using using in-house hyper-immune mouse ascites fluids directed to individual or groups of more than 70 African arboviruses (flaviviruses, bunyaviruses, orbiviruses and alphaviruses). Identifications were later confirmed by complement fixation and seroneutralization tests. All virological tests were carried out by the WHO Collaborating Center of Reference and Research on Arboviruses and Hemorrhagic Fever Viruses of the Institut Pasteur de Dakar.
Data analysis
Mosquito seasonal distribution patterns were evaluated by measuring the absolute abundance of mosquitoes, quantified as the average females per trap (CDC+CO2 light traps only) per night. The proportion (%) of each mosquito vector collected in all traps per habitat was also calculated. The Minimum Field Infection Rate (MFIR ‰) was calculated as the number of positive pools per 1000 mosquitoes. Differences of frequencies between groups were tested by contingency table analyses. All analyses were conducted using R [28].
Results
Mosquito collection
A total of 42,992 mosquitoes (41,921 females) representing 7 genera and 42 species were collected (Table 1), including 1 species of Aedomyia, 15 Aedes, 7 Anopheles, 13 Culex, 2 Mansonia, 3 Mimomyia and 1 Uranotaenia. The predominant species in both the ponds (22.9% of all mosquitoes collected) and villages (49.1%) was Ae. vexans. This species was followed by Ae. ochraceus (11.2%) and Cx. poicilipes (9.0%) in villages and by Cx. poicilipes (19.7%), Ma. uniformis (11.0%) and Cx. neavei (10.9%) in ponds.
Table 1.
Mosquito females collected in ponds and 694 villages in the Barkedji area, 2012-2013
| Species | Ponds | Villages | Total | |||
|---|---|---|---|---|---|---|
| No collected (males) | % | No collected (males) | % | No collected (males) | % | |
| Aedomya africana | 7 | 0 | 0 | 0 | 7 | 0 |
| Aedes aegypti | 110 (20) | 0.3 | 10 | 0.2 | 120 (20) | 0.3 |
| Aedes argenteopunctatus | 258 | 0.7 | 14 | 0.3 | 272 | 0.6 |
| Aedes dalzieli | 1297 (3) | 3.5 | 94 | 1.7 | 1391 (3) | 3.2 |
| Aedes fowleri | 120 | 0.3 | 73 | 1.3 | 193 | 0.4 |
| Aedes furcifer | 205 | 0.5 | 1 | 0 | 206 | 0.5 |
| Aedes hirsutus | 29 | 0.1 | 1 | 0 | 30 | 0.1 |
| Aedes luteocephalus | 22 | 0.1 | 3 | 0.1 | 25 | 0.1 |
| Aedes mcintoshi | 5 | 0 | 3 | 0.1 | 8 | 0 |
| Aedes metallicus | 18 (1) | 0 | 2 | 0 | 37 (1) | 0.1 |
| Aedes minutus | 156 | 0.4 | 54 | 1 | 210 | 0.5 |
| Aedes ochraceus | 1377 (1) | 3.7 | 627 | 11.2 | 2004 (1) | 4.7 |
| Aedes sudanensis | 659 (9) | 1.8 | 115 | 2.1 | 774 (9) | 1.8 |
| Aedes unilineatus | 21 (1) | 0.1 | 0 | 0 | 21 (1) | 0 |
| Aedes vexans | 8546 (12) | 22.9 | 2751 | 49.1 | 11297 (12) | 26.3 |
| Aedes vittatus | 0 | 0 | 1 | 0 | 1 | 0 |
| Anopheles coustani | 1 | 0 | 0 | 0 | 1 | 0 |
| Anopheles funestus | 1 | 0 | 0 | 0 | 1 | 0 |
| Anopheles gambiae | 122 (8) | 0.3 | 86 | 1.5 | 208 (8) | 0.5 |
| Anopheles pharoensis | 256 (13) | 0.7 | 37 | 0.7 | 293 (13) | 0.7 |
| Anopheles rufipes | 15 | 0 | 96 | 1.7 | 111 | 0.3 |
| Anopheles squamosus | 54 (5) | 0.1 | 44 | 0.8 | 98 (5) | 0.2 |
| Anopheles ziemanni | 2396 (4) | 6.4 | 250 | 4.5 | 2646 (4) | 6.2 |
| Culex annulioris | 0 | 0 | 17 | 0.3 | 17 | 0 |
| Culex antennatus | 1264 (10) | 3.4 | 114 | 2 | 1378 (10) | 3.2 |
| Culex bitaeniorhynchus | 533 (1) | 1.4 | 76 | 1.4 | 609 (1) | 1.4 |
| Culex decens | 18 | 0 | 0 | 0 | 18 | 0 |
| Culex ethiopicus | 1107 (12) | 3 | 55 | 1 | 1162 (12) | 2.7 |
| Culex neavei | 4077 (459) | 10.9 | 146 | 2.6 | 4223 (459) | 9.8 |
| Culex nebulosus | 2 | 0 | 10 | 0.2 | 12 | 0 |
| Culex perfuscus | 525 | 1.4 | 22 | 0.4 | 547 | 1.3 |
| Culex poicilipes | 7382 (113) | 19.7 | 502 | 9 | 7884 (113) | 18.3 |
| Culex quinquefasciatus | 13 (1) | 0 | 72 (1) | 1.3 | 84 (1) | 0.2 |
| Culex sp | 3 | 0 | 0 | 0 | 3 | 0 |
| Culex tigripes | 4 | 0 | 0 | 0 | 4 | 0 |
| Culex tritaeniorhynchus | 1470 (9) | 3.9 | 176 | 3.1 | 1646 (9) | 3.8 |
| Mansonia africana | 810 (162) | 2.2 | 32 | 0.6 | 842 (162) | 2 |
| Mansonia uniformis | 4110 (191) | 11 | 112 | 2 | 4222 (191) | 9.8 |
| Mimomyia mimomyiaformis | 57 | 0.2 | 0 | 0 | 57 | 0.1 |
| Mimomyia plumosa | 115 (1) | 0.3 | 4 | 0.1 | 119 (1) | 0.3 |
| Mimomyia splendens | 224 (34) | 0.6 | 0 | 0 | 224 (34) | 0.5 |
| Uranotaenia mayeri | 3 | 0 | 0 | 0 | 3 | 0 |
| Total collected (males) | 37392 (1070) | 100 | 5600 (1) | 100 | 42992 (1071) | 100 |
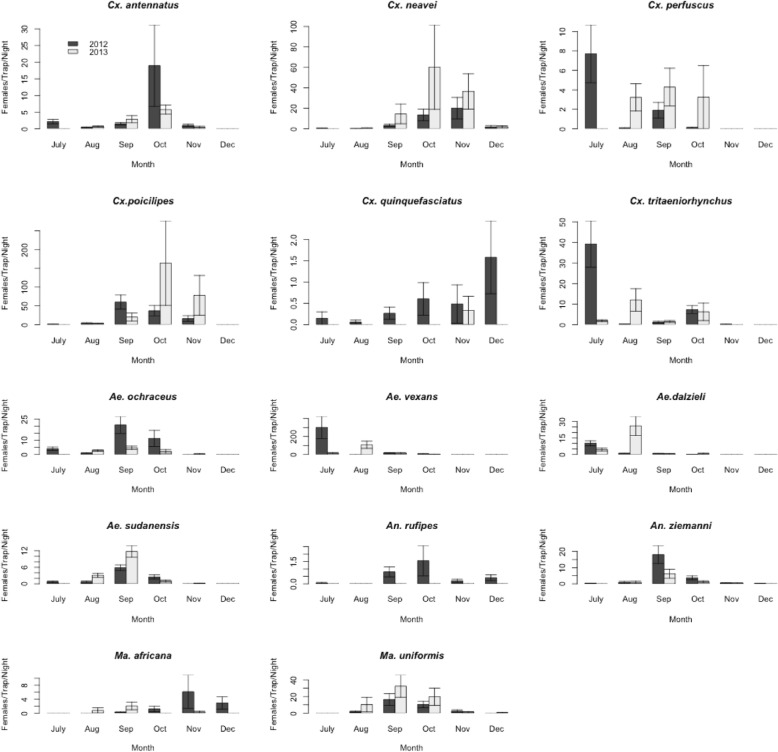
Only mosquito species (Table 2) found infected were considered in the following analyses. Proportions of each species varied significantly between ponds and villages (p < 0.0001). Seasonal patterns of mosquito females collected by CDC+CO2 light-traps are presented in Fig. 2. The largest numbers of females per CDC+CO2 trap per night were collected in October of both years for Cx. antennatus, November 2012 and October 2013 for Cx. neavei, September 2012 and October 2013 for Cx. poicilipes, July 2012 and September 2013 for Cx. perfuscus, November 2012 and September 2013 for Ma. africana, July 2012 and August 2013 for Cx. tritaeniorhynchus, July 2012 and August 2013 for Ae. vexans and Ae. dalzieli, September 2012 and 2013 for Ae. ochraceus, Ae. sudanensis, An. ziemanni and Ma. uniformis, October 2012 for An. rufipes.
Table 2.
Viruses isolated, associated mosquitoes and vertebrates, and pathogen potential, Barkedji, 2012-2013
| Virus | Year of isolation | Family | Mosquitoes found infected in this study | Vertebrates associated | Pathogen potential |
|---|---|---|---|---|---|
| Acado | 2013 | Reoviridae | Culex antennatus, Cx. neavei, Mansonia uniformis | Humans, domestic ungulates, | unknown |
| Bagaza | 2012 | Flaviviridae | Culex antennatus, Cx. neavei | Humans, wild birds, red-legged partridges and ring-necked pheasants, common wood pigeons | Unknown |
| 2013 | Cx. neavei | ||||
| Barkedji | 2012 | Flaviviridae | Cx. neavei, Ae. sudanensis | Unknown | Unknown |
| 2013 | Cx. neavei | ||||
| Ndumu | 2012 | Togaviridae | Aedes dalzieli, Ae. vexans, Anopheles rufipes, An. ziemanni, Cx. poicilipes, Ma. uniformis | Domestic pigs, Humans | Unknown |
| Rift valley fever | 2013 | Bunyaviridae | Ae. ochraceus | Yes | |
| Sanar | 2013 | Reoviridae | Cx. neavei, Ma. uniformis | Unknown | Unknown |
| Sindbis | 2012 | Togaviridae | Cx. antennatus, Cx. neavei | Humans, Birds, Orangutans, Sheeps, frogs, reed warblers, Bats | Yes |
| 2013 | Cx. neavei | ||||
| Usutu | 2012 | Flaviviridae | Culex antennatus, Cx. neavei | Human, African furred rat, blackbirds, Bats | Yes |
| 2013 | Cx. neavei | ||||
| West Nile | 2012 | Flaviviridae | Ae. dalzieli, An. rufipes, Cx. antennaus, Cx. neavei, Cx. perfuscus, Cx. poicilipes, Cx. quinquefasciatus | Horses, Birds, Human | Yes |
| 2013 | Cx. neavei, Cx. tritaeniorhynchus | ||||
| Yaounde | 2012 | Flaviviridae | Cx. tritaeniorhynchus | Bird (Bycanistes sharpii), Rodents (Praomys sp., Cavia porcellus) | Unknown |
Fig. 2.
Temporal dynamics of mosquito vectors of arboviruses in the Barkedji area, 2012-2013
Viruses
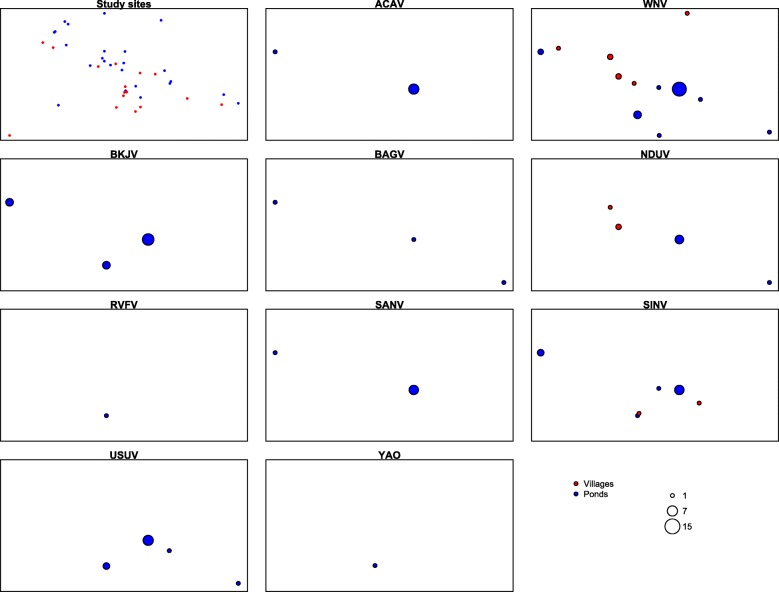
All the 42,969 mosquitoes in 4,429 pools were tested for virus isolation yielding 103 virus isolates within 10 virus species (Tables 2 and 3). WNV (30.1%), BKJV (17.5%), SINV (12.6%), and USUV (11.6%) were the most prevalent. All virus isolates were from 9 ponds (88.3 % of the virus and 50% of total ponds) and 7 villages (out of 31.8% of total villages sampled). Most of the isolates from ponds were from Kangaledji (59.3%), Fourdou (14,3%) and Niakha (13.2%). Three virus species (WNV, NDUV and SINV) were isolated from villages and 10 from ponds during the study period. The largest numbers of viruses were isolated in October (29.1% of total isolates) and November (50.5%). Viruses were isolated from 14 mosquito species. The largest numbers of viral isolations were from Cx. neavei (69.9% of the strains), Cx. antennatus (9.7%), and Ma. uniformis (4.8%). Seven different viral species were isolated from Cx. neavei (ACAV, BAGV, BKJV, SANV, SINV, USUV, and WNV), and Cx. antennatus (ACAV, BAGV, NDUV, SANV, SINV, USUV, and WNV) and 3 from Ma. uniformis (ACAV, NDUV and SANV). Viruses were detected from vectors before, during and after their peak abundances.
Table 3.
Viruses isolated and minimum field infection rates by species, Barkedji area, 2012-2013
| Species | ACAV | BAGV | BARV | NDUV | RVFV | SANV | SINV | USUV | WNV | YAOV | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P+ | MFIR | P+ | MFIR | P+ | MFIR | P+ | MFIR | P+ | MFIR | P+ | MFIR | P+ | MFIR | P+ | MFIR | P+ | MFIR | P+ | MFIR | P+ | |
| Aedes dalzieli | 1 | 0.7 | 1 | 0.7 | 2 | ||||||||||||||||
| Aedes ochraceus | 1 | 0.5 | 1 | ||||||||||||||||||
| Aedes sudanensis | 1 | 1.4 | 1 | ||||||||||||||||||
| Aedes vexans | 1 | 0.9 | 1 | ||||||||||||||||||
| Anopheles rufipes | 1 | 10.0 | 1 | 10.0 | 2 | ||||||||||||||||
| Anopheles ziemanni | 1 | 0.4 | 1 | ||||||||||||||||||
| Culex antennatus | 1 | 0.7 | 1 | 0.7 | 1 | 0.7 | 1 | 0.7 | 1 | 0.7 | 2 | 1.5 | 3 | 2.2 | 10 | ||||||
| Culex neavei | 5 | 1.3 | 2 | 0.5 | 16 | 4.3 | 4 | 1.1 | 12 | 3.2 | 9 | 2.4 | 22 | 5.8 | 70 | ||||||
| Culex perfuscus | 1 | 1.8 | 1 | ||||||||||||||||||
| Culex poicilipes | 1 | 0.1 | 1 | 0.1 | 2 | ||||||||||||||||
| Culex quinquefasciatus | 1 | 12.5 | 1 | ||||||||||||||||||
| Culex tritaeniorhynchus | 1 | 0.6 | 1 | 0.6 | 2 | ||||||||||||||||
| Mansonia africana | 1 | 1.5 | 1 | 1.5 | 2 | ||||||||||||||||
| Mansonia uniformis | 3 | 0.7 | 3 | ||||||||||||||||||
| Culex neavei male | 1 | 1 | 2 | ||||||||||||||||||
| Mansonia uniformis male | 1 | 1 | 2 | ||||||||||||||||||
| Total | 8 | 3 | 18 | 9 | 1 | 7 | 13 | 12 | 31 | 1 | 103 | ||||||||||
P+: number of positive pools; MFIR: minimum field infection rate (total number positive pools / the total number of mosquitoes tested *1000)
West Nile virus
Overall, WNV was isolated from 31 mosquito pools belonging to 3 genera and 8 species collected in 8 ponds (24/77.4% of all the WNV strains) and 5 villages (Table 4 & Fig. 3). The largest numbers of WNV isolates were from the Kangaledji ponds (54.2% of the strains detected in ponds). The virus was identified from 22 pools of Cx. neavei (71%), 3 pools of Cx. antennatus (9.7%) and 1 pool of each of the 6 other vectors (Cx. poicilipes, Cx. perfuscus, Cx. tritaeniorhunchus, Cx. quinquefasciatus, Ae. dalzieli and An. rufipes). Infected pools were collected in July (2 pools / 6.4 % of the total) September (3 pools / 9.7 %), October (12 pools / 38.7 % and November (14 pools / 45.2 %). The WNV MFIR per thousand pooled mosquitoes for all species are presented Tables 3 and 4. Mean minimum field infection rates among species (Table 3), differed significantly (χ2 = 54.8, DF = 7, p < 0.0001) among mosquito vectors, ranging from 12.5 ‰ for Cx. quinquefasciatus to 0.1 ‰ for Cx. poicilipes. The highest infection rate was observed in November for Cx. neavei and Cx. antennatus (Table 4). Differences across the season in MFIR were not statistically significant for Cx. neavei and Cx. antennatus (p > 0.2).
Table 4.
Viruses isolated in mosquito females and minimum field infection rates by month, Barkedji area, 2012-2013
| Virus | July | September | October | November | December | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | No | NP | P+ | MFIR | No | NP | P+ | MFIR | No | NP | P+ | MFIR | No | NP | P+ | MFIR | No | NP | P+ | MFIR | |
| WNV | Aedes dalzieli | 367 | 52 | 0 | 0.0 | 108 | 46 | 0 | 0.0 | 7 | 6 | 1 | 142.9 | 0 | 0 | 0 | 0.0 | 0 | 0 | 0 | 0.0 |
| Anopheles rufipes | 1 | 1 | 0 | 0.0 | 33 | 9 | 0 | 0.0 | 61 | 10 | 1 | 16.4 | 8 | 6 | 0 | 0.0 | 8 | 6 | 0 | 0.0 | |
| Culex antennatus | 46 | 15 | 0 | 0.0 | 176 | 77 | 0 | 0.0 | 990 | 71 | 2 | 2.0 | 96 | 35 | 1 | 10.4 | 0 | 0 | 0 | 0.0 | |
| Culex neavei | 12 | 6 | 0 | 0.0 | 669 | 97 | 3 | 4.5 | 1193 | 78 | 6 | 5.0 | 1721 | 128 | 13 | 7.6 | 104 | 25 | 0 | 0.0 | |
| Culex perfuscus | 155 | 16 | 1 | 6.5 | 222 | 64 | 0 | 0.0 | 64 | 21 | 0 | 0.0 | 0 | 0 | 0 | 0.0 | 0 | 0 | 0 | 0.0 | |
| Culex poicilipes | 19 | 6 | 0 | 0.0 | 3168 | 149 | 0 | 0.0 | 2401 | 114 | 1 | 0.4 | 1974 | 113 | 0 | 0.0 | 4 | 4 | 0 | 0.0 | |
| Culex quinquefasciatus | 4 | 2 | 1 | 250.0 | 13 | 6 | 0 | 0.0 | 23 | 3 | 0 | 0.0 | 20 | 5 | 0 | 0.0 | 20 | 5 | 0 | 0.0 | |
| Culex tritaeniorhynchus | 842 | 48 | 0 | 0.0 | 98 | 40 | 0 | 0.0 | 343 | 47 | 1 | 2.9 | 14 | 12 | 0 | 0.0 | 0 | 0 | 0 | 0.0 | |
| Total vectors | 1446 | 146 | 2 | 1.4 | 4487 | 488 | 3 | 0.7 | 5082 | 350 | 12 | 2.4 | 3833 | 299 | 14 | 3.7 | 136 | 40 | 0 | 0.0 | |
| USUV | Culex antennatus | 46 | 15 | 0 | 0.0 | 176 | 77 | 0 | 0.0 | 990 | 71 | 1 | 1.0 | 96 | 35 | 1 | 10.4 | 0 | 0 | 0 | 0.0 |
| Culex neavei | 12 | 6 | 0 | 0.0 | 669 | 97 | 3 | 4.5 | 1193 | 78 | 2 | 1.7 | 1721 | 128 | 4 | 2.3 | 104 | 25 | 0 | 0.0 | |
| Total vectors | 58 | 21 | 0 | 0.0 | 845 | 174 | 3 | 3.6 | 2183 | 149 | 3 | 1.4 | 1817 | 163 | 5 | 2.8 | 104 | 25 | 0 | 0.0 | |
| ACAV | Culex antennatus | 46 | 15 | 0 | 0.0 | 176 | 77 | 0 | 0.0 | 990 | 71 | 0 | 0.0 | 96 | 35 | 1 | 10.4 | 0 | 0 | 0 | 0.0 |
| Culex neavei | 12 | 6 | 0 | 0.0 | 669 | 97 | 0 | 0.0 | 1193 | 78 | 1 | 0.8 | 1721 | 128 | 4 | 2.3 | 104 | 25 | 0 | 0.0 | |
| Mansonia africana | 0 | 0 | 0 | 0.0 | 64 | 30 | 0 | 0.0 | 76 | 17 | 0 | 0.0 | 256 | 31 | 0 | 0.0 | 263 | 20 | 1 | 3.8 | |
| Total vectors | 58 | 21 | 0 | 0.0 | 909 | 204 | 0 | 0.0 | 2259 | 166 | 1 | 0.5 | 2073 | 194 | 5 | 2.8 | 367 | 45 | 1 | 2.7 | |
| BAGV | Culex antennatus | 46 | 15 | 0 | 0.0 | 176 | 77 | 0 | 0.0 | 990 | 71 | 1 | 1.0 | 96 | 35 | 0 | 0.0 | 0 | 0 | 0 | 0.0 |
| Culex neavei | 12 | 6 | 0 | 0.0 | 669 | 97 | 0 | 0.0 | 1193 | 78 | 0 | 0.0 | 1721 | 128 | 2 | 1.2 | 104 | 25 | 0 | 0.0 | |
| Total vectors | 58 | 21 | 0 | 0.0 | 845 | 0 | 0.0 | 2183 | 149 | 1 | 0.5 | 1817 | 163 | 2 | 1.1 | 104 | 25 | 0 | 0.0 | ||
| BARV | Aedes sudanensis | 15 | 8 | 0 | 0.0 | 517 | 103 | 1 | 1.9 | 111 | 36 | 0 | 0.0 | 2 | 2 | 0 | 0.0 | 0 | 0 | 0 | 0.0 |
| Culex neavei | 12 | 6 | 0 | 0.0 | 669 | 97 | 2 | 3.0 | 1193 | 78 | 4 | 3.4 | 1721 | 128 | 10 | 5.8 | 104 | 25 | 0 | 0.0 | |
| Total vectors | 27 | 14 | 0 | 0.0 | 1186 | 200 | 3 | 2.5 | 1304 | 114 | 4 | 0.0 | 1723 | 130 | 10 | 0.0 | 104 | 25 | 0 | 0.0 | |
| NDUV | Aedes dalzieli | 367 | 52 | 0 | 0.0 | 108 | 46 | 0 | 0.0 | 7 | 6 | 1 | 142.9 | 0 | 0 | 0 | 0.0 | 0 | 0 | 0 | 0.0 |
| Aedes vexans | 6603 | 187 | 0 | 0.0 | 1408 | 102 | 0 | 0.0 | 278 | 44 | 1 | 3.6 | 4 | 4 | 0 | 0.0 | 0 | 0 | 0 | 0.0 | |
| Anopheles rufipes | 1 | 1 | 0 | 0.0 | 33 | 9 | 0 | 0.0 | 61 | 10 | 1 | 16.4 | 8 | 6 | 0 | 0.0 | 8 | 6 | 0 | 0.0 | |
| Anopheles ziemanni | 6 | 6 | 0 | 0.0 | 1879 | 129 | 1 | 0.5 | 434 | 44 | 0 | 0.0 | 62 | 33 | 0 | 0.0 | 10 | 8 | 0 | 0.0 | |
| Culex antennatus | 46 | 15 | 0 | 0.0 | 176 | 77 | 0 | 0.0 | 990 | 71 | 0 | 0.0 | 96 | 35 | 0 | 0.0 | 0 | 0 | 0 | 0.0 | |
| Culex poicilipes | 19 | 6 | 0 | 0.0 | 3168 | 149 | 0 | 0.0 | 2401 | 114 | 1 | 0.4 | 1974 | 113 | 0 | 0.0 | 4 | 4 | 0 | 0.0 | |
| Mansonia uniformis | 0 | 0 | 0 | 0.0 | 2277 | 142 | 3 | 1.3 | 864 | 70 | 0 | 0.0 | 127 | 30 | 0 | 0.0 | 2 | 2 | 0 | 0.0 | |
| Total vectors | 7042 | 267 | 0 | 0.0 | 9049 | 654 | 4 | 0.4 | 5035 | 359 | 4 | 1.0 | 2271 | 221 | 0 | 0.0 | 24 | 20 | 0 | 0.0 | |
| SANV | Culex antennatus | 46 | 15 | 0 | 0.0 | 176 | 77 | 0 | 0.0 | 990 | 71 | 0 | 0.0 | 96 | 35 | 1 | 10.4 | 0 | 0 | 0 | 0.0 |
| Culex neavei | 12 | 6 | 0 | 0.0 | 669 | 97 | 0 | 0.0 | 1193 | 78 | 0 | 0.0 | 1721 | 128 | 4 | 2.3 | 104 | 25 | 0 | 0.0 | |
| Mansonia africana | 0 | 0 | 0 | 0.0 | 64 | 30 | 0 | 0.0 | 76 | 17 | 0 | 0.0 | 256 | 31 | 0 | 0.0 | 263 | 20 | 1 | 3.8 | |
| Total vectors | 58 | 21 | 0 | 0.0 | 909 | 204 | 0 | 0.0 | 2259 | 166 | 0 | 0.0 | 2073 | 194 | 5 | 2.8 | 367 | 45 | 1 | 2.7 | |
| SINV | Culex antennatus | 46 | 15 | 0 | 0.0 | 176 | 77 | 0 | 0.0 | 990 | 71 | 0 | 0.0 | 96 | 35 | 1 | 10.4 | 0 | 0 | 0 | 0.0 |
| Culex neavei | 12 | 6 | 0 | 0.0 | 669 | 97 | 1 | 1.5 | 1193 | 78 | 5 | 4.2 | 1721 | 128 | 6 | 3.5 | 104 | 25 | 0 | 0.0 | |
| Total vectors | 58 | 21 | 0 | 0.0 | 845 | 174 | 1 | 1.2 | 2183 | 149 | 5 | 2.3 | 1817 | 163 | 7 | 3.9 | 104 | 25 | 0 | 0.0 | |
| RVFV | Aedes ochraceus | 83 | 17 | 0 | 0.0 | 1250 | 110 | 1 | 0.8 | 516 | 53 | 0 | 0.0 | 5 | 4 | 0 | 0.0 | 0 | 0 | 0 | 0.0 |
| YAOV | Culex tritaeniorhynchus | 842 | 48 | 1 | 1.2 | 98 | 40 | 0 | 0.0 | 343 | 47 | 0 | 0.0 | 14 | 12 | 0 | 0.0 | 0 | 0 | 0 | 0.0 |
No: Number of female mosquitoes collected; NP: Number of pools; P+: number of positive pools; MFIR: minimum field infection rate (total number positive pools / the total number of mosquitoes tested *1000)
Fig. 3.
Spatial distribution of arboviruses detected in ponds and villages of the Barkedji area, 2012-2013. Saaka pond, from which Barkedji and West Nile viruses were isolated (1 pool each) is not included in the map because it is located about 50Km from the other sites
Usutu virus
>This virus was isolated from 10 pools (9 females and 1 male) of Cx. neavei (83.3% of the isolates) and 2 pools of Cx. antennatus collected in 4 ponds (Table 4 & Fig. 3). The largest numbers of viral isolates were from the Kangaledji pond with 7 pools (58.3% of the strains; Fig. 3). Infected pools were collected in September (3 pools), October (3) and November (5) (Table 4). The mean minimum infection rates of Cx. antennatus (1.5‰) and Cx. neavei (2.4‰) (Table 3) were comparable (p = 0.7). The highest infection rate was observed in November for Cx. antennatus and September for Cx. neavei. The differences in variation in MFIR across the season were not statistically significant for Cx. neavei and Cx. antennatus (p > 0.2).
Acado virus
This virus was isolated from 5 pools of Cx. neavei (71.4% of the isolates), 1 pool of both Cx. antennatus and Ma. africa and 1 pool of Ma. uniformis male (Table 3) collected from the Kangaledji and Niakha ponds (Fig. 3). Infected pools were collected in October (1 pool), November (5 pools) and December (1 pool) (Table 4). The mean MFIR of Cx. antennatus (0.7 ‰) and Cx. neavei (1.3 ‰) presented in Table 3 were comparable (p = 1).
The highest infection rate was observed in late November for Cx. neavei. The differences were not statistically significant (p = 0.86). The differences of the seasonal variation in MFIR were not statistically significant for Cx. neavei (p > 0.09)
Bagaza virus
This virus was isolated from 2 pools of Cx. neavei and 1 pool of Cx. antennatus (Tables 2 and 3) collected in 3 ponds (Fig. 3). Infected pools were collected in October (1 pool) and November (2 pools) (Table 4). The mean MFIR of Cx. antennatus (0.7 ‰) and Cx. neavei (0.5 ‰) were comparable (p = 1).
Barkedji virus
This virus was isolated from Cx. neavei (17 pools/16 females and 1 male), and Ae. sudanensis (1 pool) collected in 4 ponds (Table 3 & Fig. 3). The largest numbers of viral strains were from the Kangaledji pond with 9 pools (52.9 % of the strains). Positive pools were found in September (3 pools) October (4 pools), and November (10 pools) (Table 4). The highest seasonal infection rate was observed in November but the difference with the other months was not statistically significant (p = 0.6).
Ndumu virus
A total of 9 NDUV strains were isolated from mosquito pools belonging to 4 genera and 7 species (Tables 2 and 3) collected in 2 ponds (6/66.7% of all strains) and 2 villages (Fig. 3). The largest numbers of viral strains were from the Kangaledji pond (83.3% of the strains collected in ponds). The virus was identified from Ma. uniformis (3 pools/33.3% of the total), Ae. dalzieli, Ae. vexans, An. rufipes, An. ziemanni Cx. antennatus and Cx. poicilipes (1 pool each). Infected pools were collected in September (5 pools) and October (4 pools). No species showed seasonal variation of it MFIR (Table 4).
Sanar virus
This virus was isolated from Cx. neavei (4 pools; 57.1 % of the strains), Cx. antennatus, Ma. africana and Ma. uniformis male (1 pool for each species) collected from the Kangaledji (6 pools) and Niakha (1pools) pond (Table 3 & Fig. 3). Infected female pools were collected in November (5 pools) and December (1 pool) (Table 4). The mean MFIR of Cx. antennatus (0.7‰) and Cx. neavei (1.1‰) were comparable (p = 1).
Sindbis virus
SINV was isolated from 12 pools of Cx. neavei and 1 pool of Cx. antennatus (Tables 2 and 3) collected in 4 different ponds and 2 villages (Fig. 3). Infected pools were collected in September (1 pool), October (5 pools) and November (7 pools). The mean MFIR of Cx. antennatus (0.7‰) and Cx. neavei (3.2‰) were comparable (p = 0.2).
Yaounde virus
Only one isolate of this virus was recovered from a pool of Cx. tritaeniorhynchus (Tables 2 and 3) collected in a pond on July (Table 4 & Fig. 3).
Rift valley fever virus
The single strain of RVFV was isolated from a pool of Ae. ochraceus collected from a pond in September 2013 (Table 4 & Fig. 3).
Discussion
In this study we detected 10 arbovirus species from the Barkedji area, emphasizing the importance of this area as a hotspot of enzootic arboviruses transmission and a good platform for vector-borne diseases surveillance [1]. Moreover, we detected NDUV, ACAV, and SINV for the first time in the Barkedji area after more than 20 years of arbovirus surveillance, demonstrating the high biodiversity of arboviruses in the region and suggesting that our knowledge of the arboviruses present is still probably incomplete. Our study also highlights the importance of using diverse sampling methods and sites to characterize vector and arbovirus diversity. WNV, NDUV and SINV were the only three virus species isolated from villages, suggesting that these viruses pose a particular risk for transmission to human and domestic animals in the Barkedji area.
ACAV and SANV were isolated from a pool of male Ma. uniformis and USUV and BARV from a pool of male Cx. neavei, representing the first time in our knowledge, that these viruses have been detected in male mosquitoes, supporting the possibility of the maintenance of these viruses by vertical transmission in the wild. Thus, these mosquito species may be investigated as reservoirs of these viruses during the dry season.
RVFV has been isolated from several mosquito species in many African countries [1, 29]. This virus has been responsible for widespread outbreaks in both humans and domestic ungulates in Africa and the Arabian Peninsula [30–36]. The disease is characterized by mass abortions and high mortality in animals resulting in high economic losses [4]. In humans also, high mortality rates and severe complications have been observed [37]. Detection of a single isolate of RVFV in September 2013 in our study, was followed by several outbreaks of RVFV in human and ungulates in others parts of Senegal (http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?reportid=14211). This very low amplification of RVF did not involve Ae. vexans and Cx. poicilipes, known as the most abundant species and the main vectors of RVFV in West Africa [13, 18]. This single isolation of RVFV may also suggest that the amplification was too low to spill over to human and domestic ungulates in the Barkedji area.
This study provides the first documented evidence of ACAV in Senegal. ACAV, like SANV, is a member of the Corriparta virus species/serogroup (family: Reoviridae, genus: Orbivirus). This virus was first isolated in 1963 from a mixed Culex antennatus and Cx. neavei pool collected in the Acado Baro River, Ilubabor Pro in Ethiopia [38]. ACAV has never been associated to any vertebrates but mosquito species infected by ACAV are ornithophagic and have been associated with pathogenic viruses. Thus, ACAV may be potentially pathogenic to human and animals in Barkedji. This virus was detected in only 2 of the ponds investigated and mainly in November indicating a highly focal spatio-temporal amplification pattern.
SANV was isolated for the first time in 1990 from Cx. poicilipes in Dakar-Bango near Saint Louis, Senegal, and later several other mosquitoes in the Senegal River Basin, Kedougou, and Barkedji area [1, 18]. In this study, the virus was isolated mainly from Cx. neavei, which should be considered as the main vector. Cx. antennatus, Ma. africana, and Ma. uniformis are potential secondary vectors. Its vertebrate hosts and disease association are unknown. However, the ornitophagic tendency of the SANV vectors suggest that birds may play a potential role in the natural history of this virus. Viruses belonging to Corriparta virus serogroup detected in Australia, Africa and South America have been detected in a wide range of vertebrate hosts [39–42].
Five members of the genus Flavivirus, family Flaviviridae (WNV, USU, BAGV, BARV, and YAOV) were isolated in our study. WNV was isolated from 31 mosquito pools (belonging to 8 species), 8 ponds and 5 villages confirming the high and wide enzootic activity of the virus in the area [1, 43]. Indeed, this virus was continuously isolated from mosquitoes collected in the area and antibodies directed against the virus were detected in more than 78% of horses, predicted in up to 39% of resident birds from Barkedji [44, 45]. Antibodies directed against WNV were detected in 80% of a human sample collected in a village located at 80 km from Barkedji [1]. Human disease attributable to WNV in Africa has most often been categorized as mild febrile illness with rash, although more severe symptoms including hepatitis and encephalitis have been reported [46, 47]. WNV was isolated mainly from Cx. neavei and from Cx. antennatus, Cx. poicilipes, Cx. perfuscus, Cx. tritaeniorhunchus, Cx. quinquefasciatus, Ae. dalzieli and An. rufipes. WNV was isolated previously from Cx. neavei, Cx. perfuscus and Cx. tritaeniorhynchus in Barkedji and from Cx. antennatus and Ma. uniformis in the Senegal River Basin [18, 43] and Ae. dalzieli in Kedougou [43]. The virus was detected for the first time from Cx. antennatus, Ae. dalzieli and Ma. uniformis in Barkedji and from Cx. quinquefasciatus in Senegal. Vector competence of Cx. neavei, Cx. quinquefasciatus, Cx. tritaeniorhunchus and Ae. vexans for WNV has been demonstrated [48–50]. Moreover, Cx. neavei is mainly ornithophagic but feed also to a lesser extent on humans, cattle and horses [51–53], and Cx. antennatus is zoophagic but feeds occasionally on humans, cattle and birds [17, 54]. These facts suggest that Cx. neavei should be considered as the principal enzootic vector and Cx. antennatus an epizootic vector of WNV in Barkedji. Contrary to our results, most of the WNV strains isolated in previous studies were from Cx. poicilipes in the Barkedji area [1, 43] and from Ma. uniformis in the Senegal River Basin [18], suggesting that the importance a given species as a vector could vary in space and time.
USUV was detected mainly from Cx. neavei in temporary ponds in concordance with previous isolation of USUV in Senegal and South Africa, and the enzootic and sylvatic pattern of its transmission in Africa [55]. USUV been isolated from an African furred rat, and a human with fever and rash as clinical symptoms. Considering its bionomics and the virus isolations, we should consider Cx. neavei as the main enzootic and bridge vector of USUV in Senegal.
BAGV was detected from Cx. neavei and Cx. antennatus in ponds in this study, suggesting that the virus circulates in an enzootic cycle between these ornithophagic mosquitoes and birds within ponds. This virus has a wide distribution and was recovered from several West African countries, South Africa, Spain, Israel and India [55–57]. It was first isolated in 1966 in the Bagaza province of the Central African Republic from Culex spp. and later from several mosquito species in Senegal and other West African countries [55, 58]. This virus has also been isolated from Cx. tritaeniorhynchus from India [57]. Vector competence of Cx. tritaeniorhynchus, Cx. quinquefasciatus and Ae. aegypti and the vertical transmission of the virus by Cx. tritaeniorhynchus have been demonstrated in India [56]. In the current study, Cx. neavei and Cx. antennatus could be considered as probable vectors of BAGV. BAGV was responsible for febrile illness in humans and high mortality to wild birds in Spain [59]. Antibodies to the virus have been also found in 15 % of human samples from India [57].
Most of BARV strains were isolated in Cx. neavei. This virus was isolated for the first time in Barkedji and then later isolated in Culex perexiguus in Israel in 2011 [59]. BARV virus has never been isolated from human or other vertebrate hosts. However, the ornithophagic tendency [17, 51, 53] of its vectors suggest that birds may play a potential role in the natural history of this virus.
YAOV was first isolated in 1968 from Cx. nebulosus in Cameroon [27]. Later, the virus was isolated from several other mosquito species in Senegal and other West African countries [60]. YAOV was isolated from Cx. tritaeniorhynchus for the first time, in our knowledge, in our study. YAOV has been isolated from one bird (Bycanistes sharpii) two rodents (Praomys sp. and Cavia porcellus) but this virus has never been detected in humans.
NDUV and SINV are the two members of the genus Alphavirus, family: Togaviridae identified in this study. NDUV was identified from mainly Ma. uniformis suggesting that it was the main vector of this virus. NDUV was first isolated in 1959 from Ma. uniformis in South Africa and later from mosquitoes and ticks in other African countries [11, 12]. NDUV was also isolated from 2 pools of Cx. pipiens females collected as larvae in Kenya, suggesting for the first time a natural vertical transmission of this virus [61]. This virus was isolated from villages in our study, suggesting the possible involvement of domestic animals and/or humans in the transmission cycle. This assertion is supported by detection of NDUV in domestic pigs in Uganda and antibodies to the virus in humans from several African countries [62–64].
SINV was originally isolated from Cx. pipiens and Cx. univittatus collected in a village named Sindbis in Egypt in 1952 [65]. Later, the virus was detected from several mosquito species in Africa, and several mosquito and tick species outside of Africa [66]. Vector competence of Cx. neavei for SINV has been demonstrated in South Africa [49]. In this study, we reported SINV mainly in Cx. neavei and a single pool of Cx. antennatus for the first time in the Barkedji Area. Infected mosquitoes have been found in ponds and villages supporting the possible involvement of birds, domestic animals and humans in the transmission cycle of this virus. Indeed, SINV seropositivity in humans has been reported in various areas of Africa, Australia, Northern Europe, and the Middle East, and antibodies to SINV have also been found from various vertebrates including birds [67] orangutans [68] and sheep [69]. The virus has been isolated from frogs [70], reed warblers [71], bats [72], and humans [67, 73]. SINV symptoms of infection in humans include fever, arthritis, rash, tenderness and persistent arthralgia [74].
Conclusion
This paper described an update of a mosquito-based arbovirus surveillance system, with more sampling methods and sites included, in the Barkedji area. Ten virus species, including 3 new for the study area, were detected, indicating a high biodiversity of viruses of medical, veterinary and unknown importance in this area. The distribution of arboviruses, detected in 14 mosquito species, was highly focal in space and time, and was highest in October and November. WNV outbreaks in the Americas have shown that arboviruses considered of little importance can emerge quickly and invade globally. Because the Barkedji area hosts a high number of arboviruses with potential to emerge, this area, and others like it, should be consistently surveilled in order to be well-prepared to deal with future invaders.
Acknowledgements
The authors would like to thank Sadio SOW and Arame BA for their technical assistance and all the population of Barkedji for their collaboration.
Funding
Not applicable.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its tables. Additional data may be available from the corresponding author upon reasonable request.
Abbreviations
- ACAV
Acado virus
- Ae
Aedes
- An
Anopheles
- AP61
Aedes pseudoscutellaris
- BAGV
Bagaza virus
- BARV
Barkedji virus
- Cx
Culex
- Er
Eretmapodites
- Ma
Mansonia
- MFIR
Minimum field Infection Rate
- Mi
Mimomyia
- NDUV
Ndumu virus
- RVFV
Rift valley fever virus
- SANV
Sanar virus
- SINV
Sindbis virus
- USUV
Usutu virus
- WHO
World Health Organization
- WNV
West Nile virus
- YAOV
Yaounde virus
Authors’ contributions
MD, DD, YB, ID, GF and OF conceived and designed the study. E N’D, YB, ID and MD carried out the field work. GF and OF performed virus tests. DD, E N’D, ID and MD analyzed the data and drafted the manuscript. All authors read, critically revised and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
El Hadji Ndiaye, Email: ndiayeelhadji135@yahoo.fr.
Diawo Diallo, Phone: +221 33 839 92 28, Email: Diawo.DIALLO@pasteur.sn.
Gamou Fall, Email: Gamou.FALL@pasteur.sn.
Yamar Ba, Email: Yamar.BA@pasteur.sn.
Ousmane Faye, Email: Ousmane.FAYE@pasteur.sn.
Ibrahima Dia, Email: Ibrahima.DIA@pasteur.sn.
Mawlouth Diallo, Email: Mawlouth.DIALLO@pasteur.sn.
References
- 1.Traore-Lamizana M, Fontenille D, Diallo M, Ba Y, Zeller HG, Mondo M, Adam F, Thonon J, Maiga A. Arbovirus surveillance from 1990 to 1995 in the Barkedji area (Ferlo) of Senegal, a possible natural focus of Rift Valley fever virus. J Med Entomol. 2001;38(4):480–492. doi: 10.1603/0022-2585-38.4.480. [DOI] [PubMed] [Google Scholar]
- 2.Wilson ML. Rift Valley fever virus ecology and the epidemiology of disease emergence. Ann N Y Acad Sci. 1994;740:169–180. doi: 10.1111/j.1749-6632.1994.tb19867.x. [DOI] [PubMed] [Google Scholar]
- 3.Campbell GL, Marfin AA, Lanciotti RS, Gubler DJ. West Nile virus. Lancet Infect Dis. 2002;2(9):519–529. doi: 10.1016/S1473-3099(02)00368-7. [DOI] [PubMed] [Google Scholar]
- 4.Gerdes GH. Rift Valley fever. Rev Sci Tech. 2004;23(2):613–623. doi: 10.20506/rst.23.2.1500. [DOI] [PubMed] [Google Scholar]
- 5.Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85(2):328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashraf U, Ye J, Ruan X, Wan S, Zhu B, Cao S. Usutu virus: an emerging flavivirus in Europe. Viruses. 2015;7(1):219–238. doi: 10.3390/v7010219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolodziejek J, Pachler K, Bin H, Mendelson E, Shulman L, Orshan L, Nowotny N. Barkedji virus, a novel mosquito-borne flavivirus identified in Culex perexiguus mosquitoes, Israel, 2011. J Gen Virol. 2013;94(11):2449–2457. doi: 10.1099/vir.0.056200-0. [DOI] [PubMed] [Google Scholar]
- 8.Gubler DJ. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res. 2002;33(4):330–342. doi: 10.1016/S0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 9.van den Hurk AF, Hall-Mendelin S, Johansen CA, Warrilow D, Ritchie SA. Evolution of mosquito-based arbovirus surveillance systems in Australia. J Biomed Biotechnol. 2012;2012:325659. doi: 10.1155/2012/325659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu W, Unnasch TR, Katholi CR, Lampman R, Novak RJ. Fundamental issues in mosquito surveillance for arboviral transmission. Trans R Soc Trop Med Hyg. 2008;102(8):817–822. doi: 10.1016/j.trstmh.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crabtree M, Sang R, Lutomiah J, Richardson J, Miller B. Arbovirus surveillance of mosquitoes collected at sites of active Rift Valley fever virus transmission: Kenya, 2006-2007. J Med Entomol. 2009;46(4):961–964. doi: 10.1603/033.046.0431. [DOI] [PubMed] [Google Scholar]
- 12.Ochieng C, Lutomiah J, Makio A, Koka H, Chepkorir E, Yalwala S, Mutisya J, Musila L, Khamadi S, Richardson J, et al. Mosquito-borne arbovirus surveillance at selected sites in diverse ecological zones of Kenya; 2007 - 2012. Virol J. 2013;10:140. doi: 10.1186/1743-422X-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ba Y, Sall AA, Diallo D, Mondo M, Girault L, Dia I, Diallo M. Re-emergence of Rift Valley fever virus in Barkedji (Senegal, West Africa) in 2002-2003: identification of new vectors and epidemiological implications. J Am Mosq Control Assoc. 2012;28(3):170–178. doi: 10.2987/12-5725.1. [DOI] [PubMed] [Google Scholar]
- 14.Fontenille D, Traore-Lamizana M, Diallo M, Thonnon J, Digoutte JP, Zeller HG. New vectors of Rift Valley fever in West Africa. Emerg Infect Dis. 1998;4(2):289–293. doi: 10.3201/eid0402.980218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diallo D, Talla C, Ba Y, Dia I, Sall AA, Diallo M. Temporal distribution and spatial pattern of abundance of the Rift Valley fever and West Nile fever vectors in Barkedji, Senegal. J Vector Ecol. 2011;36(2):426–436. doi: 10.1111/j.1948-7134.2011.00184.x. [DOI] [PubMed] [Google Scholar]
- 16.BA Y, Diallo D, Kebe CM, Dia I, Diallo M. Aspects of bioecology of two Rift Valley Fever Virus vectors in Senegal (West Africa): Aedes vexans and Culex poicilipes (Diptera: Culicidae) J Med Entomol. 2005;42(5):739–750. doi: 10.1093/jmedent/42.5.739. [DOI] [PubMed] [Google Scholar]
- 17.Ba Y, Diallo D, Dia I, Diallo M. Feeding pattern of Rift Valley Fever virus vectors in Senegal. Implications in the disease epidemiology. Bull Soc Pathol Exot. 2006;99(4):283–289. [PubMed] [Google Scholar]
- 18.Diallo M, Nabeth P, Ba K, Sall AA, Ba Y, Mondo M, Girault L, Abdalahi MO, Mathiot C. Mosquito vectors of the 1998-1999 outbreak of Rift Valley Fever and other arboviruses (Bagaza, Sanar, Wesselsbron and West Nile) in Mauritania and Senegal. Med Vet Entomol. 2005;19(2):119–126. doi: 10.1111/j.0269-283X.2005.00564.x. [DOI] [PubMed] [Google Scholar]
- 19.Sudia WD, Chamberlain RW. Battery-operated light trap, an improved model. By W. D. Sudia and R. W. Chamberlain, 1962. J Am Mosq Control Assoc. 1988;4(4):536–538. [PubMed] [Google Scholar]
- 20.Diallo D, Ba Y, Dia I, Sall AA, Diallo M. Evaluation of the efficiency of bird-baited traps for sampling potential West Nile fever mosquito vectors (Diptera: Culicidae) in Senegal. Parasite. 2010;17(2):143–147. doi: 10.1051/parasite/2010172143. [DOI] [PubMed] [Google Scholar]
- 21.Huang YM, Ward RA. A pictorial key for the identification of the mosquitoes associated with yellow fever in Africa. Mosq Syst. 1981;13:138–149. [Google Scholar]
- 22.Ferrara L, Germain M, Hervy JP. Aedes (Diceromyia) furcifer (Edwards, 1913) et Aedes (Diceromyia) taylori Edwards, 1936: le point sur la différenciation des adultes. Cah ORSTOM, Sér Ent Méd Parasitol. 1984;22(3):179–183. [Google Scholar]
- 23.Huang Y-M. Aedes (Stegomyia) bromeliae (Diptera: Culicidae), the yellow fever virus vector in East Africa. J Med Entomol. 1986;23(2):196–200. doi: 10.1093/jmedent/23.2.196. [DOI] [PubMed] [Google Scholar]
- 24.Diagne N, Fontenille D, Konate L, Faye O, Lamizana MT, Legros F, Molez JF, Trape JF. Anopheles of Senegal. An annotated and illustrated list. Bull Soc Pathol Exot. 1994;87(4):267–277. [PubMed] [Google Scholar]
- 25.Jupp PG. Mosquitoes of southern Africa: Culicinae and Toxorhynchitinae. Hartbeespoort: Ekogilde cc Publishers; 1997. [Google Scholar]
- 26.Edwards F. Mosquitoes of the Ethiopian region: III Culicine adults and pupae. London: British Museum (Natural History); 1941. [Google Scholar]
- 27.Digoutte JP, Calvo-Wilson MA, Mondo M, Traore-Lamizana M, Adam F. Continuous cell lines and immune ascitic fluid pools in arbovirus detection. Res Virol. 1992;143(6):417–422. doi: 10.1016/S0923-2516(06)80135-4. [DOI] [PubMed] [Google Scholar]
- 28.R Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 29.Turell MJ, Bailey CL. Transmission studies in mosquitoes (Diptera: Culicidae) with disseminated Rift Valley fever virus infections. J Med Entomol. 1987;24(1):11–18. doi: 10.1093/jmedent/24.1.11. [DOI] [PubMed] [Google Scholar]
- 30.Caminade C, Ndione JA, Diallo M, MacLeod DA, Faye O, Ba Y, Dia I, Morse AP. Rift Valley Fever outbreaks in Mauritania and related environmental conditions. Int J Environ Res Public Health. 2014;11(1):903–918. doi: 10.3390/ijerph110100903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll SA, Reynes JM, Khristova ML, Andriamandimby SF, Rollin PE, Nichol ST. Genetic evidence for Rift Valley fever outbreaks in Madagascar resulting from virus introductions from the East African mainland rather than enzootic maintenance. J Virol. 2011;85(13):6162–6167. doi: 10.1128/JVI.00335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de La Rocque S, Formenty P. Applying the One Health principles: a trans-sectoral coordination framework for preventing and responding to Rift Valley fever outbreaks. Rev Sci Tech. 2014;33(2):555–567. doi: 10.20506/rst.33.2.2288. [DOI] [PubMed] [Google Scholar]
- 33.Nderitu L, Lee JS, Omolo J, Omulo S, O'Guinn ML, Hightower A, Mosha F, Mohamed M, Munyua P, Nganga Z, et al. Sequential Rift Valley fever outbreaks in eastern Africa caused by multiple lineages of the virus. J Infect Dis. 2011;203(5):655–665. doi: 10.1093/infdis/jiq004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez AM, Medanic RC, Thurmond MC. Rift Valley fever outbreaks in South Africa. Vet Rec. 2010;166(25):798. doi: 10.1136/vr.c3062. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Vizcaino F, Martinez-Lopez B, Sanchez-Vizcaino JM. Identification of suitable areas for the occurrence of Rift Valley fever outbreaks in Spain using a multiple criteria decision framework. Vet Microbiol. 2013;165(1-2):71–78. doi: 10.1016/j.vetmic.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 36.Sindato C, Karimuribo ED, Pfeiffer DU, Mboera LE, Kivaria F, Dautu G, Bernard B, Paweska JT. Spatial and temporal pattern of Rift Valley fever outbreaks in Tanzania; 1930 to 2007. PloS One. 2014;9(2):e88897. doi: 10.1371/journal.pone.0088897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laughlin LW, Meegan JM, Strausbaugh LJ, Morens DM, Watten RH. Epidemic Rift Valley fever in Egypt: observations of the spectrum of human illness. Trans R Soc Trop Med Hyg. 1979;73(6):630–633. doi: 10.1016/0035-9203(79)90006-3. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez HA, Knudson DL. Genetic relatedness of corriparta serogroup viruses. J Gen Virol. 1987;68(3):661–672. doi: 10.1099/0022-1317-68-3-661. [DOI] [PubMed] [Google Scholar]
- 39.Whitehead RH, Doderty RL, Domrow R, Standfast HA, Wetters EJ. Studies of the epidemiology of arthropod-borne virus infections at Mitchell River Mission, Cape York Peninsula, North Queensland. 3. Virus studies of wild birds, 1964-1967. Trans R Soc Trop Med Hyg. 1968;62(3):439–445. doi: 10.1016/0035-9203(68)90096-5. [DOI] [PubMed] [Google Scholar]
- 40.Doherty RL, Carley JG, Mackerras MJ, Marks EN. Studies of arthropod-borne virus infections in Queensland. III. Isolation and characterization of virus strains from wild caught mosquitoes in North Queensland. Aust J Exp Biol Med Sci. 1963;41:17–39. doi: 10.1038/icb.1963.2. [DOI] [PubMed] [Google Scholar]
- 41.Doherty RL, Whitehead RH, Wetters EJ, Gorman BM, Carley JG. A survey of antibody to 10 arboviruses (Koongol group, Mapputta group and ungrouped) isolated in Queensland. Trans R Soc Trop Med Hyg. 1970;64(5):748–753. doi: 10.1016/0035-9203(70)90017-9. [DOI] [PubMed] [Google Scholar]
- 42.Boughton CR, Hawkes RA, Naim HM. Arbovirus infection in humans in NSW: seroprevalence and pathogenicity of certain Australian bunyaviruses. Aust N Z J Med. 1990;20(1):51–55. doi: 10.1111/j.1445-5994.1990.tb00371.x. [DOI] [PubMed] [Google Scholar]
- 43.Traore-Lamizana M, Zeller HG, Mondo M, Hervy JP, Adam F, Digoutte JP. Isolations of West Nile and Bagaza viruses from mosquitoes (Diptera: Culicidae) in central Senegal (Ferlo) J Med Entomol. 1994;31(6):934–938. doi: 10.1093/jmedent/31.6.934. [DOI] [PubMed] [Google Scholar]
- 44.Chevalier V, Lancelot R, Diaite A, Mondet B, Sall B, De Lamballerie X. Serological assessment of West Nile fever virus activity in the pastoral system of Ferlo, Senegal. Ann N Y Acad Sci. 2006;1081:216–225. doi: 10.1196/annals.1373.026. [DOI] [PubMed] [Google Scholar]
- 45.Chevalier V, Reynaud P, Lefrancois T, Durand B, Baillon F, Balanca G, Gaidet N, Mondet B, Lancelot R. Predicting West Nile virus seroprevalence in wild birds in Senegal. Vector Borne Zoonotic Dis. 2009;9(6):589–596. doi: 10.1089/vbz.2008.0130. [DOI] [PubMed] [Google Scholar]
- 46.Burt FJ, Grobbelaar AA, Leman PA, Anthony FS, Gibson GV, Swanepoel R. Phylogenetic relationships of southern African West Nile virus isolates. Emerg Infect Dis. 2002;8(8):820–826. doi: 10.3201/eid0808.020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kulasekera VL, Kramer L, Nasci RS, Mostashari F, Cherry B, Trock SC, Glaser C, Miller JR. West Nile virus infection in mosquitoes, birds, horses, and humans, Staten Island, New York, 2000. Emerg Infect Dis. 2001;7(4):722–725. doi: 10.3201/eid0704.017421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fall G, Diallo M, Loucoubar C, Faye O. Vector competence of Culex neavei and Culex quinquefasciatus (Diptera: Culicidae) from Senegal for lineages 1, 2, Koutango and a putative new lineage of West Nile virus. Am J Trop Med Hyg. 2014;90(4):747–754. doi: 10.4269/ajtmh.13-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jupp PG, McIntosh BM, Blackburn NK. Experimental assessment of the vector competence of Culex (Culex) neavei Theobald with West Nile and Sindbis viruses in South Africa. Trans R Soc Trop Med Hyg. 1986;80(2):226–230. doi: 10.1016/0035-9203(86)90019-2. [DOI] [PubMed] [Google Scholar]
- 50.Goddard LB, Roth AE, Reisen WK, Scott TW. Vector competence of California mosquitoes for West Nile virus. Emerg Infect Dis. 2002;8(12):1385–1391. doi: 10.3201/eid0812.020536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McIntosh BM. Rift Valley fever. 1. Vector studies in the field. J S Afr Vet Assoc. 1972;43(4):391–395. [PubMed] [Google Scholar]
- 52.Snow WF. The attractiveness of some birds and mammals for mosquitoes in The Gambia, West Africa. Ann Trop Med Parasitol. 1983;77(6):641–651. doi: 10.1080/00034983.1983.11811763. [DOI] [PubMed] [Google Scholar]
- 53.Fall AG, Diaite A, Seck MT, Bouyer J, Lefrancois T, Vachiery N, Aprelon R, Faye O, Konate L, Lancelot R. West Nile virus transmission in sentinel chickens and potential mosquito vectors, Senegal River Delta, 2008-2009. Int J Environ Res Public Health. 2013;10(10):4718–4727. doi: 10.3390/ijerph10104718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gad AM, Riad IB, Farid HA. Host-feeding patterns of Culex pipiens and Cx. antennatus (Diptera: Culicidae) from a village in Sharqiya Governorate, Egypt. J Med Entomol. 1995;32(5):573–577. doi: 10.1093/jmedent/32.5.573. [DOI] [PubMed] [Google Scholar]
- 55.Nikolay B, Diallo M, Boye CS, Sall AA. Usutu virus in Africa. Vector Borne Zoonotic Dis. 2011;11(11):1417–1423. doi: 10.1089/vbz.2011.0631. [DOI] [PubMed] [Google Scholar]
- 56.Sudeep AB, Bondre VP, Mavale MS, Ghodke YS, George RP, Aher RV, Gokhale MD. Preliminary findings on Bagaza virus (Flavivirus: Flaviviridae) growth kinetics, transmission potential & transovarial transmission in three species of mosquitoes. Indian J Med Res. 2013;138:257–261. [PMC free article] [PubMed] [Google Scholar]
- 57.Bondre VP, Sapkal GN, Yergolkar PN, Fulmali PV, Sankararaman V, Ayachit VM, Mishra AC, Gore MM. Genetic characterization of Bagaza virus (BAGV) isolated in India and evidence of anti-BAGV antibodies in sera collected from encephalitis patients. J Gen Virol. 2009;90(11):2644–2649. doi: 10.1099/vir.0.012336-0. [DOI] [PubMed] [Google Scholar]
- 58.Gordon SW, Tammariello RF, Linthicum KJ, Dohm DJ, Digoutte JP, Calvo-Wilson MA. Arbovirus isolations from mosquitoes collected during 1988 in the Senegal River basin. Am J Trop Med Hyg. 1992;47(6):742–748. doi: 10.4269/ajtmh.1992.47.742. [DOI] [PubMed] [Google Scholar]
- 59.Aguero M, Fernandez-Pinero J, Buitrago D, Sanchez A, Elizalde M, San Miguel E, Villalba R, Llorente F, Jimenez-Clavero MA. Bagaza virus in partridges and pheasants, Spain, 2010. Emerg Infect Dis. 2011;17(8):1498–1501. doi: 10.3201/eid1708.110077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.CRORA . Rapport Annuel. Centre Collaborateur OMS de Reference et de Recherche pour les Arbovirus et Virus de Fievres Hemorragiques. Senegal: Institut Pasteur de Dakar; 1998. [Google Scholar]
- 61.Lutomiah J, Ongus J, Linthicum KJ, Sang R. Natural vertical transmission of ndumu virus in Culex pipiens (Diptera: Culicidae) mosquitoes collected as larvae. J Med Entomol. 2014;51(5):1091–1095. doi: 10.1603/ME14064. [DOI] [PubMed] [Google Scholar]
- 62.Masembe C, Michuki G, Onyango M, Rumberia C, Norling M, Bishop RP, Djikeng A, Kemp SJ, Orth A, Skilton RA, et al. Viral metagenomics demonstrates that domestic pigs are a potential reservoir for Ndumu virus. Virol J. 2012;9:218. doi: 10.1186/1743-422X-9-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kokernot RH, BM MI, Worth CB. Ndumu virus, a hitherto unknown agent, isolated from culicine mosquitoes collected in northern Natal. Union of South Africa. Am J Trop Med Hyg. 1961;10:383–386. doi: 10.4269/ajtmh.1961.10.383. [DOI] [PubMed] [Google Scholar]
- 64.Karabatsos N. International catalogue of arboviruses including certain other viruses of vertebrates, 3rd ed. San Antonio: American Society of Tropical Medicine and Hygiene; 1985.
- 65.Taylor RM, Hurlbut HS, Work TH, Kingston JR, Frothingham TE. Sindbis virus: a newly recognized arthropod transmitted virus. Am J Trop Med Hyg. 1955;4(5):844–862. doi: 10.4269/ajtmh.1955.4.844. [DOI] [PubMed] [Google Scholar]
- 66.Monath TP. The Arboviruses: Epidemiology and Ecology. Boca Raton: CRC Press; 1988. [Google Scholar]
- 67.Kurkela S, Ratti O, Huhtamo E, Uzcategui NY, Nuorti JP, Laakkonen J, Manni T, Helle P, Vaheri A, Vapalahti O. Sindbis virus infection in resident birds, migratory birds, and humans. Finland. Emerg Infect Dis. 2008;14(1):41–47. doi: 10.3201/eid1401.070510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolfe ND, Kilbourn AM, Karesh WB, Rahman HA, Bosi EJ, Cropp BC, Andau M, Spielman A, Gubler DJ. Sylvatic transmission of arboviruses among Bornean orangutans. Am J Trop Med Hyg. 2001;64(5-6):310–316. doi: 10.4269/ajtmh.2001.64.310. [DOI] [PubMed] [Google Scholar]
- 69.Juricova Z, Mitterpak J, Prokopic J, Hubalek Z. Circulation of mosquito-borne viruses in large-scale sheep farms in eastern Slovakia. Folia parasitol. 1986;33(3):285–288. [PubMed] [Google Scholar]
- 70.Kozuch O, Labuda M, Nosek J. Isolation of sindbis virus from the frog Rana ridibunda. Acta virol. 1978;22(1):78. [PubMed] [Google Scholar]
- 71.Ernek E, Kozuch O, Gresikova M, Nosek J, Sekeyova M. Isolation of Sindbis virus from the reed warbler (Acrocephalus scirpaceus) in Slovakia. Acta virol. 1973;17(4):359–361. [PubMed] [Google Scholar]
- 72.Blackburn NK, Foggin CM, Searle L, Smith PN. Isolation of Sindbis virus from bat organs. Cent Afr J Med. 1982;28(8):201. [PubMed] [Google Scholar]
- 73.Malherbe H, Strickland-Cholmley M, Jackson AL. Sindbis virus infection in man. Report of a case with recovery of virus from skin lesions. S Afr Med J. 1963;37:547–552. [PubMed] [Google Scholar]
- 74.Kurkela S, Helve T, Vaheri A, Vapalahti O. Arthritis and arthralgia three years after Sindbis virus infection: clinical follow-up of a cohort of 49 patients. Scand J Infect Dis. 2008;40(2):167–173. doi: 10.1080/00365540701586996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its tables. Additional data may be available from the corresponding author upon reasonable request.