Abstract
Adoptive immunotherapy using TCR-engineered PBLs against melanocyte differentiation Ags mediates objective tumor regression but is associated with on-target toxicity. To avoid toxicity to normal tissues, we targeted cancer testis Ag (CTA) MAGE-A3, which is widely expressed in a range of epithelial malignancies but is not expressed in most normal tissues. To generate high-avidity TCRs against MAGE-A3, we employed a transgenic mouse model that expresses the human HLAA*0201 molecule. Mice were immunized with two HLA-A*0201–restricted peptides of MAGE-A3: 112–120 (KVAELVHFL) or MAGE-A3: 271–279 (FLWGPRALV), and T cell clones were generated. MAGE-A3–specific TCR α- and β-chains were isolated and cloned into a retroviral vector. Expression of both TCRs in human PBLs demonstrated Ag-specific reactivity against a range of melanoma and nonmelanoma tumor cells. The TCR against MAGE-A3: 112–120 was selected for further development based on superior reactivity against tumor target cells. Interestingly, peptide epitopes from MAGE-A3 and MAGE-A12 (and to a lesser extent, peptides from MAGE-A2 and MAGE-A6) were recognized by PBLs engineered to express this TCR. To further improve TCR function, single amino acid variants of the CDR3 α-chain were generated. Substitution of alanine to threonine at position 118 of the α-chain in the CDR3 region of the TCR improved its functional avidity in CD4 and CD8 cells. On the basis of these results, a clinical trial is planned in which patients bearing a variety of tumor histologies will receive autologous PBLs that have been transduced with this optimized anti–MAGE-A3 TCR.
In the past two decades, fundamental advances in immunology and tumor biology combined with the identification of a large number of tumor Ags have led to significant progress in the field of cell-based immunotherapy (1–4). Adoptive cellular immunotherapy involving transfer of tumor-reactive T cells has shown some notable antitumor responses in patients with meta-static melanoma. The administration of naturally occurring tumor infiltrating lymphocytes (TILs) expanded ex vivo mediated an objective response rate ranging from 50–70% in melanoma patients, including bulky invasive tumors at multiple sites involving liver, lung, soft tissue, and brain (3, 5). A major limitation to the widespread application of TIL therapy is the difficulty in generating human T cells with antitumor potential. It has been reported that approximately only half of melanomas reproducibly give rise to antitumor TILs (6). As an alternative approach, high-affinity TCRs can be introduced into normal T cells of the patients, and the adoptive transfer of these cells into lymphodepleted patients has been shown to mediate cancer regression (7, 8).
Adoptive transfer of TCR-transduced PBLs targeting melanoma differentiation Ags such as MART-1 and gp100 resulted in objective cancer regression in up to 30% of patients (7). However, patients also exhibited significant toxicity associated with destruction of normal melanocytes in the skin, eye, and ear (7). This trial revealed that T cells expressing highly reactive TCRs mediate cancer regression and also target cognate Ag-expressing cells throughout the body. Efforts to enhance adoptive immunotherapy response rates may hinge on targeting tumor Ags with little or no expression in normal tissue. In an effort to overcome the on-target toxicities associated with immunotherapies directed against Ags expressed on normal tissues, we and others have focused on generating TCRs targeting cancer testis Ags (CTAs). CTAs are immunogenic proteins, which are normally expressed in non-MHC-expressing germ cells of testis yet are aberrantly expressed in many tumors; thus, CTAs may represent ideal targets for tumor immunotherapy. More than 110 CTA genes or gene families have been identified that are expressed in multiple tumor types (9–11). These immunogenic proteins are being vigorously pursued as targets for therapeutic cancer vaccines and TCR-based adoptive immunotherapy (12–14). In theory, targeting T cells against tumor-associated CTAs might selectively eliminate tumor cells while avoiding toxicity to normal tissue.
Since the identification of the first human MAGE CTA gene in 1991, the number MAGE family genes have grown to >25 members (15, 16). MAGE-A is a multigene family consisting of 12 homologous genes MAGE-A1 to MAGE-A12 located at chromosome Xq28 (11). The precise function and biological role of MAGE proteins have not been completely elucidated. However, members of MAGE-A, MAGE-B, and MAGE-C proteins have been implicated in the suppression of p53-dependent apoptosis (17, 18), and MAGE-A3 has been attributed to mediate fibronectin-controlled progression and metastasis (19). Genomic clustering, restricted expression pattern, and single exon open reading frame of the MAGE genes are consistent with the possibility that these genes evolved from retrotransposition and subsequent duplication (20). Expression of CTAs including MAGE genes in tumor cells has been attributed to global DNA demethylation and other mechanisms that normally silence these genes in somatic cells (21). MAGE-A3 is one of the more frequently expressed CTAs in human tumors, including melanoma (22), non-small cell lung carcinoma (NSCLC) (23), head and neck squamous cell carcinoma (24), hepatocellular carcinoma, (25) and multiple myeloma (26). The expression of MAGE-A3 has been shown to be higher in more advanced stages of the disease and is associated with poor disease prognosis (27–29). Several antigenic peptides that bind to HLA class I or class II molecules on tumor cells have been reported (30–36).
Because of its high expression in a wide array of tumor types, MAGE-A3 is an attractive target for cancer immunotherapy. In an effort to generate TCRs against MAGE-A3, we immunized transgenic mice expressing the HLA-A*0201 molecule with two MAGE-A3 peptides. In this report, we describe a TCR with a high avidity for similar peptides derived from MAGE-A3 and MAGEA12 that also cross-reacts with related peptides from MAGE-A2 and MAGE-A6. Normal PBLs engineered with this TCR demonstrated potent cytolytic activity and secreted high levels of IFN-γ in response to HLA-A*0201+/MAGE+ tumor cells from multiple histologies. These findings suggest that the use of this TCR for adoptive immunotherapy may be valuable for the treatment of patients with cancers that express these gene products.
Materials and Methods
Cell lines and human PBLs
HLA-A*0201+/MAGE-A3+ melanoma cell lines 526, 624.38, 1300, and 2984 and non-HLA-A*0201 cell lines 397, 888, 938, 1359, and 1088 were established from surgically resected metastatic melanoma tumors and maintained at the Surgery Branch, National Cancer Institute, National Institutes of Health (Bethesda, MD). The HLA-A*0201+/MAGEA3− 2361R cell line was isolated from a surgically resected metastatic renal cell carcinoma. NSCLC line H1299 (HLA-A*0201−/MAGE-A3+); small cell lung carcinoma lines H2721 (HLA-A*0201+/MAGE-A3−), H2122 (HLA-A*0201−/MAGE-A3+), and H1250 (HLA-A*0201+/MAGEA3+); and esophageal cancer cell line BE-3 (HLA-A*0201+/MAGE-A3+) were obtained from the laboratory of Dr. David Schrump (Surgery Branch, National Cancer Institute, National Institutes of Health). Breast cancer cell lines MDA-MB-453S (HLA-A*0201−/MAGE-A3+) were from American Type Culture Collection (Manassas, VA). Glioma cell line U251 (HLA-A*0201+/MAGE-A3+) was obtained from the Division of Cancer Treatment and Diagnosis Tumor Repository, National Cancer Institute, National Institutes of Health (Frederick, MD). The COS7-A*0201, 293-A*0201, 397-A*0201, 1359-A*0201, MDA-MB-453S-A*0201, and H1299-A*0201 cells were retrovirally engineered to express HLA-A*0201 as described previously (14, 37). COS7-A*0201-MAGE-A3, COS7-A*0201-MAGE-A12, 293-A*0201-MAGE-A3, and 293-A*0201-MAGEA12 cells were transduced with a retroviral vector expressing the respective MAGE genes. T2 is a lymphoblastoid cell line lacking TAP function, whose HLA class I proteins can be easily loaded with exogenous peptides.
All of the PBLs used in this study were obtained from melanoma patients treated in the Surgery Branch, National Cancer Institute, National Institutes of Health, on Institutional Review Board-approved protocols. Human lymphocytes were cultured in AIM-V medium (Invitrogen, Carlsbad, CA) supplemented with 5% human serum from donors of blood group AB (Valley Biomedical, Winchester, VA), 50 U/ml penicillin, 50 μg/ml streptomycin (Invitrogen), and 300 IU/ml IL-2 and maintained at 37°C with 5% CO2.Murine lymphocytes were cultured in RPMI 1640 supplemented with 10% FBS, 2 mmol/l L-glutamine, 1 mmol/l sodium pyruvate, MEM nonessential amino acids, 55 μmol/l 2-ME, 50 U/ml penicillin, 50 μg/ml streptomycin, and 30 IU/ml recombinant human IL-2 (rhIL-2) (R10) (Invitrogen).
Quantitative real-time PCR for MAGE-A3/6 and MAGE-A12 expression
Total RNA from tumor cell lines was isolated using an RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. A total of 1 μg of RNA was then used for cDNA synthesis using oligo(dT) with the SuperScript First-Strand Synthesis kit (Invitrogen). The cDNA was used as the templates for subsequent real-time PCR with primers designed specifically for MAGE-A3/6 (38), MAGE-A12, and β-actin (TaqMan Gene Expression Assays; Applied Biosystems, Foster City, CA). Each experiment was performed in duplicate using a TaqMan 7900 (Applied Bio-systems) real-time PCR machine according to the manufacturer’s instructions. Absolute numbers of copies were estimated using standard curves with plasmid DNA expressing respective genes.
Synthetic peptides
The following peptides were used in this study: MAGE-A1: 105–113 (KVADLVGFL), MAGE-A2: 112–120 (KMVELVHFL), MAGE-A3: 112–120 (KVAELVHFL), MAGE-A3: 271–279 (FLWGPRALV), MAGE-A4: 113–121 (KVDELAHFL), MAGE-A6: 112–120 (KVAKLVHFL), MAGEA8: 115–123 (KVAELVRFL), MAGE-A12: 112–120 (KMAELVHFL), and MAGE-C2: 144–152 (KVAELVEFL). The peptides were synthesized using a solid-phase method based on Fmoc chemistry with one of two multiple peptide synthesizers (model AMS 422; Gilson, or Pioneer; Applied Biosystems). Purity of the peptides was verified by mass spectrometry. Peptides were dissolved in DMSO and diluted in RPMI 1640 medium.
Immunization of HLA-A*0201 transgenic mice and isolation of TCRs
Transgenic mice expressing the full-length human HLA-A*0201 gene were obtained from The Jackson Laboratory. Mouse studies were conducted according to the protocols approved by the National Cancer Institute Animal Care and Use Committee as described previously (37). Briefly, 8-to 12-wk-old mice were immunized s.c. at the base of the tail with 100 μg of MAGE-A3: 112–120 (KVAELVHFL) or MAGE-A3: 271–279 (FLWGPRALV) plus 120 μg of hepatitis B virus core: 128–140 helper peptide (TPPAYRPPNAPIL) emulsified in 100 μl of IFA (Montanide ISA-51). A booster immunization was given 1 wk later. One week after the booster immunization, mice were euthanized and splenocytes were harvested and stimulated in vitro with irradiated T2 cells (18,000 rad) loaded with 0.01, 0.1, or 1 μg/ml of the immunizing peptide in R10 medium containing 30 IU/ml rhIL-2. Cultures were set up in 24-well plates with 1–3 million splenocytes and 0.2–0.4 million peptide-loaded T2 cells. One week after stimulation, bulk murine T cell cultures were tested in coculture assays for peptide-specific reactivity using T2 cells and tumor cell recognition using 1300 melanoma and H1299-A*0201 cells. Ag-specific IFN-γ secretion was measured by ELISA (Thermo Scientific, Rockford,IL). Peptide-reactive bulk cultures were cloned at 10 cells per well in U-bottom 96-well plates, with 5 × 104 peptide-pulsed irradiated T2 cells and 5 × 104 irradiated (3000 rad) C57BL/6 feeder splenocytes in medium containing 30 IU/ml recombinant human IL-2. Two to three weeks later, growth-positive wells were identified, and the T lymphocytes from those wells were transferred into 48-well plates and stimulated with 2 × 105 peptide-pulsed irradiated T2 cells and 1 × 106 irradiated C57BL/6 splenocytes in medium containing 30 IU/ml rhIL-2. One to 2 wk later, these T cell cultures were evaluated for specific recognition of peptide and tumor cells by means of specific IFN-γ secretion. T cells from each of the tumor-reactive clones were expanded by restimulation as described previously, and the total RNA was extracted for TCR isolation.
Cloning of MAGE-A3–specific, HLA-A*0201–restricted TCRs
Total RNA was extracted from tumor-reactive T cell clones using an RNeasy mini kit (Qiagen). TCR α- and β-chains from each tumor-reactive T cell clone were cloned using a SMART RACE cDNA amplification kit (Clontec, Mountain View, CA). For the amplification of TCRs, gene-specific primers were made from the C region of mouse TCR α- and β-chains, TCR α C region-5′-ACTGGTACACAGCAGGTTCTGG-3′ and TCR β C region-5′-AAGGAGACCTTGGGTGGAGTC-3′. The PCR products of the 5′-RACE were cloned into the PCR2.1 TOPO vector (Invitrogen Life Technologies), and the insert DNA fragments were sequenced. The DNA sequence data were analyzed using The International Immunogenetics Information System (http://imgt.cines.fr/IMGT_vquest/vquest?livret=0&Option=mouseTcR) for the identification of mouse TCR α- and β-chains. After the identification of the variable regions of α- and β-chains and the identification of the C region of the β-chain (CB1 or CB2), specific primers were used to amplify the full-length TCR α- and β-chains from the cDNA.
Electroporation of TCRs into PBLs
The full-length α- and β-chains were individually cloned into the RNA expression vector pGEM-4Z/64A. In vitro-transcribed mRNA encoding aand β-chains were generated using mMESSAGE mMACHINE (Ambion, Austin, TX) and electroporated into OKT3-stimulated human lymphocytes with an ElectroSquarePorator ECM-830 (BTX, San Diego, CA) as described previously (39). Electroporated PBLs were tested for Ag-specific reactivity using peptide-loaded T2 cells and tumor cell lines 2361-RCC, 938 melanoma, H1299-A*0201, and 1300 melanoma. The Ag-specific re sponse of TCR-electroporated T cells was evaluated by coculture with respective MAGE-A3 peptide-loaded T2 cells and tumor cell lines, and the culture supernatants were tested for IFN-γ levels by ELISA.
Construction of retroviral vectors expressing MAGE-A3–specific HLA-A*0201–restricted TCRs
Two MSGV1-based retroviral vectors were constructed using the overlapping PCR method with the transgene construct arranged in the following order of configuration: TCR α-chain, linker peptide furin-SGSGP2A (13), and TCR β-chain. The cloned TCR inserts were verified by restriction enzyme digestion and DNA sequencing.
Transduction of PBLs
Retroviral supernatants were generated by transfecting respective MSGV1-MAGE-TCR vector DNA from each of the constructs with a plasmid encoding RD114 envelope into 293-GP cells using the Lipofectamine 2000 reagent (Invitrogen) in Opti-MEM medium (Invitrogen) (13). Retroviral vector expressing TCR against NY ESO-1 was used as a positive control in all of the experiments (12). MSGV1 vector expressing GFP was also generated. Viral supernatants were then loaded onto RetroNectin-coated (Takara Bio, Japan) non-tissue culture-treated six-well plates. PBLs were stimulated with OKT3 (50 ng/ml) and rhIL-2 (300 IU/ml) 48 h prior to transduction, and the transduction was carried out as described previously (13, 40).
Tetramer staining
HLA-A*0201–restricted MAGE-A3–derived peptides MAGE-A3: 112–120 (KVAELVHFL) and MAGE-A3: 271–279 (FLWGPRALV) were used by the National Institutes of Health Tetramer Core Facility at Emory University to produce tetramers linking PE as the fluorophore. MAGE-A3 TCR-transduced T cells were stained with a FITC-labeled anti-CD8 (BD Pharmingen, San Jose, CA) and with PE-labeled HLA-A*0201 tetramers. FITC-conjugated mAb against the C region of the murine β-chain (eBioscience, San Diego, CA) and PE-conjugated anti-CD8 Abs were also used to detect the expression of MAGE-A3 TCRs in the human PBLs. Cells were analyzed using a FACScan flow cytometer with CellQuest software (BD Biosciences) or FlowJo software (Tree Star, Ashland, OR).
Intracellular cytokine staining
Intracellular cytokine staining was performed using a BD Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions. Briefly, cells were first stained with cell surface markers CD3 and CD8 and then stained with FITC-conjugated anti–IFN-γ and allophycocyanin-conjugated anti–IL-2 Abs for intracellular detection of the cytokines. All of the Abs as well as isotype controls were purchased from BD Biosciences. Cells were analyzed using a FACSCanto II flow cytometer with CellQuest software (BD Biosciences) or FlowJo software (Tree Star).
CD107a mobilization assay
The cell surface mobilization of the CD107a molecule was determined as a measure of degranulation and functional reactivity after Ag recognition by the TCR. In these assays, 1 × 105 H1299-A2 or H1299 cells were cocultured with an equal number of PBLs at 37°C for 2 h. The cells were then stained with mouse anti-human Abs against CD107a and CD8 (BD Biosciences) and analyzed by FACS.
Cytokine release assay
TCR-engineered PBLs were tested for Ag-specific reactivity in cytokine release assays using peptide-loaded T2 cells and tumor cells. In these assays, effector cells (1 × 105) were cocultured with an equal number of target cells in AIM-V medium in a final volume of 0.2 ml in duplicate wells of a 96-well U-bottom microplate. Culture supernatants were harvested 18–24 h after the initiation of coculture and assayed for IFN-γ and GM-CSF by ELISA (Thermo Scientific).
[51Cr] release assay
The ability of the transduced PBLs to lyse HLA-A*0201+/MAGE-A3+ tumor cells was measured using a [51Cr] release assay as described previously (12,41). In these assays, TCR-engineered PBLs were coincubated with decreasing ratios of 51Cr-labeled target cells (E:T ratio) in AIM-V medium in 96-well U-bottom plates at 37°C for 4 h. Lysis was measured by [51Cr] release in the medium: percent lysis = (sample release − minimum release)/(maximum release − minimum release) × 100%, average of duplicate samples.
CD4/CD8 separation
MAGE TCR-engineered CD4+ and CD8+ populations were separated using magnetic bead-based BD IMag human CD4 or CD8 T lymphocyte enrichment set DM kit for negative selection of those subsets (BD Biosciences).
Lymphocyte proliferation assay
TCR-transduced PBLs were tested for Ag-specific proliferation using the [3H]thymidine incorporation assay. Briefly, effector cells (1 × 105) were cocultured with equal number of irradiated (18,000 rad, cesium source) H1299 or H1299-A2 target cells in AIM-V medium in a final volume of 0.2 ml in triplicate wells of a 96-well U-bottom microplate. The cells were cocultured for 3 d and pulsed with 1 μCi [3H]thymidine (DuPont, New England Nuclear, Shelton, CT) per well and cultured for an additional 18 h. The cells were then harvested onto a glass fiber filter (Wallac Oy, Turku, Finland), and radionucleotide incorporation was measured using a Perkin-Elmer Microbeta Trilux counter (Shelton, CT). Results expressed as cpm.
Generation of single amino acid variants of the CDR3 α-chain MAGE-A3: 112–120 TCR
We generated 85 single amino acid variant TCRs in four stages. 1) Site-directed mutagenesis using a QuikChange Lightning Site-Directed Muta-genesis kit (Stratagene, La Jolla, CA) was used to substitute all of the other 19 aa at position D115 introducing appropriate nucleotide changes in the PCR primer. 2) Alanine substitutions at eight of the residues F114, D115, T116, N117, Y119, K120, V121, and I122 were introduced except at position A118 that already had an alanine residue. 3) Conservative amino acid substitutions were introduced at positions N117 to Q/K/R, A118 to V/L/I, and Y119 to R/K/Q/N by site-directed mutagenesis. 4) Single amino acid substitutions at these three residues were synthesized (GENEART, Regensburg, Germany) to produce a 16 aa substitution library at each position. Retroviral vector supernatants expressing all of the above-mentioned single amino acid variant TCRs were generated by transient transfection into 293-GP cells; PBLs from donors were transduced and tested for tetramer binding and IFN-γ production in coculture assays with tumor cell lines.
Results
Generation of MAGE-A3–reactive murine T cell clones from HLA-A*0201 transgenic mice
Transgenic mice expressing the full-length HLA-A*0201 molecule were immunized with one of the two previously identified naturally processed and presented HLA-A*0201–restricted peptides from MAGE-A3 [MAGE-A3: 112–120 (KVAELVHFL) (35) or MAGE-A3: 271–279 (FLWGPRALV) (42)] along with a helper peptide, hepatitis B virus core: 128–140. After two immunizations, murine T cells were harvested from spleen and lymph nodes and stimulated in vitro with the respective peptide and IL-2. Bulk T cell cultures from mice immunized with the MAGE-A3 peptides demonstrated specific reactivity against T2 cells pulsed with the relevant peptide and the HLA-A*0201+/MAGE-A3+ tumor cell lines H1299-A*0201, 1300 melanoma, and 624 melanoma after two in vitro stimulations (data not shown). Reactive T cells from positive wells were cloned by limiting dilution and tested for Ag-specific reactivity. Five clones derived from the mice immunized with the MAGE-A3: 112–120 peptide and six clones derived from the mice immunized with the MAGE-A3: 271–279 peptide that secreted high levels of IFN-γ in response to tumor cells and peptide-loaded T2 cells were expanded and further characterized.
Cloning of MAGE-reactive TCRs
TCR α- and β-chains from each tumor-reactive T cell clone were cloned using a SMART RACE cDNA amplification kit with gene-specific primers in the C region of mouse TCR α- and β-chains. After the identification of the variable regions of the α- and β-chains and the specific C region of the β-chain, specific primers were used to amplify the full-length TCR α- and β-chains from the cDNA. The TCR α- and β-chains were then cloned into the RNA expression vector pGEM. In vitro-transcribed RNA of TCR α- and β-chains were electroporated into human PBLs and tested for Ag-specific reactivity as described previously (39) using peptide-loaded T2 cells, H1299-A*0201, and 1300 melanoma tumor cell lines. On the basis of the specific reactivity, we selected a TCR against MAGE-A3: 112–120 peptide (TCR α-TRAV12D-3, TCR β-TRBV29*01, and CB1) and a TCR against MAGE-A3: 271–279 peptide (TRAV17*02, TRBV15*01, and CB2) for further evaluation.
Construction of a MAGE-A3 TCR-expressing retroviral vector and transduction of PBLs
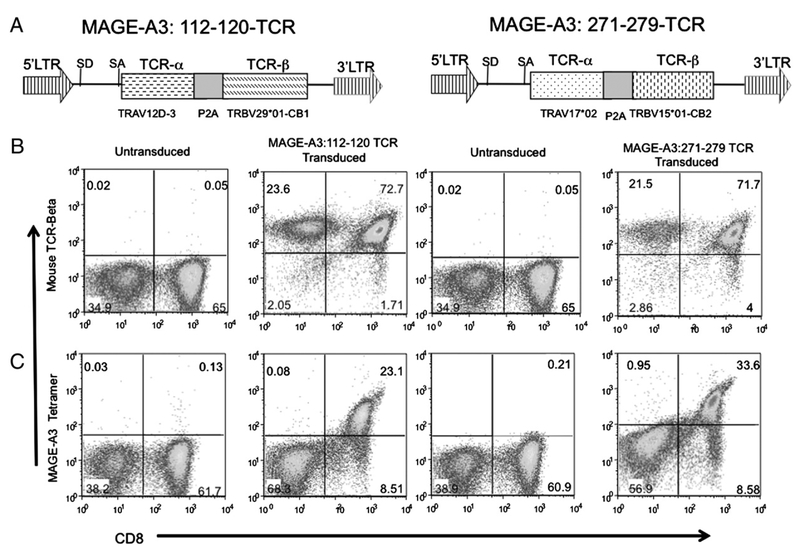
Two MSGV1-based retroviral vectors with expression cassettes consisting of TCR α-TRAV12D-3 and TCR β-TRBV29*01-CB1 and TRAV17*02 and TRBV15*01-CB2 were constructed (Fig. 1A). The TCR expression in these vectors is driven by the viral long terminal repeat, and α- and β-chains are expressed as a single open reading frame using the 2A linker peptide (13, 43). Human PBLs were stimulated for 2 d and then transduced. FACS analysis of transduced PBLs using the anti-mouse TCR β-chain revealed that both CD8+ and CD4+ cells had been transduced with these TCR vectors (Fig. 1B); however, specific tetramer binding was observed only in transduced CD8+ and not CD4+ T cells (Fig. 1C).
FIGURE 1.
A, Schematic illustration of the MSGV1-based retroviral vector encoding anti–MAGE-A3 TCR expression cassette. TCR α- and β-chains are linked with furin-spacer (SGSG)-P2A ribosomal skip peptide sequence. B and C, Flow cytometric analysis of MAGE-A3 TCR-transduced PBLs. B, Dot plots showing the FACS profile of PBLs stained with anti–human-CD8-PE and anti–mouse-TCR β-FITC Abs. C, Dot plots showing the FACS profile of PBLs stained with anti–human-CD8-FITC Ab and PE-conjugated MAGE-A3: 112–120 or MAGE-A3: 271–279 HLA-A*0201 tetramers.
Evaluation of the function of MAGE TCR-engineered PBLs
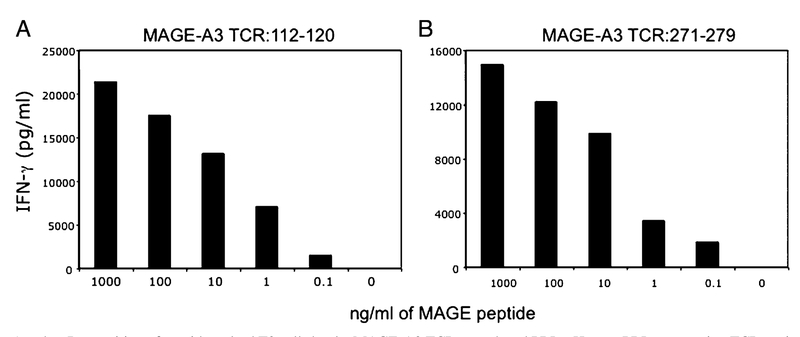
To evaluate the recognition of the respective MAGE-A3 TCRs, transduced PBLs were subjected to coculture assay with peptide-pulsed T2 cells. TCR-transduced PBLs specifically secreted IFN-γ upon encounter with the antigenic peptide in a dose-dependent manner (Fig. 2). PBLs transduced with either MAGE-A3: 112–120 or MAGE-A3: 271–279 TCRs recognized T2 cells pulsed with as little as 0.1 ng/ml MAGE-A3 peptides, indicating that both of the TCRs were relatively high-avidity receptors. Coculture of PBLs expressing TCRs against MAGE-A3: 112–120 or MAGEA3: 271–279 with control T2 cells that were not pulsed with any peptides produced background levels of IFN-γ. To assess the specific recognition of tumor cells, TCR-engineered PBLs were co-cultured with a panel of HLA-A*0201+ and HLA-A*0201− melanoma- and lung tumor-derived cell lines. Specific release of IFN-γ was observed when the TCR-engineered PBLs were co-cultured with HLA-A*0201+/MAGE-A3+ cell lines but not HLAA*0201−/MAGE-A3+ or HLA-A*0201+/MAGE-A3− cell lines (Table I). A comparison of the two TCRs revealed that T cells transduced with the MAGE-A3: 112–120 TCR released ~10-fold higher levels of IFN-γ in response to HLA-A*0201+/MAGE-A3+ tumor cell targets (Table I). These responses were specific because low levels of IFN-γ were released in response to MAGE+/HLAA*0201− cell lines and MAGE−/HLA-A*0201+ cell lines. Although MAGE-A3: 271–279 TCR-transduced PBLs efficiently recognized the peptide loaded on T2 cells, the recognition of MAGE-A3+/HLA-A*0201+ tumor cells as measured by the release of IFN-γ production was relatively weak (Table I).
FIGURE 2.
A and B, Recognition of peptide-pulsed T2 cells by the MAGE-A3 TCR-transduced PBLs. Human PBLs expressing TCR against MAGE-A3: 112–120 or MAGE-A3: 271–279 were cocultured for 16 h with T2 cells that were previously pulsed with different concentrations of the respective peptides. Coculture of PBLs expressing TCR against MAGE-A3: 112–120 or MAGE-A3: 271–279 with control T2 cells that were not pulsed with peptides produced background levels of IFN-γ. The concentration of IFN-γ secreted into the culture medium was measured by ELISA.
Table I.
IFN-γ production by the TCR-transduced PBLs after coculture with tumor cell lines
| Cell Line | Histology | mRNA Copies/106 β-Actin | HLA-A2+/ | IFN-γ (pg/ml) | |||
|---|---|---|---|---|---|---|---|
| MAGE-A3/A6 | MAGE-A12 | Untransduced | MAGE-A3: 112–120 | MAGE-A3: 271–279 | |||
| 2361 | RCC | A<100 | <100 | A2+ | <30 | <30 | <30 |
| H2721 | SCLC | <100 | <100 | A2+ | <30 | <30 | <30 |
| 1088 | Melanoma | <100 | <100 | A2- | <30 | <30 | <30 |
| 888 | Melanoma | <100 | <100 | A2- | <30 | <30 | <30 |
| H1299 | NSCLC | 32,346 | 7,862 | A2- | <30 | <30 | <30 |
| H2122 | SCLC | <100 | <100 | A2- | <30 | <30 | <30 |
| 938 | Melanoma | 19,871 | 6,623 | A2- | <30 | <30 | <30 |
| 397-A*0201 | Melanoma | 33,039 | 8,781 | A2+ | <30 | 25,077 | 1,992 |
| 526 | Melanoma | 8,030 | 6,966 | A2+ | <30 | 17,142 | 1,996 |
| 1300 | Melanoma | 19,401 | 7,463 | A2+ | <30 | 68,746 | 7,212 |
| 2984 | Melanoma | 31,137 | 7,087 | A2+ | <30 | 36,330 | 4,605 |
| 624.38 | Melanoma | 41,541 | 1,040 | A2+ | 55 | 29,090 | 4,131 |
| 1359-A*0201 | Melanoma | 20,766 | 10,977 | A2+ | 37 | 32,166 | 5,759 |
| H1299-A*0201 | NSCLC | 57,045 | 18,875 | A2+ | <30 | 26,229 | 475 |
| HI 250 | SCLC | 136 | 932 | A2+ | <30 | 7,730 | 231 |
PBLs transduced with a retroviral vector expressing TCRs were cocultured with tumor cell lines. Ag-specific IFN-γ secretion was measured by ELISA. MAGE-A3/A6 and MAGE-A12 mRNA expression levels were quantified using a quantitative real-time PCR assay. A coculture assay was done with PBLs from three donors, and the result of a representative donor is presented. Values are means of duplicate samples. Results are presented as a representative of three experiments of PBLs from separate donors.
RCC, renal cell carcinoma; SCLC, small cell lung carcinoma.
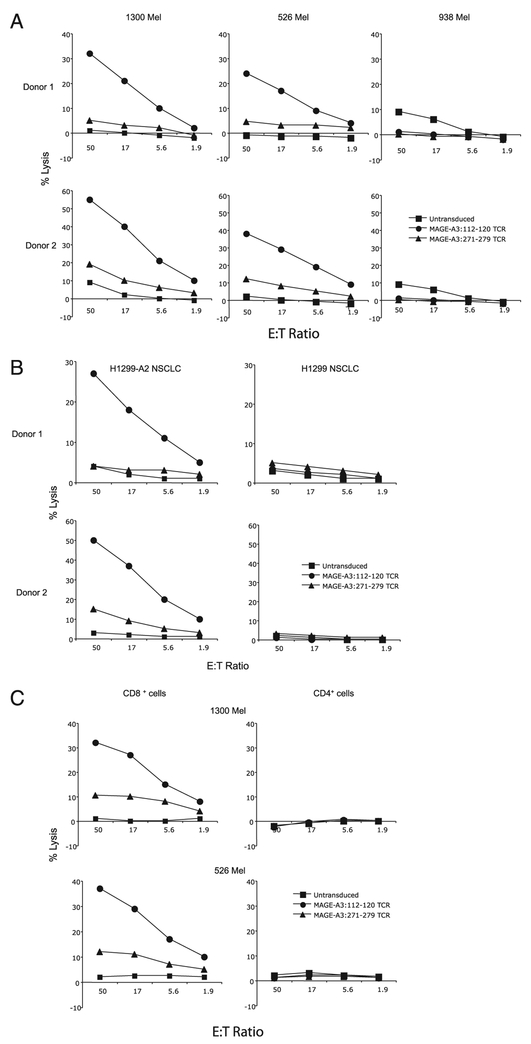
We next measured the specific lysis of melanoma cell lines by the TCR-engineered PBLs. MAGE-A3: 112–120 TCR-transduced PBLs demonstrated superior lytic function against MAGE-A3+/HLA-A*0201+ tumor cell lines 1300 melanoma and 526 melanoma cells compared with that of MAGE-A3: 271–279 TCR-transduced PBLs (Fig. 3A). There was little or no lysis of the HLA-A*0201− cell line 938 melanoma, and the untransduced PBLs showed little reactivity against any of the target cells (Fig. 3A). MAGE-A3: 112–120 TCR-transduced PBLs also showed superior lytic function against NSCLC cell line H1299-A*0201+ and did not recognize the parental non-HLA-A*0201 cell line H1299 (Fig. 3B). Because tetramer binding was observed only in MAGE-A3 TCR-transduced CD8+ cells and not in CD4+ cells, we investigated IFN-γ production and cytolytic activity using purified lymphocytes. As shown in Fig. 3C, Ag-specific lysis of melanoma cell lines 1300 melanoma and 526 melanoma was evident in CD8+ cells but not seen in CD4+ cells. Similarly, IFN-γ production after coculture with tumor cells was observed only in CD8+ cells and not seen in CD4+ cells (data not shown). On the basis of these data, the MAGE-A3: 112–120 TCR was chosen for further analysis.
FIGURE 3.
Specific killing of tumor cell lines by MAGE-A3 TCR-transduced PBLs. TCR-transduced human PBLs were cocultured for 4 h with the indicated 51Cr-labeled target tumor cell lines. Specific lysis of tumor cells was measured at the given E:T ratio using the formula: ([specific release − spontaneous release]/[total release − spontaneous release]). Specific lysis of untransduced, MAGE-A3: 112–120- or MAGE-A3: 271–279-specific TCR-transduced human PBLs are plotted on the graph as indicated. A, Specific lysis of melanoma tumor cell lines 1300 melanoma (HLAA*0201+/MAGE-A3+), 526 melanoma (HLA-A*0201+/MAGE-A3+), and 938 melanoma (HLA-A*0201−/MAGE-A3+) by the TCR-engineered PBLs. B, Specific lysis of NSCLC cell lines H1299 (MAGE+/HLA-A2+) and H1299-A*0201 (MAGE+/HLA-A2−). C, The cytolytic activities of TCR-engineered CD8+ and CD4+ T cells were evaluated using purified lymphocytes.
Single amino acid substitution variants within CDR3 of the TCR α-chain enhance the function of TCR-engineered PBLs
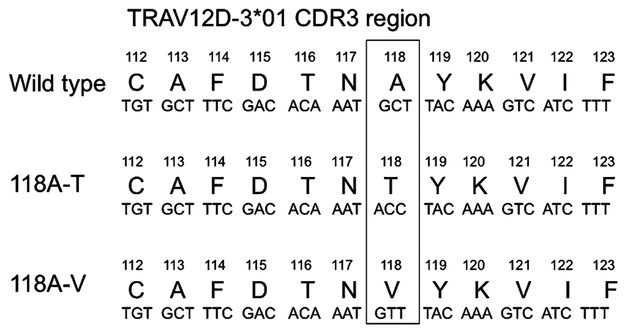
Previous studies by our group have shown that it is possible to improve the function of the TCR by engineering single or multiple amino acid changes in the CDR3 region of the TCR α-chain (14, 37, 44). We generated retroviral vectors expressing 85 single amino acid variants in the CDR3 region of the TCR α-chain and tested their function in PBLs. Preliminary screening experiments were carried out by single amino acid residue alanine substitution in the CDR3 of the α-chain. Alanine substitution at F114, D115, T116, N117, Y119, K120, and V121 completely abolished the activity of the TCR. This was seen by the complete loss of tetramer binding as well as a lack of production of IFN-γ after co-culture with peptide-pulsed T2 cells or MAGE+/HLA-A*0201+ tumor cells. We next focused on position 115 and created 19 aa substitutions at this at position. Complete loss of TCR activity, as seen by the complete lack of tetramer binding and loss of IFN-γ production, was observed when aspartic acid at position 115 was substituted with any of the other amino acids. Finally, we created a retroviral vector library of single amino acid variants at positions 117, 118, and 119 of the α-chain. During this screening, we found that a substitution of valine or threonine for the alanine residue present at position 118 in the wild-type α-chain retained TCR function (Fig. 4).
FIGURE 4.
Single amino acid substitutions at the CDR3 region of the MAGE-A3 TCR α-chain.
Table II shows the IFN-γ secretion results of a coculture assay with tumor cells of different histologies using the modified TCRs. Ag-specific HLA-A*0201–restricted recognition of tumor cells from diverse histologies were observed after coculture of TCR-engineered PBLs with breast cancer line MDA-454S-A2, glioma line U251, melanoma lines 624 and 526, esophageal line BE-3, and NSCLC line H1299-A*0201. The results demonstrated that T cells transduced with the A118V and A118T TCR variants secreted higher levels of IFN-γ than cells transduced with the wild-type TCR. T cells transduced with the A118T variant TCR secreted higher levels of IFN-γ than the A118V variant when tested against multiple MAGE+/HLA-A*0201+ cells. The HLA-A*0201− cell lines H1299 and 888 as well as MAGE−/HLA-A*0201+ cell line 2361-RCC were not recognized by the PBLs engineered to express either the wild-type or the A118T variant, indicating that this amino acid alteration did not alter the specificity of this TCR.
Table II.
IFN-γ production by the modified MAGE-A3 A118T TCR-transduced PBLs after coculture with tumor cell lines of different histologies
| Cell Lines | Histology | mRNA Copies/106 β-Actin | IFN-γ (pg/ml) | ||||
|---|---|---|---|---|---|---|---|
| MAGE-A3/A6 | MAGE-A12 | Untransduced | MAGE-WT | MAGE-A118V | MAGE-A118T | ||
| H1299 | NSCLC | 32,346 | 7,862 | <30 | <30 | <30 | <30 |
| 2361 | RCC | <100 | <100 | <30 | <30 | <30 | <30 |
| 888 | Melanoma | <100 | <100 | 356 | <30 | <30 | 48 |
| MDA-453S-A*0201 | Breast | 4,437 | 629 | 325 | 4,030 | 5,496 | 6,083 |
| U251 | CNS-glioma | 3,643 | 1,087 | <30 | 3,102 | 13,253 | 25,597 |
| 624.38 | Melanoma | 41,541 | 1,040 | 46 | 3,303 | 14,581 | 31,049 |
| BE-3 | Esophageal | 2,554 | 1,228 | <30 | 1,791 | 6,117 | 23,242 |
| 526 | Melanoma | 3,002 | 6,966 | 125 | 874 | 3,809 | 16,920 |
| H1299-A*0201 | NSCLC | 57,045 | 18,875 | 48 | 27,191 | 45,158 | 83,294 |
Donor PBLs transduced with a retroviral vector expressing wild-type or single amino acid variants of MAGE-A3: 112–120 TCR were cocultured with tumor cell lines. Untransduced PBLs were used as controls. Ag-specific IFN-γ secretion was measured by ELISA. MAGE-A3/A6 and MAGE-A12 mRNA expression levels were estimated using a quantitative real-time PCR assays. Values are means of duplicate samples. Results are presented as a representative of two experiments using PBLs from separate donors.
RCC, renal cell carcinoma.
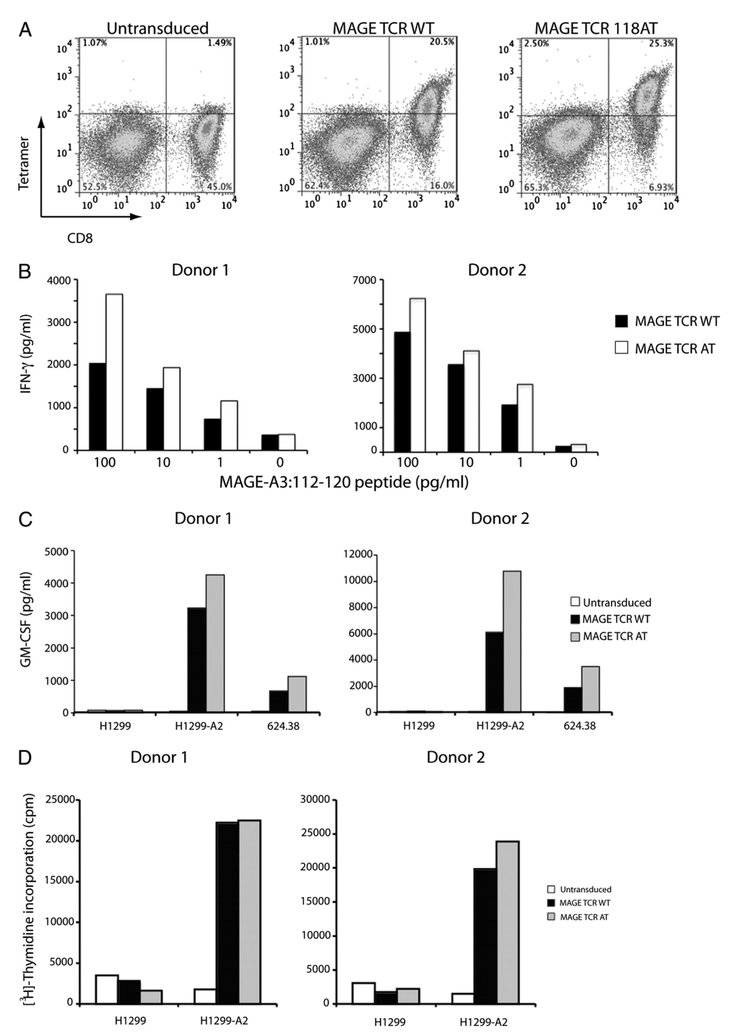
To further test the function of this improved MAGE TCR, we purified the CD4+ cells and tested these cells in a coculture assay with 1300 melanoma, H1299-A*0201 NSCLC, U251 glioma, and peptide-loaded T2 cells. CD4+ cells engineered with the MAGEA3: 112–120 TCR A118T variant specifically secreted IFN-γ in response to MAGE-A3+/HLA-A*0201+ tumor cells, whereas no response was observed in CD4+ T cells transduced with the wild-type TCR (Table III). In addition, the A118T variant led to enhanced recognition of peptide-pulsed target cells. Tetramer analysis of MAGE-A3: 112–120 A118T variant TCR-transduced PBLs showed MAGE-A3: 112–120/HLA-A*0201-specific tetramer binding in CD4+ cells, though with lesser intensity than that in CD8+ cells (Fig. 5A).
Table III.
IFN-γ production by the CD4+ cells expressing wild-type or modified CDR3 amino acid variant MAGE-A3 TCRs after coculture with tumor cell lines
| Cell Lines | IFN-03B3 (pg/ml) | ||
|---|---|---|---|
| MAGE WT | MAGE Al18V | MAGE All8T | |
| 1300 melanoma | <30 | 145 | 263 |
| H1299-A*0201 NSCLC | <30 | 132 | 589 |
| U251 glioma | <30 | 110 | 666 |
| T2 cells pulsed with MAGE peptide | 395 | 2727 | 9710 |
CD4+ cells were purified from the donor PBLs transduced with a retroviral vector expressing wild-type or single amino acid variants of MAGE-A3: 112–120 TCR and were cocultured with tumor cell lines. Ag-specific IFN-γ secretion was measured by ELISA. Values are means of duplicate samples. Results are presented as a representative of two independent experiments using PBLs from separate donors.
FIGURE 5.
A, Flow cytometric analysis of wild-type and modified MAGE-A3 TCR-A118T-transduced PBLs. Dot blot showing the FACS profile of PBLs stained with anti–human-CD8-FITC Ab and PE-conjugated MAGE-A3: 112–120-HLA-A*0201 tetramer. B, Recognition of MAGE-A3: 112–120 peptide-pulsed T2 cells by the MAGE-A3 TCR-transduced PBLs. Human PBLs expressing MAGE-A3: 112–120 wild-type or A118T variant TCR were cocultured for 16 h with T2 cells that were previously pulsed with different concentrations of MAGE-A3: 112–120 peptide. The concentration of IFN-γ secreted into the culture medium was measured by ELISA, and values indicate the mean of duplicate samples. C, A representative assay showing GM-CSF release data from two donors after overnight coculture with tumor cell targets. The concentration of GM-CSF secreted into the culture medium was measured by ELISA, and values indicate the mean of duplicate samples. D, Proliferation of PBLs after coculture with H1299 or H1299-A2 cells measured by [3H]thymidine incorporation after 3 d in culture. Values indicate average counts of triplicate wells.
In an effort to compare the function of MAGE-A3: 112–120 wild-type and 118AT variant TCR-expressing PBLs, we performed coculture assays with MAGE-A3: 112–120 peptide-pulsed T2 cells. MAGE-A3: 112–120 A118T variant TCR-expressing PBLs produced higher levels of IFN-γ than the wild-type TCR-transduced PBLs (Fig. 5B). We also measured secretion of GMCSF by the MAGE-A3 TCR-transduced PBLs after coculture with H1299, H1299-A2, and 624.38 cells. MAGE-A3: 112–120 118AT variant TCR-expressing PBLs produced higher levels of GM-CSF than the wild-type TCR-transduced PBLs (Fig. 5C). We then performed a cell proliferation assay to measure the Ag-specific proliferation of MAGE TCR-engineered PBLs. When cocultured with H1299-A2 tumor cell lines both MAGE-A3: 112–120 wild-type as well as 118AT variant TCR-transduced PBLs proliferated extensively as measured by the radiolabeled thymidine incorporation but not with H1299 cells (Fig. 5D). Untransduced PBLs exhibited a background level of proliferation in response to both H1299 and H1299-A2 cells.
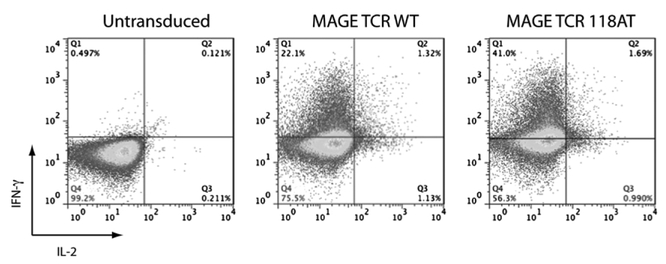
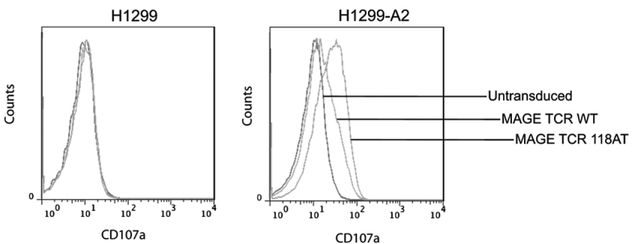
The percentage of MAGE-A3: 112–120 wild-type or 118AT variant TCR-transduced PBLs that produced IFN-γ and IL-2 after overnight coculture with H1299-A2 cells were determined by intracellular cytokine staining. The percentage of cells that produced IFN-γ in response to Ag exposure was almost twice in number in the case of A118T variant TCR-transduced PBLs than that of wild-type TCR-expressing cells, whereas a marginal increase in the number of IL-2–producing cells was observed (Fig.6). As a marker of degranulation, we compared the ability of MAGE-A3 wild-type and 118AT TCR-engineered PBLs to mobilize CD107a to the cell surface. MAGE TCR-transduced PBLs were cocultured with H1299 or H1299-A2 cells for 2 h and analyzed for CD107a Ag expression. A significantly higher number of MAGE-A3 A118T TCR-transduced cells stained positive for CD107a expression compared with wild-type TCR-transduced PBLs after coculture with H1299-A2 cells. Untransduced cells showed background levels of staining (Fig. 7).
FIGURE 6.
Intracellular cytokine staining for IFN-γ and IL-2 of MAGE-A3 TCR-transduced PBLs after Ag-specific stimulation. Ten days after transduction with MAGE-A3: 112–120 wild-type or A118T, TCR-transduced PBLs were subjected to overnight coculture with H1299-A2 cells. Cells were then stained for the detection of intracellular IFN-γ and IL-2 and cell surface expression of CD3 molecule and analyzed by FACS. The transduced PBLs produced IFN-γ in an Ag-specific manner. The plots are gated on CD3+ lymphocytes, and the percentage on each quadrant is shown on the plot. A representative of three experiments is shown.
FIGURE 7.
Expression of degranulation marker CD107a on MAGE-A3 TCR-transduced PBLs after Ag exposure. Expression of CD107a on MAGE-A3: 112–120 wild-type or A118T TCR-transduced PBLs was detected by FACS analysis after coculture with H1299-A2 cells. The percentages of CD107a-positive cells are shown. Results from a representative of three experiments are presented.
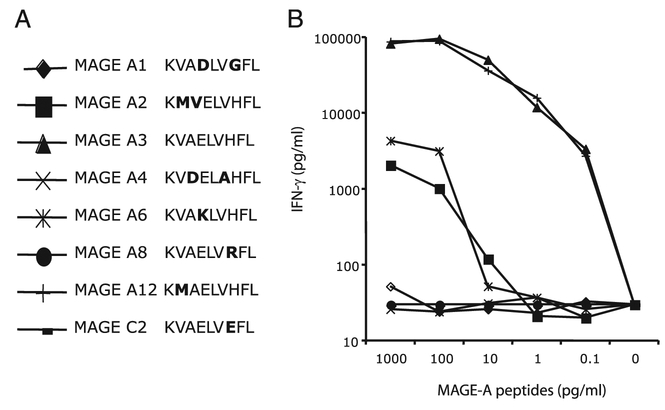
Recognition of other members of the MAGE-A family by the MAGE-A3: 112–120 TCR
The MAGE-A gene family consists of 12 genes encoding proteins with 50–99% sequence homology to each other. Several other members of the MAGE-A gene family encode amino acid sequences that are similar to that of the MAGE-A3: 112–120 peptide, differing only in 1 or 2 aa residues (Fig. 8A). We tested MAGE-A3 TCR-transduced PBLs for the recognition of peptides derived from other members of the MAGE-A family by loading T2 cells with synthetic peptides and performing coculture assays to test the peptide recognition and secretion of IFN-γ. As shown in Fig. 8B, in addition to the MAGE-A3: 112–120 (KVAELVHFL) peptide, MAGE-A12: 112–120 (KMAELVHFL) was also recognized efficiently by the MAGE-A3 TCR-engineered PBLs. Peptides derived from MAGE-A2 and MAGE-A6 were also recognized by the TCR-engineered PBLs albeit with lower avidity (Fig. 8B). We then asked whether TCR-engineered PBLs were able to recognize endogenously processed peptides from MAGEA3– and MAGE-A12–expressing cells. The recognition of COS7-A*0201 and 293-A*0201 cells that were retrovirally transduced with full-length MAGE-A3 or MAGE-A12 genes was tested in coculture assays using PBLs transduced with the wild-type and TCR variants. As seen in Table IV, superior recognition of MAGE-A12 was evident in both COS7-A*0201 and 293-A*0201 cells, and again the A118T TCR demonstrated the most activity.
FIGURE 8.
Recognition of other related MAGE peptides by the MAGE-A3: 112–120 TCR-transduced PBLs. PBLs expressing TCRs against MAGE-A3: 112–120 were cocultured for 16 h with T2 cells that were previously pulsed with different concentrations of respective peptides. The amount of IFN-γ secreted into the medium was measured by ELISA. A, Amino acid sequence alignment of related MAGE peptides. All of the peptides were 9-mers, and the amino acid variation from the MAGEA3 peptide is indicated in bold. B, Peptide recognition titration was measured as the IFN-γ production after coculture with peptide-pulsed T2 cells.
Table IV.
IFN-γ production by the TCR-transduced PBLs after coculture with COS-A2 and 293-A2 cell lines expressing full-length MAGE-A3 or MAGE-A12 genes
| Cell Lines | mRNA Copies/106 β-Actin | IFN-γ (pg/ml) | ||||
|---|---|---|---|---|---|---|
| MAGE-A3/6 | MAGE-A12 | Untransduced | MAGE-WT | MAGE-A118V | MAGE-A118T | |
| H1299-A2 | 57,045 | 18,875 | 48 | 27,191 | 45,158 | 83,294 |
| COS7-A2 | <100 | <100 | <30 | <30 | <30 | 69 |
| 293-A2 | <100 | <100 | <30 | <30 | <30 | <30 |
| COS7-A2-MAGE A3 | 602,975 | <100 | 93 | 3,644 | 6,063 | 25,920 |
| COS7-A2-MAGE A12 | 257 | 335,943 | 102 | 27,375 | 33,884 | 72,582 |
| 293-A2-MAGE A3 | 547,966 | 673 | 108 | 446 | 829 | 3,199 |
| 293-A2-MAGE A12 | 334 | 285,454 | 88 | 4,456 | 8,291 | 25,015 |
PBLs transduced with a retroviral vector expressing TCRs were cocultured with COS7-A2 or 293-A2 cells expressing MAGE-A3 or MAGE-A12 genes. Ag-specific IFN-γ secretion was measured by ELISA. MAGE-A3/A6 and MAGE-A12 mRNA expression levels were estimated using a quantitative real-time PCR assay. Values are means of duplicate samples. Results are a presented as a representative of two independent experiments using PBLs from separate donors.
Discussion
Toxicity to normal tissues is a potential negative side effect in TCR-based adoptive immunotherapy targeting shared tumor Ags that are also expressed in some normal tissue (7). Ideally, adoptive immunotherapy should strive to target T cells toward tumor-specific Ags that are not expressed in normal tissue. Toward this goal, CTAs are ideal candidates due to their overexpression in multiple tumor types and limited expression in normal tissue. MAGE-A3 is a CTA that belongs to the MAGE-A gene family and is expressed in tumors of different histologies, including melanoma, lung, ovarian, hepatocellular, head and neck, many types of sarcomas, and multiple myeloma but not found in normal tissue with the exception of testis and placenta. Because germ line cells do not present Ags due to the lack of expression of MHC molecules and can therefore not be targeted by the TCR-engineered T cells, immune responses directed against CTAs such as MAGE-A3 are not likely to lead to the recognition of these tissues.
In an attempt to derive multiple TCRs that would recognize MAGE-A3, we immunized HLA-A*0201 transgenic mice with two of the peptide epitopes that had been shown previously to be endogenously processed and presented in the context of this HLAA2 restriction element. Expression of both TCRs in human PBLs demonstrated Ag-specific reactivity in the form of tumor cell lysis and cytokine secretion against a range of melanoma and nonmelanoma tumor cell targets. Both of the TCRs equally recognized target cells pulsed with their respective MAGE-A3–derived synthetic peptides 112–120 (KVAELVHFL) and 271–279 (FLWGPRALV); however, recognition of the tumor cells was relatively poor in the case of the TCR that recognized the 271–279 epitope. This weak recognition was evident by the relatively lower levels of IFN-γ secretion and cytolytic activity by the MAGE-A3: 271–279 TCR-engineered T cells upon coculture with MAGE-A3–positive tumor cells. However, MAGE-A3: 112–120 TCR-engineered PBLs secreted high levels of proinflammatory cytokine IFN-γ and exhibited cytolytic activity against a variety of tumor cells, including melanoma, lung, breast, esophageal, and glioma. This discrepancy could be due to the possible differences between the synthetic peptide loaded externally on T2 cells and the relatively poor efficiency in MAGE-A3 Ag processing of the 271–279 epitope observed in tumor cells (30, 45). It has been previously shown that the MAGE-A3: 271–279 epitope is not efficiently presented on the surface of tumor cell lines (46). In addition, Miconnet et al. (45) demonstrated that MAGE-A3 amino acid residues 278 and 280 are major proteasome cleavage sites but not 279. In the analysis of any TCR generated by immunization, it is critical to verify the recognition of multiple tumors and not rely on a limited number of cell lines or peptide-pulsed cells because this can result in the study of pseudo-tumor Ags, which are of no clinical benefit (47).
Because the MAGE-A family of genes have 50–99% sequence homology, we made synthetic peptides of other closely related MAGE gene family peptides that differed by one or two amino acids from the MAGE-A3: 112–120 peptide. Peptides from MAGE-A3 as well as MAGE-A12 presented by the tumor cells in the context of the HLA-A*0201 allele are efficiently recognized by this TCR. The recognition of MAGE-A12 was expected based on the fact that there was a single amino acid difference between the two peptides, a conservative substitution of methionine for the valine residue present at the second position anchor residue in the MAGE-A3 peptide, both of which represent consensus amino acids (48). This might potentially broaden the number of tumors that can be targeted by this TCR. It has been reported that MAGEA genes are coordinately expressed in tumors (49), leading to the possibility that MAGE-A3/MAGE-A12 TCR-engineered T cells may recognize peptides presented on the tumor cell surface from both the MAGE-A3 and MAGE-A12 genes, resulting in more efficient elimination of tumors with heterogeneous MAGE-A gene expression.
High-avidity recognition of tumor Ags has been shown to be important for the in vivo antitumor response (50). In an effort to enhance the function of the MAGE TCR, we introduced single amino acid substitutions in the CDR3 region of the α-chain as described previously (14, 37). Modifications in the CDR3 regions can increase the Ag-specific recognition in CD4 and CD8 cells (14, 37). A TCR variant with a single amino acid substitution at position 118 from alanine to threonine of the α-chain most notably enhanced CD8 T cell function and induced Ag-specific reactivity in CD4 cells that was not apparent for the wild-type TCR. Although the wild-type TCR was derived from HLA-A*0201 transgenic mice, it was not CD8-independent, a finding that we have previously reported on for an anti-CEA TCR (37). In multiple independent assays, the A118T modified TCR functionally outperformed the wild-type TCR (Figs. 5–7, Tables II–IV). Importantly, this high-avidity modified receptor retained its specificity and did not introduce any nonspecific recognition of MAGE− or non-HLA-A*0201 cells. Preservation of TCR specificity is important to avoid potential off-target toxicity, as previously observed in some modified TCRs (14, 37, 44).
A potential advantage in targeting CTAs such as MAGE-A3 and NY ESO-1 is that the expression of these Ags can be induced selectively in tumor cells by pharmacological agents (13, 51). CTA expression on tumor cells can be induced by demethylating agents such as 5-aza-2′-deoxycytidine and histone deacetylase inhibitors such as depsipeptide. It may be an attractive clinical strategy to induce MAGE-A3 expression in the patient’s tumor before the infusion of MAGE TCR-engineered PBLs. MAGE-A3 has been the target of several clinical studies, for example, using dendritic cells loaded with MAGE-A3 peptides alone or in a pool of other melanoma Ag peptides or PBMCs pulsed with a MAGE-A3 peptide and coadministered with IL-12 (52–57). In the largest study to date, GlaxoSmithKline biologics has undertaken a MAGE-A3 protein-based tumor vaccine multinational Phase III clinical trial to treat NSCLC patients in the adjuvant setting to prevent disease relapse (58, 59). MAGE-A3 recombinant protein vaccines have been shown to be generally well tolerated in patients, though only modest clinical responses were reported (60). Unlike peptide vaccines, recombinant protein vaccines have the potential to induce a broad array of immune responses. No significant toxicity has been reported in any of the patients treated with protein vaccines (58–61). In animal models of adoptive cell therapy, vaccination was found to be essential for effective tumor treatment (62). As we move forward with the TCR-based adoptive immunotherapy targeting MAGE-A3, combining it with a MAGE-A3 vaccine may be a useful clinical strategy to stimulate the transferred T cells in vivo after adoptive cell transfer therapy.
The MAGE-A3 TCR may be an ideal candidate for tumor immunotherapy for several reasons. First, TCR-engineered T cells can be directed against a very large variety of tumor types. Second, considering the high percentage of tumors that express MAGE genes, (for example >60% of melanomas and >50% of NSCLCs), a large number of patients can be eligible for treatment. Third, coordinated expression of MAGE genes are reported, and the TCR-engineered PBLs can recognize MAGE-A3 and MAGE-A12 epitopes. Finally, because MAGE is not expressed in normal tissue except testis, this may limit the risk of on-target toxicity to normal tissue. In summary, the data presented in this article have significant implications to MAGE TCR-based adoptive immunotherapy for melanoma and nonmelanoma epithelial tumors, where we can potentially administer large numbers of Ag-specific T cells without toxicity. In addition, MAGE TCR-engineered T cells could be combined with TCRs targeted against other CTAs, such as NYESO-1, to enhance tumor killing.
Acknowledgments
We thank Arnold Mixon and Shawn Farid for technical support for FACS analysis and Yong Li for technical help.
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Abbreviations used in this article:
- CTA
cancer testis Ag
- NSCLC
non-small cell lung carcinoma
- rhIL-2
recombinant human IL-2
- TIL
tumor infiltrating lymphocyte
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Blattman JN, and Greenberg PD. 2004. Cancer immunotherapy: a treatment for the masses. Science 305: 200–205. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg SA 1999. A new era for cancer immunotherapy based on the genes that encode cancer antigens. Immunity 10: 281–287. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, and Dudley ME. 2008. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat. Rev. Cancer 8: 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg SA 2001. Progress in human tumour immunology and immuno-therapy. Nature 411: 380–384. [DOI] [PubMed] [Google Scholar]
- 5.Dudley ME, Yang JC, Sherry R, Hughes MS, Royal R, Kammula U,Robbins PF, Huang J, Citrin DE, Leitman SF, et al. 2008. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J. Clin. Oncol 26: 5233–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudley ME, Wunderlich JR, Shelton TE, Even J, and Rosenberg SA. 2003. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J. Immunother 26: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC,Hughes MS, Kammula US, Royal RE, Sherry RM, Wunderlich JR, et al. 2009. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC,Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, et al. 2006. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 314: 126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suri A 2006. Cancer testis antigens—their importance in immunotherapy and in the early detection of cancer. Expert Opin. Biol. Ther 6: 379–389. [DOI] [PubMed] [Google Scholar]
- 10.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, and Old LJ. 2005. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer 5: 615–625. [DOI] [PubMed] [Google Scholar]
- 11.Caballero OL, and Chen YT. 2009. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 100: 2014–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Zheng Z, Robbins PF, Khong HT, Rosenberg SA, andMorgan RA. 2005. Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J. Immunol 174: 4415–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wargo JA, Robbins PF, Li Y, Zhao Y, El-Gamil M, Caragacianu D,Zheng Z, Hong JA, Downey S, Schrump DS, et al. 2009. Recognition of NY-ESO-1+ tumor cells by engineered lymphocytes is enhanced by improved vector design and epigenetic modulation of tumor antigen expression. Cancer Immunol. Immunother 58: 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robbins PF, Li YF, El-Gamil M, Zhao Y, Wargo JA, Zheng Z, Xu H,Morgan RA, Feldman SA, Johnson LA, et al. 2008. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J. Immunol 180: 6116–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker PA, and Salehi A. 2002. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci. Res 67: 705–712. [DOI] [PubMed] [Google Scholar]
- 16.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, and Boon T. 1991. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254: 1643–1647. [DOI] [PubMed] [Google Scholar]
- 17.Yang B, O’Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM,Gravekamp C, Setaluri V, Peters N, Hoffmann FM, et al. 2007. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 67: 9954–9962. [DOI] [PubMed] [Google Scholar]
- 18.Monte M, Simonatto M, Peche LY, Bublik DR, Gobessi S, Pierotti MA,Rodolfo M, and Schneider C. 2006. MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc. Natl. Acad. Sci. USA 103: 11160–11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Cheng S, Asa SL, and Ezzat S. 2008. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer Res. 68: 8104–8112. [DOI] [PubMed] [Google Scholar]
- 20.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, and Lucas S. 2001. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 61: 5544–5551. [PubMed] [Google Scholar]
- 21.De Smet C, Lurquin C, Lethé B, Martelange V, and Boon T. 1999. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol. Cell. Biol 19: 7327–7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roeder C, Schuler-Thurner B, Berchtold S, Vieth G, Driesch P, Schuler G, and Lüftl M. 2005. MAGE-A3 is a frequent tumor antigen of metastasized melanoma. Arch. Dermatol. Res 296: 314–319. [DOI] [PubMed] [Google Scholar]
- 23.Tajima K, Obata Y, Tamaki H, Yoshida M, Chen YT, Scanlan MJ,Old LJ, Kuwano H, Takahashi T, Takahashi T, and Mitsudomi T. 2003. Expression of cancer/testis (CT) antigens in lung cancer. Lung Cancer 42: 23–33. [DOI] [PubMed] [Google Scholar]
- 24.Filho PA, López-Albaitero A, Xi L, Gooding W, Godfrey T, andFerris RL. 2009. Quantitative expression and immunogenicity of MAGE-3 and −6 in upper aerodigestive tract cancer. Int. J. Cancer 125: 1912–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo G, Huang S, Xie X, Stockert E, Chen YT, Kubuschok B, andPfreundschuh M. 2002. Expression of cancer-testis genes in human hepatocellular carcinomas. Cancer Immun. 2: 11. [PubMed] [Google Scholar]
- 26.Jungbluth AA, Ely S, DiLiberto M, Niesvizky R, Williamson B, Frosina D,Chen YT, Bhardwaj N, Chen-Kiang S, Old LJ, and Cho HJ. 2005. The cancer-testis antigens CT7 (MAGE-C1) and MAGE-A3/6 are commonly expressed in multiple myeloma and correlate with plasma-cell proliferation. Blood 106: 167–174. [DOI] [PubMed] [Google Scholar]
- 27.Bolli M, Kocher T, Adamina M, Guller U, Dalquen P, Haas P, Mirlacher M,Gambazzi F, Harder F, Heberer M, et al. 2002. Tissue microarray evaluation of Melanoma antigen E (MAGE) tumor-associated antigen expression: potential indications for specific immunotherapy and prognostic relevance in squamous cell lung carcinoma. Ann. Surg 236: 785–793, discussion 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gure AO, Chua R, Williamson B, Gonen M, Ferrera CA, Gnjatic S,Ritter G, Simpson AJ, Chen YT, Old LJ, and Altorki NK. 2005. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin. Cancer Res 11: 8055–8062. [DOI] [PubMed] [Google Scholar]
- 29.Dhodapkar MV, Osman K, Teruya-Feldstein J, Filippa D, Hedvat CV,Iversen K, Kolb D, Geller MD, Hassoun H, Kewalramani T, et al. 2003. Expression of cancer/testis (CT) antigens MAGE-A1, MAGE-A3, MAGE-A4, CT-7, and NY-ESO-1 in malignant gammopathies is heterogeneous and correlates with site, stage and risk status of disease. Cancer Immun. 3: 9. [PubMed] [Google Scholar]
- 30.Valmori D, Liénard D, Waanders G, Rimoldi D, Cerottini JC, andRomero P. 1997. Analysis of MAGE-3-specific cytolytic T lymphocytes in human leukocyte antigen-A2 melanoma patients. Cancer Res. 57: 735–741. [PubMed] [Google Scholar]
- 31.Chaux P, Vantomme V, Stroobant V, Thielemans K, Corthals J, Luiten R,Eggermont AM, Boon T, and van der Bruggen P. 1999. Identification of MAGE-3 epitopes presented by HLA-DR molecules to CD4(+) T lymphocytes.J. Exp. Med 189: 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manici S, Sturniolo T, Imro MA, Hammer J, Sinigaglia F, Noppen C,Spagnoli G, Mazzi B, Bellone M, Dellabona P, and Protti MP. 1999. Melanoma cells present a MAGE-3 epitope to CD4(+) cytotoxic T cells in association with histocompatibility leukocyte antigen DR11. J. Exp. Med 189: 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultz ES, Lethé B, Cambiaso CL, Van Snick J, Chaux P, Corthals J,Heirman C, Thielemans K, Boon T, and van der Bruggen P. 2000. A MAGEA3 peptide presented by HLA-DP4 is recognized on tumor cells by CD4+ cytolytic T lymphocytes. Cancer Res. 60: 6272–6275. [PubMed] [Google Scholar]
- 34.Kobayashi H, Song Y, Hoon DS, Appella E, and Celis E. 2001. Tumor-reactive T helper lymphocytes recognize a promiscuous MAGE-A3 epitope presented by various major histocompatibility complex class II alleles. Cancer Res. 61: 4773–4778. [PubMed] [Google Scholar]
- 35.Kawashima I, Hudson SJ, Tsai V, Southwood S, Takesako K, Appella E,Sette A, and Celis E. 1998. The multi-epitope approach for immunotherapy for cancer: identification of several CTL epitopes from various tumor-associated antigens expressed on solid epithelial tumors. Hum. Immunol 59: 1–14. [DOI] [PubMed] [Google Scholar]
- 36.Graff-Dubois S, Faure O, Gross DA, Alves P, Scardino A, Chouaib S,Lemonnier FA, and Kosmatopoulos K. 2002. Generation of CTL recognizing an HLA-A*0201-restricted epitope shared by MAGE-A1, -A2, -A3, -A4, -A6, -A10, and -A12 tumor antigens: implication in a broad-spectrum tumor immunotherapy. J. Immunol 169: 575–580. [DOI] [PubMed] [Google Scholar]
- 37.Parkhurst MR, Joo J, Riley JP, Yu Z, Li Y, Robbins PF, andRosenberg SA. 2009. Characterization of genetically modified T-cell receptors that recognize the CEA:691–699 peptide in the context of HLA-A2.1 on human colorectal cancer cells. Clin. Cancer Res 15: 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riker AI, Kammula US, Panelli MC, Wang E, Ohnmacht GA,Steinberg SM, Rosenberg SA, and Marincola FM. 2000. Threshold levels of gene expression of the melanoma antigen gp100 correlate with tumor cell recognition by cytotoxic T lymphocytes. Int. J. Cancer 86: 818–826. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y, Zheng Z, Cohen CJ, Gattinoni L, Palmer DC, Restifo NP,Rosenberg SA, and Morgan RA. 2006. High-efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol. Ther 13: 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes MS, Yu YY, Dudley ME, Zheng Z, Robbins PF, Li Y,Wunderlich J, Hawley RG, Moayeri M, Rosenberg SA, and Morgan RA. 2005. Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum. Gene Ther 16: 457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan RA, Dudley ME, Yu YY, Zheng Z, Robbins PF, Theoret MR,Wunderlich JR, Hughes MS, Restifo NP, and Rosenberg SA. 2003. High efficiency TCR gene transfer into primary human lymphocytes affords avid recognition of melanoma tumor antigen glycoprotein 100 and does not alter the recognition of autologous melanoma antigens. J. Immunol 171: 3287–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Bruggen P, Bastin J, Gajewski T, Coulie PG, Boël P, De Smet C,Traversari C, Townsend A, and Boon T. 1994. A peptide encoded by human gene MAGE-3 and presented by HLA-A2 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE-3. Eur. J. Immunol 24: 3038–3043. [DOI] [PubMed] [Google Scholar]
- 43.Yang S, Cohen CJ, Peng PD, Zhao Y, Cassard L, Yu Z, Zheng Z, Jones S,Restifo NP, Rosenberg SA, and Morgan RA. 2008. Development of optimal bicistronic lentiviral vectors facilitates high-level TCR gene expression and robust tumor cell recognition. Gene Ther. 15: 1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, Bennett AD, Zheng Z, Wang QJ, Robbins PF, Yu LY, Li Y,Molloy PE, Dunn SM, Jakobsen BK, et al. 2007. High-affinity TCRs generated by phage display provide CD4+ T cells with the ability to recognize and kill tumor cell lines. J. Immunol 179: 5845–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miconnet I, Servis C, Cerottini JC, Romero P, and Lévy F. 2000. Amino acid identity and/or position determines the proteasomal cleavage of the HLAA*0201-restricted peptide tumor antigen MAGE-3271–279. J. Biol. Chem 275: 26892–26897. [DOI] [PubMed] [Google Scholar]
- 46.Valmori D, Gileadi U, Servis C, Dunbar PR, Cerottini JC, Romero P,Cerundolo V, and Lévy F. 1999. Modulation of proteasomal activity required for the generation of a cytotoxic T lymphocyte-defined peptide derived from the tumor antigen MAGE-3. J. Exp. Med 189: 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkhurst MR, Riley JP, Igarashi T, Li Y, Robbins PF, andRosenberg SA. 2004. Immunization of patients with the hTERT:540–548 peptide induces peptide-reactive T lymphocytes that do not recognize tumors endogenously expressing telomerase. Clin. Cancer Res 10: 4688–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker KC, Bednarek MA, and Coligan JE. 1994. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol 152: 163–175. [PubMed] [Google Scholar]
- 49.Bredenbeck A, Hollstein VM, Trefzer U, Sterry W, Walden P, andLosch FO. 2008. Coordinated expression of clustered cancer/testis genes encoded in a large inverted repeat DNA structure. Gene 415: 68–73. [DOI] [PubMed] [Google Scholar]
- 50.Zeh HJ III, Perry-Lalley D, Dudley ME, Rosenberg SA, and Yang JC. 1999. High avidity CTLs for two self-antigens demonstrate superior in vitro and in vivo antitumor efficacy. J. Immunol 162: 989–994. [PubMed] [Google Scholar]
- 51.Weiser TS, Ohnmacht GA, Guo ZS, Fischette MR, Chen GA,Hong JA, Nguyen DM, and Schrump DS. 2001. Induction of MAGE-3 expression in lung and esophageal cancer cells. Ann. Thorac. Surg 71: 295–301, discussion 301–302. [DOI] [PubMed] [Google Scholar]
- 52.Schuler-Thurner B, Schultz ES, Berger TG, Weinlich G, Ebner S, Woerl P,Bender A, Feuerstein B, Fritsch PO, Romani N, and Schuler G. 2002. Rapid induction of tumor-specific type 1 T helper cells in metastatic melanoma patients by vaccination with mature, cryopreserved, peptide-loaded monocyte-derived dendritic cells. J. Exp. Med 195: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadanaga N, Nagashima H, Mashino K, Tahara K, Yamaguchi H, Ohta M,Fujie T, Tanaka F, Inoue H, Takesako K, et al. 2001. Dendritic cell vaccination with MAGE peptide is a novel therapeutic approach for gastrointestinal carcinomas. Clin. Cancer Res 7: 2277–2284. [PubMed] [Google Scholar]
- 54.Mackensen A, Herbst B, Chen JL, Köhler G, Noppen C, Herr W,Spagnoli GC, Cerundolo V, and Lindemann A. 2000. Phase I study in melanoma patients of a vaccine with peptide-pulsed dendritic cells generated in vitro from CD34(+) hematopoietic progenitor cells. Int. J. Cancer 86: 385–392. [DOI] [PubMed] [Google Scholar]
- 55.Gajewski TF, Fallarino F, Ashikari A, and Sherman M. 2001. Immunization of HLA-A2+ melanoma patients with MAGE-3 or MelanA peptide-pulsed autologous peripheral blood mononuclear cells plus recombinant human interleukin 12. Clin. Cancer Res 7(3, Suppl.): 895s–901s. [PubMed] [Google Scholar]
- 56.Carrasco J, Van Pel A, Neyns B, Lethé B, Brasseur F, Renkvist N, van der Bruggen P, van Baren N, Paulus R, Thielemans K, et al. 2008. Vaccination of a melanoma patient with mature dendritic cells pulsed with MAGE-3 peptides triggers the activity of nonvaccine anti-tumor cells. J. Immunol 180: 3585–3593. [DOI] [PubMed] [Google Scholar]
- 57.Banchereau J, Palucka AK, Dhodapkar M, Burkeholder S, Taquet N,Rolland A, Taquet S, Coquery S, Wittkowski KM, Bhardwaj N, et al. 2001. Immune and clinical responses in patients with metastatic melanoma to CD34(+) progenitor-derived dendritic cell vaccine. Cancer Res. 61: 6451–6458. [PubMed] [Google Scholar]
- 58.Tyagi P, and Mirakhur B. 2009. MAGRIT: the largest-ever phase III lung cancer trial aims to establish a novel tumor-specific approach to therapy. Clin. Lung Cancer 10: 371–374. [DOI] [PubMed] [Google Scholar]
- 59.Brichard VG, and Lejeune D. 2007. GSK’s antigen-specific cancer immunotherapy programme: pilot results leading to Phase III clinical development. Vaccine 25(Suppl. 2): B61–B71. [DOI] [PubMed] [Google Scholar]
- 60.Marchand M, Punt CJ, Aamdal S, Escudier B, Kruit WH, Keilholz U,Håkansson L, van Baren N, Humblet Y, Mulders P, et al. 2003. Immunisation of metastatic cancer patients with MAGE-3 protein combined with adjuvant SBAS-2: a clinical report. Eur. J. Cancer 39: 70–77. [DOI] [PubMed] [Google Scholar]
- 61.Atanackovic D, Altorki NK, Cao Y, Ritter E, Ferrara CA, Ritter G,Hoffman EW, Bokemeyer C, Old LJ, and Gnjatic S. 2008. Booster vacci-nation of cancer patients with MAGE-A3 protein reveals long-term immuno-logical memory or tolerance depending on priming. Proc. Natl. Acad. Sci. USA 105: 1650–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Overwijk WW, Theoret MR, Finkelstein SE, Surman DR, de Jong LA,Vyth-Dreese FA, Dellemijn TA, Antony PA, Spiess PJ, Palmer DC, et al. 2003. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J. Exp. Med 198: 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]