Abstract
Accurate chromosome segregation requires the proper assembly of kinetochore proteins. A key step in this process is the recognition of the histone H3 variant CENP-A in the centromeric nucleosome by the kinetochore protein CENP-N. We report cryo–electron microscopy (cryo-EM), biophysical, biochemical, and cell biological studies of the interaction between the CENP-A nucleosome and CENP-N. We show that human CENP-N confers binding specificity through interactions with the L1 loop of CENP-A, stabilized by electrostatic interactions with the nucleosomal DNA. Mutational analyses demonstrate analogous interactions in Xenopus, which are further supported by residue-swapping experiments involving the L1 loop of CENP-A. Our results are consistent with the coevolution of CENP-N and CENP-A and establish the structural basis for recognition of the CENP-A nucleosome to enable kinetochore assembly and centromeric chromatin organization.
The correct transfer of genetic material from mother to daughter cells require the accurate segregation of chromosomes during mitosis. Mis-segregation of chromosomes can lead to aneuploidy, a hallmark of cancer (1). Chromosome segregation requires the assembly of kinetochore proteins at the centro-mere, which is epigenetically marked by nucleosomes containing the histone H3 variant CENP-A (2). The centromeric nucleosome associates with a group of 16 inner kinetochore proteins, collectively known as the constitutive centromere-associated network (CCAN) (3). Among the CCAN proteins, CENP-C and CENP-N can directly recognize the CENP-A nucleosome core region and are essential for kinetochore assembly and chromosome segregation (4). Two regions of CENP-C, the central region and the CENP-C motif, bind the CENP-A nucleosome by interacting with the C-terminal tail of CENP-A and the acidic patch on histones H2A and H2B in the centromeric nucleosome (5, 6). The CENP-N N-terminal region (residues 1 to 286 in humans) is responsible for recognition of the CENP-A nucleosome (7, 8), whereas its C-terminal region (residues 287 to 339) interacts with the CCAN through CENP-L (9, 10). The recruitment of CENP-N to the centromere requires recognition of the L1 loop of CENP-A (11).
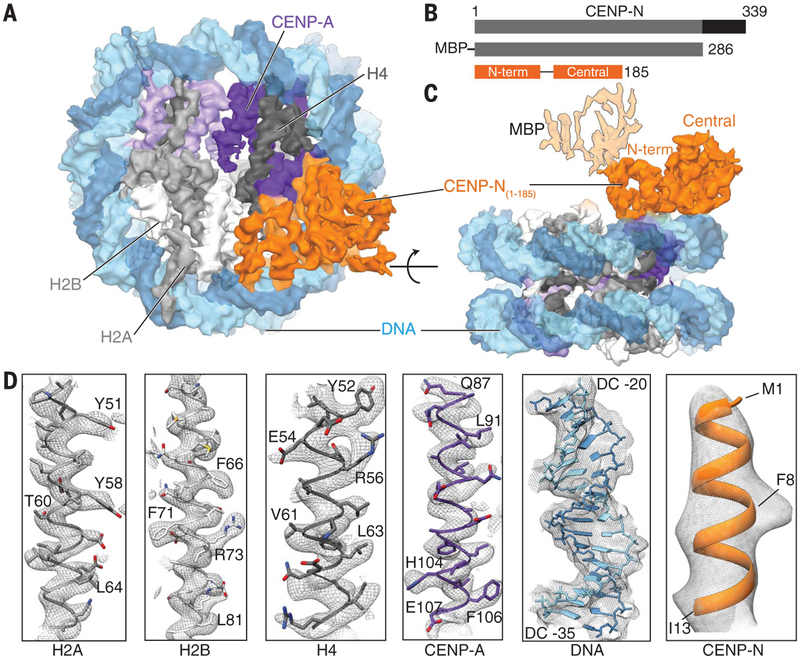
To determine structural aspects of the binding between CENP-A and CENP-N, we carried out cryo–electron microscopy (cryo-EM) analyses of the complexes formed between residues 1 to 286 of human CENP-N [tagged at its N terminus with His8 and maltose binding protein (MBP); hereafter referred to as hCENP-N1–286] and the CENP-A nucleosome containing the “601” DNA sequence (figs. S1 and S2). CENP-N binding to the CENP-A nucleosome is independent of DNA sequence (8), and we observed that CENP-A nucleosomes containing either “601” DNA or human centromeric α-satellite DNA bound hCENP-N1–286 with similar affinity and stoichiometry (fig. S3). When excess hCENP-N1–286 was used, CENP-N/CENP-A nucleosome complexes with 1:1 or 2:1 stoichiometry were observed in the cryo-EM two-dimensional (2D) class averages (fig. S4 and table S1, data set 1). The complex with 2:1 stoichiometry is a smaller fraction of the overall population, and the density observed for the second site is weaker than at the first site. In the absence of excess hCENP-N1–286, most of the 2D class averages displayed only one molecule of hCENP-N1–286 bound to the CENP-A nucleosome (fig. S5C). We refined this population to obtain a 3D reconstruction at an overall resolution of 3.9 Å for the complex formed between hCENP-N1–286 and the CENP-A nucleosome (Fig. 1; figs. S5 and S6; and table S1, data set 2).
Fig. 1. Structure of the human CENP-N/CENP-A nucleosome complex.
(A) Cryo-EM density map of the hCENP-N1–286/CENP-A nucleosome complex viewed down theaxis of the DNA supercoil. (B) Schematicof the functional domains of CENP-N known to bind the CENP-A nucleosome (gray) and CENP-L (black) (top panel). The CENP-N construct used for the present structural analysis (hCENP-N1–286) and the regions of the sequence whose structure we report here [N-terminal domain: residues 1 to 81, and central domain: residues 101 to 185; hCENP-N(1–185)] are shown in the middle and bottom panels, respectively. (C) Cryo-EM density mapof the hCENP-N1–286/CENP-A nucleosome complex as viewed from the side, at an orientation 90° to the view shownin (A). This view also depicts the extra density connected to the N-terminal domain that we assign to MBP, shown with lighter shading. (D) Representative regions of the cryo-EM density mapto illustrate map quality (from left to right) for canonical histones H2A, H2B, and H4, centromere-specific H3 variant CENP-A, nucleosomal DNA, and CENP-N.
The density map shows an identifiable nucleo-some and a bi-lobed assembly corresponding to hCENP-N1–286 (Fig. 1, A to C). The resolution of this map is highest in the core of the CENP-A nucleosome and lower in the periphery and in the bound CENP-N (Fig. 1D and fig. S6). The resolution (~3.5 Å) of the CENP-A nucleosome core is adequate for direct interpretation in terms of an atomic model, as side-chain densities are clearly visualized (Fig. 1D and fig. S7). The final structure of the CENP-A nucleosome core is similar to the crystal structure of the free CENP-A nucleosome [Protein Data Bank (PDB) ID 3AN2] (12). Although the DNA near the entry and exit sites can be visualized (fig. S8) and is displaced farther from the core histones in comparison with the canonical nucleosome structure (PDB 3LZ0) (13), the importance of these minor differences with previously reported structures is currently unclear.
The map of the CENP-N region is at a lower resolution (~4.5 Å) (fig. S6, C to F). At this resolution, densities for several α-helical regions and β strands are well resolved, but because side-chain densities are not delineated, it is not possible to thread the sequence unambiguously into the density map. To identify CENP-N residues that may form the interface with the CENP-A nucleosome, we carried out an extensive, iterative process to predict and fit various stretches of secondary structure elements and subdomains into the density map (see materials and methods). These efforts resulted in a predicted molecular model with a sequence assignment based on the modeled subdomains (figs. S9 and S10 and table S1). From this model, we identified residues likely to form the interface with the CENP-A nucleosome.
In our model, residues 1 to 185 of hCENP-N are organized into distinct N-terminal and central domains (Fig. 1 and figs. S9 and S10). On the basis of binding studies, the MBP moiety, which is attached to the N terminus of hCENP-N1–286 through a dipeptide (alanine-alanine) linker, does not interfere with the hCENP-N/CENP-A nucleo-some interaction (fig. S3, C to G). The apparent dissociation constant (KD) values for MBP-tagged and untagged hCENP-N1–286 are 0.042 ± 0.007 μM and 0.046 ± 0.008 μM, respectively. This is consistent with our assignment of the location of MBP to the weak density on the side of hCENPN1–286 that is distal from the core (Fig. 1C and figs. S5E and S6A). The density marked “unaccounted” in fig. S9E likely reflects an ordered polypeptide segment beyond residue 185 (fig. S9, A and B). The N-terminal hCENP-N domain, comprising residues 1 to 81, is composed of five antiparallel α helices that show structural conservation to proteins of the death domain super-family, whereas the central domain, comprising residues 101 to 185, contains both α helices and β strands with similarity to regions of the ap pendage domain of β2 adaptin (fig. S9, F to H) (14, 15).
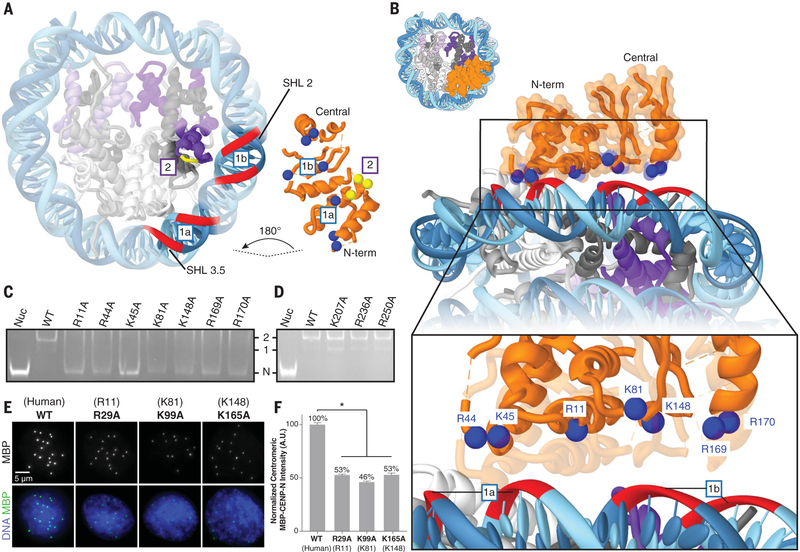
The structure of the complex shows that hCENP-N1–286 has direct interactions with the DNA of the nucleosome, as well as with the L1 loop of CENP-A (Fig. 2A and fig. S11, A and B). We identified several positively charged residues of hCENP-N proximal to nucleosomal DNA that likely form stabilizing interactions with the backbone phosphates of DNA (Fig. 2B). Consistent with our structural model, mutations of hCENP-N residues R11, K15, and K45 have already been shown to reduce this protein’s ability to bind the CENP-A nucleosome, and further, mutation of R11 disrupts centromeric localization and the recruitment of other CCAN proteins in human cells (8). To further verify the role of these and other residues identified at the interface, we constructed hCENP-N1–286 mutants in the N-terminal domain [R11→A11 (R11A), R44A, K45A, and K81A] and in the central domain (K148A, R169A, and R170A) and analyzed the binding by electro-phoretic mobility shift analysis; these mutations impair hCENP-N1–286 binding to the CENP-A nucleosome (Fig. 2C). Mutation of residues H77, H79, or K90, which are located on the linker connecting the N-terminal and central domains, also resulted in various extents of decreased binding affinity (fig. S12A), likely due to perturbations of CENP-N tertiary structure. As controls, we also tested the effects of mutating several positively charged residues predicted to be distal from the nucleosomal DNA (R60A, K109A/K110A, and K143A) or residues beyond the central domain (K207A, R236A, and R250A). All of these mutations had minimal effects on hCENP-N1–286 binding affinity (Fig. 2D and fig. S12A) (7).
Fig. 2. Interaction of CENP-N with nucleosomal DNA.
(A) Cut-away view of the hCENP-N1–286/CENP-A nucleosome model to highlight interfaces involved in complex formation (see also fig. S11, A and B). For the CENP-N/DNA interface (labeled “1a” and “1b”), nucleosomal DNAis shown as a red ribbon, whereas positively charged residues of CENP-N that are proposed to interact with it are shown as blue spheres. Forthe CENP-N/CENP-A interface (labeled “2”), CENP-A residues (R80, G81, and V82) are marked by the short yellow ribbon, whereas interacting CENP-N residues (E3, T4, and E7) are shown as yellow spheres. (B) View of the CENP-N/DNA interface at different magnifications to highlight details of interactions between the nucleosomal DNA and positively charged residues of CENP-N. (C) Gel mobility shift experiment to examine the effects of CENP-N mutations (indicated atop the gel) on binding to the CENP-A nucleosome. Impaired binding is reflected by increased intensity of the free nucleosome (Nuc) band, concomitant with the disappearance of defined 1:1 and 2:1 bands. “N” indicates the migration position of the free CENP-A nucleosome; “1” and “2” denote the migration positions of CENP-A nucleosomes bound with either one or two molecules of CENP-N, respectively. WT, wild type. (D) Similar analysis to that in (C), carried out with a set of CENP-N mutations involving residues distal from the binding interface. (E) Images of interphase nuclei in Xenopus egg extracts with exogenous MBP-xCENP-N and xCENP-L proteins containing the indicated mutations (with analogous human mutations in parentheses), stained with an antibody forMBP (green) and Hoechst (blue). (F) Centromeric MBP fluorescence intensity normalized as a percentage of that observed for wild-type MBP-xCENP-N. Error bars represent SEM (n > 200 centromeres). A.U., arbitrary units.
To examine the physiological importance of CENP-N interactions with the nucleosomal DNA, we mutated conserved and analogous residues within Xenopus laevis CENP-N (xCENP-N) (fig. S13A) and tested their effects on the centromeric localization of the exogenously expressed MBP-xCENP-N/CENP-L complex in interphase Xenopus egg extracts (fig. S14). Mutation of xCENP-N residues R29, K99, and K165 (which correspond to residues R11, K81, and K148 in hCENP-N) led to a significant reduction in the localization of the xCENP-N/L complex to centromeres (Fig. 2, E and F). Mutation of K108 (K90 in hCENP-N) in the linker region caused minor defects in localization (fig. S14, E and F). Furthermore, mutation of K223 (K207 in hCENP-N), which is beyond the central domain, did not diminish centromeric localization (fig. S14, E and F). These results are in agreement with our findings on hCENP-N (Fig. 2 and fig. S14I) and suggest that the interactions with nucleosomal DNA identified in the structure of the hCENP-N1–286/CENP-A nucleosome complex are conserved across Xenopus and human centromeres.
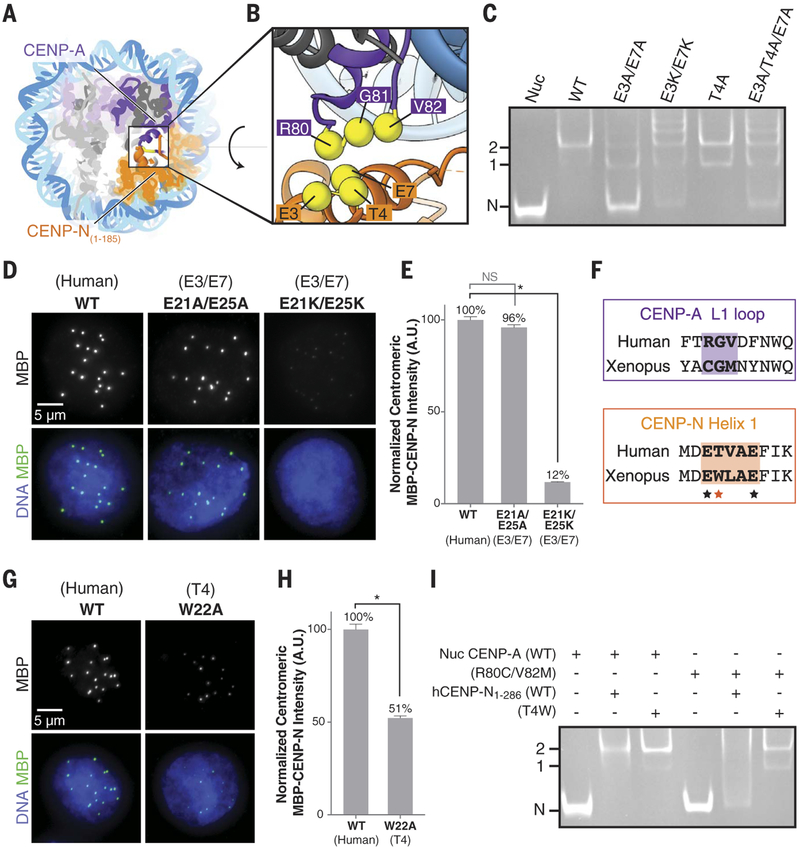
In our structural model, the N-terminal helix of hCENP-N1–286 is in contact with the L1 loop of CENP-A, in which residues R80 and G81 constitute an insertion compared to canonical H3.1 (fig. S11B) (12). Previous studies have shown that hCENP-N fails to localize to the centromere upon a double mutation of R80A and G81A in CENP-A (11), and substitution of the CENP-A targeting domain (CATD), including the L1 loop and α2 helix, to the corresponding region in H3 (H3CATD) is sufficient for recruitment of CENP-N (8, 16). We further corroborated these findings by in vitro mutational analysis (fig. S12B) and cryo-EM structure determination of the hCENP-N CATD 1–286/H3.1 chimeric nucleosome complex (fig. S15 and table S1), which revealed that hCENP-N1–286 binding to the chimeric nucleosome is structurally similar to its binding to CENP-A containing nucleo-some (fig. S15E). In our structural model of the hCENP-N1–286/CENP-A nucleosome complex, conserved residues E3 and E7 from helix 1 of hCENP-N are proximal to R80 of CENP-A, and residue T4 of hCENP-N is close to V82 of CENP-A (Fig. 3, A and B, and fig. S11). The structure thus suggests that both electrostatic and hydrophobic interactions are involved at the interface. Gel shift assays indicate that the E3A/E7A and T4A mutants of hCENP-N1–286 bind to the CENP-A nucleosome with reduced affinity, whereas the E3K, E7K, E3K/E7K, and E3A/T4A/E7A mutants show aberrant binding (Fig. 3C and fig. S12C). Further, the E3K/E7K and E3A/T4A/E7A mutants exhibit loss of specificity for CENP-A over H3.1 nucleosomes (fig. S12, D and E). Together, these results are consistent with a model in which a small number of residues of hCENP-N that interact with the L1 loop of CENP-A provide interaction specificity, whereas broad electrostatic interactions with the backbone phosphates of DNA enhance binding affinity.
Fig. 3. Interaction between theL1 loop of CENP-A and helix 1 of CENP-N.
(A and B) Overall (A) and close-up (B) view of the hCENP-N1–286/CENP-A interface formed betweenR80, G81, and V82 on the L1 loop of CENP-A and E3, T4, and E7 on helix 1of CENP-N. (C) Gel mobility shift experiment to examine the effects of CENP-N mutations (indicated atopthe gel) on binding to the CENP-A nucleosome. (D) Images of interphase nuclei in Xenopus egg extracts with exogenous MBP-xCENP-L andxCENP-L proteins containing the indicated mutations of xCENP-N residues E21 and E25 (correspondingto residues E3 and E7 in hCENP-N), stained with an antibody forMBP (green) and Hoechst (blue).(E) Centromeric MBP fluorescence intensity normalized as a percentageof that observed for wild-type MBP-xCENP-N. Error bars representSEM (n > 200 centromeres). (F) Alignment of human and Xenopus laevis sequences corresponding to the L1 loop of CENP-A and helix 1 of CENP-N. Closely interacting segments of the L1 loop of CENP-A and helix 1 of CENP-N are highlighted bythe shaded areas. The asterisks indicate conserved glutamic acid residues(black asterisks) and variability in the hydrophobic residue correspondingto position T4 (red asterisk) of human CENP-N. (G) Images of interphase nuclei in Xenopus egg extracts with exogenous MBP-xCENP-N and xCENP-L proteins containing the indicated mutations of xCENP-N, as in (D).(H) Centromeric MBP fluorescence intensity, determined as in (E). (I) Gel mobility shift experiment to examine the effects of correlated amino acid substitutions between the L1 loop of CENP-A and helix 1 of CENP-N.
To further evaluate the physiological importance of CENP-N interactions with the L1 loop of CENP-A, we examined the effects of the xCENP-N double mutants E21A/E25A and E21K/E25K (which correspond to mutations E3A/E7A and E3K/E7K in hCENP-N) on centromeric localization in Xenopus egg extracts (Fig. 3, D and E, and fig. S13A). Complete loss of centromeric localization was observed in the E21K/E25K mutant but not in the E21A/E25A mutant (Fig. 3, D and E, and fig. S14C). Furthermore, the W22A mu tation in xCENP-N (corresponding to mutation of T4 in hCENP-N) severely disrupted centromeric localization (Fig. 3, F to H, and fig. S14D). Thus, the localization of xCENP-N appears to depend more on hydrophobic rather than electrostatic interactions as compared with that of hCENP-N, where the E3A/E7A mutation leads to a total loss of centromeric localization in human cells (fig. S14, G to I). Our results support a model in which residues within helix 1 of hCENP-N (E3, T4, and E7) are critical for the recognition of the L1 loop within the CENP-A nucleosome.
CENP-A and centromeric DNA sequences show evidence for rapid evolution (17, 18), and alignments of the CENP-N N-terminal region and the CENP-A L1 loop across eukaryotes show a high degree of variability (fig. S13, B to D). To test whether CENP-N may have coevolved with CENP-A to maintain kinetochore assembly, we created a chimeric CENP-A nucleosome in which R80 and V82 of hCENP-A were respectively replaced by Cys and Met to mimic the Xenopus CENP-A L1 loop sequence (Fig. 3F). Wild-type hCENP-N does not interact with the chimeric nucleosome (Fig. 3I), in agreement with decreased centromeric localization of a W22T xCENP-N mutant (fig. S14, E and F). However, chimeric nucleosome binding could be enabled by a complementary hCENP-N T4W mutant, which also retained its affinity for wild-type hCENP-A nucleosomes (Fig. 3I). Together, these results suggest that CENP-N has likely coevolved with CENP-A to enable kinetochore assembly using hydrophobic and electrostatic interactions to recognize centromeric chromatin across different species.
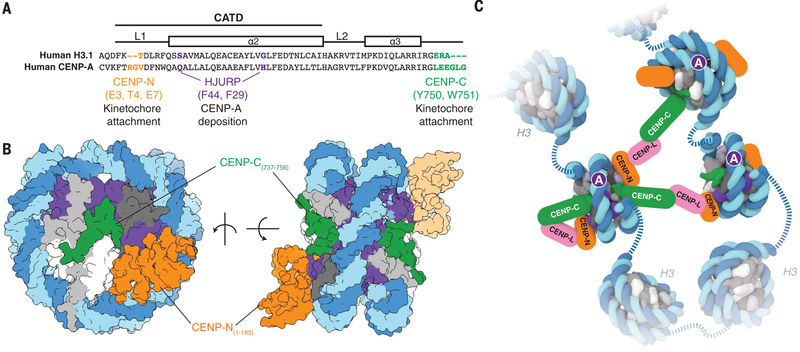
Centromere identity and function are specified by a two-step mechanism wherein the deposition of CENP-A at the centromere by the CENP-A chaperone HJURP allows for the recognition of CENP-A by the kinetochore proteins CENP-C and CENP-N (19). Structures of HJURP (20–22), CENP-C fragments (5), and CENP-N (this study) in complex with different forms of CENP-A show that, in all three cases, only a small number of residues are involved in the specific recognition of CENP-A, whereas additional interactions are used for further stabilization (Fig. 4A, and fig. S11, C and D). These residues on CENP-A have both conserved and necessary functions, despite the variability of their amino acid sequence across eukaryotes. Our data suggest that the CENP-A–interacting residues of CENP-N coevolve with CENP-A, which could reflect a common strategy for maintaining CENP-A specificity (17). Previous studies (7, 23) and our results (Fig. 4B and fig. S2, A and D) show that CENP-N and fragments of CENP-C that include one of its two CENP-A nucleosome binding domains [fig. S2E, central domain (7, 23), or CENP-C motif (this work)] can simultaneously bind to the same face of the CENP-A nucleosome. Furthermore, the kinetochore protein CENP-L has been shown to mediate interactions between CENP-C and CENP-N (9, 23–25). Thus, the valency of CENP-C/CENP-N/CENP-L interactions could facilitate clustering of sparse and nonadjacent CENP-A nucleosomes (Fig. 4C) (9, 26), which might help to establish the folding of centromeric chromatin (27, 28) and/or the integrity of the kinetochore (29, 30).
Fig. 4. Structural determinants of kinetochore assembly on the CENP-A nucleosome.
(A) Sequence alignment between humanH3.1 and CENP-A to highlight distinct CENP-A motifs involvedin deposition and recognition of CENP-A at centromeric chromatin (see also fig. S11, C and D). Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. (B) Two different views of the CENP-A nucleosome bound to hCENP-N and a modeled CENP-C motif peptide (5) to highlight potential dual binding of full-length CENP-C and CENP-N proteins on the CENP-A nucleosome. The second CENP-N (shown with lighter shading) is modeled on the basis of the cryo-EM density map obtained in the presence of excess hCENP-N1–286 (fig. S4), whereas the CENP-C motif peptides (human numbering shown for clarity) on each face of the nucleosome are positioned according to the crystal structure of the nucleosome in complex with the rat CENP-C motif (5). (C) Schematic view to highlight recognition and possible enrichment of CENP-A nucleosomes by the CCAN proteins CENP-C, CENP-N, and CENP-L. Other kinetochore proteins and the dimerization of CENP-C have been omitted for clarity.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the intramural programs of the NCI and NIDDK of the NIH. We thank T. Fox and U. Baxa atthe Center for Molecular Microscopy for help with cryo-EM data collection, A. Bartesaghi for advice with cryo-EM analysis,L. Jenkins for mass spectrometry, S. Li for DNA plasmid purifications, and V. Falconieri for assistance with figure preparation. This study utilized the computational resources of the High-Performance Computing Biowulf cluster at the NIH (http://hpc.nih.gov). The cryo-EM density map and atomic coordinates for the hCENP-N1–286/CENP-A nucleosome complex have been deposited in the Electron Microscopy Data Bank and the PDB under accession codes EMD-7293 and 6BUZ, respectively. S.C., J.H., A.E.K., Y.B., and S.S. were involved in all stages of experiment design and interpretation of results; J.H. prepared nucleosome complex samples and performed biochemical analysis; H.F. and J.H. performed nuclearmagnetic resonance experiments; R.G. and J.H. performed analytical ultracentrifugation experiments; H.S. andA.E.K. performed in vivo experiments; S.C. carried outcryo-EM grid preparation, data collection, and image processing; S.C. built the structural model, with input fromJ.H.; S.C., Y.B., A.E.K., and J.H. carried out structural analysis; and S.C., J.H., A.E.K., Y.B., and S.S. integrated all of thedata and wrote the manuscript, with help from all authors. We declare no competing financial interests.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/359/6373/339/suppl/DC1
Materials and Methods
Figs. S1 to S15
Table S1
References (31–55)
REFERENCES AND NOTES
- 1.Kops GJ, Weaver BA, Cleveland DW, Nat. Rev. Cancer 5, 773–785 (2005). [DOI] [PubMed] [Google Scholar]
- 2.McKinley KL, Cheeseman IM, Nat. Rev. Mol. Cell Biol. 17, 16–29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdaasdonk JS, Bloom K, Nat. Rev. Mol. Cell Biol. 12, 320–332 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll CW, Milks KJ, Straight AF, Cell Biol J. 189, 1143–1155 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato H et al. , Science 340, 1110–1113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF, Nature 477, 354–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo LY et al. , Nat. Commun 8, 15775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll CW, Silva MC, Godek KM, Jansen LE,Straight AF, Nat. Cell Biol. 11, 896–902 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKinley KL et al. , Mol. Cell 60, 886–898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinshaw SM, Harrison SC, Cell Reports 5, 29–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang J et al. , Genes Dev. 29, 1058–1073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tachiwana H et al. , Nature 476, 232–235 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Vasudevan D, Chua EY, Davey CA, J. Mol. Biol 403, 1–10 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Sukits SF et al. , J. Mol. Biol 310, 895–906 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, Albrecht TR, Baillat D, Wagner EJ, Tong L, Proc. Natl. Acad. Sci. U.S.A. 114, 4394–4399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black BE et al. , Nature 430, 578–582 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Malik HS, Henikoff S, Genetics 157, 1293–1298 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henikoff S, Trends Genet. 17, 689–690 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Fachinetti D et al. , Nat. Cell Biol. 15, 1056–1066 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z et al. , Nature 472, 234–237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu H et al. , Genes Dev. 25, 901–906 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho US, Harrison SC, Proc. Natl. Acad. Sci. U.S.A. 108, 9367–9371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weir JR et al. , Nature 537, 249–253 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Nagpal H et al. , Mol. Biol. Cell 26, 3768–3776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klare K et al. , J. Cell Biol. 210, 11–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodor DL et al. , eLife 3, e02137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blower MD, Sullivan BA, Karpen GH, Dev. Cell 2, 319–330 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro SA et al. , Proc. Natl. Acad. Sci. U.S.A. 107, 10484–10489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki A, Badger BL, Wan X, DeLuca JG, Salmon ED, Dev. Cell 30, 717–730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vargiu G et al. , Proc. Natl. Acad. Sci. U.S.A. 114, 3133–3138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.