Abstract
Background
A multidrug-resistant (MDR) RR2 gene cluster was identified by whole-genome sequencing in several highly virulent (ST-17) Group B streptococcus (GBS) isolates, which caused neonatal invasive infections in southern China in 2016. Tracing the transmission and distribution of MDR isolates in this area is important for the effective management of future infections. The aim of this study was to obtain longitudinal data of MDR isolates to monitor epidemiological trends of general common isolates in southern China, and provide evidence for future characterization of antimicrobial resistance mechanisms.
Methods
Clinical information and antimicrobial susceptibility of GBS isolates were acquired from electronic information management system databases of the hospital under study between January 2011 and December 2017. To confirm the presence of intact RR2, the tetO, ant6, lnuB, and ant9 genes located upstream, midstream, and downstream of RR2 were detected by PCR and DNA sequencing.
Results
A total of 149 cases of neonatal invasive GBS infection were identified during the period 2011–2017. Among them, 119 cases (79.9%) were caused by MDR isolates, with a general increasing trend over the past 7 years. Further characterization of 11 isolates showed that six isolates causing late-onset disease (LOD) carry the tetO, ant6, and lnuB genes, which are located on RR2. Moreover, lnuB and ant9 consistently co-occurred in GBS isolates, which suggests their close proximity to one another in the RR2 gene cluster.
Conclusion
The MDR GBS is responsible for a large number of neonatal invasive infections and occurs with increasing frequency over time. Particularly, the MDR GBS isolates that cause LOD are more likely to carry the RR2 gene cluster, compared with those that cause early-onset disease. The rise in number of MDR GBS isolates emphasizes the pressing need for continuous surveillance to monitor their antibiotic susceptibility and epidemiology.
Keywords: invasive infection, gene cluster, neonates, multidrug-resistance
Introduction
Group B streptococcus (GBS or Streptococcus agalactiae) is a leading cause of neonatal morbidity and mortality. It is commonly found in the genital and gastrointestinal tracts of women. Approximately 10%–30% of pregnant women are colonized with GBS worldwide,1 and neonates can be affected by vertical transmission of the bacterium through contaminated amniotic or vaginal secretions of a colonized mother before or during delivery.2–4 An overall incidence of 0.55 per 1,000 live births in the urban areas of southern China was reported in our previous study.5 Following neonatal exposure to GBS during delivery, the bacteria can quickly spread into the bloodstream with rapid onset of illnesses diagnosed in hospitals, such as pneumonia, sepsis, or meningitis.6–8 The overall fatality ratio in such cases is 9.6% (95% CI: 7.5–11.8).9 Among the infected infants who survive meningitis, some are affected by permanent neurological sequelae due to the absence of specific clinical symptoms for a proper diagnosis, and therefore, receive delayed treatment.10–14 Overall, GBS infection affects both the neonate and the public health system.15,16
Neonatal GBS infections are of two types: early-onset disease (EOD) and late-onset disease (LOD). The EOD occurs during the first week of life (age 0–6 days) and accounts for 30% of all GBS disease in neonates.1 The LOD occurs in newborns and infants aged 7–89 days. The EOD presents with sepsis (80%–85%), meningitis (7%), or pneumonia (10%), whereas the LOD primarily presents with bacteremia (65%), meningitis (25%–30%), or other signs (9%).17 The recommended universal screening of pregnant women for GBS carriage and the use of intrapartum antibiotic prophylaxis during delivery have significantly reduced the incidence of EOD, but not the incidence of LOD.7
Two groups of antibiotics, penicillins and aminopenicillins, are recommended as first-line therapy against GBS infections; macrolides (erythromycin) and lincosamide (clindamycin) represent the second-line antibiotics that are usually prescribed for those with an allergy to beta-lactams.2,4 Within recent years, the emerging antimicrobial resistance in clinical settings has become a threat to public health worldwide.3,5,18 The GBS with reduced penicillin susceptibility (PRGBS) in isolated strains from clinical settings rose from 2.3% to 14.7% between 2005 and 2013 in Japan.18 In the USA, clindamycin-resistant GBS and erythromycin-resistant isolates increased from 10.5% to 15.0% and 15.8% to 32.8%, respectively.19 China is even more severely affected, as clindamycin and erythromycin resistance occurs at very high levels of 87.5% and 92.5%, respectively.20 In 2014, the situation worsened owing to the appearance of a vancomycin-resistant strain; vancomycin is the last-line drug for treatment against GBS infections.21
A multidrug-resistant (MDR) isolate refers to a strain that is resistant to three or more types of antimicrobial drugs simultaneously. Several infections caused by MDR GBS have been reported.18,22 Moreover, an MDR gene cluster, which is harbored in integrative and conjugative elements (ICEs), named RR2, has been identified in several MDR clinical isolates from southern China.23 The RR2 gene cluster carries several antibiotic resistance genes, including tetO for tetracyclines, lnuB for lincosamides, ant6, ant9, and aphA for aminoglycosides, and ermB for macrolides. Despite this marked increase in antibiotic resistance, the isolation rate of MDR GBS has not been widely investigated in the region of southern China, and the roles of RR2 in MDR GBS infection remain unclear.
In this study, a longitudinal survey of population-based surveillance of neonatal invasive GBS infection in southern China from 2011 to 2017 was undertaken to assess trends in the epidemiology of MDR isolates. Furthermore, 11 isolates were selected to probe for tetO, ant6, lnuB, and ant9 genes located upstream, midstream, and downstream of the RR2 gene cluster to investigate whether partial or intact RR2 is present in these isolates. This study may shed light on the trends of MDR GBS infection in southern China and may provide new directions for the study of the mechanisms of antimicrobial resistance in GBS.
Materials and methods
Bacterial strains
Routine laboratory reports of neonatal invasive GBS disease submitted by microbiology laboratories of Guangzhou Women and Children’s Medical Center were reviewed for the period 1 January 2011 to 31 December 2017. Laboratory surveillance is subject to internal quality control and external quality assessment. An invasive infection was defined as yielding positive GBS cultured from blood or non-sterile sites and clinical symptoms.24 Records were considered as the same episode if specimens were taken within 7 days and showed the same biological response; they were thus merged accordingly to form a single record. A portion of the cases and isolates from 2011 to 2014 described here have also been analyzed and reported in a previous study conducted by our group.5 These data have been included for analysis in this study to provide a clearer perspective of the trends in GBS infections. This study was approved by the Ethics Committee of Guangzhou Women and Children’s Medical Center.
Antimicrobial susceptibility testing
Identification of GBS is based on the following tests: Gram stain, catalase, automated bacterial identification system CAMP, hippurate hydrolysis, and a commercial automatic bacterial identification system (VITEK 2 COMPACT, bio-Mérieux, Marcy L’Étoile, France). An MDR isolate refers to an isolate, which is non-susceptible to aminoglycosides, macrolides/lincosamides, and/or tetracyclines.6,18,22 The test results of GBS isolates were collected from electronic databases of the hospital information management system. Antibacterial susceptibility testing of all 11 isolates in which resistance genes were detected was performed by disk diffusion, according to the Clinical and Laboratory Standards Institute methods and interpretation criteria for Streptococcus spp. β-Hemolytic Group.25
Molecular analysis by PCR-based assays
Detection of tetO, ant6, lnuB, and ant9 genes was achieved by PCR. The PCR was conducted in a final volume of 50 µL of 0.8× PCR buffer (50 mM KCl, 10 mM Tris-HCl [pH 9.0], 0.1% Triton X-100, 0.01% [wt/vol] stabilizer, 1.5 mM MgCl2) containing 300 µM deoxynucleoside triphosphates, 3.5 mM MgCl2, 2 U Taq polymerase (Takara, Kusatsu, Japan), and 10 ng of template DNA, extracted by Lysis Buffer for Microorganism for direct PCR (Takara); primer concentrations were all 0.5 µM. The PCR was conducted on a DNA thermal cycler (Bio-Rad, Hercules, CA, USA) with the following cycling conditions: an initial cycle of 3 minutes at 94°C (5 minutes for tetO); 30 cycles of 30 seconds of denaturation at 94°C, 30 seconds of annealing at 55°C, and 50 seconds of extension at 72°C (45 seconds for tetO); followed by 33 cycles of elongation at 72°C. The PCR products were analyzed by electrophoresis on a 2% agarose gel at 220 V for 45 minutes in 0.5× Tris-borate-EDTA buffer.26,27 Visualization and image acquisition were performed with gold view (SBS, Beijing, China) and ultra-violet transillumination. The results are shown in the accompanying figures. Primers used for the detection of genes were as follows: tetO_F: 5′-AACTTAGGCATTCTG-GCTCAC-3′ and tetO_R: 5′-TCCCACTGTTCCATATC-GTCA-3′; ant6_F: 5′-CCAATGATGCTGATTGTATCG-3′ and ant6_R: 5′-CTCTTCTATATCAGCGGCATA-3′; lnuB_F: 5′-CATGAAAGGGTGAAGAAATG-3′ and lnuB_R: 5′-ACCCAATACTGTGAATAACG-3′; ant9_F: 5′-GGGTTGGCTACTATTGGGATT-3′ and ant9_R: 5′-AACGTGGCTCTAGTTGATGG-3′. The PCR products were extracted and sequenced by BGI (Guangdong, China).
Statistical analysis
Statistical analysis was performed using the IBM SPSS 17.0 software. The chi-squared test was used for comparisons between two groups. Data from the drug susceptibility tests were analyzed by the WHONET 5.6 software, and graphs were created by the GraphPad Prism 5.0 software. A P-value≤0.05 was considered significant.
Results
Clinical data of specimens
To evaluate the trends in GBS infection in southern China, we integrated previously reported data5 with newly acquired data from the present study. From January 2001 to December 2017, 149 cases of neonatal invasive GBS infections were reported at Guangzhou Women and Children’s Medical Center. Among those cases, 119 (79.9%) were caused by MDR isolates, including 95 (79.8%) that were diagnosed as sepsis, and 24 (20.2%) as meningitis (Table 1). Of the 30 infections caused by non-MDR isolates, 25 (83.3%) had diagnoses of sepsis and 5 (16.7%) had diagnoses of meningitis. No significant differences were noted in the proportions of sepsis and meningitis cases between MDR and non-MDR isolates. Among the 119 MDR isolates, 34 (28.6%) were from EOD cases. The number of EOD and LOD cases caused by non-MDR isolates were 11 (36.7%) and 19 (63.3%), respectively. The proportions of MDR and non-MDR cases between EOD and LOD cases were not significantly different. The MDR and non-MDR isolates showed no differences in clinical characteristics of the neonatal disease by age (Table 1). However, most of the isolates from female cases were reported as MDR infections, whereas those from male cases were predominantly caused by non-MDR isolates (P=0.019).
Table 1.
Clinical characteristics of GBS isolates in neonatal invasive infections from 2011 to 2017
| Specimens | All cases (n=149) | MDR (n=119) | Non-MDR (n=30) | P-value | ||
|---|---|---|---|---|---|---|
| Cases | Age (days) | Mean ± SD | 20±19.3 | 20±19.6 | 17±18.1 | 0.543 |
| Median | 15 | 16 | 12 | – | ||
| Sex ratio (male/female) | 0.80 | 0.65 | 1.72 | 0.019 | ||
| Symptom | Sepsis only | 120 (80.5) | 95 (79.8) | 25 (83.3) | 0.665 | |
| Meningitis | 29 (19.5) | 24 (20.2) | 5 (16.7) | |||
| Diseases | EOD (0–6 days) | 45 (30.2) | 34 (28.6) | 11 (36.7) | 0.388 | |
| LOD (7–89 days) | 104 (69.8) | 85 (71.4) | 19 (63.3) | |||
Abbreviations: EOD, early-onset disease; GBS, Group B streptococcus; LOD, late-onset disease; MDR, multidrug-resistant.
Trends in the epidemiology of neonatal invasive GBS infection from 2011 to 2017
The overall proportion of MDR isolates among all GBS isolates under study from 2011 to 2017 was 79.9%. Between 2011 and 2013, the number of MDR cases rose from 7 to 11. From 2014 onward, the annual number of MDR cases was maintained at >20. The annual percentages of MDR isolates from 2011 to 2013 were 70.0%, 78.6%, and 73.3%, respectively, which were all less than the overall mean value. In contrast to the period 2011–2013, an increasing trend in the percentages of MDR isolates was observed from 2014 to 2017, which were all higher than the overall average. Overall, there was an increasing trend in the epidemiology of neonatal invasive GBS infections from 2011 to 2017 (Table 2).
Table 2.
MDR isolates in neonatal invasive infection from 2011 to 2017
| Years | MDR isolates (n) | All isolates (n) | Percentage (%) |
|---|---|---|---|
| 2011 | 7 | 10 | 70.0 |
| 2012 | 11 | 14 | 78.6 |
| 2013 | 11 | 15 | 73.3 |
| 2014 | 23 | 28 | 82.1 |
| 2015 | 24 | 30 | 80.0 |
| 2016 | 22 | 26 | 84.6 |
| 2017 | 21 | 26 | 80.8 |
| Total | 119 | 149 | 79.9 |
Abbreviations: MDR, multidrug-resistant.
MDR isolates partially or fully carried by the RR2 gene cluster
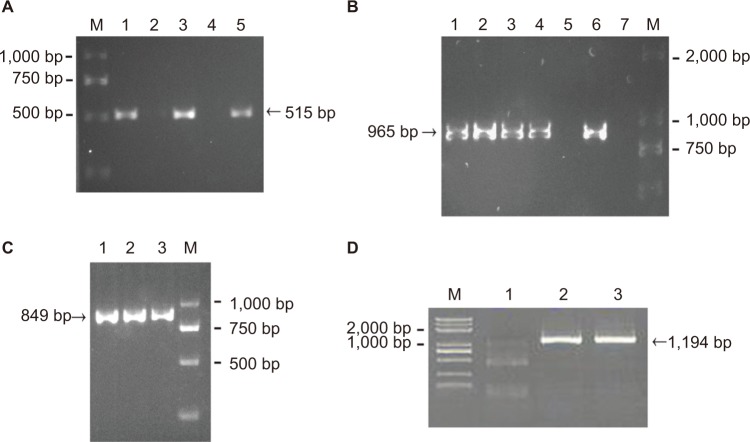
A previous study identified an RR2 gene cluster that bears multiple antibiotic resistant genes from five MDR strains isolated in southern China.23 Thus, it is logical to hypothesize that the rise in the number of MDR isolates may be due to the spread of the RR2 gene cluster among the GBS population. To verify this hypothesis, eleven isolates (three EOD isolates and eight LOD isolates, according to the ratio of MDR cases from EOD and LOD) were selected for PCR analysis. This analysis was designed to detect the presence of tetO, lnuB, and ant6 genes located upstream, midstream, and downstream, respectively, of the RR2 cluster, and which confer resistance to tetracycline, clindamycin, and streptomycin, respectively. The PCR products of tetO, ant6, and lnuB were detected as 515, 965, and 849 bp, respectively (Figure 1A–C). Basic Local Alignment Search Tool (BLAST) analysis revealed that tetO, ant6, and lnuB sequences showed 97%, 99%, and 99% nucleotide identity, respectively. The sequencing results are shown in Figures S1–S4. The genotypic and phenotypic details of the selected three EOD cases and eight LOD cases are listed in Table 3.
Figure 1.
Representative PCR results of tetO (A), ant6 (B), lnuB (C), and ant9 (D) genes.
Notes: M: DNA ladder. (A) tetO, lane 1: strains 1–1; lane 3: strains 1–2; lane 5: strains 1–3; lanes 2 and 4: no specimen. (B) ant6, lane 1: strains 2–3; lane 2: strains 2–4; lane 3: strains 2–5; lane 4: strains 2–6; lane 5: strains 2–7; lane 6: strains 2–8; lane 7: no specimen. (C) lnuB, lane 1: strains 2–1; lane 2: strains 2–2; lane 3: strains 2–3; (D) ant9, lane 1: strains 1–3; lane 2: strains 2–6; lane 3: strains 2–7.
Table 3.
Genotype and phenotype characteristics of the eleven GBS isolates from neonatal invasive infections
| Strain | Stage | Resistance gene | Resistance pattern | ||||
|---|---|---|---|---|---|---|---|
| tetO | ant6 | lnuB | Tetracycline | Streptomycin | Clindamycin | ||
| 1–1 | EOD | + | + | − | + | + | + |
| 1–2 | EOD | + | + | − | + | + | + |
| 1–3 | EOD | + | + | − | + | + | + |
| 2–1 | LOD | + | + | + | + | + | + |
| 2–2 | LOD | + | + | + | + | + | + |
| 2–3 | LOD | + | + | + | + | + | + |
| 2–4 | LOD | + | + | − | + | + | + |
| 2–5 | LOD | + | + | + | + | + | + |
| 2–6 | LOD | + | + | + | + | + | + |
| 2–7 | LOD | + | − | + | + | + | + |
| 2–8 | LOD | + | + | + | + | + | + |
Abbreviations: EOD, early-onset disease; GBS, Group B streptococcus; LOD, late-onset disease.
The PCR results showed that all three EOD cases carried tetO and ant6, but not lnuB, whereas six of the eight LOD cases carried tetO, ant6, and lnuB. One of the LOD cases had no ant6, and the other had no lnuB sequences.
The absence of lnuB and ant9
Therefore, it can be concluded that the lnuB gene played an important role in EOD and LOD. To further analyze the deletion of lnuB genes and determine the presence of the ant9 gene near to the lnuB gene, we selected three strains, the numbers of which were 1–3, 2–6, and 2–7, with genotypes tetO (+)/ant6 (+)/lnuB (−); tetO (+)/ant6 (+)/lnuB (+); and tetO (+)/ant6 (−)/lnuB (+), respectively. The PCR product of ant9 was 1,194 bp in length (Figure 1D), and BLAST analysis revealed that the ant9 sequences showed 100% nucleotide identity. The sequencing results are shown in the supplementary figures. As shown in Table 4, lnuB and ant9 consistently appeared together. This suggests that the relative positions of these two genes on RR2 are either close to each other or located in the same transmissible element.
Table 4.
Presence of resistance genes in three selected strains
| Strains | tetO | ant6 | lnuB | ant9 |
|---|---|---|---|---|
| 1–3 | + | + | − | − |
| 2–6 | + | + | + | + |
| 2–7 | + | − | + | + |
Discussion
As GBS has now become an issue of global public health concern, a number of cases of MDR GBS have been reported worldwide within recent years, especially in Asia.18 The epidemiological data of GBS in southern China have historically been limited. A previous study suggested that antimicrobial resistance of GBS in this area is worsening and more attention is needed.16 To oversee the trend of MDR GBS infections in this area, we combined the data from 2011 to 2014 with epidemiological data from 2015 to 2017. Thus, the present study provides a broad view of neonatal invasive GBS infections in southern China, and included 149 cases during the 7-year study period. The isolation rate of MDR strains from neonatal invasive GBS infections between January 2011 and December 2017 was 79.9% (119/149), with a significant increasing trend in resistance from 2014 to 2017.
The increasing rate at which MDR GBS has been isolated from neonates has been critical over the past few years. The latest report showed that all strains were sensitive to vancomycin, but not to penicillin, ceftriaxone, or cefotaxime, with susceptibility of 1.1%, 1%, and 1%, respectively, among 181 cases of bloodstream infection.28 In Japan, PRGBS has also been reported within the past 5 years.29–31 In Iran, 93.5% (n=58) of erythromycin-resistant GBS showed an increased minimum inhibitory concentration in response to penicillin.32 Surveillance in Taiwan showed that levofloxacin-resistant GBS isolates increased from 2.2% during 2002–2006 to 6.2% during 2008–2012.33 Furthermore, a new class of resistant GBS has been reported, of which MDR GBS with PI-1 (CC17/PI-2b) lost has shown resistance to macrolides, lincosamides, tetracycline, high-level streptomycin, and kanamycin.34 Chloramphenicol-resistant MDR GBS has also been reported, despite the rare occurrence of chloramphenicol resistance in GBS.22 The MDR GBS strains that are resistant to penicillin form relatively small, less hemolytic colonies.35 This may be a strategy of GBS to enhance its survival and spread in hospital settings, resulting in nosocomial infections, and leading to global spread and increased prevalence.36 These studies have demonstrated the emerging threat of MDR GBS isolates to global public health. It is, therefore, essential to examine the epidemiology of MDR GBS and characterize MDR mechanisms.
In this study, eleven isolates were selected to detect the tetO, lnuB, and ant6 genes located upstream, midstream, and downstream, respectively, of the RR2 gene cluster, which has been previously identified.23 Of these eleven isolates, six were from LOD cases and they all carried the tetO, ant6, and lnuB genes. None of the three EOD cases carried lnuB genes. Therefore, we speculated that MDR GBS isolates of EOD mainly carried fragments of the RR2 cluster rather than the complete set. Moreover, a considerable proportion of the LOD isolates most likely carried the full range of the RR2 cluster.
Furthermore, three strains (1–3 for EOD, and 2–6 and 2–7 for LOD) were selected to detect the ant9 gene close to the lnuB gene. The lnuB gene was observed to co-occur with the ant9 gene in strains causing LOD. In addition, sequences of the lnuB and ant9 genes were highly consistent with the partial sequence of ICE SGB76 (GenBank accession number KF772204) reported by Montilla (Figure 2).37 This suggests that the partial RR2 in strain 1–3 might have been deleted (Figure 3). Further whole-genome sequencing will be required to clarify gene structure and composition of RR2. The BLAST analysis revealed that this sequence (aadE, ant9, and lnuB) exhibited similarity to the lnu(B)-containing structures previously identified in Staphylococcus aureus (JQ861959 and JX560992).37 Thus, whether the lnuB and ant9 genes in RR2 are acquired from S. aureus and whether this acquisition leads to the emergence of MDR isolates require further investigation.
Figure 2.
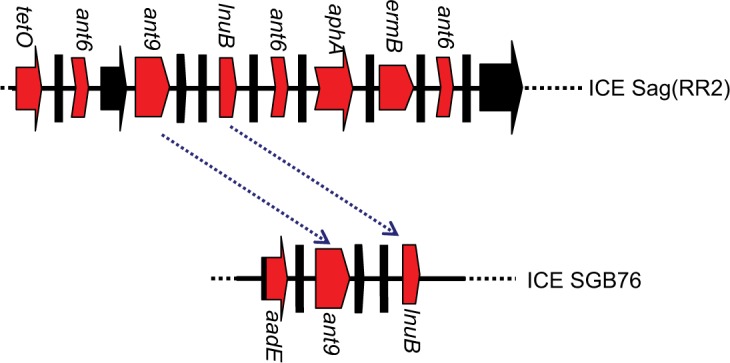
Relationship between RR2 and SGB 76.
Note: Red arrows represent the region carrying antibiotic resistance genes. Black arrows indicate genes duplicated in RR2. The region between blue dotted arrows indicates 99% of sequence identity. The diagram does not represent the actual size.
Figure 3.
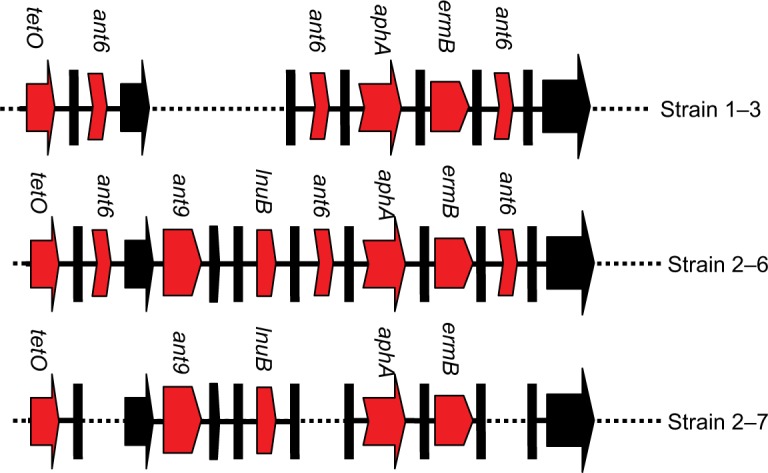
The diagram of proposed gene structures in three selected strains.
Note: Red arrows represent the region carrying antibiotic resistance genes. Black arrows indicate genes duplicated. The diagram does not represent the actual size.
Conclusion
Here, we reported on the epidemiological characteristics of neonatal invasive MDR GBS infections in southern China over a 7-year study period. We showed that the proportion of MDR isolates has been increasing over time and speculated that LOD isolates most likely carry the MDR gene cluster. This study provides perspective for further research on the stability of the MDR gene cluster and its role in LOD.
Supplementary materials
ant6 sequencing map.
ant9 sequencing map.
tetO sequencing map.
lnuB sequencing map.
Acknowledgments
We thank Wenjing Ji for her professional assistance in format and proofreading and David J McIver for his assistance in language editing in earnest. They offered and shared plenty of good experiences and opinions unselfishly as well.
This work was supported by grants from the Department of Science and Technology of Guangdong Province (2015A030401007) and Guangzhou Science Technology and Innovation Commission (201804010447), and CYC thanks the support from the Research Development Fund from University of Bradford.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Morgan JA, Cooper DB. Pregnancy, Group B Streptococcus. StatPearls. edn. Treasure Island FL: StatPearls Publishing LLC; 2018. [Google Scholar]
- 2.Wilson CB, Nizet V, Maldonado YA. Remington and Klein’s infectious diseases of the fetus and newborn infant. Seminars in Fetal & Neonatal Medicine. 2015;20(6):442. [Google Scholar]
- 3.Verani JR, McGee L, Schrag SJ, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention (CDC) Prevention of perinatal group B streptococcal disease – revised guidelines from CDC, 2010. MMWR Recomm Rep. 2010;59(RR-10):1–36. [PubMed] [Google Scholar]
- 4.Madhi SA, Cutland CL, Jose L, et al. Safety and immunogenicity of an investigational maternal trivalent group B streptococcus vaccine in healthy women and their infants: a randomised phase 1b/2 trial. Lancet Infect Dis. 2016;16(8):923–934. doi: 10.1016/S1473-3099(16)00152-3. [DOI] [PubMed] [Google Scholar]
- 5.Guan X, Mu X, Ji W, et al. Epidemiology of invasive group B streptococcal disease in infants from urban area of South China, 2011–2014. BMC Infect Dis. 2018;18(1):14. doi: 10.1186/s12879-017-2811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotten CM. Antibiotic stewardship: reassessment of guidelines for management of neonatal sepsis. Clin Perinatol. 2015;42(1):195–206. doi: 10.1016/j.clp.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, Huang J, Chen Z, Guo D, Yao Z, Ye X. Antibiotic prevention for maternal group B streptococcal colonization on neonatal GBS-related adverse outcomes: a meta-analysis. Front Microbiol. 2017;8:374. doi: 10.3389/fmicb.2017.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi M, Schrag SJ, Alderson MR, et al. WHO consultation on group B Streptococcus vaccine development: report from a meeting held on 27–28 April 2016. Vaccine. 2016 doi: 10.1016/j.vaccine.2016.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edmond KM, Kortsalioudaki C, Scott S, et al. Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet. 2012;379(9815):547–556. doi: 10.1016/S0140-6736(11)61651-6. [DOI] [PubMed] [Google Scholar]
- 10.Quan V, Verani JR, Cohen C, et al. Invasive group B streptococcal disease in South Africa: importance of surveillance methodology. PLoS One. 2016;11(4):e0152524. doi: 10.1371/journal.pone.0152524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie D, Rashid H, El-Bashir H, et al. Impact of meningitis on intelligence and development: a systematic review and meta-analysis. PLoS One. 2017;12(8):e0175024. doi: 10.1371/journal.pone.0175024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romain AS, Cohen R, Plainvert C, et al. Clinical and laboratory features of group B streptococcus meningitis in infants and newborns: study of 848 cases in France, 2001–2014. Clin Infect Dis. 2018;66(6):857–864. doi: 10.1093/cid/cix896. [DOI] [PubMed] [Google Scholar]
- 13.Arfi A, Cohen R, Varon E, Béchet S, Bonacorsi S, Levy C. Case-control study shows that neonatal pneumococcal meningitis cannot be distinguished from group B Streptococcus cases. Acta Paediatr. 2017;106(12):1915–1918. doi: 10.1111/apa.13919. [DOI] [PubMed] [Google Scholar]
- 14.Zhu M, Hu Q, Mai J, Lin Z. Analysis of pathogenic bacteria and drug resistance in neonatal purulent meningitis. Zhonghua Er Ke Za Zhi. 2015;53(1):51–56. [PubMed] [Google Scholar]
- 15.White A, Madhi SA. Ethical considerations for designing GBS maternal vaccine efficacy trials in low-middle income countries. Vaccine. 2015;33(47):6396–6400. doi: 10.1016/j.vaccine.2015.07.108. [DOI] [PubMed] [Google Scholar]
- 16.Ji W, Liu H, Jin Z, et al. Disease burden and antimicrobial resistance of invasive group B streptococcus among infants in China: a protocol for a national prospective observational study. BMC Infect Dis. 2017;17(1):377. doi: 10.1186/s12879-017-2475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song JY, Lim JH, Lim S, Yong Z, Seo HS. Progress toward a group B streptococcal vaccine. Hum Vaccin Immunother. 2018;41(85):1–13. doi: 10.1080/21645515.2018.1493326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki T, Kimura K, Reid ME, et al. High isolation rate of MDR group B streptococci with reduced penicillin susceptibility in Japan. J Antimicrob Chemother. 2015;70(10):2725–2728. doi: 10.1093/jac/dkv203. [DOI] [PubMed] [Google Scholar]
- 19.Castor ML, Whitney CG, Como-Sabetti K, et al. Antibiotic resistance patterns in invasive group B streptococcal isolates. Infect Dis Obstet Gynecol. 2008;2008:727505–5. doi: 10.1155/2008/727505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P, Ma Z, Tong J, et al. Serotype distribution, antimicrobial resistance, and molecular characterization of invasive group B Streptococcus isolates recovered from Chinese neonates. Int J Infect Dis. 2015;37:115–118. doi: 10.1016/j.ijid.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 21.Park C, Nichols M, Schrag SJ. Two cases of invasive vancomycin-resistant group B streptococcus infection. N Engl J Med. 2014;370(9):885–886. doi: 10.1056/NEJMc1308504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simoni S, Vincenzi C, Brenciani A, et al. Molecular characterization of Italian isolates of fluoroquinolone-resistant streptococcus agalactiae and relationships with chloramphenicol resistance. Microb Drug Resist. 2018;24(3):225–231. 225–231. doi: 10.1089/mdr.2017.0139. [DOI] [PubMed] [Google Scholar]
- 23.Campisi E, Rosini R, Ji W, et al. Genomic analysis reveals multi-drug resistance clusters in group B streptococcus CC17 hypervirulent isolates causing neonatal invasive disease in Southern Mainland China. Front Microbiol. 2016;7(1265):1265. doi: 10.3389/fmicb.2016.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie S, Lu X, Jin Z, et al. Characterization of group B Streptococcus isolated from sterile and non-sterile specimens in China. Diagn Microbiol Infect Dis. 2018;92(1):56–61. doi: 10.1016/j.diagmicrobio.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute M100-S24: Performance Standards for Antimicrobial Susceptibility Testing - Twenty-Fourth Informational Supplement Wayne: Clinical and Laboratory Standards Institute. 2014. [Accessed November 26, 2018]. Available from https://microbiolab-bg.com/wp-content/uploads/2015/05/CLSI-2014.pdf.
- 26.Malhotra-Kumar S, Lammens C, Piessens J, Goossens H. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob Agents Chemother. 2005;49(11):4798–4800. doi: 10.1128/AAC.49.11.4798-4800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou L, Yu SJ, Gao W, Yao KH, Shen AD, Yang YH. Serotype distribution and antibiotic resistance of 140 pneumococcal isolates from pediatric patients with upper respiratory infections in Beijing, 2010. Vaccine. 2011;29(44):7704–7710. doi: 10.1016/j.vaccine.2011.07.137. [DOI] [PubMed] [Google Scholar]
- 28.Hua CZ, Yu H, Zhuang JQ, et al. An analysis of 181 cases with blood stream infection caused by Streptococcus agalactiae in children from 2011 to 2015: a multi-center retrospective study. Zhonghua Er Ke Za Zhi. 2016;54(8):577–581. doi: 10.3760/cma.j.issn.0578-1310.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Yamada R, Kimura K, Nagano N, et al. Comparative analysis of penicillin-susceptible and non-susceptible isolates of group B streptococci by multilocus sequence typing. Jpn J Infect Dis. 2015;68(4):326–329. doi: 10.7883/yoken.JJID.2014.387. [DOI] [PubMed] [Google Scholar]
- 30.Kamiya C, Kimura K, Doyama Y, et al. Ceftibuten-containing agar plate for detecting group B streptococci with reduced penicillin susceptibility (PRGBS) Diagn Microbiol Infect Dis. 2015;82(4):269–273. doi: 10.1016/j.diagmicrobio.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Fukigai S, Morimoto M, Kimura K, et al. Effectual detection of group B streptococci with reduced penicillin susceptibility (PRGBS) by commercially available methicillin-resistant-Staphylococcus aureus (MRSA)-selective agar. Diagn Microbiol Infect Dis. 2016;85(3):309–312. doi: 10.1016/j.diagmicrobio.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 32.Mousavi SM, Nasaj M, Hosseini SM, Arabestani MR. Survey of strain distribution and antibiotic resistance pattern of group B streptococci (Streptococcus agalactiae) isolated from clinical specimens. GMS Hyg Infect Control. 11:Doc18. doi: 10.3205/dgkh000278. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu CJ, Lai JF, Huang IW, et al. Multiclonal emergence of levofloxacin-resistant group B Streptococcus, Taiwan. J Antimicrob Chemother. 2017;72(12):3263–3271. doi: 10.1093/jac/dkx297. [DOI] [PubMed] [Google Scholar]
- 34.Martins ER, Pedroso-Roussado C, Melo-Cristino J, Ramirez M, The Portuguese Group for the Study of Streptococcal Infections Streptococcus agalactiae causing neonatal infections in Portugal (2005–2015): diversification and emergence of a CC17/PI-2b multidrug resistant sublineage. Front Microbiol. 2017;8:499. doi: 10.3389/fmicb.2017.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banno H, Kimura K, Tanaka Y, et al. Analysis of multidrug resistant group B streptococci with reduced penicillin susceptibility forming small, less hemolytic colonies. PLoS One. 2017;12(8):e0183453. doi: 10.1371/journal.pone.0183453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagano N, Nagano Y, Toyama M, et al. Nosocomial spread of multidrug-resistant group B streptococci with reduced penicillin susceptibility belonging to clonal complex 1. J Antimicrob Chemother. 2012;67(4):849–856. doi: 10.1093/jac/dkr546. [DOI] [PubMed] [Google Scholar]
- 37.Montilla A, Zavala A, Cáceres Cáceres R, et al. Genetic environment of the lnu(B) gene in a Streptococcus agalactiae clinical isolate. Antimicrob Agents Chemother. 2014;58(9):5636–5637. doi: 10.1128/AAC.02630-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ant6 sequencing map.
ant9 sequencing map.
tetO sequencing map.
lnuB sequencing map.