Abstract
Genetic parameters are required to evaluate carcass merit using correlated real-time ultrasound (RTU) measurements. Many registered bulls and heifers are measured using RTU before consideration for selection as parents, whereas few animals are recorded for carcass traits and those are often crossbred steers. The objective of this study was to estimate genetic parameters required for evaluating carcass merit in the American Hereford Association (AHA) and the American Simmental Association (ASA) using multivariate models and to assess accuracy of carcass trait estimated breeding values (EBV) for selection candidates. All available carcass data including carcass weight (CWT), fat thickness (FAT), longissimus muscle area (LMA), and marbling score (MRB) were provided by the AHA and the ASA along with RTU data including fat thickness (UFAT), longissimus muscle area (ULMA), and percentage of intramuscular fat (UIMF). Carcass data comprised 6,054 AHA and 9,056 ASA cattle, while RTU data in comparable numbers from close relatives comprised 6,074 AHA and 7,753 ASA cattle. Pedigrees included 33,226 AHA and 37,665 ASA animals. Fixed effects for carcass and RTU data included contemporary group, age at scan/slaughter, and major breed percentages. Restricted maximum likelihood procedures were applied to all the carcass and RTU measurements, along with birth weight to account for selection, fitting 8-trait multivariate models separately for each breed association. Heritability estimates for AHA and ASA carcass traits were 0.41 ± 0.04 and 0.25 ± 0.03 for FAT, 0.47 ± 0.04 and 0.32 ± 0.03 for LMA, 0.48 ± 0.04 and 0.43 ± 0.04 for MRB, 0.51 ± 0.04 and 0.34 ± 0.03 for CWT, and for RTU traits were 0.29 ± 0.04 and 0.37 ± 0.03 for UFAT, 0.31 ± 0.04 and 0.44 ± 0.03 for ULMA, and 0.45 ± 0.04 and 0.42 ± 0.03 for UIMF. Genetic correlations for AHA and ASA analyses between FAT and UFAT were 0.74 ± 0.08 and 0.28 ± 0.13, between LMA and ULMA were 0.81 ± 0.07 and 0.57 ± 0.10, and between MRB and UIMF were 0.54 ± 0.08 and 0.73 ± 0.07. Predictions of carcass merit using RTU measurements in Hereford cattle would be more reliable for FAT and LMA than MRB, but the reverse would be true for admixed Simmental cattle. Genetic correlations for MRB in AHA and for FAT and LMA in ASA are less than currently assumed in their national evaluations. Collection of greater numbers of carcass measurements would improve the accuracy of genetic evaluations for carcass traits in both breeds.
Keywords: beef cattle, carcass, genetic parameters, Hereford, indirect selection, Simmental, ultrasound
INTRODUCTION
Variance parameters including heritabilities, genetic and residual correlations are required for national cattle evaluation. There are more than adequate amounts of data available for estimating such variance parameters for growth and real-time ultrasound (RTU) traits, but much less data are available in North America for carcass traits. Like many other breed associations, American Hereford Association (AHA) and American Simmental Association (ASA) have long been using RTU data in addition to carcass measurements to enhance national cattle evaluations for carcass traits. Evaluations combining ultrasound and carcass data will be more accurate than those based on carcass data alone, provided realistic variance parameters are used in the evaluations. Combining RTU and carcass data provides for estimation of expected progeny differences (EPD) using larger and more random samples than could be achieved using carcass data alone (Crews and Kemp, 2002). Most published reports of parameters have been based on bivariate rather than multivariate animal model analyses (Crews et al., 2003; Eriksson et al., 2004). The objective of this study was to separately estimate genetic parameters required for evaluating carcass merit in AHA and ASA using some RTU along with all available carcass data, fitting multivariate models with major breed percentages as fixed effects, and hence evaluate the accuracy of carcass traits of living individuals by assessing theoretical prediction accuracy of estimated breeding values (EBV), obtained from traditional best linear unbiased prediction (BLUP) analysis that uses various phenotypic information sources and the variance components estimated in this study.
MATERIALS AND METHODS
Data sets were constructed using all reliable carcass data from structured progeny test programs managed by AHA and ASA. A subset of the available RTU data and ancestral pedigrees augmented that carcass data to produce the data sets described below. Animal Care and Use Committee approval was not obtained for this study because the data were extracted from existing industry databases.
Carcass and Ultrasound Data
All available carcass data including carcass weight (CWT), back fat thickness (FAT), longissimus muscle area (LMA), and marbling score (MRB) were obtained from AHA and ASA. A small number of records that were missing 1 or 2 of these 4 traits was also included. Due to computing limitations, only close relatives such as half-siblings of animals with carcass records were extracted from the complete database, along with their contemporaries, if they had at least 2 of the 3 RTU records including ultrasound subcutaneous fat (UFAT), ultrasound longissimus muscle area (ULMA), and intramuscular fat percentage (IMF). RTU measurements on animals with carcass data were ignored as slaughtered animals are typically scanned a short time prior to harvest rather than using the same protocols as applied to the seed-stock animals. Extended pedigrees based on ancestors of animals with data were used to construct relationship matrices linking animals with ultrasound data to those with carcass data. This resulted in CWT, FAT, LMA, and MRB being available from 6,054 AHA and 9,056 ASA cattle, while UFAT, ULMA, and UIMF were available on 6,074 AHA and 7,753 ASA cattle (Table 1). Birth weight (BWT) records were available for all individuals with carcass or RTU measurements.
Table 1.
Summary statistics for carcass traits and their real-time ultrasound live indicators
| Data source1 | Carcass2 | Ultrasound3 | Weight4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Item | FAT (mm)5 | LMA (cm2) | MRB6 | UFAT (mm) | ULMA (cm2) | UIMF (%) | BWT (kg) | CWT (kg) | |
| AHA | Minimum | 1.78 | 53.16 | 0.61 | 1.02 | 25.94 | 0.25 | 22.07 | 200.99 |
| Mean | 14.55 | 81.81 | 4.09 | 5.46 | 61.55 | 3.38 | 38.99 | 362.24 | |
| Maximum | 35.31 | 131.03 | 8.54 | 20.32 | 106.77 | 8.72 | 59.60 | 495.78 | |
| SD | 4.60 | 8.90 | 1.00 | 2.49 | 14.32 | 0.96 | 4.42 | 40.89 | |
| No. of siresg | 423 | ||||||||
| No. of records | 6,054 | 6,074 | 12,128 | 6,054 | |||||
| No. of Contemporary Groups | 348 | 528 | 679 | 348 | |||||
| ASA | Minimum | 1.02 | 11.61 | 0.21 | 1.02 | 40.00 | 0.50 | 13.61 | 171.91 |
| Mean | 12.02 | 83.87 | 5.32 | 5.01 | 78.32 | 3.27 | 39.14 | 359.06 | |
| Maximum | 35.56 | 150.79 | 10.80 | 21.59 | 131.61 | 7.85 | 64.41 | 553.38 | |
| SD | 4.06 | 10.32 | 1.10 | 1.85 | 12.84 | 0.90 | 5.59 | 47.04 | |
| No. of sires7 | 1358 | ||||||||
| No. of records | 9,056 | 7,753 | 16,809 | 9,056 | |||||
| No. of Contemporary Groups | 819 | 324 | 741 | 819 | |||||
Data Source: AHA, American Hereford Association; ASA, American Simmental Association.
Carcass traits: FAT, back fat thickness; LMA, longissimus muscle area; MRB, marbling score.
Ultrasound traits: UFAT, real time ultrasound (RTU) fat thickness; ULMA, RTU longissimus muscle area; UIMF, RTU intramuscular fat percentage.
Weights: BWT, birth weight; CWT, carcass weight.
Unit conversions were undertaken from imperial to metric for all the traits except MRB and UIMF.
MRB for AHA was scored on the graders scale and divided by 100, whereas for ASA marbling was scored according to BIF guidelines.
Number of sires of individuals with phenotypic records.
Pedigree
Extended pedigree files were constructed for all the animals with carcass or RTU phenotypes for AHA data (33,226 animals in the pedigree) and ASA data (37,665 animals in the pedigree).
Analysis Models
Analyses on AHA and ASA data were done separately, both using 8-trait multivariate models. The multivariate model is represented in matrix notation as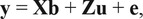 where X and Z are design matrices relating observations (y) to their respective fixed (b) and random (u) effects, and e is a vector of random residuals. Random effects were assumed to have null means, with Var(u) = G0 ⊗ A where G0 represents the genetic (co)variance matrix of order 8 being the number of traits analyzed and A represents the pedigree-based numerator relationship matrix (NRM) of order equal to the number of animals in the pedigree. The residuals have Var(e) = R, where R would be R0 ⊗ I if all animals had all traits observed, but when some traits are missing the corresponding rows and columns of R0 ⊗ I they are deleted to obtain R which has order equal to the number of observed phenotypes across the 8 traits, in which R0 represents the residual (co)variance matrix of order 8 and I is an identity matrix of order equal to the number of animals with at least one phenotypic observation. No animal with carcass data had its RTU data included in the analysis; therefore, residual covariances were not estimated between carcass and ultrasound traits.
where X and Z are design matrices relating observations (y) to their respective fixed (b) and random (u) effects, and e is a vector of random residuals. Random effects were assumed to have null means, with Var(u) = G0 ⊗ A where G0 represents the genetic (co)variance matrix of order 8 being the number of traits analyzed and A represents the pedigree-based numerator relationship matrix (NRM) of order equal to the number of animals in the pedigree. The residuals have Var(e) = R, where R would be R0 ⊗ I if all animals had all traits observed, but when some traits are missing the corresponding rows and columns of R0 ⊗ I they are deleted to obtain R which has order equal to the number of observed phenotypes across the 8 traits, in which R0 represents the residual (co)variance matrix of order 8 and I is an identity matrix of order equal to the number of animals with at least one phenotypic observation. No animal with carcass data had its RTU data included in the analysis; therefore, residual covariances were not estimated between carcass and ultrasound traits.
In the model fitted to AHA data, fixed effects for carcass traits included: linear regression of breed percentages comprising Hereford and Angus, linear regression of heterosis effect between Hereford and Angus, harvest contemporary group (CG; harvest date × sex), and the linear regression of age at harvest. Fixed effects for RTU traits that were only measured on purebred Herefords included: scan CG (herd × scan date × sex), and the linear regression of age at scanning. Fixed effects for birth weight included birth CG (herd × sex × birth date window, the birth date windows span 60 d from the first calf born in the herd × sex contemporary group). These contemporary groups are the same as used in AHA national cattle evaluation. Individuals with carcass data represented 348 harvest CG, while animals with ultrasound data represented 528 scan CG, whereas all animals with carcass or RTU measures were represented in 679 birth CG.
In ASA data, admixed composite cattle are routinely registered and very few animals with carcass or RTU data are purebred. The average pedigree estimated breed composition was 42% Angus and 47% Simmental. Fixed effects for all traits included: linear regression on Angus and Simmental breed percentages, linear regression on heterosis fraction, and linear regression of age at harvest or scan. Fixed effects for carcass traits included harvest CG (herd × harvest date × sex). Fixed effects for RTU traits included scan CG (herd × scan date × sex). Fixed effects for birth weight included birth CG (herd × birth date window × sex). Individuals with carcass data represented 819 harvest CG, while animals with ultrasound data represented 324 scan CG. All animals with carcass or RTU measures were represented in 741 birth CG.
All the phenotypic data were measured and recorded in imperial units, i.e., inches rather than millimeters (mm) for back fat thickness, square inches rather than square centimeters for longissimus muscle area, and U.S. pounds (lbs) rather than kilograms (kg) for birth and carcass weights. More details regarding collection and national evaluation of these traits are available in the Beef Improvement Federation guidelines (https://beefimprovement.org/wp-content/uploads/2013/07/BIFGuidelinesFinal_updated0916.pdf). Phenotypes were standardized which leads to better convergence of variance components. Preliminary bivariate and trivariate animal models for combinations of traits were performed to obtain initial values of (co)variance parameters that were then used as starting values in subsequent multivariate analyses. Heritabilities and genetic correlations reported here were obtained using estimates of the variance components obtained from the 8-trait multivariate analyses. Estimates of genetic variance/covariances and their associated standard errors were obtained using average information restricted maximum likelihood (REML) as implemented in ASREML version 4 (Gilmour et al., 2014). For reporting results, variance component estimates were rescaled back, and unit conversions were undertaken from imperial to metric for all the traits besides MRB and UIMF.
Assessing Accuracy of Carcass Trait Evaluations
Multitrait evaluations were undertaken separately for each breed association using the estimated variance parameters as if they represented the true parameters. The EBV and accuracies were obtained on all the animals, but only those that were considered to be typical of selection candidates (SC) were further characterized.
Animals with offspring must have been selected to be parents, and their accuracies would be higher than would have been the case at the time of their selection, so those animals did not comprise SC. To represent a typical SC, the accuracies of those animals without offspring in the smallest and the largest scan CG were not considered in the scenario analysis described below. The sizes of selected scan CG were between 20 and 80 individuals for the AHA evaluation and between 75 and 125 for the ASA evaluation.
In scenario 1 (carcass only) RTU measurements were treated as missing, so that the carcass EBV of SC were predicted using their relatives' carcass phenotypes. In scenario 2 (RTU only) all carcass trait measurements from relatives were treated as missing so that the carcass EBV of the SC were predicted using their own yearling RTU phenotypic records. All available RTU and carcass phenotypic information was used in scenario 3, representing the same analysis as done to estimate variance components, except that the variance parameters were assumed known.
The multitrait Mixed Model Equations (MME) were built separately for each scenario and each data set from AHA and ASA, using the same models as previously described in this study. A single-site Gibbs sampling method implemented in BOLT software package (www.ThetaSolutionsLLC.com) was used to solve the MME with preconditioned conjugate gradient (PCG) solutions as starting values and a chain length of 50,000, with samples from the first 10,000 iterations discarded as burn-in. The sample variance of Markov Chain Monte Carlo (MCMC) samples for each animal was used as the prediction error variance (PEV) to calculate the theoretical prediction accuracy as 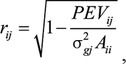 where subscripts i and j represent the ith individual and the jth trait, σ2gj is the genetic variance corresponding to the jth trait, and Aii is the ith diagonal element in the NRM corresponding to the ith individual calculated to account for inbreeding. The value of using carcass and/or RTU phenotypes to predict carcass EBV was assessed by comparing distributions of the theoretical prediction accuracies for each trait in each scenario, plotted using custom Python scripts.
where subscripts i and j represent the ith individual and the jth trait, σ2gj is the genetic variance corresponding to the jth trait, and Aii is the ith diagonal element in the NRM corresponding to the ith individual calculated to account for inbreeding. The value of using carcass and/or RTU phenotypes to predict carcass EBV was assessed by comparing distributions of the theoretical prediction accuracies for each trait in each scenario, plotted using custom Python scripts.
RESULTS AND DISCUSSION
Summary statistics for carcass traits, their corresponding RTU measurements, and birth weight are provided in Table 1. Mean FAT was 14.55 mm for AHA data and 12.02 mm for ASA data, comparable to 13.2 mm reported for Hereford (Arnold et al., 1991) and 12.42 mm for multibreed beef cattle including Angus, Charolais, Hereford, and Simmental composites (Miar et al., 2014). Mean UFAT for AHA and ASA cattle were 5.46 mm and 5.01 mm, both higher than reported values of 4.8 mm (Arnold et al., 1991) and 3.27 mm (Kause et al., 2015) for Hereford, 4.06 mm (Crews et al., 2003) and 4.07 (Crews and Kemp, 2001) for Simmental, but lower than the 8.80 mm reported (Miar et al., 2014) for Canadian composite cattle. The latter may be due to the fact that the Canadian results are based on the 12th and 13th ribs, whereas U.S. measurements are based on the 12th rib alone. Average MRB were 4.09 for AHA, which is smaller than the reported 5.21 (Arnold et al., 1991) for Hereford, and 5.32 for ASA data which is larger than the reported 5.01 (Crews et al., 2003) for Simmental, and both are larger than the 2.19 (Miar et al., 2014) for Canadian composite beef cattle.
Estimates of heritability were moderate, ranging between 0.25 and 0.48 for carcass and RTU traits, and moderate to high ranging between 0.34 and 0.61 for birth weight and carcass weight, as presented by diagonal elements in Table 2 for AHA data and Table 3 for ASA data. The highest heritability was 0.61 for birth weight in AHA data, while the lowest heritability was 0.25 for FAT in ASA data. Heritability estimates may be influenced by several factors including sampling, population size (Benyshek 1981; Koots et al., 1994), pedigree depth, breed, and heterosis effects in crossbred populations, and the endpoint defined by age, back fat, weight, or marbling (Rumph et al., 2007). Carcass and RTU traits generally have been reported in previous studies as being traits with moderate to high heritability. These results confirm that there are good opportunities for genetic improvement of carcass merit traits based on their direct measurements in these populations since the heritability estimates for these traits ranged from moderate to high.
Table 2.
Estimates of genetic (below diagonal) and residual (above diagonal) correlations, heritabilities (diagonal), and their standard error of estimates among carcass and ultrasound traits for the American Hereford Association
| UFAT1 | FAT2 | ULMA3 | LMA4 | UIMF5 | MRB6 | BWT7 | CWT8 | |
|---|---|---|---|---|---|---|---|---|
| Heritability, genetic correlation, and residual correlation | ||||||||
| UFAT | 0.29 ± 0.04 | N/A | 0.25 ± 0.03 | N/A | 0.18 ± 0.04 | N/A | -0.08 ± 0.04 | N/A |
| FAT | 0.74 ± 0.08 | 0.41 ± 0.04 | N/A | -0.01 ± 0.05 | N/A | 0.22 ± 0.04 | -0.00 ± 0.05 | 0.41 ± 0.04 |
| ULMA | 0.19 ± 0.09 | -0.15 ± 0.10 | 0.31 ± 0.04 | N/A | -0.01 ± 0.04 | N/A | -0.02 ± 0.04 | N/A |
| LMA | -0.11 ± 0.10 | -0.23 ± 0.07 | 0.81 ± 0.07 | 0.47 ± 0.04 | N/A | -0.09 ± 0.05 | 0.05 ± 0.05 | 0.34 ± 0.04 |
| IMF | 0.44 ± 0.07 | 0.34 ± 0.08 | -0.03 ± 0.08 | -0.18 ± 0.08 | 0.45 ± 0.04 | N/A | -0.07 ± 0.05 | N/A |
| MRB | 0.14 ± 0.10 | 0.23 ± 0.07 | -0.10 ± 0.10 | 0.02 ± 0.07 | 0.54 ± 0.08 | 0.48 ± 0.04 | 0.09 ± 0.06 | 0.20 ± 0.05 |
| BWT | -0.08 ± 0.07 | -0.10 ± 0.06 | 0.23 ± 0.07 | 0.11 ± 0.06 | 0.02 ± 0.06 | -0.19 ± 0.06 | 0.61 ± 0.03 | 0.16 ± 0.06 |
| CWT | 0.19 ± 0.10 | 0.24 ± 0.07 | 0.45 ± 0.09 | 0.51 ± 0.05 | 0.09 ± 0.09 | 0.12 ± 0.07 | 0.31 ± 0.06 | 0.51 ± 0.04 |
| Additive genetic variance (Vg) and phenotypic variance (Vp) | ||||||||
| Unit | mm2 | mm2 | (cm2)2 | (cm2)2 | - | - | kg2 | kg2 |
| Vg | 0.69 ± 0.09 | 7.23 ± 0.79 | 13.54 ± 1.86 | 28.09 ± 2.81 | 0.19 ± 0.02 | 0.23 ± 0.02 | 8.58 ± 0.52 | 498.27 ± 48.80 |
| Vp | 2.37 ± 0.05 | 17.54 ± 0.38 | 44.15 ± 0.97 | 59.43 ± 1.31 | 0.43 ± 0.01 | 0.48 ± 0.01 | 14.06 ± 0.25 | 969.56 ± 21.85 |
UFAT is real-time ultrasound (RTU) back fat thickness.
FAT is carcass back fat thickness.
ULMA is RTU longissimus dorsi muscle area.
LMA is carcass longissimus dorsi muscle area.
UIMF is intramuscular fat percentage, i.e., the RTU indicator of MRB.
MRB is carcass marbling score.
BWT is birth weight.
CWT is carcass weight.
Table 3.
Estimates of genetic (below diagonal) and residual (above diagonal) correlations, heritabilities (diagonal), and their standard error of estimates among carcass and ultrasound traits for the American Simmental Association
| UFAT1 | FAT2 | ULMA3 | LMA4 | UIMF5 | MRB6 | BWT7 | CWT8 | |
|---|---|---|---|---|---|---|---|---|
| Heritability, genetic correlation and residual correlation | ||||||||
| UFAT | 0.37 ± 0.03 | N/A | 0.18 ± 0.03 | N/A | 0.18 ± 0.03 | N/A | -0.08 ± 0.04 | N/A |
| FAT | 0.28 ± 0.13 | 0.25 ± 0.03 | N/A | -0.09 ± 0.03 | N/A | 0.23 ± 0.03 | -0.06 ± 0.03 | 0.30 ± 0.03 |
| ULMA | -0.01 ± 0.06 | 0.22 ± 0.12 | 0.44 ± 0.03 | N/A | -0.04 ± 0.03 | N/A | 0.08 ± 0.04 | N/A |
| LMA | -0.04 ± 0.12 | -0.27 ± 0.08 | 0.57 ± 0.10 | 0.32 ± 0.03 | N/A | 0.01 ± 0.03 | 0.09 ± 0.03 | 0.38 ± 0.03 |
| IMF | 0.33 ± 0.06 | -0.01 ± 0.11 | -0.24 ± 0.06 | -0.22 ± 0.10 | 0.42 ± 0.03 | N/A | -0.08 ± 0.04 | N/A |
| MRB | 0.13 ± 0.11 | 0.22 ± 0.07 | 0.03 ± 0.10 | -0.23 ± 0.07 | 0.73 ± 0.07 | 0.43 ± 0.04 | -0.01 ± 0.04 | 0.18 ± 0.04 |
| BWT | -0.11 ± 0.06 | -0.16 ± 0.07 | 0.22 ± 0.06 | 0.23 ± 0.06 | -0.16 ± 0.06 | -0.11 ± 0.06 | 0.47 ± 0.02 | 0.22 ± 0.03 |
| CWT | -0.03 ± 0.12 | 0.08 ± 0.08 | 0.52 ± 0.10 | 0.51 ± 0.06 | -0.17 ± 0.10 | -0.00 ± 0.07 | 0.42 ± 0.05 | 0.34 ± 0.03 |
| Additive genetic variance (Vg) and phenotypic variance (Vp) | ||||||||
| Unit | mm2 | mm2 | (cm2)2 | (cm2)2 | - | - | kg2 | kg2 |
| Vg | 0.71 ± 0.07 | 2.81 ± 0.04 | 26.36 ± 2.23 | 19.71 ± 2.13 | 0.20 ± 0.02 | 0.29 ± 0.03 | 9.20 ± 0.54 | 278.21 ± 29.10 |
| Vp | 1.92 ± 0.04 | 11.12 ± 0.18 | 60.09 ± 1.16 | 62.49 ± 1.02 | 0.46 ± 0.01 | 0.67 ± 0.01 | 19.54 ± 0.25 | 817.31 ± 13.50 |
UFAT is real-time ultrasound (RTU) back fat thickness.
FAT is carcass back fat thickness.
ULMA is RTU longissimus dorsi muscle area.
LMA is carcass longissimus dorsi muscle area.
UIMF is intramuscular fat percentage, i.e., the RTU indicator of MRB.
MRB is carcass marbling score.
BWT is birth weight.
CWT is carcass weight.
Carcass versus Ultrasound Measures of Fat Thickness
The phenotypic standard deviations (SD) for FAT (UFAT) from AHA and ASA data were 4.19 (1.54) and 3.33 (1.39) mm, comparable to reported estimates of 3.7 (0.85) mm (Arnold et al., 1991) for Hereford and 3.44 (1.28) mm (Crews et al., 2003) for Simmental. The relatively smaller phenotypic SD for UFAT were not surprising as RTU measures are taken on breeding stock at younger ages and lesser degrees of finish than is the case for carcass measures.
Additive genetic SD estimates for FAT (UFAT) were 2.69 (0.83) and 1.68 (0.84) mm for AHA and ASA, which were comparable to those previously reported values of 2.59 (0.43) mm for Hereford (Arnold et al., 1991), and 2.02 (0.93 and 1.05) mm for Simmental (yearling bulls and heifers; Crews et al., 2003). Crews et al. (2003) used a subset of the data that was available to our study but did not fit breed and heterosis effects in the manner done in this analysis.
Heritability of FAT (UFAT) were 0.41 (0.29) and 0.25 (0.37) for AHA and ASA cattle, similar to reported values of 0.49 (0.26; Arnold et al., 1991) for Hereford, 0.35 (0.53 and 0.69; Crews et al., 2003) for Simmental (yearling bulls and heifers), and 0.27 (0.31; Miar et al., 2014) for Canadian composite cattle. The relatively low heritability for UFAT from AHA and FAT from ASA relative to literature reports may be due to these cattle having been harvested at a constant finish endpoint, differences in breeds, and the fact that the model included a covariate for age, whereas some other literature analyses include a covariate for harvest weight.
The genetic correlation between FAT and UFAT was high at 0.74 for AHA data but relatively low at 0.28 for ASA data, both estimates being lower than previously reported values of 0.88 (Devitt and Wilton, 2001) for crossbred beef cattle, 0.79 and 0.83 (Crews et al., 2003) for Simmental bulls and heifers, and 0.82 and 0.96 (Reverter et al., 2000) for Angus bulls and heifers.
Carcass versus Ultrasound Measures of Longissimus Muscle Area
Phenotypic SD of LMA (ULMA) for AHA and ASA cattle were 7.71 (6.64) and 7.90 (7.75) cm2, which were within the range of reported estimates of 6.14 (5.43) cm2 (Arnold et al., 1991) for Hereford and 8.4 (7.55 and 6.94) cm2 (Crews et al., 2003) for Simmental (yearling bulls and heifers).
Additive genetic SD estimates of LMA (ULMA) for AHA and ASA were 5.30 (3.68) and 4.44 (5.13) cm2, which were similar to reported estimates of 4.16 (2.72) cm2 (Arnold et al., 1991) for Hereford, and 5.6 (4.6 and 4.9) cm2 (Crews et al., 2003) for Simmental (yearling bulls and heifers).
Heritability estimates were 0.47 (0.31) and 0.32 (0.44) for AHA and ASA LMA (ULMA), similar to reported values of 0.46 (0.25; Arnold et al., 1991) for Hereford, 0.46 (0.37; Crews et al., 2003) for Simmental, and higher than estimates of 0.24 (0.17; Miar et al., 2014) for Canadian composite cattle.
Genetic correlations between LMA and ULMA were 0.81 for AHA and 0.57 for ASA, both within the range of reported estimates of 0.29 for Angus, 0.94 for Hereford (Reverter et al., 2000), 0.66 (Moser et al., 1998; Devitt and Wilton, 2001) for Simmental, 0.71 and 0.67 for Simmental between LMA and ULMA from yearling bulls and heifers (Crews and Kemp, 2001), and 0.80 (Crews et al., 2003) for Simmental.
Carcass Marbling Score versus Ultrasound Intramuscular Fat Percentage Measures
Phenotypic SD of MRB (UIMF) for AHA and ASA cattle were 0.69 (0.65) and 0.82 (0.68) cm2, which were similar to reported estimates of 0.88 (0.52 and 0.65; Crews et al., 2003) for Simmental (yearling bulls and heifers) and 0.59 (0.54; McAllister et al., 2011) for Red Angus.
Additive genetic SD estimates of MRB (UIMF) for AHA and ASA were 0.48 (0.44) and 0.54 (0.44), which were within the range of reported estimates of 0.64 (0.36 and 0.47; Crews et al., 2003) for Simmental (yearling bulls and heifers).
Heritability estimates were 0.48 (0.45) and 0.43 (0.42) for MRB (UIMF) from AHA and ASA cattle, similar to reported estimates of 0.54 (0.47 and 0.52 for yearling bulls and heifers; Crews et al., 2003) for Simmental cattle, 0.35 (0.29; McAllister et al., 2011) for Red Angus cattle, and 0.38 (0.37; Miar et al., 2014) for Canadian composite cattle.
Genetic correlation estimates of MRB with UIMF were 0.54 for AHA and 0.73 for ASA cattle, lower than the reported value of 0.80 (Devitt and Wilton, 2001) for Simmental cattle, while Wilson et al. (1999) found this correlation to be 0.77. Crews et al. (2003) reported that genetic correlations of MRB with bull and heifer UIMF were 0.74 and 0.69 for Simmental cattle.
Birth and Carcass Weights
Heritability estimates for birth weight were 0.61 for AHA and 0.47 for ASA cattle, higher than the reported estimate of 0.43 (Eriksson et al., 2004) for direct birth weight for Hereford, and estimates of 0.40 and 0.45 (Garrick et al., 1989) for male and female Simmental. Heritability estimates of CWT were 0.51 for AHA and 0.34 for ASA, which were within the ranges of reported values of 0.24 (Arnold et al., 1991), 0.54 (Reverter et al., 2000), 0.50 (Eriksson et al., 2004), and 0.48 (Kause et al., 2015) for Hereford, and 0.38 (Crews and Kemp, 2001), 0.47 (Devitt and Wilton, 2001), 0.32 (Shanks et al., 2001), and 0.48 (Crews et al., 2003) for Simmental. Three reviews reported average CWT heritabilities of 0.41 (Marshall, 1994), 0.42 (Utrera and van Vleck, 2004), and 0.45 (Koots et al., 1994), while a range of 0.17 to 0.65 was reported for 8 different European beef cattle breeds (Hickey et al., 2007) including Hereford and Simmental.
Genetic correlations between BWT and CWT were 0.31 for AHA and 0.42 for ASA. Eriksson et al. (2004) reported a similar genetic correlation of 0.30 for Hereford between CWT and maternal BWT, but a lower value of 0.11 was found between CWT and direct BWT.
Considerations using Crossbred Data
It is difficult to obtain carcass data from purebred pedigree herds, and such data tend to only be recorded on inferior animals. It is much easier to obtain carcass data from commercial rather than bull-breeding herds, and these are commonly crossbred animals, for both Hereford and Simmental breeds. The genetic correlation between pure- and crossbred performance may be less than unity (Newman et al., 2002; Núñez-Dominguez et al., 1993), which would make equivalent amounts of crossbred data less effective for ranking purebreds than would be the case if purebred data were used. However, if Hereford and Simmental bulls are commonly outcrossed, purebred selection should be for crossbred performance.
Theoretical Prediction Accuracies for Carcass Traits using Ultrasound and/or Carcass Data
A total of 1,788 and 1,083 RTU measured individuals from 54 AHA and 11 ASA scan CG were chosen to represent typical SC according to descriptions in Materials and Methods. These SC were sired by 117 and 176 bulls, respectively, and these bulls also produced 1,612 and 1,345 harvested individuals that had carcass trait measurements.
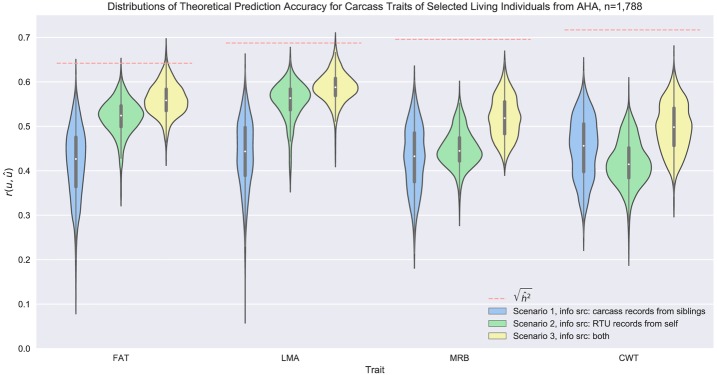
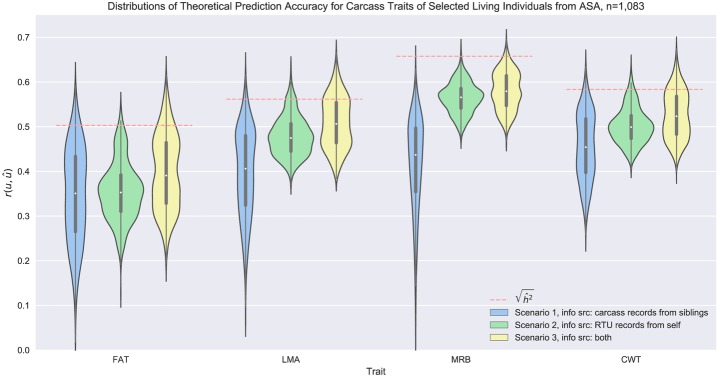
Predicting carcass EBV for SC ignoring RTU but using their relatives' carcass measurements, i.e., Scenario 1, yielded the lowest mean theoretical accuracy from the 3 scenarios, except for CWT from AHA data (Fig. 1 and 2). That low accuracy was due to the fact that none of the SC had their own phenotypic information besides BWT; therefore, their carcass EBV accuracies were critically determined by (1) the genetic correlation between carcass traits and BWT, which were as low as -0.10, 0.11, and -0.19 for AHA data and -0.16, 0.23, and -0.11 for ASA data; and (2) carcass EBV accuracies of their sires, which was dramatically affected by their own RTU measurements and the number of their harvested offspring. Huge variations in the accuracies of the sires' carcass EBV contributed to wider distributions of theoretical prediction accuracy of carcass EBV for the SC, as observed in Scenario 1.
Figure 1.
Distribution of theoretical prediction accuracy of EBV for carcass traits based on various sources of phenotypic information from the American Hereford Association. The boxes inside the violins showed the quartiles of the data set while the whiskers extended to show the rest of the distribution, and the orange dashed lines represent square roots of estimated heritabilities, which are the theoretical upper bounds of prediction accuracies. Abbreviations: AHA, American Hereford Association; FAT, back fat thickness; LMA, longissimus muscle area; MRB, marbling score; CWT, carcass weight; RTU, real-time ultrasound.
Figure 2.
Distribution of theoretical prediction accuracy of EBV for carcass traits based on various sources of phenotypic information from the American Simmental Association. The boxes inside the violins showed the quartiles of the data set while the whiskers extended to show the rest of the distribution, and the orange dashed lines represent square roots of estimated heritabilities, which are the theoretical upper bounds of prediction accuracies. Abbreviations: ASA, American Simmental Association; FAT, back fat thickness; LMA, longissimus muscle area; MRB, marbling score; CWT, carcass weight; RTU, real-time ultrasound.
In Scenario 2 carcass EBV of SC were predicted primarily from the individual's own RTU phenotypic records and were influenced by the genetic correlations between carcass traits and their corresponding RTU live indicators. A tendency of higher means and narrower distributions of prediction accuracies were observed for this scenario compared to Scenario 1, which could be inferred from the fact that genetic correlations between carcass traits and their corresponding RTU measurements (0.74, 0.81, 0.54 and 0.28, 0.57, 0.73 as shown in Tables 2 and 3) were higher than the genetic correlations between carcass traits and BWT (showed previously) and the expected half-sib coefficient of kinship (0.25), and that prediction accuracies were less affected by the sires' EBV accuracies if the SC had their own measurements. However, theoretical prediction accuracy of CWT from the AHA evaluation was lower than that of Scenario 1. Since no corresponding RTU trait was present for CWT, the lower accuracy could be explained by relatively small genetic correlations between CWT and RTU traits (0.19, 0.45, 0.09) and between CWT and BWT (0.31) in the AHA evaluation.
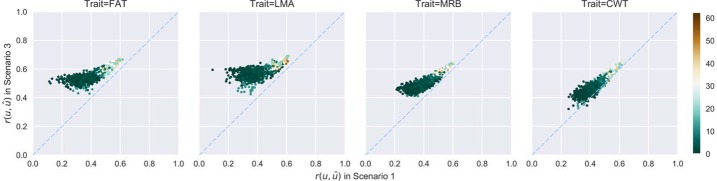
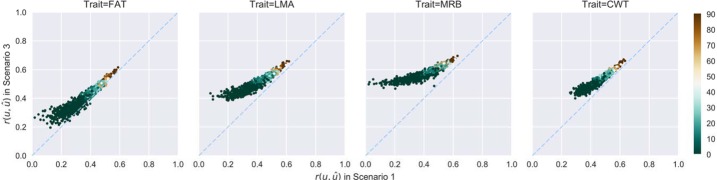
Using all the yearling RTU measurements of SC along with their relatives' carcass measurements (Scenario 3), the mean theoretical prediction accuracies of carcass EBV for SC were increased by 0.05∼0.16 (Table 4) compared to those that were obtained from Scenario 1. Increases were greater for individuals with the lowest accuracies, which were those with small numbers of carcass phenotyped relatives (Fig. 3 and 4). A few SC had no carcass siblings, and their carcass EBV from Scenario 1 were predicted through their carcass phenotyped relatives that share common grand sires or more distant relatives, which caused low prediction accuracies as expected. Adding the SC RTU information into the model narrowed the distributions of theoretical prediction accuracies by 9.8%∼62.3% (Table 5), indicative of decreased prediction uncertainty and increased prediction reliability.
Table 4.
Mean theoretical prediction accuracy of EBV for carcass traits based on various scenarios varying in terms of sources of phenotypic information
| Data Source1 | Trait2 | Scenario 1 Carcass only | Scenario 2 RTU3 only | Scenario 3 All data | Delta4: S3-S1 |
|---|---|---|---|---|---|
| AHA | CWT | 0.45 | 0.42 | 0.50 | 0.05 |
| FAT | 0.42 | 0.52 | 0.56 | 0.14 | |
| LMA | 0.44 | 0.56 | 0.59 | 0.15 | |
| MRB | 0.43 | 0.45 | 0.52 | 0.09 | |
| ASA | CWT | 0.46 | 0.50 | 0.53 | 0.07 |
| FAT | 0.35 | 0.36 | 0.40 | 0.05 | |
| LMA | 0.40 | 0.48 | 0.51 | 0.11 | |
| MRB | 0.42 | 0.56 | 0.58 | >0.16 |
Data Source: AHA, American Hereford Association; ASA, American Simmental Association.
Trait: CWT is carcass weight; FAT is back fat thickness; LMA is longissimus dorsi muscle area; MRB is marbling score.
RTU refers to real-time ultrasound measures UFAT, ULMA, and ultrasound intramuscular fat percentage (UIMF).
Delta: S3 is short for Scenario 3; S1 is short for Scenario 1.
Figure 3.
Pair-wise scatter plot of theoretical prediction accuracies of EBV from Scenario 1 to Scenario 3 for the American Hereford Association evaluation. Each point stands for an individual, and the color represents the number of its carcass phenotyped siblings. Dashed line is y = x. Abbreviations: FAT, back fat thickness; LMA, longissimus muscle area; MRB, marbling score; CWT, carcass weight.
Figure 4.
Pair-wise scatter plot of theoretical prediction accuracies of EBV from Scenario 1 to Scenario 3 for the American Simmental Association evaluation. Each point stands for an individual, and the color represents the number of its carcass phenotyped siblings. Dashed line is y = x. Abbreviations: FAT, back fat thickness; LMA, longissimus muscle area; MRB, marbling score; CWT, carcass weight.
Table 5.
Distribution range of theoretical prediction accuracy of EBV for carcass traits based on various sources of phenotypic information
| Data Source1 | Trait2 | Scenario 1 Carcass only | Scenario 2 RTU3 only | Scenario 3 All data | Reduced4: (S1-S3)/S1% |
|---|---|---|---|---|---|
| AHA | CWT | 0.37 | 0.38 | 0.34 | 9.8 |
| FAT | 0.50 | 0.30 | 0.25 | 49.7 | |
| LMA | 0.54 | 0.29 | 0.27 | 49.6 | |
| MRB | 0.39 | 0.29 | 0.24 | 38.9 | |
| ASA | CWT | 0.38 | 0.24 | 0.28 | 26.8 |
| FAT | 0.57 | 0.42 | 0.42 | 26.6 | |
| LMA | 0.54 | 0.26 | 0.28 | 47.6 | |
| MRB | 0.61 | 0.21 | 0.23 | 62.3 |
Data Source: AHA, American Hereford Association; ASA, American Simmental Association.
Trait: CWT is carcass weight; FAT is back fat thickness; LMA is longissimus dorsi muscle area; MRB is marbling score.
RTU refers to real-time ultrasound measures UFAT, ULMA and ultrasound intramuscular fat percentage (UIMF).
Reduced: S3 is short for Scenario 3; S1 is short for Scenario 1.
Square roots of heritabilities determined the theoretical upper bounds of prediction accuracies measured as correlations between EBV and phenotypic records (orange dashed lines in Fig. 1 and 2). In contrast, we assessed the correlations between EBV and the true breeding values (TBV) from the prediction error variance obtained by MCMC sampling using the MME. Results showed that the 8-trait animal model explained phenotypic variations well from the ASA data (Fig. 2), while a promising increase in prediction accuracy for AHA's MRB and CWT can be expected if more data representing direct measurements of the target trait and measurements from its genetically correlated traits are considered into the model (Fig. 1).
These results indicated that the accuracy of genetic evaluation of carcass traits is significantly enhanced by inclusion of RTU data. However, the paucity of actual carcass data currently being collected by either AHA or ASA would argue that selection would likely be more effective if there was a greater extent of collection of actual carcass data. In the past, collection of carcass data has been problematic due to the lack of pedigree recording in commercial herds with multisire mating, but the relatively low cost of genotyping panels for parentage or genomic prediction now allows much easier access to carcass data than was the case in the past.
Conclusion
In conclusion, genetic parameter estimates indicated that carcass traits were moderately to highly heritable for both AHA and ASA data. Furthermore, similarly moderate to high heritability estimates were obtained for fat thickness, longissimus muscle area, and percentage of intramuscular fat measured using ultrasound in yearling replacement animals, for both AHA and ASA data. Genetic correlations between carcass traits and their ultrasound indicators were favorable but sometimes less than currently assumed in national evaluations. Predictions of carcass merit on potential replacements using RTU measurements would increase prediction accuracy by 0.05∼0.16 and reduce the uncertainty of prediction by 9.8%∼62.3%. Although ultrasound measurements provide for genetic evaluation of a more representative sample of cattle within populations and an earlier estimate of carcass merit evaluation was possible when data are available only from organized carcass progeny tests, collection of greater numbers of carcass measurements would improve the accuracy of genetic evaluations in both breeds.
CONFLICT OF INTEREST
DJG and BLG disclose that they are partners in ThetaSolutionsLLC, a company that licenses BOLT software that is used in the genetic and genomic evaluations published by American Simmental Association and American Hereford Association, but believe this does not represent a conflict of interest in this study.
LITERATURE CITED
- Arnold J. W., Bertrand J. K., Benyshek L. L., Ludwig C. 1991. Estimates of genetic parameters for live animal ultrasound, actual carcass data, and growth traits in beef cattle. J. Anim. Sci. 69:985–992. doi: 10.2527/1991.693985x [DOI] [PubMed] [Google Scholar]
- Benyshek L. L. 1981. Heritabilities for growth and carcass traits estimated from data on Herefords under commercial conditions. J. Anim. Sci. 53:49–56. [Google Scholar]
- Crews D. H., Kemp R. A. 2002. Genetic evaluation of carcass yield using ultrasound measures on young replacement beef cattle. J. Anim. Sci. 80:1809–1818. doi: 10.2527/2002.8071809x [DOI] [PubMed] [Google Scholar]
- Crews D. H., Pollak E. J., Weaber R. L., Quaas R. L., Lipsey R. J. 2003. Genetic parameters for carcass traits and their live animal indicators in Simmental cattle. J. Anim. Sci. 81:1427–1433. doi: 10.2527/2003.8161427x [DOI] [PubMed] [Google Scholar]
- Crews D. H., Kemp R. A. 2001. Genetic parameters for ultrasound and carcass measures of yield and quality among replacement and slaughter beef cattle. J. Anim. Sci. 79:3008–3020. doi: 10.2527/2001.79123008x [DOI] [PubMed] [Google Scholar]
- Devitt C. J., Wilton J. W. 2001. Genetic correlation estimates between ultrasound measurements on yearling bulls and carcass measurements on finished steers. J. Anim. Sci. 79:2790–2797. doi: 10.2527/2001.79112790x [DOI] [PubMed] [Google Scholar]
- Eriksson S., Näsholm A., Johansson K., Philipsson J. 2004. Genetic relationships between calving and carcass traits for Charolais and Hereford cattle in Sweden. J. Anim. Sci. 82:2269–2276. doi: 10.2527/2004.8282269x [DOI] [PubMed] [Google Scholar]
- Garrick D. J., Pollak E. J., Quaas R. L., Van Vleck L. D. 1989. Variance Heterogeneity in Direct and Maternal Weight Traits by Sex and Percent Purebred for Simmental-Sired Calves1. J. Anim. Sci. 67:2515–2528. doi: 10.2527/jas1989.67102515x [DOI] [PubMed] [Google Scholar]
- Gilmour A. R., Gogel B. J., Cullis B. R., Thompson R. 2014. ASReml User Guide Release 4.0. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK: www.vsni.co.uk [Google Scholar]
- Hickey J. M., Keane M. G., Kenny D. A., Cromie A. R., Veerkamp R. F. 2007. Genetic parameters for EUROP carcass traits within different groups of cattle in Ireland. J. Anim. Sci. 85:314–321. doi: 10.2527/jas.2006-263 [DOI] [PubMed] [Google Scholar]
- Kause A., Mikkola L., Strandén I., Sirkko K. 2015. Genetic parameters for carcass weight, conformation and fat in five beef cattle breeds. Animal 9:35–42. doi: 10.1017/S1751731114001992 [DOI] [PubMed] [Google Scholar]
- Koots K. R., Wade K. M., Kennedy B. W., Dekkers J. 1994. Method and effect of adjustment for heterogeneous variance of Holstein conformation traits. J. Dairy Sci. 77(1):294–302. doi: 10.3168/jds.S0022-0302(94)76954-X [DOI] [PubMed] [Google Scholar]
- Marshall D. M. 1994. Breed differences and genetic parameters for body composition traits in beef cattle. J. Anim. Sci. 72:2745–2755. doi: 10.2527/1994.72102745x [DOI] [PubMed] [Google Scholar]
- McAllister C., Speidel S., Crews D., Enns R. 2011. Genetic parameters for intramuscular fat percentage, marbling score, scrotal circumference, and heifer pregnancy in Red Angus cattle. J. Anim. Sci. 89:2068–2072. doi: 10.2527/jas.2010-3538 [DOI] [PubMed] [Google Scholar]
- Miar Y., Plastow G., Bruce H., Moore S., Durunna O., Nkrumah J., Wang Z. 2014. Estimation of genetic and phenotypic parameters for ultrasound and carcass merit traits in crossbred beef cattle. Can. J. Anim. Sci. 94:273–280. doi: 10.4141/cjas2013-115 [DOI] [Google Scholar]
- Moser D. W., Bertrand J. K., Misztal I., Kriese L. A., Benyshek L. L. 1998. Genetic parameter estimates for carcass and yearling ultrasound measurements in Brangus cattle. J. Anim. Sci. 76:2542–2548. [DOI] [PubMed] [Google Scholar]
- Newman S., Reverter A., Johnston D. J. 2002. Purebred-crossbred performance and genetic evaluation of postweaning growth and carcass traits in Bos indicus × Bos taurus crosses in Australia. J. Anim. Sci. 80:1801–1808. doi: 10.2527/2002.8071801x [DOI] [PubMed] [Google Scholar]
- Núñez-Dominguez R., Vleck D. L., Boldman K. G., Cundiff L. V. 1993. Correlations for genetic expression for growth of calves of Hereford and Angus dams using a multivariate animal model. J. Anim. Sci. 71:2330–2340. doi: 10.2527/1993.7192330x [DOI] [PubMed] [Google Scholar]
- Reverter A., Johnston D. J., Graser H. U., Wolcott M. L., Upton W. H. 2000. Genetic analyses of live-animal ultrasound and abattoir carcass traits in Australian Angus and Hereford cattle. J. Anim. Sci. 78:1786–1795. doi: 10.2527/2000.7871786x [DOI] [PubMed] [Google Scholar]
- Rumph J., Shafer W., Crews D., Enns R., Lipsey R., Quaas R., Pollak E. 2007. Genetic evaluation of beef carcass data using different endpoint adjustments. J. Anim. Sci. 85:1120–1125. doi: 10.2527/jas.2006-694 [DOI] [PubMed] [Google Scholar]
- Shanks B. C., Tess M. W., Kress D. D., Cunningham B. E. 2001. Genetic evaluation of carcass traits in Simmental-sired cattle at different end points. J. Anim. Sci. 79:595–604. [DOI] [PubMed] [Google Scholar]
- Wilson D. E., Rouse G. H., Hays C., Hassen A. 1999. Genetic evaluation of ultrasound measures: Angus. In Proc. 31st Beef Improv. Fed. Ann. Res. Symp. Annu. Mtg., Roanoke, VA: p. 197–198. [Google Scholar]
- Utrera R. A., Vleck V. L. 2004. Heritability estimates for carcass traits of cattle: a review. Genet. Mol. Res. 3(3): 380–394. [PubMed] [Google Scholar]