ABSTRACT
Alterations in progesterone (P4) catabolism due to high feed intake underlie some effects of nutrition on reproduction. Based on previous research, we hypothesized that high feed intake could potentially increase P4 catabolism, likely due to increased liver blood flow. However, there could also be an opposing action due to increased circulating insulin, which has been shown to inhibit hepatic expression of key enzymes involved in P4 catabolism. To test which effect would have the greatest impact on circulating P4 during a 1- and 2 -mo time frame, we used a noncyclic ewe model. The plane of nutrition was controlled, and effects on circulating insulin, P4 catabolism in response to exogenous P4, and steady state mRNA for key hepatic enzymes were evaluated. Twenty-four F1 Dorper × Santa Inês ewe lambs (5 mo old and approximately 25 kg BW) were used. After 14 d of adaptation, ewes were randomized into 2 groups: ad libitum fed (Ad), with intake of 3.8% DM/kg BW, or restricted feed intake (R), with 2% DM/kg BW, for 8 wk. At wk 4 and 8, ewes received an intravaginal P4 implant to evaluate P4 catabolism. As designed, Ad ewes had greater daily feed intake than R ewes (means of 1.8 [SE 0.03] and 0.6 kg/ewe [SE 0.01]; P < 0.001) and greater weekly gain in BW (means of 1.7 [SE 0.12] vs. −0.1 kg/ewe [SE 0.03]; P < 0.001). Mean circulating insulin of samples collected from −0.5 to 7 h after the start of feeding was over 5-fold greater in Ad ewes than in R ewes (least squares means of 8.2 [SE 0.93] vs. 1.5 μIU/mL [SE 0.16], respectively, at wk 4 and 12.0 [SE 1.02] vs. 2.2 μIU/mL [SE 0.18], respectively, at wk 8; P < 0.001). Although both groups received the same P4 treatment, mean circulating P4 of samples collected from −0.5 to 7 h after feeding was much lower in Ad ewes than in R ewes (least squares means of 3.2 [SE 0.32] vs. 5.5 ng/mL [SE 0.32], respectively, at wk 4 and 2.8 [SE 0.28] vs. 5.2 ng/mL [SE 0.28], respectively, at wk 8; P < 0.001) indicating much greater P4 catabolism in ewes with high feed intake. Unexpectedly, there was no effect of diet on hepatic mRNA concentrations for CYP2C, CYP3A, AKR1C, or AKR1D at wk 4 or 8 in spite of dramatically elevated insulin. Therefore, high energy/feed intake primarily increased P4 catabolism with no evidence for offsetting effects due to insulin-induced changes in hepatic P4 metabolizing enzymes.
Keywords: hepatic gene expression, metabolism, nutrition, progesterone
INTRODUCTION
Among the interactions of nutrition with reproduction in mammalian species, increased DMI in sheep was found to reduce circulating progesterone (P4) concentrations and reduce embryo survival (Parr et al., 1987, 1993a, 1993b; Parr, 1992). This negative effect is particularly pronounced in lactating dairy cows, in which dramatic increases in energy intake to meet the demands of high milk production can result in major increases in catabolism of P4 and estradiol (Sangsritavong et al., 2002; Vasconcelos et al., 2003), resulting in substantial changes in reproductive physiology (Wiltbank et al., 2006).
Progesterone is primarily inactivated or catabolized in the liver (Parr et al., 1993b; Freetly and Ferrell, 1994). The enzymes involved in Phase 1 of hepatic P4 catabolism include enzymes of the cytochrome P450 (CYP) family such as CYP2C and CYP3A (Murray, 1991, 1992) and members of the aldo-keto reductase (AKR) family including AKR1C and AKR1D (Jin et al., 2011). One intriguing concept that has been pursued during the last few years is that circulating insulin may regulate P4 catabolism due to regulation of hepatic expression of the key enzymes involved in P4 inactivation in the liver (Lemley et al., 2010a,b). For example, in both sheep and cattle, in vivo studies have reported that alterations in diets that increased circulating insulin or directed changes in circulating insulin led to a reduction in expression of CYP2C and CYP3A and reduced the clearance rate of P4 (Lemley et al., 2008a,b, 2010b,c). In addition, studies with bovine hepatocytes have found that in vitro treatment with insulin produced an acute and dramatic decrease in expression of CYP2C and CYP3A (Lemley et al., 2009).
Therefore, there are 2 simultaneous and potentially opposing processes involved in hepatic P4 catabolism that could be altered by high energy intake. First, high energy intake could increase liver blood flow, thus increasing P4 catabolism due to greater delivery of P4 molecules to the enzymes involved in P4 catabolism. Conversely, high energy intake could increase circulating insulin and elevated insulin could reduce expression of hepatic enzymes involved in P4 catabolism. A reduction in P4 metabolizing enzymes due to elevated insulin could potentially increase circulating P4 concentrations, assuming all other components involved in P4 production and catabolism were constant.
The objective of this experiment was to use a noncyclic ewe model to analyze the longer-term effects, 4 to 8 wk, of high vs. low energy intake on P4 catabolism. We hypothesized that high energy intake would increase circulating insulin and that high insulin would reduce expression of hepatic enzymes involved in P4 catabolism. Furthermore, we hypothesized that the increase in hepatic P4 catabolism, due to an expected increase in liver blood flow due to high DMI, would be offset by the reduction in hepatic expression of P4 metabolizing enzymes. To test these hypotheses, we provided ewes with restricted vs. ad libitum DMI for 4 and 8 wk and measured the effects on circulating insulin and P4 concentrations and hepatic mRNA abundance for key P4 metabolizing enzymes: CYP2C, CYP3A, AKR1C, and AKR1D.
MATERIALS AND METHODS
All procedures and protocols with animals were approved by the Ethics Committee on the Use of Animals in Research (University of São Paulo, Piracicaba, Brazil, Protocol no.: 2010-4) and by the Ethics Committee on Animal Use (University of São Paulo State, Botucatu, Brazil, Protocol no.: 104/2010). This study was conducted in the Intensive Production System of Sheep and Goats, Department of Animal Science, Luiz de Queiroz College of Agriculture, Piracicaba, São Paulo, Brazil.
Animals and Nutritional Management
Twenty-four prepubertal ewe lambs with an average age of 5 mo, F1 Dorper × Santa Inês, were used in this experiment. Ewes were housed in 24 individual pens. During the adaptation period (14 d), the animals received the same diet (Table 1), with a daily intake of 3% DM/kg BW. After adaptation, the ewes were divided into blocks using BW and, within the initial block, randomly assigned to the 2 experimental groups: ad libitum–fed (Ad) group or restricted feed intake (R) group.
Table 1.
Ingredients and composition of the diet offered to sheep
| Ingredient | Percent of DM |
|---|---|
| Coastcross hay | 10.0 |
| Ground corn | 70.4 |
| Soybean meal | 16.0 |
| Ammonium chloride | 0.5 |
| Limestone | 1.5 |
| Mineral mixture1 | 1.6 |
| Chemical composition, % | |
| DM | 90.4 |
| CP | 18.0 |
| Ether extract | 4.0 |
7.5% P, 13.4% Ca, 1.0% Mg, 7.0% S, 14.5% Na, 21.8% Cl, 500 mg/kg Fe, 300 mg/kg Cu, 4,600 mg/kg Zn, 1,100 mg/kg Mn, 55 mg/kg I, 40 mg/kg Co, and 30 mg/kg Se.
The groups were fed the diets for 8 wk. The ewes received the same diet, based on recommendations of the NRC (2007), consisting of 90% concentrate and 10% forage (Table 1), with the only difference being the quantity of diet that was offered to the ewes. Half of the ewes were offered feed with a daily intake of 2% DM/kg BW (R group) and the other half received an unlimited diet, with an expected daily intake of 3.8% DM/kg BW (Ad group). All animals had free access to water. The DMI was calculated on a daily basis by weighing the quantity of feed offered minus the amount remaining in each trough.
On a weekly basis, ewes were weighed and had other measurements (height, from ground to the top of shoulder or withers, and length, the distance between the sternum and the ischiatic tuberosity) taken to estimate body mass index (BMI) using the formula BMI = BW/([height/100] × [length/100]). Body weight was measured in kilograms and height and length were measured in centimeters.
Blood Sampling and Hormone Assays
In the last week of the adaptation period, blood samples were taken before feeding (0 h) and at 1, 2, 3, 4, 5, 6, and 7 h after offering feed. These samples were used to determine the dynamic profile for circulating insulin and P4.
At 4 and 8 wk after initiation of the diets, all ewes had a blood sample taken before inserting an intravaginal P4 implant (controlled internal drug release [CIDR] device; 0.33 g P4; Zoetis, São Paulo, Brazil; Day 0) to determine if ewes were prepubertal (circulating P4 concentration < 1 ng/mL). In addition, all ewes received PGF2α (intramuscular, 5 mg Lutalyse; Zoetis) at time of P4 implant insertion and on Day 6 after CIDR device insertion to assure regression of any corpus luteum that might be present. These actions were done to preclude the confounding effects of a corpus luteum on the circulating P4 concentrations during the measurement period. On Day 8 after CIDR device insertion, blood samples were collected before feeding and at 1, 2, 3, 4, 5, 6, and 7 h after feeding to determine the profiles of circulating insulin and P4 concentrations (Fig. 1). Blood samples were collected by jugular venipuncture into Vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ) with anticoagulant (sodium heparin) and centrifuged at 2,000× g for 15 min at room temperature. Plasma was stored at −20°C until hormones were measured using RIA. Circulating insulin was evaluated using a solid-phase RIA using a commercial kit (Coat-A-Count Insulin; Siemens Medical Solutions Diagnostics, Inc., Los Angeles, CA). The assay sensitivity was 1.2 μIU/mL and the intra- and interassay CV were 8.5 and 9.9%, respectively. Circulating P4 was evaluated using a solid-phase RIA using a commercial kit (Coat-A-Count Progesterone; Siemens Medical Solutions Diagnostics, Inc.). The assay sensitivity was 0.02 ng/mL and the intra- and interassay CV were 2.9 and 7.4%, respectively.
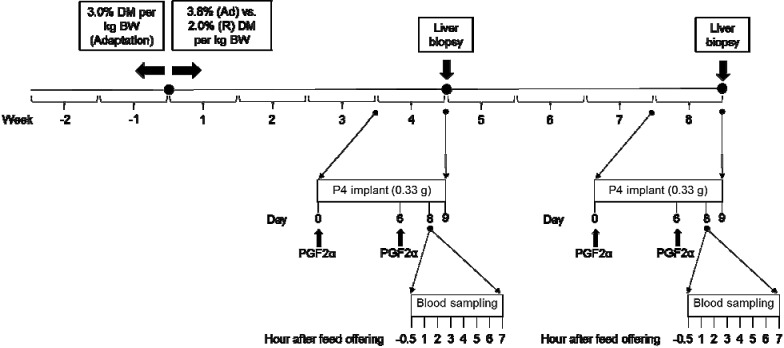
Figure 1.
Experimental design. During the adaptation period (14 d), ewe lambs received the same diet, with a daily intake of 3.0% DM/kg BW. After that, ewes were randomly assigned to 2 groups based on the amount of feed offered: ad libitum fed (Ad; daily intake of 3.8% DM/kg BW) or restricted feed intake (R; daily intake of 2.0% DM/kg BW). During the fourth and eighth weeks, after initiation of the diets on Day 0, ewes were treated with PGF2α (to assure the absence of corpus luteum) and received an intravaginal progesterone (P4) controlled internal drug release device (0.33 g of P4) kept for 9 d. On Day 6, another treatment with PGF2α was performed, and on Day 8, blood samples were collected at the following intervals: −0.5, 1, 2, 3, 4, 5, 6, and 7 h relative to the time the diet was offered (0 h). On Day 9, liver biopsies were performed.
Liver Biopsies
Liver biopsies were performed on Day 9 after CIDR device insertion, 4 h after offering the diet, and before removing the CIDR device. After hair removal and skin disinfection of the surgical area, subcutaneous and intramuscular anesthesia was done with 2% lidocaine infusion at the 11th intercostal space, approximately 12 cm below the spinal vertebrae on the right side of the ewes. An incision of approximately 1 cm was made in the skin, and a 14 gauge by 15-cm biopsy needle (TRU-CUT; Biocompany, São Paulo, Brazil) was used to collect liver fragments. Samples were stored in RNAlater (Ambion Inc., Austin, TX) at 4°C overnight before long-term storage at −80°C.
CYP2C, CYP3A, AKR1C, and AKR1D mRNA Measurements
Liver biopsies were individually homogenized (Ika Ultra Turrax T10 basic; Merck, Darmstadt, Germany) in TRIzol reagent (Invitrogen Corp., Carlsbad, CA) for total RNA extraction according to the protocol described by Chomczynski and Sacchi (1987). Concentration of RNA in each sample was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Purity of RNA was verified using the ratio of the optical density at 260 nm to the optical density at 280 nm, and integrity was verified using 1% agarose gel electrophoresis. Total RNA was treated with RNase-free DNase I (QIAGEN Inc., Valencia, CA) before cDNA synthesis to eliminate any potential genomic DNA.
One microgram of total RNA from each sample was reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen Corp.) following the protocol of the manufacturer. The cDNA was stored at −20°C until used.
Primer pairs specific for each of the genes studied were designed on different exons using Primer3 primer design software (Koressaar and Remm, 2007) based on the sheep-specific GenBank database (https://www.ncbi.nlm.nih.gov/gene) sequences (Table 2). Aldo-keto reductase 1 primer pairs were based on bovine-specific sequences due to the lack of sheep sequences in the GenBank database. Primers were analyzed using Premier Biosoft Net Primer, Palo Alto, CA (http://www.premierbiosoft.com/netprimer/), to avoid secondary structures such as hairpins and loops. All primers were optimized to amplify at 58°C and purchased from Sigma-Aldrich Brasil Ltda and Life Technologies Brasil Ltda (São Paulo, Brazil). Specific methods used for real-time quantitative PCR (RT-qPCR) were previously described (Udvardi et al., 2008; Bustin et al., 2009).
Table 2.
Sequence of primers and size of the amplified fragments for evaluation of target genes (cytocrome P450 2C [CYP2C19], cytocrome P450 3A [CYP3A24], aldo-keto reductase 1C4 [AKR1C4], and aldo-keto reductase 1D1 [AKR1D1]) and housekeeping genes (β-actin [ACTB], cyclophilin A [PPIA], glyceraldehyde-3-phosphate dehydrogenase [GAPDH], ATPase, and ribosomal protein L19 [RPL19])
| Gene | Sequence1 (5′–3′) | Product size, bp | GenBank2 access number | |
|---|---|---|---|---|
| CYP2C19 | F | CAAGAATCCCTGGACCTCAA | 193 | NM_001205152.1 |
| R | GCTTCAGCAGGAGCAGGAG | |||
| CYP3A24 | F | GGCTCTGTAAGAAGGATGTGG | 189 | NM_001129904.1 |
| R | CCAGTTCCAAAAGGCAGGTA | |||
| AKR1C4 | F | ACCAGCCTTGGAAAAGTCAC | 183 | NM_181027.2 |
| R | TGCGTCCTTACACTTCTCCA | |||
| AKR1D1 | F | AGGACACTCAAGGACCTCCA | 186 | NM_001192358.1 |
| R | ACTTCACCAAGCCAGCATCT | |||
| ACTB | F | ACTGGGACGACATGGAGAAG | 167 | NM_001009784.1 |
| R | GTACATGGCAGGGGTGTTG | |||
| PPIA | F | TCTGAGCACTGGAGAGAAAGG | 178 | AY251270.1 |
| R | GATGCCAGGACCTGTATGCT | |||
| GAPDH | F | GGCGTGAACCACGAGAAGTATAA | 394 | NM_001190390.1 |
| R | CCCTCCACGATGCCAAAGT | |||
| ATPase | F | TTCCTACTGCCCTGGAATG | 98 | NM_001009360.1 |
| R | CACGAAGATGAGAAGCGAGT | |||
| RPL19 | F | GAAATCGCCAATGCCAAC | 361 | AY158223.1 |
| R | GAGCCTTGTCTGCCTTCA | |||
F = Forward; R = Reverse.
Five housekeeping genes were chosen and used to normalize results for the target genes, based on previous reliability information using geNorm software (Goossens et al., 2005): β-actin (ACTB), cyclophilin A (PPIA), ribosomal protein L19 (RPL19), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and ATPase. Reverse transcribed products were diluted 1:8 in ribonuclease-free water. Real-time quantitative PCR was performed using a LightCycler 480 (Roche Diagnostics GmbH, Mannheim, Germany). For each gene, the reaction was performed in duplicate wells using optical 96-well reaction plates using the SYBR Green I Master Mix (Roche Diagnostics GmbH). Positive and negative (no cDNA) controls were included for each primer set on each plate. The RT-qPCR conditions underwent initial denaturation at 95°C for 5 min followed by 40 cycles of amplification at 95°C for 5 s, 58°C for 30 s, and 72°C for 20 s. Specificity of RT-qPCR products were analyzed by comparing the dissociation melting curves with the positive and negative controls and by agarose gel electrophoresis.
The amplification efficiency for each sample was calculated using the program LingRegPCR (http://www.hartfaalcentrum.nl/index.php?main=files&fileName=LinRegPCR.zip&description=LinRegPCR:%20qPCR%20data%20analysis&sub=LinRegPCR). Four points on the exponential curve were determined during the exponential phase of amplification. Fluorescence data obtained from the RT-qPCR program were exported to LingRegPCR, which uses a linear regression analysis of the fluorescence data of the exponential phase of PCR to determine the amount of mRNA and the efficiency of amplification. Using LingRegPCR, threshold cycle (Ct) values were determined for each sample. Based on the amplification efficiency for each individual sample, obtained from the software, procedures were adopted to correct the Ct values for the optimal efficiency of amplification.
Statistical Analysis
Data were analyzed with the methodology of generalized linear models, using PROC GLIMMIX of SAS (SAS Inst. Inc., Cary, NC). The statistical model used to analyze feed consumption included the fixed effects block, diet (ad libitum vs. restricted diet), week of evaluation, and interaction between diet and week. For analysis of weight gain, the model included effects of block and diet (ad libitum vs. restricted). Analysis of repeated measurements of insulin concentrations during the adaptation period contained the effects of block and week and the random effects of animal and experimental error, using the best correlation matrix between measurements in the same animal. Concentrations of insulin and P4 in the fourth and eighth weeks were independently analyzed with a model that included the effects of block, diet (ad libitum vs. restricted), time in relation to feeding, the interaction between diet and time, and random effects of animal and experimental error, using the best correlation matrix between the measurements of the animal.
All statistical comparisons were performed on the adjusted means using the least squares method and are presented as means and SE, to aid in interpretation of results. Least squares mean separation was performed using the Tukey test, and the differences between treatments were considered significant for values of P < 0.05.
For relative quantification, the Ct values for each sample were determined using the program LingRegPCR (Ramakers et al., 2003) using at least 4 points of the amplification curve during the exponential phase of PCR. A procedure was adopted from the individual efficiencies obtained through LinRegPCR to correct values for the ideal efficiency of 2. The adjust was calculated using the following equation: Ctij = log2[E_geneijCt_gene(ij)], in which Ctij is the Ct of the jth sample from the ith gene corrected for efficiency of 2 and Ct_gene(ij) is the original threshold cycle on E_geneij with the actual efficiency calculated by the program.
For data normalization, a general linear mixed model that considers the fixed and random effects originating from the experimental design was used, according to the following equation: Yij = μ + Gi + Sj + eij, in which Yij is the Ct for ith target gene of the jth sample, μ is the average of Ct, Gi is the fixed effect for the ith gene, Sj is the random effect associated with the sample considering Sj ∼ normally and independently distributed (NID; 0, σa2), where σa2 is additive genetic variance, and eij is the random residual effect, with eij ∼ NID(0, σe2) where σe2 is residual variance.
From this model we obtained the values of the BLUP for the experimental animals, and these were used as adjustment factors for Ct values of target genes (CYP2C, CYP3A, AKR1C, and AKR1D). The effects of different treatments on mRNA for each liver enzyme were analyzed using the model
in which Yij is the phenotypic value for concentrations of each mRNA (in Ct), μ is a constant inherent to each mRNA, Mi is the fixed effect of treatments for each mRNA (CYP2C, CYP3A, AKR1C, and AKR1D), and eij = random residual effect associated with the trait Yij, with eij ∼ NID(0, σe2).
The equations were used for quantification of mRNA as well as to calculate the delta-delta Ct values for each mRNA.
RESULTS
There were no differences in BW between the experimental groups during wk −1 and 0, before initiation of the experimental diets (Fig. 2). As designed for the experiment, ewes that received the ad libitum diet had greater (P < 0.001) average daily DMI during the experimental period compared with the group that received the restricted diet (means of 1.8 [SE 0.03] vs. 0.6 kg/ewe per day [SE 0.01], respectively). By 1 wk after initiation of the experimental diets, Ad ewes had greater BW and this difference was present (P < 0.001) throughout the remainder of the experimental period (Fig. 2), resulting in greater (P < 0.001) rate of BW gain than R ewes (1.7 ± 0.12 vs. −0.1 ± 0.03 kg/wk, respectively). Moreover, Ad ewes had increasing BMI during the study (from 74.5 at wk 0 to 80.8 at wk 4 and 83.4 at wk 8) whereas R ewes lost BMI during the experiment (from 78.5 at wk 0 to 69.9 at wk 4 and 63.4 at wk 8; P < 0.001).
Figure 2.
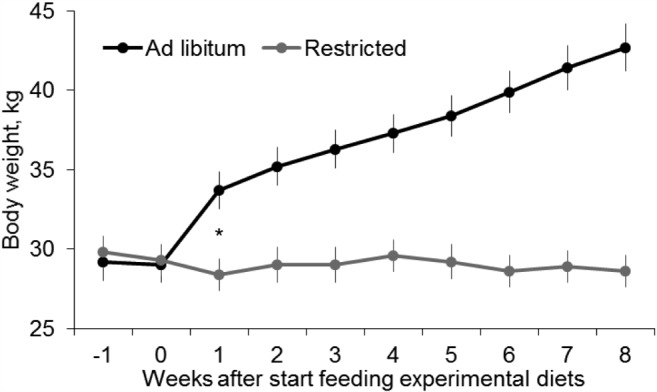
Body weight (mean ± SE) of ewes fed ad libitum (3.8% DM/kg BW; n = 12) or restricted (2.0% DM/kg BW; n = 12) diets throughout the experimental period. *P < 0.001 for wk 1 to 8.
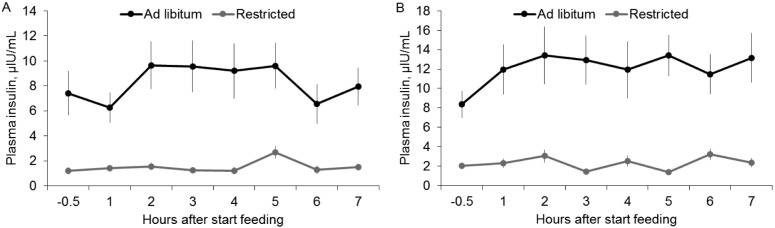
Prior to initiation of the experimental diets, basal circulating insulin concentrations were least squares mean of 3.5 μIU/mL (SE 0.69). Also during this adaptation period, the peak postprandial circulating concentrations of insulin were least squares mean of 13.9 μIU/mL (SE 2.33), and the peak was observed at 1 h after feed was offered. There was no difference (P > 0.10) in circulating insulin during the adaptation period between Ad ewe lambs and R ewe lambs (data not shown). During the experimental period, average circulating insulin concentrations were much greater (P < 0.001) for the Ad ewes than for the R ewes, both at the fourth week (8.2 ± 0.93 vs. 1.5 ± 0.16 μIU/mL, respectively) and at the eighth week (12.0 ± 1.02 vs. 2.2 ± 0.18 μIU/mL, respectively). Moreover, there was an effect of treatment (P < 0.001) and time (P < 0.05) on circulating insulin at the end of the fourth and eighth weeks (Fig. 3). There was, however, no interaction between treatment and time at any of the evaluated periods (P > 0.10).
Figure 3.
Least squares means ± SE of plasma concentrations of insulin (μIU/mL) before (−0.5 h) and after (1, 2, 3, 4, 5, 6, and 7 h) offering feed to ewes submitted to the ad libitum diet (3.8% DM/kg BW; n = 12) or the restricted diet (2% DM/kg BW; n = 12) at the end of the fourth (A) and eighth (B) experimental week. For the fourth week: treatment (P < 0.001), time (P < 0.01), and treatment × time (P = 0.30). For the eighth week: treatment (P < 0.001), time (P < 0.05), and treatment × time (P = 0.11).
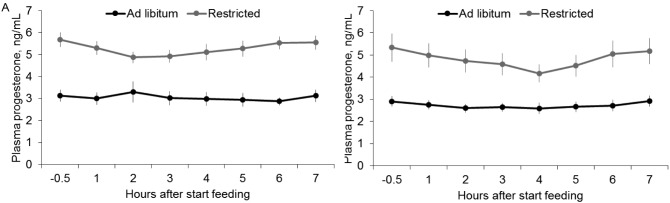
Prior to PGF2α treatment/CIDR device insertion, 1 ewe from the Ad group at the fourth week and another one from the same treatment at the eighth week had circulating P4 above 1 ng/mL. Despite that, during the subsequent evaluations (in the presence of a P4 implant), their concentrations of circulating P4 were not different (P > 0.10) from the others within the same treatment group, indicating that PGF2α treatment was effective. Average circulating P4 concentrations during the experimental periods, in which ewes were implanted with an intravaginal P4 device, were lower (P < 0.001) for the Ad group than for the R group both at the fourth week (least squares means of 3.2 [SE 0.32] vs. 5.5 ng/mL [SE 0.32], respectively) and at the eighth week (least squares means of 2.8 [SE 0.28] vs. 5.2 ng/mL [SE 0.28], respectively). Moreover, there was an effect of treatment and time (P < 0.001) on circulating P4 but no interaction between treatment and time at the end of the fourth week (P = 0.11) or at the end of the eighth week (P = 0.31; Fig. 4). There was no difference between the fourth and eighth weeks for circulating P4 for either the R ewes or the Ad ewes (P > 0.15). Despite significant differences in BW among ewes within experimental groups considering evaluations performed at the fourth and eighth weeks (restricted: ranging from 21.5 to 35.5 kg; ad libitum: ranging from 30.3 to 52.3 kg), there was no correlation (P > 0.15) within groups between BW and circulating P4 for either R ewes (R = 0.24) or Ad diet ewes (R = 0.08).
Figure 4.
Least squares means ± SE of plasma progesterone concentrations (ng/mL) before (−0.5 h) and after (1, 2, 3, 4, 5, 6, and 7 h) offering feed to ewes submitted to the ad libitum diet (3.8% DM/kg BW; n = 12) or the restricted diet (2% DM/kg BW; n = 12) at the end of the fourth (A) and eighth (B) experimental week. For the fourth week: treatment (P < 0.001), time (P < 0.001), and treatment × time (P = 0.11). For the eighth week: treatment (P < 0.001), time (P < 0.001), and treatment × time (P = 0.31).
The expression of mRNA for the steroid metabolizing enzymes was evaluated in liver biopsies from ewes in the 2 treatment groups at the fourth and eighth weeks of the study. There were no effects of dietary treatments at the end of the experimental week on concentrations of mRNA for CYP2C19 (P = 0.86 in the fourth week and P = 0.07 in the eighth week), CYP3A4 (P = 0.91 in the fourth week and P = 0.39 in the eighth week), AKR1C4 (P = 0.88 in the fourth week and P = 0.91 in the eighth week), or AKR1D1 (P = 0.74 in the fourth week and P = 0.19 in the eighth week; Table 3).
Table 3.
Expression of mRNA (ΔCT1) and confidence interval (in parentheses) for the liver enzymes cytochrome P450 2C19 (CYP2C19), CYP3A4, aldo-keto reductase 1C4 (AKR1C4), and AKR1D1 in noncyclic ewes at 4 and 8 wk after onset of treatment diets (ad libitum vs. restricted diets; 3.8 vs. 2.0% DM/kg BW per day, respectively). Reference genes: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), cyclophilin A (PPIA), ATPase, ribosomal protein L19 (RPL19), and β-actin (ACTB).
| 4 wk | 8 wk | |||||
|---|---|---|---|---|---|---|
| Enzymes | Ad libitum n = 10 | Restricted n = 11 | P-value | Ad libitum n = 10 | Restricted n = 11 | P value |
| CYP2C19 | 0.88 (0.33–2.36) | 0.79 (0.30–2.06) | 0.86 | 2.39 (1.55–3.69) | 1.43 (1.07–2.18) | 0.07 |
| CYP3A4 | 3.88 (1.84–8.16) | 4.08 (1.98–8.44) | 0.91 | 8.89 (6.04–13.08) | 7.23 (4.96–10.53) | 0.39 |
| AKR1C4 | 1.45 (1.29–2.72) | 1.38 (1.35–2.58) | 0.88 | 4.54 (2.81–7.33) | 4.70 (2.91–7.60) | 0.91 |
| AKR1D1 | 0.13 (0.07–0.27) | 0.16 (0.08–0.32) | 0.74 | 0.33 (0.21–0.52) | 0.49 (0.31–0.78) | 0.19 |
CT = cycle threshold.
DISCUSSION
Plasma insulin concentrations were much greater for ewes that received ad libitum feed than for ewes that received the restricted diet. This was expected due to the high DMI of the ewe lambs in the ad libitum Ad group. It is well known that greater secretion of insulin occurs during greater energy intake and, therefore, circulating concentrations can be used as an indicator of the energy “status” of the animal (Schwartz et al., 2000; Butler et al., 2004). However, the magnitude of the effect in the present experiment was noteworthy, with >5-fold greater circulating insulin at both the fourth and eighth weeks of the experimental treatment. This dramatic difference in insulin allowed us to unambiguously test whether the longer-term effect of elevated insulin could regulate hepatic expression of P4 metabolizing enzymes and whether these changes could override the direct effects of high feed intake to increase P4 catabolism, presumably through elevated gastrointestinal and liver blood flow. Our first hypothesis was that high energy intake would increase insulin and that increased insulin would produce a decrease in expression of mRNA for the liver enzymes involved in P4 catabolism. Our results clearly indicated an increase in insulin but did not provide support for the hypothesis that increased insulin would decrease mRNA for liver enzymes involved in P4 catabolism. This result was based on an analysis of 10 or 11 ewes per treatment group and clearly demonstrated no detectable change in liver mRNA for any of the enzymes, in spite of the dramatic increase in circulating insulin either in ewes that had not been previously exposed to P4 (fourth week) or in ewes pre-exposed (eighth week). Our second hypothesis was that there would be no change in circulating P4 in the 2 groups because an increased hepatic P4 catabolism, which was expected in the group with increased feed consumption possibly due to increased liver blood flow, would be offset by an insulin-induced decrease in hepatic expression of the P4 metabolizing enzymes. However, again we found no evidence for any reduction in P4 catabolism due to the elevated circulating insulin and observed only the distinct increase in P4 catabolism that would be expected in animals with high feed consumption. This produced a clear and striking decrease in circulating P4 in the ewes that were fed the ad libitum diet compared with the ewes on the restricted diet. Therefore, our results are clearly consistent with the idea that P4 catabolism is increased by increased DMI but are surprisingly contradictory to the idea that elevated insulin causes a reduction in hepatic gene expression of P4 metabolizing enzymes.
Various studies have shown that in ruminants, high-carbohydrate diets will generate greater quantities of acetate and propionate in the rumen, and this can induce dramatic increases in circulating insulin (Bergman, 1990; Gong et al., 2002), as observed in the present study. Carbohydrates are converted into short-chain fatty acids as a result of microbial fermentation, and these fatty acids are the primary stimulus for secretion of insulin by the pancreas in ruminants (De Jong, 1982). In addition, propionate is an important precursor for gluconeogenesis in the liver. Increased availability of energy, in the form of glucose or fatty acids, has been associated with earlier time to first ovulation in cattle (Gong et al., 2002; Butler et al., 2004) due to increased GnRH/LH pulses driving follicular development (Arias et al., 1992; Beam and Butler, 1999). Similar to our work, several studies have shown the effects of increased DMI or greater energy consumption on circulating concentrations of insulin (Butler et al., 2004; Lemley et al., 2008b, 2010b; Moriel et al., 2008).
There was a small but significant increase in circulating insulin between 2 and 5 h after feeding in both experimental groups, similar to a previous study in nonlactating cows that reported an increase in circulating insulin at 2 h after feed consumption (Moriel et al., 2008). However, the major differences in circulating insulin were due to effects of diet and not the time of feeding, with over 5-fold greater circulating insulin in ewes receiving an ad libitum diet compared with ewes receiving a restricted diet at all times before and after feeding.
On the other hand, there was an inverse relationship between the level of feed intake and plasma concentrations of P4, as observed in our study and in other studies in sheep and cattle (Parr et al., 1993a,b; Vasconcelos et al., 2003). In previous studies, an increase in clearance rate for P4 has been clearly associated with greater hepatic blood flow in sheep and cattle (Parr et al., 1993b; Sangsritavong et al., 2002; Wiltbank et al., 2014). In this study, an exogenous source of P4 was used in an attempt to standardize the input of P4 to the ewe so that increased clearance would be the primary reason for any changes in circulating P4 concentrations. Differences in BW might be expected to alter steady state circulating P4 concentrations, probably due to greater liver size and liver blood flow and, therefore, greater P4 metabolism. However, it seems likely that greater P4 metabolism due to greater feed intake was primarily responsible for the decrease in circulating P4 and not just the increase in BW. This is evidenced by the similar circulating P4 concentrations in Ad ewes at the fourth and eighth weeks of treatment, despite continuing increases in BW between the fourth and eighth weeks of ad libitum feeding. Furthermore, the difference in circulating P4 between Ad ewes and R ewes (71.9% greater at the fourth week) was much greater than the difference in BW (29.3% greater at the fourth week). Therefore, a dramatic decrease in circulating P4, despite similar P4 delivery, indicates that P4 metabolism was increased in Ad ewes compared with R ewes.
Circulating P4 is inactivated or catabolized in hepatocytes by CYP2C and CYP3A and AKR, mainly AKR1C and AKR1D (Lemley and Wilson, 2010; Jin et al., 2011). Several studies have reported that insulin is an inhibitor of expression of the mRNA or of the protein for these enzymes in the liver of sheep and cattle (Lemley et al., 2008a,b; Vieira et al., 2013). For instance, in the study by Lemley et al. (2008a), treatment of cows with propylene glycol produced a 30% increase in circulating insulin with no change in CYP2C mRNA but a 40% decrease in the amount of CYP3A mRNA. In this same study (Lemley et al., 2008a), as insulin was infused (1 g insulin ∙ kg–1 BW ∙ h–1), there was a decrease in the expression of both enzymes CYP2C (88%) and CYP3A (45%). However, in another study (Lemley et al., 2010b), comparing high-grain vs. high-fiber diets, a 22% increase in circulating insulin at 10 h after feeding was associated with no change in expression of CYP2C or CYP3A. The present study produced a more definitive increase in circulating insulin and for longer periods of time with our 4- and 8-wk dietary interventions; however, we observed no change in mRNA expression of CYP2C, CYP3A, AKR1C, or AKR1D. Therefore, our results indicate that elevated insulin for extended periods of time (up to 8 wk) do not alter hepatic expression of mRNA for any of the steroid-metabolizing enzymes that were measured in this study. In addition, our results indicating a reduction in circulating P4 concentrations in ewes that had high feed consumption demonstrate that there was increased P4 catabolism in response to increased energy intake. This latter suggestion is consistent with the idea that any compensatory decrease in liver enzymes was not sufficient to offset the dramatic increase in P4 catabolism that is caused by high feed intake.
In conclusion, high energy intake decreased circulating P4 concentrations, probably through a primary effect on increased liver blood flow. In addition, we found no evidence for insulin-induced changes in mRNA for some of the major hepatic P4 metabolizing enzymes in this ewe model. It seems likely that although high DMI can produce dramatic increases in circulating insulin, the major factor responsible for alterations in circulating P4 caused by high DMI is the previously demonstrated increase in hepatic blood flow that elevates steroid catabolism.
FOOTNOTES
The first author and coauthors were supported by scholarships from the Coordination for the Improvement of Higher Education Personnel (CAPES) of Brazil and the National Council for Scientific and Technological Development (CNPq) of Brazil. This project was funded by the grant 2010/20704-4 from the São Paulo Research Foundation (FAPESP). We thank Prof. Eunice Oba of UNESP, Botucatu, for her assistance with hormone assays and Zoetis for donating hormones.
LITERATURE CITED
- Arias P., Rodríguez M., Szwarcfarb B., Sinay I. R., Moguilevsky J. A. 1992. Effect of insulin on LHRH release by perifused hypothalamic fragments. Neuroendocrinology 56:415–418. doi: 10.1159/000126257 [DOI] [PubMed] [Google Scholar]
- Beam S. W., Butler W. R. 1999. Effects of energy balance on follicular development and first ovulation in postpartum dairy cows. J. Reprod. Fertil. 54:411–424. [PubMed] [Google Scholar]
- Bergman E. N. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70:567–590. [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., Wittwer C. T. 2009. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55(4):611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Butler S. T., Pelton S. H., Butler W. R. 2004. Insulin increases 17 beta-estradiol production by the dominant follicle of the first postpartum follicle wave in dairy cows. Reproduction 127:537–545. doi: 10.1530/rep.1.00079 [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal. Biochem. 162:156–159. doi: 10.1016/0003-2697(87)90021-2 [DOI] [PubMed] [Google Scholar]
- De Jong A. 1982. Patterns of plasma-concentrations of insulin and glucagon after intravascular and intra-ruminal administration of volatile fatty-acids in the goat. J. Endocrinol. 92:357–370. doi: 10.1677/joe.0.0920357 [DOI] [PubMed] [Google Scholar]
- Freetly H. C., Ferrell C. L. 1994. Net uptakes of estradiol 17-beta and progesterone across the portal-drained viscera and liver of ewes. J. Endocrinol. 141:353–358. doi: 10.1677/joe.0.1410353 [DOI] [PubMed] [Google Scholar]
- Gong J. G., Lee W. J., Garnsworthy P. C., Webb R. 2002. Effect of dietary-induced increases in circulating insulin concentrations during the early postpartum period on reproductive function in dairy cows. Reproduction 123:419–427. doi: 10.1530/rep.0.1230419 [DOI] [PubMed] [Google Scholar]
- Goossens K., Van Poucke M., Van Soom A., Vandesompele J., Van Zeveren A., Peelman L. J. 2005. Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev. Biol. 5:27. doi: 10.1186/1471-213X-5-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Mesaros A. C., Blair I. A., Penning T. M. 2011. Stereospecific reduction of 5 beta-reduced steroids by human ketosteroid reductases of the AKR (aldo-keto reductase) superfamily: Role of AKR1C1-AKR1C4 in the metabolism of testosterone and progesterone via the 5 beta-reductase pathway. Biochem. J. 437:53–61. doi: 10.1042/BJ20101804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T., Remm M. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. doi: 10.1093/bioinformatics/btm091 [DOI] [PubMed] [Google Scholar]
- Lemley C. O., Butler S. T., Butler W. R., Wilson M. E. 2008a. Short communication: Insulin alters hepatic progesterone catabolic enzymes cytochrome P4502C and 3A in dairy cows. J. Dairy Sci. 91:641–645. doi: 10.3168/jds.2007-0636 [DOI] [PubMed] [Google Scholar]
- Lemley C. O., Koch J. M., Blemings K. P., Krause K. M., Wilson M. E. 2008b. Concomitant changes in progesterone catabolic enzymes, cytochrome P4502C and 3A, with plasma insulin concentrations in ewes supplemented with sodium acetate or sodium propionate. Animal 2:1223–1229. doi: 10.1017/S1751731108002462 [DOI] [PubMed] [Google Scholar]
- Lemley C. O., Koch J. M., Blemings K. P., Wilson M. E. 2009. Alterations in progesterone catabolic enzymes, CYP2C and CYP3A, in hepatocytes challenged with insulin and glucagon. J. Anim. Vet. Adv. 8:39–46. [Google Scholar]
- Lemley C. O., Tager L. R., Wilmoth T. A., Krause K. M., Vonnahme K. A., Wilson M. E. 2010a. Progesterone clearance in dairy cows fed an insulin stimulating diet. J. Dairy Sci. 93:2304–2305. [DOI] [PubMed] [Google Scholar]
- Lemley C. O., Vonnahme K. A., Tager L. R., Krause K. M., Wilson M. E. 2010b. Diet-induced alterations in hepatic progesterone (P-4) catabolic enzyme activity and P-4 clearance rate in lactating dairy cows. J. Endocrinol. 205:233–241. doi: 10.1677/JOE-10-0042 [DOI] [PubMed] [Google Scholar]
- Lemley C. O., Wilmoth T. A., Tager L. R., Krause K. M., Wilson M. E. 2010c. Effect of a high cornstarch diet on hepatic cytochrome P450 2C and 3A activity and progesterone half-life in dairy cows. J. Dairy Sci. 93:1012–1021. doi: 10.3168/jds.2009-2539 [DOI] [PubMed] [Google Scholar]
- Lemley C. O., Wilson M. E. 2010. Effect of cytochrome P450 and aldo-keto reductase inhibitors on progesterone inactivation in primary bovine hepatic cell cultures. J. Dairy Sci. 93:4613–4624. doi: 10.3168/jds.2010-3165 [DOI] [PubMed] [Google Scholar]
- Moriel P., Scatena T. S., Sa O. G., Cooke R. F., Vasconcelos J. L. M. 2008. Concentrations of progesterone and insulin in serum of nonlactating dairy cows in response to carbohydrate source and processing. J. Dairy Sci. 91:4616–4621. doi: 10.3168/jds.2008-1286 [DOI] [PubMed] [Google Scholar]
- Murray M. 1991. Microsomal cytochrome-P450-dependent steroid metabolism in male sheep liver – Quantitative importance of 6-beta-hydroxylation and evidence for the involvement of a P450 from the IIA subfamily in the pathway. J. Steroid Biochem. Mol. Biol. 38:611–619. doi: 10.1016/0960-0760(91)90320-5 [DOI] [PubMed] [Google Scholar]
- Murray M. 1992. Participation of the a cytochrome P450 enzyme from the 2C subfamily in progesterone 21-hydroxylation in sheep liver. J. Steroid Biochem. Mol. Biol. 43:591–593. doi: 10.1016/0960-0760(92)90248-H [DOI] [PubMed] [Google Scholar]
- NRC 2007. Nutrient requirements of small ruminants: Sheep, goats, cervids and New World camelids. Natl. Acad. Press, Washington, DC. [Google Scholar]
- Parr R. A. 1992. Nutrition-progesterone interactions during early pregnancy in sheep. Reprod. Fertil. Dev. 4:297–300. doi: 10.1071/RD9920297 [DOI] [PubMed] [Google Scholar]
- Parr R. A., Davis I. F., Fairclough R. J., Miles M. A. 1987. Overfeeding during early pregnancy reduces periperpheral progesterone concentration and pregnancy in sheep. J. Reprod. Fertil. 80:317–320. doi: 10.1530/jrf.0.0800317 [DOI] [PubMed] [Google Scholar]
- Parr R. A., Davis I. F., Miles M. A., Squires T. J. 1993a. Feed intake affects metabolic clearance rate of progesterone in sheep. Res. Vet. Sci. 55:306–310. doi: 10.1016/0034-5288(93)90099-2 [DOI] [PubMed] [Google Scholar]
- Parr R. A., Davis I. F., Miles M. A., Squires T. J. 1993b. Liver blood flow and metabolic clearance rate of progesterone in sheep. Res. Vet. Sci. 55:311–316. doi: 10.1016/0034-5288(93)90100-T [DOI] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J. M., Deprez R. H. L., Moorman A. F. M. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62–66. doi: 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- Sangsritavong S., Combs D. K., Sartori R., Armentano L. E., Wiltbank M. C. 2002. High feed intake increases liver blood flow and metabolism of progesterone and estradiol-17 beta in dairy cattle. J. Dairy Sci. 85:2831–2842. doi: 10.3168/jds.S0022-0302(02)74370-1 [DOI] [PubMed] [Google Scholar]
- Schwartz M. W., Woods S. C., Porte D., Seeley R. J., Baskin D. G. 2000. Central nervous system control of food intake. Nature 404:661–671. [DOI] [PubMed] [Google Scholar]
- Udvardi M. K., Czechowski T., Scheible W. R. 2008. Eleven golden rules of quantitative RT-PCR. Plant Cell 20:1736–1737. doi: 10.1105/tpc.108.061143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos J. L. M., Sangsritavong S., Tsai S. J., Wiltbank M. C. 2003. Acute reduction in serum progesterone concentrations after feed intake in dairy cows. Theriogenology 60:795–807. doi: 10.1016/S0093-691X(03)00102-X [DOI] [PubMed] [Google Scholar]
- Vieira F. V. R., Cooke R. F., Aboin A. C., Lima P., Vasconcelos J. L. M. 2013. Short communication: Acute but transient increase in serum insulin reduces messenger RNA expression of hepatic enzymes associated with progesterone catabolism in dairy cows. J. Dairy Sci. 96:1085–1089. doi: 10.3168/jds.2012-5783 [DOI] [PubMed] [Google Scholar]
- Wiltbank M., Lopez H., Sartori R., Sangsritavong S., Gumen A. 2006. Changes in reproductive physiology of lactating dairy cows due to elevated steroid metabolism. Theriogenology 65:17–29. doi: 10.1016/j.theriogenology.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Wiltbank M. C., Souza A. H., Carvalho P. D., Cunha A. P., Giordano J. O., Fricke P. M., Baez G. M., Diskin M. G. 2014. Physiological and practical effects of progesterone on reproduction in dairy cattle. Animal 8:70–81. doi: 10.1017/S1751731114000585 [DOI] [PubMed] [Google Scholar]