Abstract
Zebu animals (Bos indicus) are known to take longer to reach puberty compared with taurine animals (Bos taurus), limiting the supply of animals for harvest or breeding and impacting profitability. Genomic information can be a helpful tool to better understand complex traits and improve genetic gains. In this study, we performed a genomewide association study (GWAS) to identify genetic variants associated with reproductive traits in Nelore beef cattle. Heifer pregnancy (HP) was recorded for 1,267 genotyped animals distributed in 12 contemporary groups (CG) with an average pregnancy rate of 0.35 (±0.01). Disregarding one of these CG, the number of antral follicles (NF) was also collected for 937 of these animals, with an average of 11.53 (±4.43). The animals were organized in CG: 12 and 11 for HP and NF, respectively. Genes in linkage disequilibrium (LD) with the associated variants can be considered in a functional enrichment analysis to identify biological mechanisms involved in fertility. Medical Subject Headings (MeSH) were detected using the MESHR package, allowing the extraction of broad meanings from the gene lists provided by the GWAS. The estimated heritability for HP was 0.28 ± 0.07 and for NF was 0.49 ± 0.09, with the genomic correlation being −0.21 ± 0.29. The average LD between adjacent markers was 0.23 ± 0.01, and GWAS identified genomic windows that accounted for >1% of total genetic variance on chromosomes 5, 14, and 18 for HP and on chromosomes 2, 8, 11, 14, 15, 16, and 22 for NF. The MeSH enrichment analyses revealed significant (P < 0.05) terms associated with HP—“Munc18 Proteins,” “Fucose,” and “Hemoglobins”—and with NF—“Cathepsin B,” “Receptors, Neuropeptide,” and “Palmitic Acid.” This is the first study in Nelore cattle introducing the concept of MeSH analysis. The genomic analyses contributed to a better understanding of the genetic control of the reproductive traits HP and NF and provide new selection strategies to improve beef production.
Keywords: beef cattle, enrichment analysis, genomewide association study, genomics, linkage disequilibrium, Medical Subject Headings terms
INTRODUCTION
Reproductive traits are economically important in beef cattle production, especially for zebu animals (Bos indicus), where heifers often take longer to reach puberty (Abeygunawardena and Dematawewa, 2004; Eler et al., 2014) compared with taurine animals (Bos taurus). Reduced heifer fertility impacts the profitability of a given size herd in the production system by decreasing the number of weaned calves and increasing the lifetime costs of herd replacements (Eler et al., 2014).
The inclusion of genomic information in genetic evaluations can be a helpful tool to analyze polygenic traits, such as those for reproduction, shortening the time it takes to obtain reliable breeding value estimates, which can lead to reduced generation intervals and improving genetic gains. It positively affects accuracies by including a more precise measure of genetic similarity between individuals compared with traditional pedigree-based evaluations (Berry et al., 2012; Hayes et al., 2013). Genomic markers such as SNP can be used in genomewide association studies (GWAS), a well-established strategy that has identified thousands of markers spread across the cattle genome related to important economic traits (Hu et al., 2016). Genomewide association study relies on linkage disequilibrium (LD) between at least 1 marker and the causal mutation or quantitative trait nucleotides responsible for the observed phenotypic variation (Khatkar et al., 2014).
Female fertility can be measured in a number of different ways (Khatkar et al., 2014). Heifer pregnancy (HP), defined by the determination of whether or not a heifer exposed to breeding has become pregnant, is a directly measured trait for which data collection is an easy and inexpensive process (Eler et al., 2002). This allows its implementation as a selection criterion in large herds.
Because reproductive technologies, such as ovum pick up and in vitro fertilization (IVF), are widely adopted in zebu herds, the numbers of antral follicles (NF) has become a relevant trait, due to its association with female performance for ovum pick up and IVF (Baruselli et al., 2015).
A better understanding of genetic factors that affect reproduction could lead to substantial improvement in genetic selection to improve reproductive rates (Kappes et al., 2000). Identification of genomic regions associated with reproductive traits could expand our understanding of reproductive processes and be used to improve reproductive efficiency in cattle, especially in zebu animals.
Medical Subject Headings (MeSH) is a collection of comprehensive life sciences terms (Nelson et al., 2004) organized by the U.S. National Library of Medicine (Bethesda, MD); it is the annotation used for PubMed (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3013746/pdf/gkq1237.pdf) documents. These terms are clustered into 16 categories, and the size of MeSh's vocabulary library is approximately twice as large as that of Gene Ontology (Nakazato et al., 2007). They have been recently used in overrepresentation analysis (Morota et al., 2015), permitting the extraction of broad meaning from the gene lists provided by GWAS.
In this study, phenotypic information for HP and NF were collected from Nelore heifers on commercial farms in Brazil. Following whole-genome genotyping, GWAS was performed to identify genomic regions associated with these traits followed by MeSH enrichment analyses of these regions, aiming to increase knowledge about the genetic influence and biological role of genes related to HP. This is the first study in Nelore cattle introducing the concept of MeSH analysis.
MATERIAL AND METHODS
Data Set
The data set consisted of the pregnancy status on 2,283 Nelore heifers exposed to breeding, and around 48% of these (1,099 animals) also had NF measured. The data were from 3 commercial beef cattle farms called Segredo (Farm 1), Engano (Farm 2) and CFM (Farm 3), located in the state of Mato Grosso do Sul in midwestern Brazil and were stored and analyzed by the Animal Breeding and Biotechnology Group of the College of Animal Science and Food Engineering, Pirassununga, São Paulo, Brazil. The animals were about 16 mo old when data was collected, having been raised under similar environmental conditions and receiving salt and mineral supplementation on a pasture-based system, with ad libitum water.
The animals were allocated into 12 contemporary groups (CG), formed by the combination of the animal's herd of origin, management group, and year of birth. Records from CG without variability (e.g., all animals had the same pregnancy status) were not considered in the analyses. In addition, animals with an age or weight exceeding 3.5 SD above or below the overall CG mean were excluded.
Traits
Heifer Pregnancy
The diagnosis of pregnancy was performed using ultrasound (Chison 8200VET with 7.5 MHz transducer; Kylumax, Wuxi, China) or transrectal palpation 40 d after AI. Phenotypic records were treated as categorical, assigning the value of 1 (success) to heifers that were diagnosed pregnant and 0 (failure) to those that were not pregnant at the time of diagnosis.
In tropical countries, calving is normally spread from July to November, with heifers entering the October breeding season at between 12 (born in November) and 16 mo old (born in July). So challenging animals around 15 mo old is a strategy to select for sexual precocity that fits in with the routine management of the farms (Eler et al., 2002).
Number of Antral Follicles
Ovarian ultrasound (7.5 MHz transrectal linear transducer, Mindray M5Vet; Mindray, Mahwah, NJ) was performed on those Nelore heifers submitted to fixed-time AI. Visible follicles (≥3 mm of diameter) were counted and recorded on Day 4 of the protocol to establish the total NF recruited in the synchronized follicular wave. This procedure was performed in 2 (Engano and Segredo) of the 3 farms.
Genotypic Data
Hair samples of 1,267 young heifers (about 16 mo old) from 2,283 available animals were collected for genomic DNA extraction and subsequent genotyping analysis using the GeneSeek GGP Bos indicus HD array (Neogen, Lincoln, NE) with 74,677 SNP, specially developed for B. indicus cattle. All 1,267 animals had phenotypic records for HP and, of those, 938 also had NF records.
Quality control procedures were performed using PREGSF90 version 1.10 software (Misztal et al., 2014) to reduce spurious associations and, consequently, increase the accuracy of the genomic analyses. The quality control parameters used herein to delete loci were minor allele frequency < 2% and SNP call rate < 95% and when the observed percentage of heterozygous markers differed from expected (Hardy–Weinberg equilibrium) by >15%. Samples with call rate < 90% were also excluded. Genotypes from loci on the Y chromosome, the mitochondria, and markers not assigned to chromosomes were eliminated from the data. After editing, a total of 64,753 SNP and 1,255 genotyped heifers remained in the data set. The frequency of animals among farms and their phenotypic information are summarized in Table 1.
Table 1.
Average values (SD) for age, weight, heifer pregnancy (HP) rate and number of antral follicles (NF) of the genotyped (after quality control) Nelore heifers by farm
| Item | Farm 1 | Farm 2 | Farm 3 |
|---|---|---|---|
| No. of animals | 579 | 299 | 377 |
| Age of animals, mo. | 16.1 (1.20) | 14.1 (0.87) | 17.2 (0.52) |
| Weight of animals, kg | 283.2 (28.92) | 260.6 (19.20) | 276.5 (16.73) |
| HP | 0.33 | 0.35 | 0.36 |
| NF | 12.07 | 10.99 | – |
Variance Components Estimation
Genetic and residual variances or covariances for HP and NF were estimated in a bivariate model using the single-step methodology proposed by Legarra et al. (2014) under a Bayesian approach. The statistical model used was
in which y is the vector of the dependent variable (observed phenotypes) for genotyped and nongenotyped animals; b is the vector of fixed effects, including the CG and linear covariate for heifer age at pregnancy diagnosis; a is the vector of random additive genetic effects; X and Z are incidence matrices relating b and a with the dependent variable y; and e is a vector of random residual errors. Variance components for HP were estimated using a threshold model, which related the observed trait on a categorical scale to an underlying continuous normal scale, whereas NF was modeled as a continuous variable.
The covariance matrix of a and e assumed
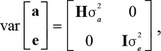 |
in which σa2 and σe2 are the variance components of the additive genetic and residual effects, respectively; I is an identity matrix of order equal to the number of animals with phenotypes; and H is the relationship matrix that combines information from the genotyped and nongenotyped animals considering pedigree information as in Aguilar et al. (2010), with its inverse calculated as
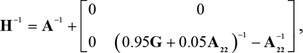 |
in which A is the additive relationship matrix (pedigree), A22−1 is the inverse of the conventional relationship matrix including just the genotyped animals, and G is the genomic relationship matrix estimated according to (VanRaden, 2008):
in which M is a transformed incidence matrix of marker alleles whose elements in the ith column are 0 − 2pi, 1 − 2pi, and 2 − 2pi for genotypes AA, AB, and BB, respectively; M′ is the transpose of M; and pi is the frequency of allele B in the ith marker.
A total of 2,200,000 chains were generated for this variance component analysis using THRGIBBS1F90 (Misztal et al., 2014), discarding the first 200,000 iterations (burn-in). The convergence of Markov chain Monte Carlo (MCMC) chains was verified by Geweke's convergence test and visual inspection of trace plots using the boa (Bayesian output analysis) package (Smith, 2007) in R software (R Development Core Team, 2013). The genetic and residual variance estimates were used as prior values in the subsequent Bayesian analysis.
Genomewide Association Study
The LD between markers was estimated using the square of the correlation (r2; Hill and Robertson, 1968), calculated as
in which A, a, B, and b are the frequencies of each allele in the studied population. The analyses were conducted with PLINK version 1.9 (Chang et al., 2015).
The associations between SNP markers and the phenotypic information were performed using BayesB methodology, which simultaneously analyzes all SNP data and assumes a different genetic variance for each SNP locus with scaled inverse χ2 prior distributions and that a fraction (1 − π) of the markers have nonzero effects (Meuwissen et al., 2001; Habier et al., 2011). The traits were analyzed separately using GenSel software (Garrick and Fernando, 2013), and the posterior distribution of marker effects was predicted under the statistical model
in which y, X, and b were defined in model [1]; k is the number of SNP; xji is the column vector representing the SNP covariate at locus i for animal j, coded as the number of the minor allele frequency allele; βi is the substitution effect for locus i, assuming βi|π, σi2 ∼ δiN(0, σi2) when δi = 1 and βi = (1 − δi)N(0, σβi2 = 0) when δi = 0; δi is an indicator variable; and ej|σe2 ∼ N(0, σe2). The prior for δi was
in which π was assumed to be 0.999, which results in about 0.1% of the SNP fitted in the model at each iteration. The marker genetic prior for 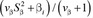 χ2 and the random residual prior
χ2 and the random residual prior 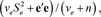 with νβ = 4 and νe = 10 df, and scale parameters estimated as
with νβ = 4 and νe = 10 df, and scale parameters estimated as 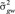 and
and 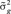 , respectively, in which n is the number of individuals (Cheng et al., 2015).
, respectively, in which n is the number of individuals (Cheng et al., 2015).
A total of 90,000 iterations was considered, where the first 2,000 were discarded (burn-in) and the following 88,000 were used to predict the posterior mean effect of each SNP marker. Rather than using effects of individual markers, the proportion of variance explained by nonoverlapping 1-Mb genomic windows comprising all the SNP effects in that region were used for inference in the genomewide association. This proportion was sampled by
in which, at every 100th post-burn-in MCMC iteration, 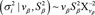 is the sampled genetic variance explained by SNP inside the genomic window w and
is the sampled genetic variance explained by SNP inside the genomic window w and 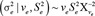 is the sampled total genetic variance. For more details on sampling
is the sampled total genetic variance. For more details on sampling 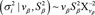 and
and 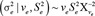 see Fernando and Garrick (2013). Genomic windows that explain at least 1% of total genetic variance based on their posterior means were considered as important regions associated with the traits, with higher chances of being associated with QTL regions (Peters et al., 2012).
see Fernando and Garrick (2013). Genomic windows that explain at least 1% of total genetic variance based on their posterior means were considered as important regions associated with the traits, with higher chances of being associated with QTL regions (Peters et al., 2012).
Gene Search and Functional Enrichment
The important regions identified by the GWAS were extended on either side (±500 kb) for the functional enrichment analyses and gene search. Annotated genes in these regions were retrieved from the Ensembl Genes 87 database using Biomart software (Aken et al., 2016).
Medical Subject Headings terms were detected using the MESHR package (Tsuyuzaki et al., 2015) for enrichment analyses considering the genes that were associated with HP (127) and NF (91). Medical subject headings identification associated with each Entrez Gene (Maglott et al., 2011)Identifications were obtained from the MESH.BTA.EG.DB annotation package (Tsuyuzaki et al., 2015), assuming a universe of all annotated genes with a unique corresponding Entrez Gene identifications (17,093). The R package uses a hypergeometric test to assess the significance of the enrichment as described by Morota et al. (2015). The terms with a P-value < 0.05 were considered significant, applying the Benjamini and Hochberg (1995) procedure to account for multiple testing.
RESULTS AND DISCUSSION
The genomic estimates of heritability on the liability scale by the single-step method were 0.28 (SD 0.07) for HP and 0.49 (SD 0.09) for NF. The estimated genomic correlation was −0.21 (SD 0.29). This high SD makes the confidence interval straddle 0, suggesting weak or no genetic association between the traits (Table 2). The highest posterior density (0.025–0.975%) interval estimated for the genomic correlation between HP and NF was remarkably wide, varying between −0.781 and 0.329. As the proposed convergence criteria were well met, the width of this interval may be attributed to the limited number of observations included in the present study.
Table 2.
Posterior mean, median, SD, and highest posterior density interval 95% (HPD 95%) of heritability (h2) and genomic correlation for early pregnancy (HP) and number of antral follicles (NF) of the Nelore heifers
| Parameter | Mean | Median | SD | HPD 95% |
|---|---|---|---|---|
| h 2 HP | 0.28 | 0.28 | 0.07 | 0.15 to 0.43 |
| h 2 NF | 0.49 | 0.49 | 0.09 | 0.32 to 0.68 |
| r HP×NF 1 | −0.21 | −0.20 | 0.29 | −0.78 to 0.33 |
r HP×NF = the genomic correlation between heifer pregnancy and number of antral follicles.
Genomewide Association Study
The average LD between adjacent markers was 0.23 ± 0.01, ranging from 0.20 on chromosome 27 to 0.25 on chromosome 5 (Fig. 1), and the overall LD on the same chromosome varied from 0.09 on chromosome 27 to 0.13 on chromosome 6, with an average of 0.11 (SD 0.01; Supplemental Fig. S1 [see the online version of the article at http://journalofanimalscience.org]). Espigolan et al. (2013) reported an r2 averaging 0.17 for overall SNP in a population of 795 Nelore bulls with approximately 447,000 markers. This observed difference is expected because LD is inversely proportional to the distance between markers, with denser genotypes generally having high overall LD. Biegelmeyer et al. (2016) found average r2 of 0.21 ± 0.27 and 0.16 ± 0.20 in 391 Hereford and 2,019 Braford animals, respectively, with approximately 41,000 SNP. As noted by those authors, LD can be influenced by several factors and is, therefore, population specific.
Figure 1.
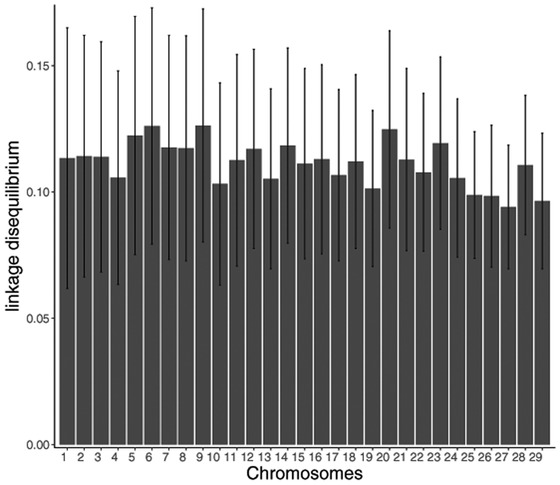
Mean values (±SD) of linkage disequilibrium (r2) of adjacent markers by chromosomes in the studied population of Nelore heifers.
A total of 2,673 genomic windows were constructed for 30 chromosomes (29 autosomes and the X chromosome) including, on average, 25.5 SNP distributed between flanking SNP that were 934 kb apart. The results identified significant regions on chromosomes 5, 14, and 18 for HP and 2, 8, 11, 14, 15, 16, and 22 for NF that each explained >1% of the total genetic variance (Fig. 2 and 3). These genomic regions were used to locate candidate genes and are presented on Tables 3 and 4 for HP and NF, respectively. Other genomic regions on chromosomes 1, 2, 3, 5, 14, 24, 29, and X demonstrated an important influence on HP (Table 5) and genomic regions on chromosomes 2, 12, 15, 19, 24, 25, and 29 demonstrated an important influence on NF (Table 6), explaining >0.5% of the total additive genetic variance. Nonetheless, these regions were not considered in the further analyses.
Figure 2.
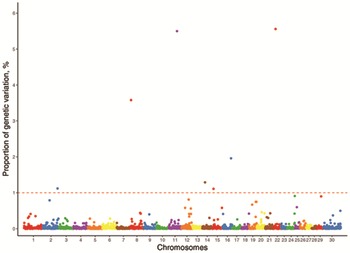
Manhattan plot of the genomewide association study for pregnancy in Nellore heifers. The dashed red line represents the threshold of the proportion of the explained genetic variance.
Figure 3.
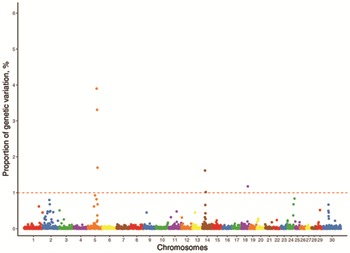
Manhattan plot of the genomewide association study for the number of antral follicles in Nellore heifers. The dashed red line represents the threshold of the proportion of the explained genetic variance.
Table 3.
Genes harbored in genomic windows that explained more than 1% of the additive genetic variance for early pregnancy in Nelore heifers
| CHR1 | Position (UMD 3.1 bovine assembly2) | Genes | Var,3 % | No. of SNP4 |
|---|---|---|---|---|
| 5 | 72,521,518–74,466,890 | APOL6, HMGXB4, HMOX1, ISX, LARGE1, MB, MCM5, RASD2, RBFOX2, SNORA76, and TOM1 | 3.90 | 22 |
| 76,522,303–78,484,850 | BICD1, DNM1L, FGD4, KIAA1551, PKP2, SYT10, and YARS2 | 3.31 | 27 | |
| 80,633,605–82,455,158 | CCDC91, COX7ALP1, ERGIC2, FAR2, KLHL42, and PTHLH | 1.70 | 22 | |
| 14 | 22,507,464–24,484,847 | ATP6V1H, FAM150A, LYPLA1, MRPL15, NPBWR1, OPRK1, PCMTD1, RB1CC1, RGS20, RP1, SOX17, ST18, TCEA1, U2, U6, and XKR4 | 1.62 | 29 |
| 18 | 54,518,279–56,456,772 | ALDH16A1, BAX, BCAT2, BOSTAUV1R403, BOSTAUV1R404, C19orf68, C5AR1, C5AR2, CA11, CABP5, CCDC114, CCDC155, CCDC9, CD37, CRX, DBP, DHDH, DHX34, DKKL1, ELSPBP1, EMP3, FAM38E, FCGRT, FGF21, FLT3LG, FTL, FUT1, FUT2, GLTSCR1, GLTSCR2, GRIN2D, GRWD1, GYS1, HRC, HSD17B14, IZUMO1, KCNA7, KCNJ14, KDELR1, KPTN, LHB, LIG1, LIN7B, LMTK3, MAMSTR, MEIS3, NAPA, NOSIP, NPAS1, NTF4, NTN5, NUCB1, PIH1D1, PLEKHA4, PPFIA3, PPP1R15A, PRRG2, PTH2, RASIP1, RCN3, RPL18, RPS11, RUVBL2, SAE1, SEC1, SEPW1, SLC17A7, SLC6A16, SLC8A2, SNRNP70, SPACA4, SPHK2, SULT2A1, SULT2B1, SYNGR4, TEAD2, TMEM143, TMEM160, TRPM4, TULP2, ZC3H4, ZNF114, and ZNF541 | 1.18 | 27 |
| 14 | 28,561,608–30,475,350 | GGH, NKAIN3, TTPA, and YTHDF3 | 1.02 | 23 |
CHR = Bovine autosome.
Var = genetic variance explained by the window.
No. of SNP is the number of SNP in the window.
Table 4.
Genes harbored in genomic windows that explained more than 1% of the additive genetic variance for number of antral follicles in Nelore heifers
| CHR1 | Position (UMD 3.1bovine assembly2) | Genes | Var,3 % | No. of SNP4 |
|---|---|---|---|---|
| 22 | 14,507,849–16,474,506 | ACKR2, ANO10, CCDC13, CCK, HHATL, HIGD1A, KLHL40, LYZL4, NKTR, SEC22C, SNRK, SS18L2, TCAIM, TOPAZ1, TRAK1, VIPR1, ZBTB47, ZNF197, ZNF445, and ULK4 | 5.56 | 25 |
| 11 | 69,520,906–71,472,289 | ALK, BRE, C11H2orf71, CLIP4, FAM179A, FOSL2, LBH, PLB1, PPP1CB, SNORD53_SNORD92, SPDYA, TRMT61B, WDR43, and YPEL5 | 5.50 | 27 |
| 8 | 6519,096–8474,290 | ADAM29, BLK, C8orf74, CTSB, DEFB134, FAM167A, FDFT1, GATA4, GLRA3, HPGD, MTMR9, NEIL2, PINX1, SOX7, TDH, and XKR6 | 3.58 | 22 |
| 16 | 70,502,173–72,455,202 | ARL8A, ELF3, GPR37L1, LGR6, PPP1R12B, PROX, PTPN14, PTPN7, RNPEP, RPS6KC1, SMYD2, SYT2, and UBE2T | 1.96 | 25 |
| 14 | 22,507,464–24,484,847 | ATP6V1H, FAM150A, LYPLA1, MRPL15, NPBWR1, OPRK1, PCMTD1, RB1CC1, RGS20, RP1, SOX17, ST18, TCEA1, and XKR4 | 1.29 | 29 |
| 2 | 122,533,476–124,485,696 | COL16A1, FABP3, HCRTR1, LAPTM5, MATN1, NKAIN1, PEF1, PUM1, SDC3, SERINC2, SNRNP40, and TINAGL1 | 1.12 | 22 |
| 15 | 8507,403–10,464,870 | ARHGAP42 and CNTN5 | 1.11 | 24 |
CHR = Bovine autosome.
Var = genetic variance explained by the window.
No. of SNP is the number of SNP in the window.
Table 5.
Description of genomic windows that explain more than 0.5% of the total genetic variation of heifer pregnancy in Nelore heifers
| CHR1 | SNP_start2 | SNP_end3 | Pos_start,4 bp | Pos_end,5 bp | Size,6 bp | No. of SNP7 | %Var8 |
|---|---|---|---|---|---|---|---|
| 5 | rs109437025 | rs110687761 | 73,021,518 | 73,966,890 | 945,372 | 22 | 3.90 |
| 5 | rs42561706 | rs137385583 | 77,022,303 | 77,984,850 | 962,547 | 27 | 3.31 |
| 5 | rs110496647 | rs136544553 | 81,133,605 | 81,955,158 | 821,553 | 22 | 1.70 |
| 14 | rs41724652 | rs133297141 | 23,007,464 | 23,984,847 | 977,383 | 29 | 1.62 |
| 18 | rs136460244 | rs41891085 | 55,018,279 | 55,956,772 | 938,493 | 27 | 1.18 |
| 14 | rs41624840 | rs136805030 | 29,061,608 | 29,975,350 | 913,742 | 23 | 1.02 |
| 5 | rs42917128 | rs136339681 | 60,131,115 | 60,941,344 | 810,229 | 23 | 0.93 |
| 24 | rs136828522 | rs137238317 | 60,015,982 | 60,980,620 | 964,638 | 43 | 0.84 |
| 5 | rs137127461 | rs109435449 | 72,059,606 | 72,982,750 | 923,144 | 28 | 0.82 |
| 2 | rs42509691 | rs134051905 | 54,015,771 | 54,997,281 | 981,510 | 23 | 0.80 |
| 24 | rs109329309 | rs135881583 | 52,121,696 | 52,957,990 | 836,294 | 23 | 0.68 |
| 5 | rs110450288 | rs133794376 | 82,043,465 | 82,961,509 | 918,044 | 26 | 0.68 |
| 2 | rs133912634 | rs134084039 | 58,011,152 | 58,971,614 | 960,462 | 25 | 0.68 |
| X | rs134685381 | rs137716652 | 44,037,579 | 44,972,292 | 934,713 | 21 | 0.67 |
| 14 | rs135852767 | rs42298467 | 25,021,594 | 25,986,431 | 964,837 | 22 | 0.66 |
| 5 | rs110797637 | rs137576699 | 51,113,065 | 51,990,170 | 877,105 | 25 | 0.62 |
| 1 | rs136647907 | rs133111309 | 125,026,356 | 125,988,072 | 961,716 | 24 | 0.62 |
| 29 | rs134769207 | rs42172278 | 24,031,236 | 24,999,881 | 968,645 | 24 | 0.52 |
| 3 | rs109945234 | rs42368646 | 3017,659 | 3968,604 | 950,945 | 42 | 0.51 |
| X | rs134673004 | rs134676523 | 43,104,725 | 43,892,074 | 787,349 | 22 | 0.50 |
CHR = Bovine autosome.
SNP_start = first SNP of the window.
SNP_end = last SNP of the window.
Pos_start = position of the first SNP.
Pos_end = position of the last SNP.
Size is the size of the window.
No. of SNP is the number of SNP in the window.
%Var = percentage of the additive genetic variance explained by the window.
Table 6.
Description of genomic windows that explain more than 0.5% of the total genetic variation of number of antral follicles in Nelore heifers
| CHR1 | SNP_start2 | SNP_end3 | Pos_start,4 bp | Pos_end,5 bp | Size,6 bp | No. of SNP7 | %Var8 |
|---|---|---|---|---|---|---|---|
| 24 | rs133021126 | rs109305902 | 62,008,417 | 62,594,751 | 586,334 | 24 | 0.91 |
| 29 | rs42183484 | rs135436243 | 33,000,052 | 33,914,381 | 914,329 | 23 | 0.90 |
| 12 | rs109641299 | rs134942057 | 60,015,627 | 60,994,461 | 978,834 | 28 | 0.81 |
| 2 | rs42321794 | rs134609840 | 56,028,202 | 56,979,919 | 951,717 | 21 | 0.79 |
| 19 | rs109030622 | rs135605613 | 61,000,109 | 61,990,875 | 990,766 | 45 | 0.75 |
| 19 | rs136160175 | rs41919438 | 56,008,922 | 56,945,797 | 936,875 | 55 | 0.75 |
| 19 | rs137834616 | rs137557574 | 26,010,131 | 26,962,473 | 952,342 | 29 | 0.67 |
| 25 | rs134031092 | rs134416406 | 16,006,913 | 16,986,653 | 979,740 | 21 | 0.60 |
| 12 | rs110539435 | rs109044869 | 30,015,791 | 30,992,759 | 976,968 | 30 | 0.58 |
| 15 | rs137836273 | rs110334648 | 81,018,176 | 81,864,846 | 846,670 | 32 | 0.58 |
| 12 | rs137815663 | rs109275028 | 79,019,126 | 79,976,465 | 957,339 | 27 | 0.56 |
| X | rs133820946 | rs135451827 | 144,008,894 | 144,988,891 | 979,997 | 35 | 0.50 |
CHR = Bovine autosome.
SNP_start = first SNP of the window.
SNP_end = last SNP of the window.
Pos_start = position of the first SNP.
Pos_end = position of the last SNP.
Size is the size of the window.
No. of SNP is the number of SNP in the window.
%Var = percentage of the additive genetic variance explained by the window.
Six and 7 genomic windows explained at least 1% each of the additive genetic variance for HP and NF, respectively. The cumulative genetic variance explained by these windows was 12.73% for HP and 20.12% for NF. The highest proportion of genetic variance explained by a window for HP was 3.90% in a window located at 73 Mb in chromosome 5, whereas for NF, the most relevant window explained 5.56% of the genetic variance; this window is located at 15 Mb in chromosome 22. Half of the genetic variance was explained by 237 windows for HP and 116 windows for NF, whereas to achieve 90% of explained variance, it would be a similar function of 1,704 windows for HP and 2,673 windows for NF.
Medical Subject Headings Enrichment Analyses
The MeSH enrichment analyses revealed 74 terms related to HP (Supplemental Table S1; see the online version of the article at http://journalofanimalscience.org) and 48 terms related to NF (Supplemental Table S2; see the online version of the article at http://journalofanimalscience.org). After the Benjamini and Hochberg correction, the terms that were significantly (P < 0.05) associated with HP were “Munc-18 Proteins,” “Fucose,” and “Hemoglobins” and the terms that were significantly (P < 0.05) associated with NF were “Cathepsin B,” “Receptors-Neuropeptide,” and “Palmitic Acid.”
The mammalian homolog of unc-18 (munc-18) has an important association with hormone secretion in the pituitary cells in mice. Weiss et al. (2007) argued that estradiol affects the production of munc-18, with a central role in regulation of LH secretion. Korteweg et al. (2005) concluded that munc-18 is involved in peptide hormone secretion from the anterior pituitary.
“Fucose” is a hexose deoxy sugar naturally found on mammalian cell surfaces and has been detected in the zona pellucida and on the spermatozoal surface. Romero-Aguirregomezcorta et al. (2015) observed increased levels of spermatozoon–zona pellucida binding and penetration when fucose was added to IVF medium, indicating a key role in fertilization. Studies in the bovine (Lefebvre et al., 1997; Suarez et al., 1998; Tanghe et al., 2004) confirmed that fucose mediates a specific interaction between sperm and oviductal epithelium, thus playing an important role prior in fertilization.
The “Hemoglobins” term was also significant, suggesting a possible association with reproduction. Hemoglobins are the oxygen-carrying proteins of erythrocytes, and vascularity within the ovary has been associated with oocyte health (Van Blerkom, 2000). Brown et al. (2014) concluded that hemoglobin is expressed by the follicular cells of the ovary, suggesting an influence on endocrine regulation through induction of a hypoxic stage that interferes with the LH surge.
“Cathepsin B,” a lysosomal cysteine proteinase, was negatively correlated with the quality of bovine oocytes and embryos (Balboula et al., 2013). According to Balboula et al. (2013), heat stress, a common condition in tropical countries, could lead to an increase in the expression of cathepsin B, with consequent impairment of oocyte development and female fertility.
The “Receptor, Neuropeptide” term refers to cell surface receptors that bind specific neuropeptides initiating intracellular modifications that can change the comportment of cells. Many of these neuropeptides are hormones when outside of the nervous system. The neuropeptide kisspeptin has been positively associated with GnRH and gonadotropin secretion (Amstalden et al., 2014), and the function of kisspeptin neurons may be influenced by postweaning weight gain in heifers. Redmond et al. (2011) related this neuropeptide to the onset of puberty in lambs through its role in activation of the hypothalamic–adenohypophyseal axis.
Palmitic acid is one of the most common SFA present in the follicular fluids of cattle, sheep, and pigs (McEvoy et al., 2000). Mu et al. (2001) associated the plasma concentration of SFA with granulosa cell apoptosis in humans, which may influence reproductive disorders. In cattle, a high concentration of palmitic acid in follicular fluid has negative effects on oocyte maturation, fertilization, and cleavage rate (Leroy et al., 2005). Marei et al. (2010) found that dietary supplementation with fatty acids may influence oocyte maturation and early embryonic development. Zeron et al. (2001) reported high follicular fluid concentrations of SFA in high temperatures (summer), which is associated with negative impacts on fertility (García-Ispierto et al., 2007).
Genes Associated with Heifer Pregnancy
Chromosome 5
Twenty-four genes in 3 genomic windows on chromosome 5 had large effects on HP. A study by Kappes et al. (2000) found a significant peak on this chromosome (approximately 40 cM) for ovulation rate and suggested that this chromosome may harbor important genes for this trait in cattle. The results of Khatkar et al. (2014) found highest meta-GWAS scores on chromosome 1, 5, 13, and 16 for fertility traits.
The genes TOM1 and HO-1 are associated with pregnancy in humans (Dumesic et al., 2015). Assidi et al. (2011) noted that TOM1 downregulates the transcription factor AP1, which is involved in the LH pathway in preovulatory bovine granulosa cells. They also confirmed that TOM1 is a negative biomarker of oocyte competence. Heme-oxygenase 1 (HO-1) is an oxidative stress-related gene, and high levels of expression in cumulus cells is associated with reduced oocyte fertilization in humans (Bergandi et al., 2014). These results support the hypothesis that this genomic region (72–74 Mb) is involved with HP outcomes.
In a study by Hawken et al. (2012), chromosome 5 harbored the most important markers associated with reproductive traits, such as age at puberty, in Tropical Composite cattle. The most significant SNP was located at around 96 Mb, which is close to the regions reported in this study (72–83 Mb). Other studies (Schulman et al., 2008; Allan et al., 2009; Kim et al., 2009; Sahana et al., 2010; Luna-Nevarez et al., 2011) also related this chromosome to reproductive traits. Insulin-like growth factor 1 (IGF1) is located at 66 Mb and is frequently associated with reproductive traits in cattle (Fortes et al., 2013). The average LD of adjacent markers between the locations of the middle of this gene (66,564,289 bp) and the middle of the closest genomic window reported in this study (73,494,204 bp) was 0.34 (95% confidence interval 0.30–0.39), suggesting a possible linkage between these markers.
Chromosome 14
Eighteen genes were associated with both HP and NF in a window (22–24 Mb) on chromosome 14. Hawken et al. (2012) associated multiple clusters of SNP related to reproductive traits in 843 Brahman and 866 Tropical Composites beef heifers, with the most significant markers located between 22 and 25 Mb. A meta-GWAS study (Khatkar et al., 2014) found the highest score on chromosome 14 for sexual maturity in cattle.
These same regions had large effects in the current study for both HP and NF, emphasizing their importance in reproductive traits and suggesting a possible pleiotropic effect. This hypothesis was reinforced by comparing the correlation of the genomewide estimated genomic values of HP and NF with the estimated genomic values when considering only the markers present in referenced genomic window (Cole et al., 2009). The estimates were −0.14 (95% confidence interval −0.21 to −0.08) when considering all SNP and −0.30 (95% confidence interval −0.36 to −0.24) considering only SNP in the referened window. A Fisher r-to-Z transformation test was performed to assess significant differences between the estimates, and the obtained result (P < 0.01) supports the pleiotropic effect previously hypothesized for this region.
SOX17 is a well-known gene linked with primordial germ cells, which are the precursors of spermatozoa and oocytes (Irie et al., 2015). Irie et al. (2015) suggested that SOX17 plays a key role in the maintenance of early human germline cells and also supports human uterine adenogenesis and glandular functions (Guimarães-Young et al., 2016). Mice with a null allele of SOX17 show decreased expression of the progesterone receptor promoter gene compared with the wild-type, with a consequent impairment of female fertility.
The RGS20 gene belongs to the regulator of G-coupled protein signaling (RGS) family of proteins. It is a regulator of G protein α subunits, inducing them into the inactive, guanosine diphosphate (GDP)–bound form (The UniProt Consortium, 2015). This gene has an important function in cell division, where it is essential for the first mitotic division of the embryo. Mouse zygotes with a loss of RGS14 expression were not able to progress to the 2-cell stage (Martin-McCaffrey et al., 2004). Feuerstein et al. (2012) identified RGS2 as biomarker of oocyte development competence, with significantly (P < 0.05) different levels of expression in pregnant and nonpregnant patients.
The PLAG1 gene, located at 25 Mb of chromosome 14 in cattle, is often associated with reproductive traits in different species, with reported functions related to cell proliferation by regulation of growth factors (Juma et al., 2016). Chen et al. (2007) concluded that the expression of this gene in the hypothalamus and pituitary glands affects egg production. Hensen et al. (2004) identified significant associations with growth rate and fertility reduction in both males and females using PLAG1 knockout mice. In cattle, variants of this gene have also been linked to growth and fertility traits as well as changes in IGF1 gene expression (Fortes et al., 2012a,b; Hawken et al., 2012). Although the exact gene location was not reported in our results, the average LD of adjacent markers between the location of the middle of this gene (25,026,431 bp) and the middle of the genomic window (23,496,156 bp) reported in this study was 0.32 (95% confidence interval 0.21–0.44), suggesting possible linkage between these markers.
Chromosome 18
The genomic window (54–56 Mb) on chromosome 18 included 83 different genes. The galactoside 2-α-L-fucosyltransferase (FUT2) gene appears to be stimulated by estrogen, and the correlated expression of fucosylated H-typed-1 carbohydrate epitope in the endometrial epithelia may be involved with embryo implantation (White and Kimber, 1994). The FUT1 gene was associated with daughter pregnancy rate as well as total number of piglets born (HongYan et al., 2009; Cochran et al., 2013), and Ortega et al. (2016) also found significant associations between this gene and cow conception rate and daughter pregnancy rate in Holstein cattle.
The proapoptotic Bcl-2 associated X protein (BAX) gene accelerates programmed cell death, and it has been shown that the absence of this gene results in infertility in males (Gaddis et al., 2016). However, in females, Bcl-2 knockout mice exhibit increased oocytes and follicle numbers (Greenfeld et al., 2007), although te Velde and Pearson (2002) found that upregulation of this gene resulted in enhanced follicle damage and premature ovarian failure in mice.
Luteinizing hormone β polypeptide (LHB) is known to promote ovulation by stimulating the ovaries to synthesize steroids (The UniProt Consortium, 2015). Luteinizing hormone is secreted by the pituitary gland and plays an important role in gonadal functions. Weiss et al. (1992) associated a homozygous mutation in the LHB gene with delayed puberty in humans. Normal pulsatile release of this hormone is crucial for successful induction of LH receptors on the granulosa cells of the developing dominant follicle (Luo et al., 2011). Bender et al. (2014) reported a positive correlation between feed intake and LH concentration, with both high and low values being harmful to embryo development.
The genes SLC17A7, SLC6A16, and SLC8A2 are part of the solute carrier family. SLC17A7 is regulated by estrogen and encodes the vesicular glutamate transporter 1 (Vglut1) protein, which is expressed in cells of the hippocampus (Bellocchio et al., 2000) and may be associated with estrogen regulation (Sárvári et al., 2015). SLC6 specifically transports neurotransmitters (e.g., dopamine and serotonin), AA (e.g., gamma 1-aminobutyric acid [GABA]), and osmolytes (e.g., betaine, taurine, and creatine; Höglund et al., 2005). SLC8A2 mediates the electrogenic exchange of Ca2+ against Na+ ions across the cell membrane. Choi et al. (2014) reported that SLC8A is expressed in the uterine endometrium of pregnant pigs and concluded that calcium extrusion molecules may be associated with the establishment and maintenance of pregnancy.
The sphingosine kinase (SPHK) gene family is part of the sphingolipid metabolic pathway, which is highly activated in decidua during pregnancy (Mizugishi et al., 2007). The isoform genes SPHK1 and SPHK2 participate in the pathway of the bioactive lipid sphingosine 1-phosphate (S1P) and have been associated with vasoconstriction in human uterine arteries during pregnancy (Hudson et al., 2007). Mizugishi et al. (2007) found that mutant mice for SPHK1 and SPHK2 produced infertile females, with reduced production of S1P.
Genes Associated with the Number of Antral Follicles
Chromosome 2
The associated genomic window on chromosome 2 (122–124 Mb) harbored 12 genes. The intracellular fatty acid–binding proteins (FBP) are associated with metabolism and transport of long-chain fatty acids that have import roles in oocyte meiotic maturation prior to ovulation (Sanchez-Lazo et al., 2014). The small nuclear ribonucleoproteins have been related to oocyte maturation, fertilization, and early embryogenesis in mouse (Prather et al., 1990).
Parisi and Lin (1999) associated the Pumilio 1 (PUM1) gene in Drosophila with embryogenesis, primordial germ cell proliferation, germline stem cell division, and the oogenic process. PUM1 is also involved in oocyte maturation in Xenopus (Nakahata et al., 2001). The presence of anti-Pum antibody, or the overexpression of Pum1, has significant impacts on oocyte maturation (Nakahata et al., 2003). Mak et al. (2016) reported that this gene plays a critical role in the establishment of primordial folliculogenesis, meiosis, and female fertility in mice.
Chromosome 8
Sixteen genes were reported in the associated genomic region on chromosome 8 (6–8 Mb). Tenghe et al. (2016) identified associations with endocrine fertility traits, such as luteal activity, in Holstein cows. The protein GATA4 is part of the erythroid transcription factor (GATA) family of zinc finger transcription factors, with possible involvement in early gonadal development in mice and humans (Bouma et al., 2007; Lourenço et al., 2011), and a mutation in GATA4 can compromise the cycle of anti-Müllerian hormone. Lourenço et al. (2011) also concluded that GATA4 is related to cases of sex development disorders and congenital heart defects in humans.
Balboula et al. (2010) found that inhibition of the cathepsin B (CTSB) protein positively affected oocyte production, resulting in an increased number of good quality embryos in bovines. They concluded that CTSB could be used as a marker for low-quality oocytes, in agreement with Bettegowda et al. (2008), who also reported negative correlations between CTSB and oocyte competence. However, Warzych et al. (2016) found favorable associations of CTSB in bovine cumulus cells with oocyte quality, suggesting that the relationship of the gene to oocyte quality is more complex that initially thought.
Members of the a disintegrin and metalloprotease domain (ADAM) gene family are known to play key roles in follicular development and ovarian organization and functioning (Brown et al., 2005; Shozu et al., 2005; Feng et al., 2016) in humans (Gao et al., 2007; Pyun et al., 2014) and mammals in general (Russell et al., 2015). Members of the ADAM with thrombospondin (ADAMTS) gene family have been extensively studied, and it is associated with follicular development through control of extracellular matrix remodeling (Russell et al., 2015). Brown et al. (2005) reported that early antral follicles were significantly reduced in ovaries of ADAMST-1 knockout mice. Gene expression has also been observed in germ and somatic cells in mice testis, suggesting association with reproduction (Choi et al., 2004; Han et al., 2009).
Chromosome 11
Eleven genes were reported in the genomic window at chromosome 11 (69–71 Mb). SPDYA is a protein-coding gene expressed in pathways related to oocyte meiosis (The UniProt Consortium, 2015). Additionally, it was also associated with oocyte maturation in humans (Barnes et al., 2003) and Xenopus (Cheng et al., 2005). Porter et al. (2002) concluded that this class of genes is an essential component of cell proliferation pathways.
FOS-like antigen 2 (FOSL2) participates in the regulation of steroidogenesis, which includes the estrogen hormone (Hatzirodos et al., 2014b). This gene encodes components of activator protein 1 (AP-1), which participates in the terminal differentiation of granulosa cells to luteal cells (Sharma and Richards, 2000). Przygrodzka et al. (2015) suggested that elements from the FOS gene family are activated in porcine corpora lutea with acquired luteolytic sensibility, playing an important role in structural luteolysis in pigs.
The protein yippee-like 5 (YPEL5) is part of the yippee like (YPEL) gene family involved with cell cycle progression and growth, with high expression in oocytes (Hosono et al., 2010; Rajput et al., 2013). Hatzirodos et al. (2014a) reported significant upregulation of this gene in large follicles (>10 mm) compared with small follicles (<5 mm) in taurine cattle.
Chromosome 15
Only 2 genes were present in the associated genomic window on chromosome 15 (8–10 Mb). The protein-coding gene Rho GTPase activating protein 42 (ARHGAP42) influences blood pressure in vascular smooth muscle (Bai et al., 2013). ARHGAP42 also is involved in biological processes related to the regulation of Rho protein, which is part of a family of proteins involved in regulation and timing of cell division (Yoshizaki et al., 2003). Spencer et al. (2013) associated this gene with age at menarche and age at natural menopause in African American women.
Chromosome 16
The associated genomic window on chromosome 16 (70–72 Mb) harbored 13 genes. La Sala et al. (2015) discussed the association of the G protein-coupled receptor 37 (GPR37) with gonadal differentiation in mice. Mutant animals (GPR37-null) had impaired testis development, affecting postnatal Sertoli cell proliferation and maturation, resulting in significant reduction of sperm count. This gene may have similar functions in females, affecting granulosa cell proliferation and animal fertility.
Although the precise function of leucine-rich repeat-containing G protein coupled receptor 6 (LGR6) is still unknown, other leucine-rich repeat-containing G-protein coupled receptor (LGR) genes have been characterized as part of the insulin gene family, which also includes important reproductive genes including LH, FSH, and TSH (Lu et al., 2005). LGR7 and LGR8 are receptors for relaxin hormone, which is primarily produced by the corpus luteum. LGR4 knockout mice were infertile and exhibited abnormal development of the reproductive tract (Mendive et al., 2006).
Protein phosphatase 1 regulatory subunit 12B (PPP1R12B) may be involved with insulin signaling (Pham et al., 2012). This gene is associated with smooth muscle contractibility and regulation of uterine cell hypertrophy in the early stages of gestation (Lartey et al., 2016). Ryu et al. (2016) found that PPP1R12B participates in protein phosphorylation, which may influence reproduction in humans.
Protein tyrosine phosphatases (PTP) regulate many cellular processes, such as metabolism, cell–cell adhesion, cell migration, cell growth, and the expression of transformation growth factor beta, which is a key regulator of follicle development in mammals (Knight and Glister, 2006; Bastian et al., 2015). van Eekelen et al. (2010) reported that 48 PTP genes are expressed in zebra fish embryos, with the majority of them being maternally derived.
Chromosome 22
Twenty genes were reported in the associated genomic window on chromosome 22 (14–16 Mb). Baillet et al. (2011) showed that testis and ovary-specific PAZ domain gene 1 (TOPAZ1) is expressed in the gonads of sheep and mice, particularly in pregnant females during fetal gonad development. They concluded that the gene has an important function in germ cell meiosis. Supporting these findings, Luangpraseuth-Prosper et al. (2015) found that this gene also plays an important role in the progression of meiosis in oocytes and spermatogenesis in mice.
Lysozyme-like (LYZL) genes belong to the class of c-type lysozymes, which are widely distributed in the animal species and which have protective bacteriolytic functions in host defense (Zhang et al., 2005). LYZL4 is expressed in the epithelium of the human epididymis, and it has also been observed on the surface of human embryonic stem cells (Gu et al., 2011). When mouse spermatozoa were incubated with anti-LYZL4 antibodies, there was a concomitant loss of fertilizing ability (Sun et al., 2011). Some studies (McReynolds et al., 2014; Kwon et al., 2015) have suggested that this gene could be used as a biomarker for male fertility. The LYZL genes may have similar functions in females, participating in the process of germ cell development
In summary, this study provides evidence of the genomic complexity involved in reproductive traits. Genomic regions on chromosomes 5, 14, and 18 showed important associations (that explained >1% of the total additive genetic variance) with HP, whereas regions on chromosomes 2, 8, 11, 14, 15, 16, and 22 had large associations with NF. Although the same genomic window on chromosome 14 was associated with both traits, their genetic correlation was not relevant, suggesting that the selection for one trait has no interference in the other trait. The MeSH terms “Munc18 Proteins,” “Fucose,” and “Hemoglobins” were significantly related to HP, and the MeSH terms “Cathepsin B,” “Receptors, Neuropeptide,” and “Palmitic Acid” were related to NF. Increasing the knowledge about associated genomic regions and genes may be useful for enhancing genomic selection, increasing evaluation accuracy, and making better selection decisions (Boichard et al., 2016).
Conclusions
Genomewide association studies allowed us to identify genomic regions associated with the reproductive traits heifer pregnancy and number of antral follicles. Medical Subject Headings enrichment analyses identified important biological processes that are related to the expression of the phenotypes, providing useful information about the genetic components of these traits. The gene search suggested that some genomic regions harbor important genes related to the traits studied, which could be used in genomic prediction to improve reproductive performance.
LITERATURE CITED
- Abeygunawardena H., Dematawewa C. M. B. 2004. Pre-pubertal and postpartum anestrus in tropical Zebu cattle. Anim. Reprod. Sci. 82–83:373–387. doi: 10.1016/j.anireprosci.2004.05.006 [DOI] [PubMed] [Google Scholar]
- Aguilar I., Misztal I., Johnson D. L., Legarra A., Tsuruta S., Lawlor T. J. 2010. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 93:743–52. doi: 10.3168/jds.2009-2730 [DOI] [PubMed] [Google Scholar]
- Aken B. L., Ayling S., Barrell D., Clarke L., Curwen V., Fairley S., Fernandez-Banet J., Billis K., Garcia-Giron C., Hourlier T., Howe K. L., Kahari A. K., Kokocinski F., Martin F. J., Murphy D. N., Nag R., Ruffier M., Schuster M., Tang Y. A., Vogel J.-H., White S., Zadissa A., Flicek P., Searle S. M. J. 2016. The Ensembl gene annotation system. Database 2016:baw093. doi: 10.1093/database/baw093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan M. F., Kuehn L. A., Cushman R. A., Snelling W. M., Echternkamp S. E., Thallman R. M. 2009. Confirmation of quantitative trait loci using a low-density single nucleotide polymorphism map for twinning and ovulation rate on bovine chromosome 5. J. Anim. Sci. 87:46–56. doi: 10.2527/jas.2008-0959 [DOI] [PubMed] [Google Scholar]
- Amstalden M., Cardoso R. C., Alves B. R. C., Williams G. L. 2014. Reproduction Symposium: Hypothalamic neuropeptides and the nutritional programming of puberty in heifers. J. Anim. Sci. 92:3211–3222. doi: 10.2527/jas.2014-7808 [DOI] [PubMed] [Google Scholar]
- Assidi M., Montag M., Van Der Ven K., Sirard M. A. 2011. Biomarkers of human oocyte developmental competence expressed in cumulus cells before ICSI: A preliminary study. J. Assist. Reprod. Genet. 28:173–188. doi: 10.1007/s10815-010-9491-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Lenhart K. C., Bird K. E., Suen A. A., Rojas M., Kakoki M., Li F., Smithies O., Mack C. P., Taylor J. M. 2013. The smooth muscle-selective RhoGAP GRAF3 is a critical regulator of vascular tone and hypertension. Nat. Commun. 4:2910. doi: 10.1038/ncomms3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillet A., Bouffant R., Volff J. N., Luangpraseuth A., Poumerol E., Thépot D., Pailhoux E., Livera G., Cotinot C., Mandon-Pépin B. 2011. TOPAZ1, a novel germ cell-specific expressed gene conserved during evolution across vertebrates. PLoS One 6:e26950. doi: 10.1371/journal.pone.0026950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboula A. Z., Yamanaka K., Sakatani M., Hegab A. O., Zaabel S. M., Takahashi M. 2010. Cathepsin B activity is related to the quality of bovine cumulus oocyte complexes and its inhibition can improve their developmental competence. Mol. Reprod. Dev. 77:439–448. doi: 10.1002/mrd.21164 [DOI] [PubMed] [Google Scholar]
- Balboula A. Z., Yamanaka K., Sakatani M., Kawahara M., Hegab A. O., Zaabel S. M., Takahashi M. 2013. Cathepsin B activity has a crucial role in the developmental competence of bovine cumulus-oocyte complexes exposed to heat shock during in vitro maturation. Reproduction 146:407–417. doi: 10.1530/REP-13-0179 [DOI] [PubMed] [Google Scholar]
- Barnes E. A., Porter L. A., Lenormand J. L., Dellinger R. W., Donoghue D. J. 2003. Human Spy1 promotes survival of mammalian cells following DNA damage. Cancer Res. 63:3701–3707. [PubMed] [Google Scholar]
- Baruselli P. S., Batista E. O. S., Vieira L. M., Souza A. H. 2015. Relationship between follicle population, AMH concentration and fertility in cattle. Anim. Reprod. 12:487–497. [Google Scholar]
- Bastian F. B., Chibucos M. C., Gaudet P., Giglio M., Holliday G. L., Huang H., Lewis S. E., Niknejad A., Orchard S., Poux S., Skunca N., Robinson-Rechavi M. 2015. The confidence information ontology: A step towards a standard for asserting confidence in annotations. Database 2015:bav043. doi: 10.1093/database/bav043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellocchio E. E., Reimer R. J., Fremeaur R. T., Jr, Edwards R. H. 2000. Uptake of glutamate into synaptic vesicles by an inorganic phosphate transporter. Science 289:957–960. [DOI] [PubMed] [Google Scholar]
- Bender R. W., Hackbart K. S., Dresch A. R., Carvalho P. D., Vieira L. M., Crump P. M., Guenther J. N., Fricke P. M., Shaver R. D., Combs D. K., Wiltbank M. C. 2014. Effects of acute feed restriction combined with targeted use of increasing luteinizing hormone content of follicle-stimulating hormone preparations on ovarian superstimulation, fertilization, and embryo quality in lactating dairy cows. J. Dairy Sci. 97:764–778. doi: 10.3168/jds.2013-6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57:289–300. [Google Scholar]
- Bergandi L., Basso G., Evangelista F., Canosa S., Dalmasso P., Aldieri E., Revelli A., Benedetto C., Ghigo D. 2014. Inducible nitric oxide synthase and heme oxygenase 1 are expressed in human cumulus cells and may be used as biomarkers of oocyte competence. Reprod. Sci. 21:1370–1377. doi: 10.1177/1933719114525268 [DOI] [PubMed] [Google Scholar]
- Berry D. P., Bastiaansen J. W. M., Veerkamp R. F., Wijga S., Wall E., Berglund B., Calus M. P. L. 2012. Genome-wide associations for fertility traits in Holstein–Friesian dairy cows using data from experimental research herds in four European countries. Animal 6:1206–1215. doi: 10.1017/S1751731112000067 [DOI] [PubMed] [Google Scholar]
- Bettegowda A., Patel O. V., Lee K.-B., Park K.-E., Salem M., Yao J., Ireland J. J., Smith G. W. 2008. Identification of novel bovine cumulus cell molecular markers predictive of oocyte competence: Functional and diagnostic implications. Biol. Reprod. 79:301–309. doi: 10.1095/biolreprod.107.067223 [DOI] [PubMed] [Google Scholar]
- Biegelmeyer P., Gulias-Gomes C. C., Caetano A. R., Steibel J. P., Cardoso F. F. 2016. Linkage disequilibrium, persistence of phase and effective population size estimates in Hereford and Braford cattle. BMC Genet. 17:32. doi: 10.1186/s12863-016-0339-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boichard D., Ducrocq V., Croiseau P., Fritz S. 2016. Genomic selection in domestic animals: Principles, applications and perspectives. C. R. Biol. 339:274–277. doi: 10.1016/j.crvi.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Bouma G. J., Washburn L. L., Albrecht K. H., Eicher E. M. 2007. Correct dosage of Fog2 and Gata4 transcription factors is critical for fetal testis development in mice. Proc. Natl. Acad. Sci. U. S. A. 104:14994–14999. doi: 10.1073/pnas.0701677104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H. M., Anastasi M. R., Frank L. A., Kind K. L., Richani D., Robker R. L., Russell D. L., Gilchrist R. B., Thompson J. G. 2014. Hemoglobin: A gas-transport molecule that is hormonally regulated in the ovarian follicle in mice and humans. Biol. Reprod. 92:26. doi: 10.1095/biolreprod.114.124594 [DOI] [PubMed] [Google Scholar]
- Brown H. M., Pritchard M., Russell D. L. 2005. Early ovarian follicle dysgenesis in a disintegrin and metalloproteinase with thrombospondin motifs type 1 (ADAMTS-1) null mice. Reprod. Fertil. Dev. 17:87. doi: 10.1071/SRB05Abs225 [DOI] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., Lee J. J. 2015. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4:7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. F., Shiue Y. L., Yen C. J., Tang P. C., Chang H. C., Lee Y. P. 2007. Laying traits and underlying transcripts, expressed in the hypothalamus and pituitary gland, that were associated with egg production variability in chickens. Theriogenology 68:1305–1315. doi: 10.1016/j.theriogenology.2007.08.032 [DOI] [PubMed] [Google Scholar]
- Cheng A., Xiong W., Ferrell J. E., Solomon M. J. 2005. Identification and comparative analysis of multiple mammalian speedy/ringo proteins. Cell Cycle 4:155–165. [DOI] [PubMed] [Google Scholar]
- Cheng H., Qu L., Garrick D. J., Fernando R. L. 2015. A fast and efficient Gibbs sampler for BayesB in whole-genome analyses. Genet. Sel. Evol. 47:80. doi: 10.1186/s12711-015-0157-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I., Oh J., Cho B. N., Ahnn J., Jung Y. K., Kim D. H., Cho C. 2004. Characterization and comparative genomic analysis of intronless Adams with testicular gene expression. Genomics 83:636–646. doi: 10.1016/j.ygeno.2003.10.001 [DOI] [PubMed] [Google Scholar]
- Choi Y., Seo H., Shim J., Yoo I., Ka H. 2014. Calcium extrusion regulatory molecules: Differential expression during pregnancy in the porcine uterus. Domest. Anim. Endocrinol. 47:1–10. doi: 10.1016/j.domaniend.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Cochran S. D., Cole J. B., Null D. J., Hansen P. J. 2013. Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet. 14:49. doi: 10.1186/1471-2156-14-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. B., VanRaden P. M., O'Connell J. R., Van Tassell C. P., Sonstegard T. S., Schnabel R. D., Taylor J. F., Wiggans G. R. 2009. Distribution and location of genetic effects for dairy traits. J. Dairy Sci. 92:2931–2946. doi: 10.3168/jds.2008-1762 [DOI] [PubMed] [Google Scholar]
- Dumesic D. A., Meldrum D. R., Katz-Jaffe M. G., Krisher R. L., Schoolcraft W. B. 2015. Oocyte environment: Follicular fluid and cumulus cells are critical for oocyte health. Fertil. Steril. 103:303–316. doi: 10.1016/j.fertnstert.2014.11.015 [DOI] [PubMed] [Google Scholar]
- Eler J. P., Bignardi A. B., Ferraz J. B. S., Santana M. L. 2014. Genetic relationships among traits related to reproduction and growth of Nelore females. Theriogenology 82:708–714. doi: 10.1016/j.theriogenology.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Eler J. P., Silva J. A., Ferraz J. B. S., Dias F., Oliveira H. N., Evans J. L., Golden B. L. 2002. Genetic evaluation of the probability of pregnancy at 14 months for Nellore heifers. J. Anim. Sci. 80:951–954. doi: 10.2527/2002.804951x [DOI] [PubMed] [Google Scholar]
- Espigolan R., Baldi F., Boligon A. A., Souza F. R., Gordo D. G., Tonussi R. L., Cardoso D. F., Oliveira H. N., Tonhati H., Sargolzaei M., Schenkel F. S., Carvalheiro R., Ferro J. A., Albuquerque L. G. 2013. Study of whole genome linkage disequilibrium in Nellore cattle. BMC Genomics 14:305. doi: 10.1186/1471-2164-14-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Wang Y., Cai H., Sun G., Niu W., Xin Q., Tang X., Zhang J., Wang C., Zhang H., Xia G. 2016. ADAM10–Notch signaling governs the recruitment of ovarian pregranulosa cells and controls folliculogenesis in mice. J. Cell Sci. 129:2202–2212. doi: 10.1242/jcs.184267 [DOI] [PubMed] [Google Scholar]
- Fernando R. L., Garrick D. J. 2013. Bayesian methods applied to GWAS. In: Gondro C., van der Werf J., Hayes B. editors. Genome-Wide Association Studies and Genomic Prediction. Vol. 1019. Springer, Totowa, NJ: p. 237–274. [DOI] [PubMed] [Google Scholar]
- Feuerstein P., Puard V., Chevalier C., Teusan R., Cadoret V., Guerif F., Houlgatte R., Royere D. 2012. Genomic assessment of human cumulus cell marker genes as predictors of oocyte developmental competence: Impact of various experimental factors. PLoS One 7:e40449. doi: 10.1371/journal.pone.0040449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes M. R. S., Lehnert S. A., Bolormaa S., Reich C., Fordyce G., Corbet N. J., Whan V., Hawken R. J., Reverter A. 2012a. Finding genes for economically important traits : Brahman cattle puberty. Anim. Prod. Sci. 52:143–150. doi: 10.1071/AN11165 [DOI] [Google Scholar]
- Fortes M. R. S., Li Y., Collis E., Zhang Y., Hawken R. J. 2013. The IGF1 pathway genes and their association with age of puberty in cattle. Anim. Genet. 44:91–95. doi: 10.1111/j.1365-2052.2012.02367.x [DOI] [PubMed] [Google Scholar]
- Fortes M. R. S., Reverter A., Hawken R. J., Bolormaa S., Lehnert S. A. 2012b. Candidate genes associated with testicular development, sperm quality, and hormone levels of inhibin, luteinizing hormone, and insulin-like growth factor 1 in Brahman bulls. Biol. Reprod. 87:58. doi: 10.1095/biolreprod.112.101089 [DOI] [PubMed] [Google Scholar]
- Gaddis K. L. P., Null D. J., Cole J. B. 2016. Explorations in genome-wide association studies and network analyses with dairy cattle fertility traits. J. Dairy Sci. 99:6420–6435. doi: 10.3168/jds.2015-10444 [DOI] [PubMed] [Google Scholar]
- Gao S., De Geyter C., Kossowska K., Zhang H. 2007. FSH stimulates the expression of the ADAMTS-16 protease in mature human ovarian follicles. Mol. Hum. Reprod. 13:465–471. doi: 10.1093/molehr/gam037 [DOI] [PubMed] [Google Scholar]
- García-Ispierto I., López-Gatius F., Bech-Sabat G., Santolaria P., Yániz J. L., Nogareda C., De Rensis F., López-Béjar M. 2007. Climate factors affecting conception rate of high producing dairy cows in northeastern Spain. Theriogenology 67:1379–1385. doi: 10.1016/j.theriogenology.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Garrick D. J., Fernando R. L. 2013. Implementing a QTL Detection Study (GWAS) Using Genomic Prediction Methodology. In: Methods in molecular biology, vol. 1019. Clifton, NJ: p. 275–298. [DOI] [PubMed] [Google Scholar]
- Greenfeld C. R., Pepling M. E., Babus J. K., Furth P. A., Flaws J. A. 2007. BAX regulates follicular endowment in mice. Reproduction 133:865–876. doi: 10.1530/REP-06-0270 [DOI] [PubMed] [Google Scholar]
- Gu B., Zhang J., Wu Y., Zhang X., Tan Z., Lin Y., Huang X., Chen L., Yao K., Zhang M. 2011. Proteomic analyses reveal common promiscuous patterns of cell surface proteins on human embryonic stem cells and sperms. PLoS One 6:e19386. doi: 10.1371/journal.pone.0019386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães-Young A., Neff T., Dupuy A. J., Goodheart M. J. 2016. Conditional deletion of Sox17 reveals complex effects on uterine adenogenesis and function. Dev. Biol. 414:219–227. doi: 10.1016/j.ydbio.2016.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Kizilkaya K., Garrick D. J. 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinf. 12:186. doi: 10.1186/1471-2105-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Choi E., Park I., Lee B., Jin S., Kim D. H., Nishimura H., Cho C. 2009. Comprehensive analysis of reproductive ADAMs: Relationship of ADAM4 and ADAM6 with an ADAM complex required for fertilization in mice. Biol. Reprod. 80:1001–1008. doi: 10.1095/biolreprod.108.073700 [DOI] [PubMed] [Google Scholar]
- Hatzirodos N., Irving-Rodgers H. F., Hummitzsch K., Harland M. L., Morris S. E., Rodgers R. J. 2014a. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genomics 15:24. doi: 10.1186/1471-2164-15-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzirodos N., Irving-Rodgers H. F., Hummitzsch K., Rodgers R. J. 2014b. Transcriptome profiling of the theca interna from bovine ovarian follicles during atresia. PLoS One 9:e99706. doi: 10.1371/journal.pone.0099706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawken R. J., Zhang Y. D., Fortes M. R. S., Collis E., Barris W. C., Corbet N. J., Williams P. J., Fordyce G., Holroyd R. G., Walkley J. R. W., Barendse W., Johnston D. J., Prayaga K. C., Tier B., Reverter A., Lehnert S. A. 2012. Genome-wide association studies of female reproduction in tropically adapted beef cattle. J. Anim. Sci. 90:1398–1410. doi: 10.2527/jas.2011-4410 [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Lewin H. A., Goddard M. E. 2013. The future of livestock breeding: Genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet. 29:206–214. doi: 10.1016/j.tig.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Hensen K., Braem C., Declercq J., Van Dyck F., Dewerchin M., Fiette L., Denef C., Van De Ven W. J. M. 2004. Targeted disruption of the murine Plag1 proto-oncogene causes growth retardation and reduced fertility. Dev. Growth Differ. 46:459–470. doi: 10.1111/j.1440-169x.2004.00762.x [DOI] [PubMed] [Google Scholar]
- Hill W. G., Robertson A. 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38:226–231. doi: 10.1007/BF01245622 [DOI] [PubMed] [Google Scholar]
- Höglund P. J., Adzic D., Scicluna S. J., Lindblom J., Fredriksson R. 2005. The repertoire of solute carriers of family 6: Identification of new human and rodent genes. Biochem. Biophys. Res. Commun. 336:175–189. doi: 10.1016/j.bbrc.2005.08.048 [DOI] [PubMed] [Google Scholar]
- HongYan Z., Song Z., Wei Z., YuHong S. 2009. Polymorphism of FUT1 gene and its relationship with litter size in northeast Hebao pigs. Anim. Husb. Feed Sci. (Pawtucket, RI, U. S.) 1:1–3. [Google Scholar]
- Hosono K., Noda S., Shimizu A., Nakanishi N., Ohtsubo M., Shimizu N., Minoshima S. 2010. YPEL5 protein of the YPEL gene family is involved in the cell cycle progression by interacting with two distinct proteins RanBPM and RanBP10. Genomics 96:102–111. doi: 10.1016/j.ygeno.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Hu Z. L., Park C. A., Reecy J. M. 2016. Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res. 44:D827–D833. doi: 10.1093/nar/gkv1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson N. K., O'Hara M., Lacey H. A., Corcoran J., Hemmings D. G., Wareing M., Baker P., Taggart M. J. 2007. Modulation of human arterial tone during pregnancy: The effect of the bioactive metabolite sphingosine-1-phosphate. Biol. Reprod. 77:45–52. doi: 10.1095/biolreprod.107.060681 [DOI] [PubMed] [Google Scholar]
- Irie N., Weinberger L., Tang W. W. C., Kobayashi T., Viukov S., Manor Y. S., Dietmann S., Hanna J. H., Surani M. A. 2015. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 160:253–268. doi: 10.1016/j.cell.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juma A. R., Damdimopoulou P. E., Grommen S. V. H., Van de Ven W. J. M., De Groef B. 2016. Emerging role of PLAG1 as a regulator of growth and reproduction. J. Endocrinol. 228:R45–R56. doi: 10.1530/JOE-15-0449 [DOI] [PubMed] [Google Scholar]
- Kappes S. M., Bennett G. L., Keele J. W., Echternkamp S. E., Gregory K. E., Thallman R. M. 2000. Initial results of genomic scans for ovulation rate in a cattle population selected for increased twinning rate. J. Anim. Sci. 78:3053–3059. [DOI] [PubMed] [Google Scholar]
- Khatkar M. S. S., Randhawa I. A. S. A. S., Raadsma H. W. W. 2014. Meta-assembly of genomic regions and variants associated with female reproductive efficiency in cattle. Livest. Sci. 166:144–157. doi: 10.1016/j.livsci.2014.05.015 [DOI] [Google Scholar]
- Kim E. S., Shi X., Cobanoglu O., Weigel K., Berger P. J., Kirkpatrick B. W. 2009. Refined mapping of twinning-rate quantitative trait loci on bovine chromosome 5 and analysis of insulin-like growth factor-1 as a positional candidate gene. J. Anim. Sci. 87:835–843. doi: 10.2527/jas.2008-1252 [DOI] [PubMed] [Google Scholar]
- Knight P. G., Glister C. 2006. TGF-β superfamily members and ovarian follicle development. Reproduction 132:191–206. doi: 10.1530/rep.1.01074 [DOI] [PubMed] [Google Scholar]
- Korteweg N., Maia A. S., Thompson B., Roubos E. W., Burbach J. P. H., Verhage M. 2005. The role of Munc18-1 in docking and exocytosis of peptide hormone vesicles in the anterior pituitary. Biol. Cell 97:445–455. doi: 10.1042/BC20040101 [DOI] [PubMed] [Google Scholar]
- Kwon W.-S., Rahman M. S., Lee J.-S., Yoon S.-J., Park Y.-J., Pang M.-G. 2015. Discovery of predictive biomarkers for litter size in boar spermatozoa. Mol. Cell. Proteomics 14:1230–1240. doi: 10.1074/mcp.M114.045369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartey J., Taggart J., Robson S., Taggart M. 2016. Altered expression of human smooth muscle myosin phosphatase targeting (MYPT) isovariants with pregnancy and labor. PLoS One 11:e0164352. doi: 10.1371/journal.pone.0164352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Sala G., Marazziti D., Di Pietro C., Golini E., Matteoni R., Tocchini-Valentini G. P. 2015. Modulation of Dhh signaling and altered Sertoli cell function in mice lacking the GPR37-prosaposin receptor. FASEB J. 29:2059–2069. doi: 10.1096/fj.14-269209 [DOI] [PubMed] [Google Scholar]
- Lefebvre R., Lo M. C., Suarez S. S. 1997. Bovine sperm binding to oviductal epithelium involves fucose recognition. Biol. Reprod. 56:1198–1204. doi: 10.1095/biolreprod56.5.1198 [DOI] [PubMed] [Google Scholar]
- Legarra A., Christensen O. F., Aguilar I., Misztal I. 2014. Single step, a general approach for genomic selection. Livest. Sci. 166:54–65. doi: 10.1016/j.livsci.2014.04.029 [DOI] [Google Scholar]
- Leroy J. L. M. R., Vanholder T., Mateusen B., Christophe A., Opsomer G., de Kruif A., Genicot G., Van Soom A. 2005. Non-esterified fatty acids in follicular fluid of dairy cows and their effect on developmental capacity of bovine oocytes in vitro. Reproduction 130:485–495. doi: 10.1530/rep.1.00735 [DOI] [PubMed] [Google Scholar]
- Lourenço D., Brauner R., Rybczynska M., Nihoul-Fékété C., McElreavey K., Bashamboo A. 2011. Loss-of-function mutation in GATA4 causes anomalies of human testicular development. Proc. Natl. Acad. Sci. U. S. A. 108:1597–1602. doi: 10.1073/pnas.1010257108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Lam H. N., Menon R. K. 2005. New members of the insulin family: Regulators of metabolism, growth and now reproduction. Pediatr. Res. 57:70R–73R. [DOI] [PubMed] [Google Scholar]
- Luangpraseuth-Prosper A., Lesueur E., Jouneau L., Pailhoux E., Cotinot C., Mandon-Pépin B. 2015. TOPAZ1, a germ cell specific factor, is essential for male meiotic progression. Dev. Biol. 406:158–171. doi: 10.1016/j.ydbio.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Luna-Nevarez P., Rincon G., Medrano J. F., Riley D. G., Chase C. C., Coleman S. W., van Leeuwen D. M., de Atley K. L., Islas-Trejo A., Silver G. A., Thomas M. G. 2011. Single nucleotide polymorphisms in the growth hormone-insulin-like growth factor axis in straightbred and crossbred Angus, Brahman, and Romosinuano heifers: Population genetic analyses and association of genotypes with reproductive phenotypes. J. Anim. Sci. 89:926–934. doi: 10.2527/jas.2010-3483 [DOI] [PubMed] [Google Scholar]
- Luo W., Gumen A., Haughian J. M., Wiltbank M. C. 2011. The role of luteinizing hormone in regulating gene expression during selection of a dominant follicle in cattle. Biol. Reprod. 84:369–378. doi: 10.1095/biolreprod.110.085274 [DOI] [PubMed] [Google Scholar]
- Maglott D., Ostell J., Pruitt K. D., Tatusova T. 2011. Entrez Gene : gene-centered information at NCBI. Nucleic Acids Res. 39:52–57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak W., Fang C., Holden T., Dratver M. B., Lin H. 2016. An important role of Pumilio 1 in regulating the development of the mammalian female germline. Biol. Reprod. 94:134. doi: 10.1095/biolreprod.115.137497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marei W. F., Wathes D. C., Fouladi-Nashta A. A. 2010. Impact of linoleic acid on bovine oocyte maturation and embryo development. Reproduction 139:979–988. doi: 10.1530/REP-09-0503 [DOI] [PubMed] [Google Scholar]
- Martin-McCaffrey L., Willard F. S., Oliveira-dos-Santos A. J., Natale D. R. C., Snow B. E., Kimple R. J., Pajak A., Watson A. J., Dagnino L., Penninger J. M., Siderovski D. P., D'Souza S. J. A. 2004. RGS14 is a mitotic spindle protein essential from the first division of the mammalian zygote. Dev. Cell 7:763–769. doi: 10.1016/j.devcel.2004.10.004 [DOI] [PubMed] [Google Scholar]
- McEvoy T. G., Coull G. D., Broadbent P. J., Hutchinson J. S. M., Speake B. K. 2000. Fatty acid composition of lipids in immature cattle, pig and sheep oocytes with intact zona pellucida. J. Reprod. Fertil. 118:163–170. doi: 10.1530/reprod/118.1.163 [DOI] [PubMed] [Google Scholar]
- McReynolds S., Dzieciatkowska M., Stevens J., Hansen K. C., Schoolcraft W. B., Katz-Jaffe M. G. 2014. Toward the identification of a subset of unexplained infertility: A sperm proteomic approach. Fertil. Steril. 102:692–699. doi: 10.1016/j.fertnstert.2014.05.021 [DOI] [PubMed] [Google Scholar]
- Mendive F., Laurent P., Van Schoore G., Skarnes W., Pochet R., Vassart G. 2006. Defective postnatal development of the male reproductive tract in LGR4 knockout mice. Dev. Biol. 290:421–434. doi: 10.1016/j.ydbio.2005.11.043 [DOI] [PubMed] [Google Scholar]
- Meuwissen T. H., Hayes B. J., Goddard M. E. 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misztal I., Tsuruta S., Lourenco D. A. L., Aguilar I., Legarra A., Vitezica Z. 2014. Manual for BLUPF90 family of programs. http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=blupf90_all1.pdf. (Accessed 6 October 2016.)
- Mizugishi K., Li C., Olivera A., Bielawski J., Bielawska A., Deng C., Proia R. L. 2007. Maternal disturbance in activated sphingolipid metabolism causes pregnancy loss in mice. J. Clin. Invest. 117:2993–3006. doi: 10.1172/JCI30674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morota G., Peñagaricano F., Petersen J. L., Ciobanu D. C., Tsuyuzaki K., Nikaido I. 2015. An application of MeSH enrichment analysis in livestock. Anim. Genet. 46:381–387. doi: 10.1111/age.12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y. M., Yanase T., Nishi Y., Tanaka A., Saito M., Jin C. H., Mukasa C., Okabe T., Nomura M., Goto K., Nawata H. 2001. Saturated FFAs, palmitic acid and stearic acid, induce apoptosis in human granulosa cells. Endocrinology 142:3590–3597. doi: 10.1210/endo.142.8.8293 [DOI] [PubMed] [Google Scholar]
- Nakahata S., Katsu Y., Mita K., Inoue K., Nagahama Y., Yamashita M. 2001. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 276:20945–20953. doi: 10.1074/jbc.M010528200 [DOI] [PubMed] [Google Scholar]
- Nakahata S., Kotani T., Mita K., Kawasaki T., Katsu Y., Nagahama Y., Yamashita M. 2003. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 120:865–880. doi: 10.1016/S0925-4773(03)00160-6 [DOI] [PubMed] [Google Scholar]
- Nakazato T., Takinaka T., Mizuguchi H., Matsuda H., Bono H., Asogawa M. 2007. Biocompass: A novel functional inference tool that utilizes MeSH hierarchy to analyze groups of genes. In Silico Biol. 8:53–61. [PubMed] [Google Scholar]
- Nelson S. J., Schopen M., Savage A. G., Schulman J. L., Arluk N. 2004. The MeSH translation maintenance system: Structure, interface design, and implementation. Stud. Health Technol. Inform. 107:67–69. [PubMed] [Google Scholar]
- Ortega M. S., Denicol A. C., Cole J. B., Null D. J., Hansen P. J. 2016. Use of single nucleotide polymorphisms in candidate genes associated with daughter pregnancy rate for prediction of genetic merit for reproduction in Holstein cows. Anim. Genet. 47:288–297. doi: 10.1111/age.12420 [DOI] [PubMed] [Google Scholar]
- Parisi M., Lin H. 1999. The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics 153:235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S. O., Kizilkaya K., Garrick D. J., Fernando R. L., Reecy J. M., Weaber R. L., Silver G. A., Thomas M. G. 2012. Bayesian genome-wide association analysis of growth and yearling ultrasound measures of carcass traits in Brangus heifers. J. Anim. Sci. 90:3398–3409. doi: 10.2527/jas.2011-4507 [DOI] [PubMed] [Google Scholar]
- Pham K., Langlais P., Zhang X., Chao A., Zingsheim M., Yi Z. 2012. Insulin-stimulated phosphorylation of protein phosphatase 1 regulatory subunit 12B revealed by HPLC-ESI-MS/MS. Proteome Sci. 10:52. doi: 10.1186/1477-5956-10-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter L. A., Dellinger R. W., Tynan J. A., Barnes E. A., Kong M., Lenormand J. L., Donoghue D. J. 2002. Human speedy: A novel cell cycle regulator that enhances proliferation through activation of Cdk2. J. Cell Biol. 157:357–366. doi: 10.1083/jcb.200109045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather R., Simerly C., Schatten G., Pilch D. R., Lobo S. M., Marzluff W. F., Dean W. L., Schultz G. A. 1990. U3 snRNPs and nucleolar development during oocyte maturation, fertilization and early embryogenesis in the mouse: U3 snRNA and snRNPs are not regulated coordinate with other snRNAs and snRNPs. Dev. Biol. 138:247–255. doi: 10.1016/0012-1606(90)90195-O [DOI] [PubMed] [Google Scholar]
- Przygrodzka E., Witek K. J., Kaczmarek M. M., Andronowska A., Ziecik A. J. 2015. Expression of factors associated with apoptosis in the porcine corpus luteum throughout the luteal phase of the estrous cycle and early pregnancy: Their possible involvement in acquisition of luteolytic sensitivity. Theriogenology 83:535–545. doi: 10.1016/j.theriogenology.2014.10.016 [DOI] [PubMed] [Google Scholar]
- Pyun J. A., Kim S., Kwack K. B. 2014. Interaction between thyroglobulin and ADAMTS16 in premature ovarian failure. Clin. Exp. Reprod. Med. 41:120–124. doi: 10.5653/cerm.2014.41.3.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput S. K., Kumar P., Roy B., Verma A., Pandey H. P., Singh D., De S., Datta T. K. 2013. Identification of some unknown transcripts from SSH cDNA library of buffalo follicular oocytes. Animal 7:446–454. doi: 10.1017/S1751731112001620 [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2013. R: A language and environment for statistical computing. The R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Redmond J. S., Macedo G. G., Velez I. C., Caraty A., Williams G. L., Amstalden M. 2011. Kisspeptin activates the hypothalamic-adenohypophyseal-gonadal axis in prepubertal ewe lambs. Reproduction 141:541–548. doi: 10.1530/REP-10-0467 [DOI] [PubMed] [Google Scholar]
- Romero-Aguirregomezcorta J., Matás C., Coy P. 2015. α-l-Fucosidase enhances capacitation-associated events in porcine spermatozoa. Vet. J. 203:109–114. doi: 10.1016/j.tvjl.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Russell D. L., Brown H. M., Dunning K. R. 2015. ADAMTS proteases in fertility. Matrix Biol. 44–46:54–63. doi: 10.1016/j.matbio.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Ryu D., Ryu J., Lee C. 2016. Genome-wide association study reveals sex-specific selection signals against autosomal nucleotide variants. J. Hum. Genet. 61:423–426. doi: 10.1038/jhg.2015 [DOI] [PubMed] [Google Scholar]
- Sahana G., Guldbrandtsen B., Bendixen C., Lund M. S. 2010. Genome-wide association mapping for female fertility traits in Danish and Swedish Holstein cattle. Anim. Genet. 41:579–588. doi: 10.1111/j.1365-2052.2010.02064.x [DOI] [PubMed] [Google Scholar]
- Sanchez-Lazo L., Brisard D., Elis S., Maillard V., Uzbekov R., Labas V., Desmarchais A., Papillier P., Monget P., Uzbekova S. 2014. Fatty acid synthesis and oxidation in cumulus cells support oocyte maturation in bovine. Mol. Endocrinol. 28:1502–1521. doi: 10.1210/me.2014-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sárvári M., Kalló I., Hrabovszky E., Solymosi N., Rodolosse A., Vastagh C., Auer H., Liposits Z. 2015. Hippocampal gene expression is highly responsive to estradiol replacement in middle-aged female rats. Endocrinology 156:2632–2645. doi: 10.1210/en.2015-1109 [DOI] [PubMed] [Google Scholar]
- Schulman N. F., Sahana G., Lund M. S., Viitala S. M., Vilkki J. H. 2008. Quantitative trait loci for fertility traits in Finnish Ayrshire cattle. Genet. Sel. Evol. 40:195–214. doi: 10.1051/gse:2007044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. C., Richards J. S. 2000. Regulation of AP1 (Jun/Fos) factor expression and activation in ovarian granulosa cells. Relation of JunD and Fra2 to terminal differentiation. J. Biol. Chem. 275:33718–33728. doi: 10.1074/jbc.M003555200 [DOI] [PubMed] [Google Scholar]
- Shozu M., Minami N., Yokoyama H., Inoue M., Kurihara H., Matsushima K., Kuno K. 2005. ADAMTS-1 is involved in normal follicular development, ovulatory process and organization of the medullary vascular network in the ovary. J. Mol. Endocrinol. 35:343–355. doi: 10.1677/jme.1.01735 [DOI] [PubMed] [Google Scholar]
- Smith B. J. 2007. boa: An R package for MCMC output convergence. J. Stat. Softw. 21:1–37. doi: 10.18637/jss.v021.i11 [DOI] [Google Scholar]
- Spencer K. L., Malinowski J., Carty C. L., Franceschini N., Fernández-Rhodes L., Young A., Cheng I., Ritchie M. D., Haiman C. A., Wilkens L., Wu C., Matise T. C., Carlson C. S., Brennan K., Park A., Rajkovic A., Hindorff L. A., Buyske S., Crawford D. C. 2013. Genetic variation and reproductive timing: African American women from the population architecture using genomics and epidemiology (PAGE) study. PLoS One 8:e55258. doi: 10.1371/journal.pone.0055258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez S. S., Revah I., Lo M., Kölle S. 1998. Bull sperm binding to oviductal epithelium is mediated by a Ca2+-dependent lectin on sperm that recognizes Lewis-a trisaccharide. Biol. Reprod. 59:39–44. doi: 10.1095/biolreprod59.1.39 [DOI] [PubMed] [Google Scholar]
- Sun R., Shen R., Li J., Xu G., Chi J., Li L., Ren J., Wang Z., Fei J. 2011. Lyzl4, a novel mouse sperm-related protein, is involved in fertilization. Acta Biochim. Biophys. Sin. (Shanghai) 43:346–353. doi: 10.1093/abbs/gmr017 [DOI] [PubMed] [Google Scholar]
- Tanghe S., Van Soom A., Duchateau L., Nauwynck H., De Kruif A. 2004. Carbohydrates and glycoproteins involved in bovine fertilization in vitro. Mol. Reprod. Dev. 68:492–499. doi: 10.1002/mrd.20095 [DOI] [PubMed] [Google Scholar]
- Tenghe A. M. M., Bouwman A. C., Berglund B., Strandberg E., de Koning D. J., Veerkamp R. F. 2016. Genome-wide association study for endocrine fertility traits using single nucleotide polymorphism arrays and sequence variants in dairy cattle. J. Dairy Sci. 99:5470–5485. doi: 10.3168/jds.2015-10533 [DOI] [PubMed] [Google Scholar]
- te Velde E. R., Pearson P. L. 2002. The variability of female reproductive aging. Hum. Reprod. Update 8:141–154. doi: 10.1093/humupd/8.2.141 [DOI] [PubMed] [Google Scholar]
- Tsuyuzaki K., Morota G., Ishii M., Nakazato T., Miyazaki S., Nikaido I. 2015. MeSH ORA framework: R/Bioconductor packages to support MeSH over-representation analysis. BMC Bioinf. 16:45. doi: 10.1186/s12859-015-0453-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium 2015. UniProt: A hub for protein information. Nucleic Acids Res. 43:D204–D212. doi: 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Blerkom J. 2000. Intrafollicular influences on human oocyte developmental competence: Perifollicular vascularity, oocyte metabolism and mitochondrial function. Hum. Reprod. 15(Suppl. 2):173–188. doi: 10.1093/humrep/15.suppl_2.173 [DOI] [PubMed] [Google Scholar]
- van Eekelen M., Overvoorde J., van Rooijen C., den Hertog J. 2010. Identification and expression of the family of classical protein-tyrosine phosphatases in zebrafish. PLoS One 5:e12573. doi: 10.1371/journal.pone.0012573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi: 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Warzych E., Cieslak A., Wolc A., Lechniak D. 2016. Cathepsin mRNA level in bovine cumulus cells fails to be a good marker of oocyte quality. Anim. Sci. Pap. Rep. 34:339–349. [Google Scholar]
- Weiss J., Axelrod L., Whitcomb R. W., Harris P. E., Crowley W. F., Jameson J. L. 1992. Hypogonadism caused by a single amino acid substitution in the β subunit of luteinizing hormone. N. Engl. J. Med. 326:179–183. doi: 10.1056/NEJM199201163260306 [DOI] [PubMed] [Google Scholar]
- Weiss J. M., Hüller H., Polack S., Friedrich M., Diedrich K., Treeck O., Pfeiler G., Ortmann O. 2007. Estradiol differentially modulates the exocytotic proteins SNAP-25 and munc-18 pituitary gonadotrophs. J. Mol. Endocrinol. 38:305–314. doi: 10.1677/jme.1.02114 [DOI] [PubMed] [Google Scholar]
- White S., Kimber S. J. 1994. Changes in alpha (1-2)-fucosyltransferase activity in the murine endometrial epithelium during the estrous cycle, early pregnancy, and after ovariectomy and hormone replacement. Biol. Reprod. 50:73–81. doi: 10.1095/biolreprod50.1.73 [DOI] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Kurokawa K., Itoh R. E., Nakamura T., Mochizuki N., Nagashima K., Matsuda M. 2003. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 162:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeron Y., Ocheretny A., Kedar O., Borochov A., Sklan D., Arav A. 2001. Seasonal changes in bovine fertility: Relation to developmental competence of oocytes, membrane properties and fatty acid composition of follicles. Reproduction 121:447–454. doi: 10.1530/rep.0.1210447 [DOI] [PubMed] [Google Scholar]
- Zimin A. V., Delcher A. L., Florea L., Kelley D. R., Schatz M. C., Puiu D., Hanrahan F., Pertea G., Van Tassell C. P., Sonstegard T. S., Marçais G., Roberts M., Subramanian P., Yorke J. A., Salzberg S. L. 2009. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Gao R., Zhang H., Cai X., Shen C., Wu C., Zhao S., Yu L. 2005. Molecular cloning and characterization of three novel lysozyme-like genes, predominantly expressed in the male reproductive system of humans, belonging to the C-type lysozyme/alpha-lactalbumin family. Biol. Reprod. 73:1064–1071. doi: 10.1095/biolreprod.105.041889 [DOI] [PubMed] [Google Scholar]


