Abstract
Purpose
FAM46C is known as a tumor suppressor in multiple myeloma. However, there are few studies about the expression and function of FAM46C in oral squamous cell carcinoma (OSCC), which is one of the most common oral cancers in the world.
Methods
mRNA and protein expression level were determined by real time PCR and Western blot, respectively. Cell Counting Kit-8 assay and flow cytometry analysis were used to analyze cell proliferation and apoptosis, respectively. Activity of caspase 3 and caspase 9 was determined using biochemical assays.
Results
Our results showed that the OSCC cells overexpressing FAM46C had a relatively slower cell proliferation rate and higher cell apoptosis rate compared with control groups. The results from Western blot showed that the expression levels of cleaved caspase 9 and cleaved caspase 3, which are the active forms of caspase 3 and caspase 9 in FAM46C overexpressed OSCC cells, were higher than in the control cells, while the phosphorylation of ERK1/2 together with its upstream regulators Ras and phosphorylation of MEK1/2 were relatively lower. Additionally, the results also showed that ERK1/2 agonist (EGF) or a caspase 3 inhibitor (Z-DEVD-FMK) inhibited activity of caspase 3 and caspase 9 and cell apoptosis rate. Furthermore, by analyzing FAM46C silencing OSCC cells, we found an increased proliferation rate and a reduced apoptosis rate compared with control cells. And those phenomena could be blocked by U0126, which is an ERK1/2 inhibitor.
Conclusion
Overall, our data suggest that FAM46C probably acts as a tumor suppressor gene in OSCC cells and the working mechanism of FAM46C may be involved in the caspases and ERK1/2 pathway.
Keywords: OSCC, FAM46C, caspases, ERK1/2
Introduction
As one of the most commonly diagnosed cancers, oral cancer, especially oral squamous cell carcinoma (OSCC), is a severe cancer worldwide with high rates of metastases and recurrence. Research has reported that there is a higher occurring rate of OSCC in males than in females.1 Similar to other cancers, a series of genetic changes are the primary cause of OSCC.2,3 Previous studies have shown that various types of physical, chemical or biological mutagens can increase the rate of genetic changes of OSCC.3,4 And a lot of genes involved in occurrence and development of OSCC have been studied.5–7 Even though great developments have been achieved in OSCC treatments, the main therapy is still surgical intervention with radiotherapy or chemotherapy, and the overall survival rate has not significantly improved.2,8 Therefore, it is still urgent to acquire more knowledge, especially regarding the molecular mechanisms of cell proliferation and apoptosis in OSCC, which may lead to new potential therapy.
FAM46 genes are widely expressed in animal genomes and the encoded proteins have noncanonical poly (A) polymerases activity. There are four FMA46 paralogs expressed in humans (FAM46A, FAM46B, FAM46C, and FAM46D) and among them FAM46C mutation was found in most multiple myeloma (MM) patients.9–11 Furthermore, research has also reported that overexpression of FAM46C can suppress cell migration and invasion in hepatocellular carcinoma cells.12 All those studies suggested that FAM46C may be one of the tumor suppressor genes.
However, whether FAM46C can also act as a tumor suppressor gene in OSCC is still unknown. To further understand the extra function of FAM46C gene in OSCC, we overexpressed or silenced FAM46C expression level in OSCC cell lines to analyze whether FAM46C gene is involved in regulating the cell growth in OSCC. Furthermore, the activity of caspase 3 and caspase 9 were detected to confirm that the enhanced cell apoptosis induced by FAM46C overexpression was achieved through caspases pathway. The expression level of p-ERK1/2 was also detected to investigate whether ERK1/2 acts as an inhibiting factor of caspases pathway in OSCC cells. In addition, upstream regulators of ERK1/2 were also detected to explain the regulating mechanism of FAM46C on ERK1/2. In summary, our findings demonstrated that in vitro FAM46C can regulate the proliferation and apoptosis of OSCC cells via a complex downstream signaling pathway.
Material and methods
Cell lines and transfection
HEK293T cells, human immortalized oral epithelial cell (HIOEC) line, and the adherent OSCC cell lines from human tongue tissues (HSC4, SCC25, SCC4, SCC15, and CAL27) used in the this study were purchased from JRDUN Biothech (Shanghai, China). These cells were cultured in DMEM together with 10% fetal calf serum (FCS) and 1% antibiotic (penicillin/streptomycin) at 37°C with 5% CO2.
HEK293T cells were co-transfected with a lentiviral plasmid (pLVX-puro) expressing FAM46C or containing a control vector with package plasmids. The viral supernatant was collected after 48 hours of transfection and added to SCC15 and CAL27 cells. SCC15 and CAL27 cells without any treatment were used as control. The expression level of FAM46C was evaluated by real time PCR and Western blot after 48 hours of transduction.
Three FAM46C shRNAs (siFAM46C-1 5′-CCAGGGATTGCATGTCCTT-3′, siFAM46C-2 5′-GGACGAGGC AACTTTCCAA-3′, siFAM46C-3 5′-GCAACTTCA GCAACTACTA-3′) and a control shRNA (siNC) were constructed into lentivirus (pLKO.1). The constructed lentivirus and package plasmids were co-transfected into HEK293T cells according to the manufacturer’s instructions (Lipofectamine 2000; Invitrogen™, Thermo Fisher Scientific, Waltham, MA, USA). The viral supernatant was collected after 48 hours of transfection and added to HSC4 cells. HSC4 cells without any treatment were used as control. The inhibition efficiency was evaluated by real time PCR and Western blot after 48 hours of transduction.
RNA extraction and real time PCR
Total RNA of cultured cells was isolated by using Trizol Reagent (1596-026; Invitrogen™, Thermo Fisher Scientific) according to the manufacturer’s instructions and then reverse transcribed into complementary DNA (cDNA) with Revert Aid First Strand cDNA Synthesis Kit (K1622; Fermentas, Thermo Fisher Scientific). SYBR Green PCR kit (K0223; Thermo Fisher Scientific) was used to conduct real time PCR on an ABI-7300 instrument (Thermo Fisher Scientific). FAM46C (forward primer: 5′ CGCAGGGTGGTGAACGAG 3′ and reverse primer: 5′ TACAGGGCAGCCAGGTAGG 3′) expression levels were determined with internal control, GAPDH (forward primer: 5′ AATCCCATCACCATCTTC 3′ and reverse primer: 5′ AGGCTGTTGTCATACTTC 3′).
Protein extraction and Western blot assay
Total proteins were extracted using RIPA lysis buffer containing protease and phosphatase inhibitors (BY240825; JRDun, Shanghai, China). Cell samples were washed twice with cold PBS and then lysed in RIPA buffer, after that the samples were kept at 4°C for several minutes. The samples were centrifuged at 12,000 g for 10 minutes and then supernatant was collected. The sample proteins were quantitated by BCA Protein Assay Kit (PICPI23223; Thermo Fisher Scientific). Equal amounts of protein (approximately 25 µg) were separated by SDS-PAGE, and transferred onto a nitrocellulose (NC) membrane (HATF00010; EMD Millipore, Billerica, MA, USA). The NC membrane was blocked with 5% skim milk at room temperature for 1 hour or 4°C overnight. The membranes were incubated with primary antibodies at room temperature for 2 hours or 4°C overnight (FAM46C, Abcam, Cambridge, UK, Ab169699; p-ERK1/2, CST (Cell Signalling Technology, Danvers, MA, USA), 9101; ERK1/2, CST, 9102; caspase 9, Abcam, Ab202068; caspase 3, Abcam, Ab44976; Ras, Solarbio, Beijing, China, K006385P; MEK1/2, CST, 9122; p-MEK1/2, CST, 4376; GAPDH, CST, 5174). After washing three times with Tris buffer with 1% Tween, an appropriate secondary horseradish peroxidase-conjugated antibody (A0208; Beyotime, Shanghai, China) was added to the NC membranes and incubated at 37°C for 1 hour. The signals were visualized with enhanced chemiluminescence reagents (WBKLS0100; EMD Millipore) and pictured with Tanon-5200 system (Tanon, Shanghai, China) and the bands were analyzed with ImageJ software.
Cell proliferation analysis
The cell proliferation assay was performed with Cell Counting Kit-8 (CCK-8). In short, SCC15, CAL27, and HSC4 cells were seeded onto 96-well plates (3×103 cells/well) and incubated overnight at 37°C. Then the SCC15 and CAL27 cells were transfected with lentivirus expressing FAM46C cDNA or containing control vector. SCC15 and CAL27 cells without any treatment were used as control. The HSC4 cells were transfected with FAM46C shRNAs lentivirus or an siNC lentivirus and exposed to 50 µmol/L U0126 (S1102; Selleck Chemicals, Boston, USA). At 0, 24, 48, and 72 hours after treatment, 100 µL medium without FCS with 10% CCK-8 (CP002; SAB Signalway Antibody, Bath, UK) was added to each well and incubated at 37°C for 1 hour. The optical density at 450 nm was read by a spectrophotometer (DNM-9602; Perlong, Beijing, China).
Cell apoptosis and caspase 3/9 activity assay
The cell apoptosis rate was assessed by FACS assay and activity of caspase 3 and caspase 9 was assessed by biochemical method. Amounts of 50 µg/L EGF (236-EG; R&D Systems, Inc., Minneapolis, MN, USA), 20 µM Z-DEVD-FMK (S7312; Selleck) or 50 µmol/L U0126 (S1102; Selleck) were used in this experiment.
For cell apoptosis assay, after 48 hours of transduction, 5–10×103 cells were collected and labeled with Annexin V-FITC (C1063; Beyotime) at 4°C for 15 minutes and then labeled with propidium iodide (C1063; Beyotime) at 4°C for another 5 minutes. Cell apoptosis rate was analyzed by a flow cytometer (Accuri C6; BD Biosciences, San Jose, CA, USA).
The activity of caspase 3 and caspase 9 were measured by caspase 3/caspase 9 Colorimetric Assay Kit (KeyGEN, Nanjing, China) according to the manufacturer’s instructions.
Statistical analysis
GraphPad Prism software Version 7.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used to analyze data. Data from three independent experiments were presented as mean ± SD. Results were performed by one-way or two-way ANOVA to determine the statistical significance among different groups. P<0.05 was defined as significant.
Results
The expression level of FAM46C in 5 OSCC cell lines
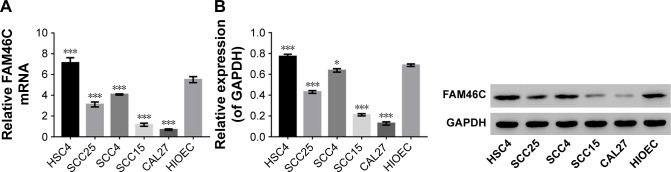
To study the biological functions of FAM46C in OSCC in vitro, we chose HIOEC cells and five different OSCC cell lines HSC4, SCC4, SCC25, SCC15, and CAL27 to detect the expression level of FAM46C. FAM46C mRNA expression level and protein expression level in HIOEC and OSCC cell lines were measured by real time PCR and Western blot respectively. As illustrated in Figure 1, a different expression level of FAM46C was shown among the five OSCC cell lines in both mRNA and protein level. The expression level of FAM46C in HSC4 was higher than in HIOEC, but in SCC25, SCC15, SCC4, and CLA27 it was lower than in HIOEC and especially lower in CAL27 and SCC15.
Figure 1.
Expression of FAM46C in HIOEC and OSCC cells.
Notes: The protein and mRNA expression levels of FAM46C in HIOEC and five OSCC cell lines were analyzed by real time PCR (A) and Western blot (B), respectively. Three independent experiments were performed and data are shown as mean ± SD. *P<0.05 vs HIOEC; ***P<0.001 vs HIOEC.
Abbreviations: HIOEC, human immortalized oral epithelial cell; OSCC, oral squamous cell carcinoma.
FAM46C overexpression in OSCC cells
To test the function of FAM46C in regulating the proliferation and apoptosis of OSCC cells, we overexpressed FAM46C in OSCC cells. Here, CAL27 and SCC15 OSCC cell lines were chosen to overexpress FAM46C because the expression level of FAM46C in these two cell lines was relatively lower (Figure 1).
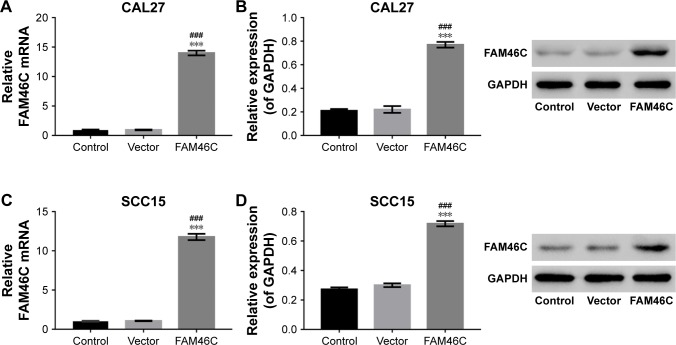
The CAL27 and SCC15 cells were transfected with the lentivirus expressing FAM46C or control vector and the cells without treatment served as control. As shown in Figure 2, the FAM46C expression level was upregulated at both mRNA and protein level compared with control and vector groups in CAL27 and SCC15 cells.
Figure 2.
Overexpression of FAM46C in OSCC cells.
Notes: mRNA and protein expression levels of FAM46C were measured by real time PCR and Western blot in CAL27 (A and B) and SCC15 (C and D) cell lines after transduction with lentivirus expressing FAM46C or control vector. SCC15 and CAL27 cells without treatment served as control. The experiments were repeated three times and presented as mean ± SD. ###P<0.001 vs control; ***P<0.001 vs vector.
Abbreviation: OSCC, oral squamous cell carcinoma.
FAM46C overexpression inhibits cell proliferation in OSCC cell lines
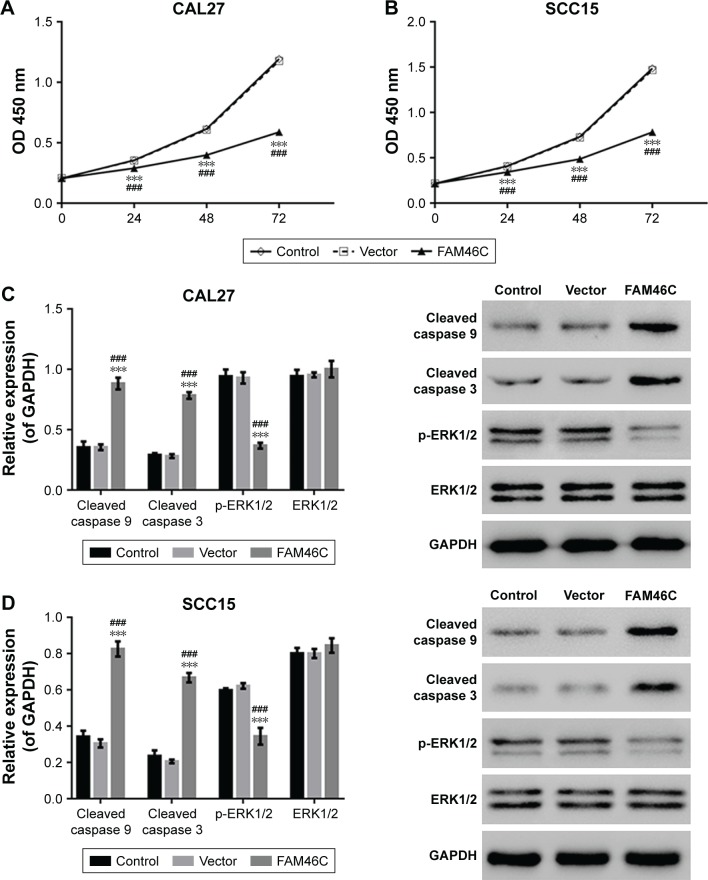
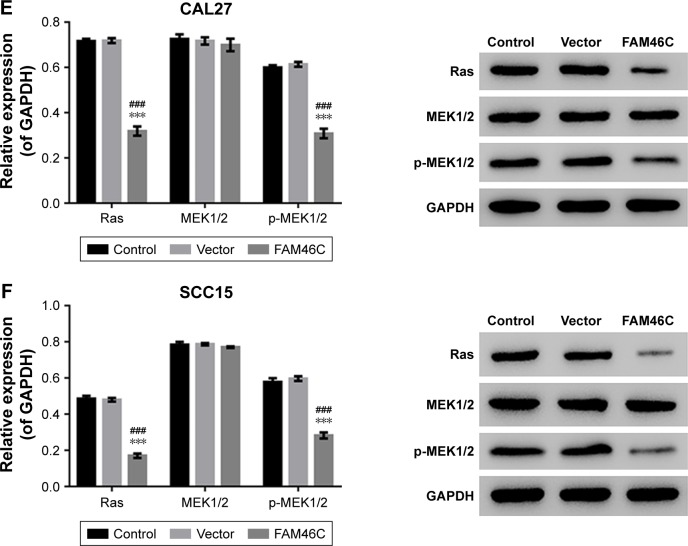
To establish whether FAM46C plays a role in regulating cell proliferation in OSCC cells, we performed CKK-8 assay with two FAM46C overexpressed OSCC cell lines (CAL27 and SCC15). The results of CCK-8 showed that, compared with control and vector groups, the cell proliferation of FAM46C overexpressed cells was significantly attenuated (Figure 3A and B). The Western blot results (Figure 3C and D) showed that, in FAM46C overexpressed cells, the expression levels of cleaved caspase 3 and cleaved caspase 9 were significantly higher but the expression level of p-ERK1/2 was lower compared with control and vector groups. Moreover, the expression level of Ras and p-MEK1/2, the upstream regulators of ERK, were also suppressed by FAM46C overexpression, as illustrated in Figure 3E and F. These data suggest that FAM46C can suppress the growth of OSCC cells and this pathway may be involved in the regulation of the activation of caspases and ERK1/2.
Figure 3.
FMA46C overexpression inhibited cell proliferation.
Notes: At 0, 24, 48, and 72 hours after transduction, cell proliferation was evaluated by CCK-8 assay in CAL27 (A) and SCC15 (B) cells. Data were based on three independent experiments and presented as mean ± SD. The protein expression levels of cleaved caspase 9, cleaved caspase 3, p-ERK1/2, and ERK1/2 in CAL27 (C) and SCC15 (D) cells after 48 hours of transduction were analyzed by Western blotting. The expression levels of Ras, MEK1/2, and p-MEK1/2 were analyzed by Western blot in CAL27 (E) and SCC15 (F). Control: original CAL27 or SCC15 cells; vector: CAL27 or SCC15 cells transduced with control lentivirus. ###P<0.001 vs control; ***P<0.001 vs vector.
Abbreviation: CCK-8, Cell Counting Kit-8.
FAM46C overexpression induces the apoptosis of OSCC cells via caspase 3 and ERK pathways
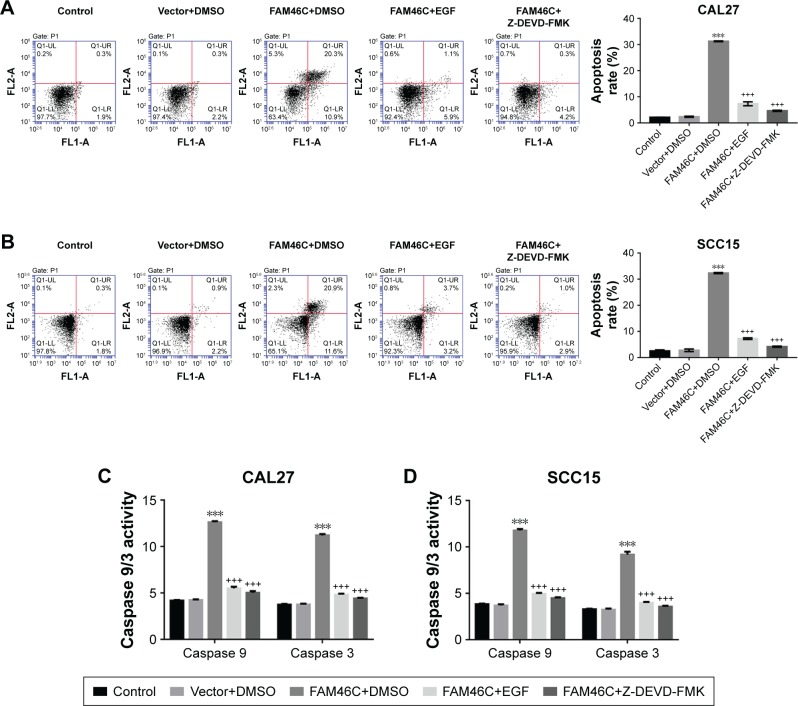
Our further study focused on whether the attenuated proliferation of OSCC cells with FAM46C overexpression was associated with an increase in apoptosis. We evaluated OSCC cell apoptosis by flow cytometry analysis with FAM46C overexpressed cells, together with treatment with ERK1/2 agonist (EGF) or a caspase 3 blocker (Z-DEVD-FMK) or vehicle (DMSO). The cells without any treatment served as control. As shown in Figure 4A and B, significantly increased cell apoptosis was shown in the FAM46C overexpressed group (CAL27 and SCC15) compared with vector group. The FAM46C overexpressed cells treated with EGF or Z-DEVD-FMK achieved an apparent decline in cell apoptosis compared with FAM46C overexpressed cells treated with vehicle. In addition, we evaluated the activity of caspase 3 and caspase 9, which are a kind of proapoptotic protein, by biochemical method. The results (Figure 4C and D) showed that both caspase 3 and caspase 9 had higher activity in FAM46C overexpressed group compared with vector group, and both EGF and Z-DEVD-FMK could reduce the activity of caspase 3 and caspase 9. These data suggest that the cell apoptosis in FAM46C overexpressed cells was regulated via caspases and ERK pathways.
Figure 4.
FAM46C overexpression induced cell apoptosis via caspase 3 and ERK1/2 pathway.
Notes: CAL27 and SCC15 cells were transduced with lentivirus expressing FAM46C (FAM46C), and then treated with 50 µg/L EGF (FAM46C+EGF) or 20 µM Z-DEVD-FMK (FAM46C+Z-DEVD-FMK) or vehicle (DMSO) for 48 hours. Cell apoptosis was evaluated by flow cytometry in CAL27 (A) and SCC15 (B) cells. Original SCC15 and CAL27 cells without any treatment (control) and the cells transduced with control vector lentivirus (vector+DMSO) were used as control. Activity of caspase 9 and caspase 3 in CAL27 (C) and SCC15 (D) cells was measured by biochemical method. ***P<0.001 vs vector+DMSO; +++P<0.001 vs FAM46C+DMSO.
Knockdown of FAM46C inhibits cell apoptosis and induces the proliferation in OSCC cells via ERK pathway
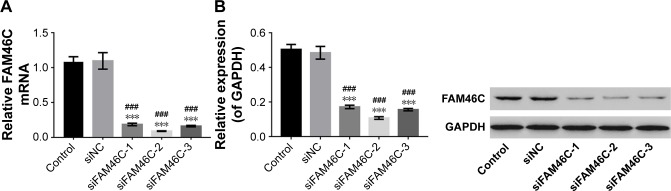
To further investigate the growth inhibition in OSCC cells, we inhibited the expression level of FAM46C in OSCC cells. As shown in Figure 1, we chose HSC4 cells to silence FAM46C because the expression level of FAM46C in HSC4 was relatively higher. The constructed lentiviruses expressing FAM46C shRNAs or control shRNA (siNC) were used to infect HSC4 cells, and HSC4 cells without any treatment served as control. In Figure 5, the results from real time PCR and Western blot showed that all three shRNAs caused a strong inhibitory effect on FAM46C at both mRNA and protein levels. siFAM46C-2 was chosen for the following experiments, because of a higher inhibitory efficiency than siFAM46C-1 and siFAM46C-3.
Figure 5.
Suppressing FAM46C expression by RNAi technology.
Notes: The efficiency of three shRNAs (siFAM46C-1, -2, and -3) or a control shRNA (siNC) in suppressing endogenous FAM46C in HSC4 cells was measured by real time PCR (A) and Western blot (B). HSC4 cells without any treatment acted as control. The experiments were repeated three times. ###P<0.001 vs control; ***P<0.001 vs siNC.
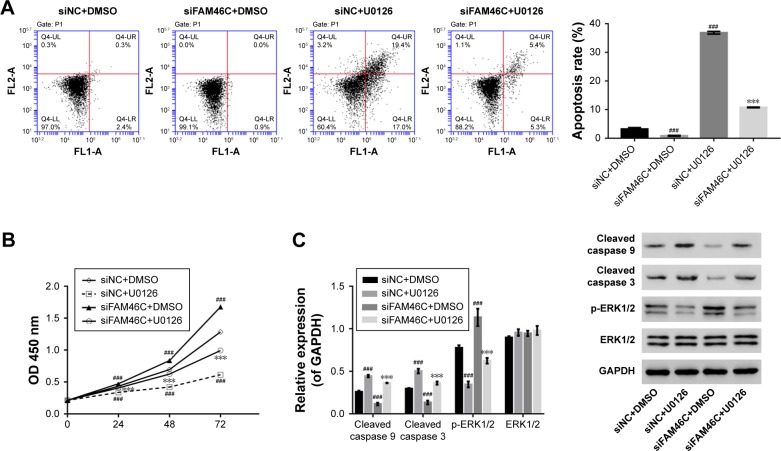
Next we analyzed the cell apoptosis and the proliferation in FAM46C knockdown cells (siFAM46C) and siNC. As shown in Figure 6, a weaker apoptosis and a stronger proliferation was exhibited in FAM46C knockdown group compared with siNC group. The Western blot results showed that the protein expression levels of cleaved caspase 3 and cleaved caspase 9 were lower but p-ERK1/2 was higher in FAM46C knockdown group compared with siNC groups. Beyond that, we treated FAM46C knockdown cells and siNC cells with an ERK1/2 inhibitor (U0126). Flow cytometry assay (Figure 6A) and CCK-8 assay (Figure 6B) showed that both FAM46C knockdown and siNC groups exhibited decreased proliferation and improved apoptosis rate compared with the cells treated by vehicle (DMSO). Western blot (Figure 6C) also showed an associative result after being treated with U0126, the protein expression level of cleaved caspase 3 and cleaved caspase 9 was upregulated in both FAM46C knockdown cells and siNC cells, but the expression level of p-ERK1/2 was reduced. These data suggest that FAM46C can regulate the growth of OSCC cells and this may be involved in activation of caspases and ERK1/2 expression pathway.
Figure 6.
Knockdown of FAM46C regulated cell apoptosis and proliferation via ERK pathway.
Notes: HSC4 cells transduced with FAM46C shRNAs (siFAM46C) lentivirus or control shRNA (siNC) lentivirus and at same time treated with U0126 or vehicle (DMSO). (A) Cell apoptosis was evaluated by flow cytometry after 48 hours of treatment. (B) at 0, 24, 48, and 72 hours after treatment, cell proliferation of these two cell groups was evaluated by ccK-8 assay. Data are based on three independent experiments and shown as mean ± SD. (C) The protein expression levels of cleaved caspase 9, cleaved caspase 3, p-ERK1/2, and ERK1/2 were analyzed by Western blotting. ###P<0.001 vs siNC+DMSO; ***P<0.001 vs siFAM46C+DMSO.
Abbreviation: CCK-8, Cell Counting Kit-8.
Discussion
The tumor-inhibiting effect of FAM46C has been studied for several years, but previous research mainly focused on MM.13,14 Here, in this research, we found that FAM46C was also related to regulating the growth of OSCC cells in vitro. First of all, we found that overexpressing FAM46C in OSCC cells could inhibit cell proliferation from CCK-8 assay. Moreover, the flow cytometry assay also showed that FAM46C induced the apoptosis of OSCC cells. Together, our findings suggest that FAM46C may inhibit the growth of OSCC cells by inducing apoptosis.
Most pathways to apoptosis involve the activation of cas-pases, which belong to the cysteinyl aspartate proteases group.15 There are approximately 14 kinds of caspases. Caspase 9, a member of caspases, usually plays a role in the early stage of cell apoptosis pathway by activating caspase 3, which is usually considered as the downstream signaling molecule in cell apoptosis pathway.16 Little is known about the pathway of FAM46C-induced apoptosis in OSCC cells. Therefore, we tested the activity of caspase 3 and caspase 9. Our results indicated that FAM46C overexpressed cell lines showed a higher activity of caspase 3 and caspase 9 and a relatively higher cell apoptosis rate compared with the vector group. Meanwhile, this activity or the higher cell apoptosis can be decreased by caspase blocker (Z-DEVD-FMK). Moreover, in FAM46C knockdown cells the protein expression level of cleaved caspase 3 and cleaved caspase 9 was downregulated compared with siNC cells. These data suggest that the pathway of FAM46C-induced apoptosis may be regulated by caspase 3/9.
A lot of research has focused on the RAF-MEK-ERK1/2 pathway, which plays a key role in cell survival.17 Moreover, it has been proven that RAS-ERK pathway can drive neoplasia, and abnormal RAS-ERK signaling exists in a lot of human malignancies.18,19 Furthermore, in this pathway, Ras acts as early signaling molecule to activate ERK1/2 and p-ERK1/2 is a final effective form which regulates cell growth.20–22 In addition, research has shown that p-ERK1/2 inhibits the activation of caspase 9 to achieve cell proliferation.18,23
Our results revealed that in FAM46C overexpressed cells, the level of p-ERK1/2 was downregulated and the level of p-ERK1/2 was relatively higher in FAM46C knockdown cells. In addition, results from Western blotting showed that the expression levels of Ras and p-MEK were also significantly suppressed by FAM46C overexpression. These data suggest that FAM46C could have an inhibitory effect on the phosphorylation of ERK1/2 via Ras/MEK pathway. In addition, ERK1/2 agonist (EGF) disturbed the activity of caspase 3 and caspase 9 in FAM46C overexpressed cells. At the same time, the Western blot results in FAM46C knockdown cells and siNC cells showed that the treatment with ERK1/2 inhibitor (U0126) reduced the level of p-ERK1/2 and increased the expression levels of cleaved caspase 3 and cleaved caspase 9 compared with the cells exposed to vehicle. These findings clearly indicate that p-ERK1/2 can really interfere with the expression level of cleaved caspase 3 and cleaved caspase 9 which can be upregulated by FAM46C overexpression in OSCC cells.
Conclusion
In summary, our results clearly illuminate that FAM46C can reduce cell proliferation and induce cell apoptosis by activating caspase 3 and caspase 9 and at the same time inhibit the phosphorylation of ERK1/2 via Ras/MEK pathway. This is the first study describing the role of FAM46C in OSCC cells and may shed light on OSCC treatment although further experiments are needed to explore how FAM46C activates caspase 3/9 and interrupts the phosphorylation of ERK1/2.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328(3):184–194. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 2.Wang XC, Ma Y, Meng PS, Han JL, Yu HY, Bi LJ. miR-433 inhibits oral squamous cell carcinoma (OSCC) cell growth and metastasis by targeting HDAC6. Oral Oncol. 2015;51(7):674–682. doi: 10.1016/j.oraloncology.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral Oncol. 2009;45(4–5):301–308. doi: 10.1016/j.oraloncology.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Petti S. Lifestyle risk factors for oral cancer. Oral Oncol. 2009;45(4–5):340–350. doi: 10.1016/j.oraloncology.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Chang JT, Wang HM, Chang KW, et al. Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int J Cancer. 2005;114(6):942–949. doi: 10.1002/ijc.20663. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh LL, Wang PF, Chen IH, et al. Characteristics of mutations in the p53 gene in oral squamous cell carcinoma associated with betel quid chewing and cigarette smoking in Taiwanese. Carcinogenesis. 2001;22(9):1497–1503. doi: 10.1093/carcin/22.9.1497. [DOI] [PubMed] [Google Scholar]
- 7.Shao Y, Qu Y, Dang S, Yao B, Ji M. MiR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6. Cancer Cell Int. 2013;13(1):51. doi: 10.1186/1475-2867-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008;87(1):14–32. doi: 10.1177/154405910808700104. [DOI] [PubMed] [Google Scholar]
- 9.Barbieri M, Manzoni M, Fabris S, et al. Compendium of FAM46C gene mutations in plasma cell dyscrasias. Br J Haematol. 2016;174(4):642–645. doi: 10.1111/bjh.13793. [DOI] [PubMed] [Google Scholar]
- 10.Kuchta K, Muszewska A, Knizewski L, et al. FAM46 proteins are novel eukaryotic non-canonical poly(A) polymerases. Nucleic Acids Res. 2016;44(8):3534–3548. doi: 10.1093/nar/gkw222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan XY, Zhai XF, Jiang YP, Han T, Zhang QY, Xin HL. Antimetastatic effects of norcantharidin on hepatocellular carcinoma cells by up-regulating FAM46C expression. Am J Transl Res. 2017;9(1):155. [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd KD, Ross FM, Walker BA, et al. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin Cancer Res. 2011;17(24):7776–7784. doi: 10.1158/1078-0432.CCR-11-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu YX, Shi CX, Bruins LA, et al. Loss of FAM46C Promotes Cell Survival in Myeloma. Cancer Res. 2017;77(16):4317–4327. doi: 10.1158/0008-5472.CAN-16-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seki K, Yoshikawa H, Shiiki K, Hamada Y, Akamatsu N, Tasaka K. Cisplatin (CDDP) specifically induces apoptosis via sequential activation of caspase-8, -3 and -6 in osteosarcoma. Cancer Chemother Pharmacol. 2000;45(3):199–206. doi: 10.1007/s002800050030. [DOI] [PubMed] [Google Scholar]
- 16.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9(1):47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 17.Ballif BA, Blenis J. Molecular mechanisms mediating mammalian mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK cell survival signals. Cell Growth Differ. 2001;12(8):397–408. [PubMed] [Google Scholar]
- 18.Balmanno K, Cook SJ. Tumour cell survival signalling by the ERK1/2 pathway. Cell Death Differ. 2009;16(3):368–377. doi: 10.1038/cdd.2008.148. [DOI] [PubMed] [Google Scholar]
- 19.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 20.Arozarena I, Calvo F, Crespo P, Ras CP. Ras, an actor on many stages: posttranslational modifications, localization, and site-specified events. Genes Cancer. 2011;2(3):182–194. doi: 10.1177/1947601911409213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrero A, Pinto A, Colón-Bolea P, et al. Small Molecule Inhibition of ERK Dimerization Prevents Tumorigenesis by RAS-ERK Pathway Oncogenes. Cancer Cell. 2015;28(2):170–182. doi: 10.1016/j.ccell.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Lefloch R, Pouysségur J, Lenormand P. Single and combined silencing of ERK1 and ERK2 reveals their positive contribution to growth signaling depending on their expression levels. Mol Cell Biol. 2008;28(1):511–527. doi: 10.1128/MCB.00800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol. 2003;5(7):647–654. doi: 10.1038/ncb1005. [DOI] [PubMed] [Google Scholar]