Abstract
African Americans (AAs) have higher incidence and mortality rates of colorectal cancer (CRC) compared with other US populations. They present with more right-sided, microsatellite stable disease and are diagnosed at earlier ages compared with non-Hispanic Whites (NHWs). To gain insight into these trends, we conducted exome sequencing (n = 45), copy number (n = 33) and methylation analysis (n = 11) of microsatellite stable AA CRCs. Results were compared with data from The Cancer Genome Atlas (TCGA). Two of the 45 tumors contained POLE mutations. In the remaining 43 tumors, only 27 (63%) contained loss-of-function mutations in APC compared with 80% of TCGA NHW CRCs. APC-mutation-negative CRCs were associated with an earlier onset of CRC (P = 0.01). They were also associated with lower overall mutation burden, fewer copy number variants and a DNA methylation signature that was distinct from the CpG island methylator phenotype characterized in microsatellite unstable disease. Three of the APC-mutation-negative CRCs had loss-of-function mutations in BCL9L. Mutations in driver genes identified by TCGA exome analysis were less frequent in AA CRC cases than TCGA NHWs. Genes that regulate the WNT signaling pathway, including SOX9, GATA6, TET1, GLIS1 and FAT1, were differentially hypermethylated in APC-mutation-negative CRCs, suggesting a novel mechanism for cancer development in these tumors. In summary, we have identified a subtype of CRC that is associated with younger age of diagnosis, lack of APC mutation, microsatellite and chromosome stability, lower mutation burden and distinctive methylation changes.
A subset of early-onset colorectal cancers is associated with lack of mutations in APC. These tumors develop by a non-canonical carcinogenic process that is characterized by lower levels of somatic mutations and driven by distinctive epigenetic changes.
Introduction
Colorectal cancer (CRC) is the third most common cancer in both men and women in the USA and the second most common cause of cancer-related deaths (1). African Americans (AAs) bear a disproportionate burden with an incidence that is approximately 20% higher than in non-Hispanic Whites (NHWs) and an even larger difference in mortality (2). Furthermore, AAs are more often diagnosed with CRCs at an earlier age, which a decade ago prompted the recommendation of CRC screening for average risk AAs to be started at age 45 instead of at age 50 (3). The early-onset CRC seen in AAs is more associated with distal location and toxic exposures (4,5).
Other biological differences have been described comparing CRCs arising in AAs and NHWs. AAs have a greater proportion of proximal cancers (6,7). Proximal CRCs have a higher percentage of microsatellite instability (MSI), increased gene promoter hypermethylation and increased gene mutation rates in comparison to distal cancers (8). Interestingly, the excess of proximal cancers in AAs in comparison to NHWs is mostly made of microsatellite stable (MSS) tumors that commonly present with lymphocytic infiltrate and are less often associated with toxic exposures or a higher body mass index compared with the distal cancers (4). In terms of hereditary risk, some CRC risk alleles are shared between NHWs and AAs but others are population-specific (9–12).
Large exome sequencing studies have been performed on CRC, but they have included few tumors from AA patients (8,13). One recent study sought to characterize somatic mutations in a significant number of MSS, non-hypermutated CRCs from AA patients (14). In that study, although genes that are commonly mutated in NHW CRCs were also found mutated in AA CRCs, mutations in 15 distinct genes were identified that were associated with CRCs in AAs but had not been previously associated with CRC in NHWs. Some of these somatic mutations, such as those in ephrin type A receptor 6 (EPHA6), foliculin (FLCN) and 5-hydroxytryptamine (serotonin) receptor 1F(HTR1F), were detected exclusively in AA CRCs.
To further characterize tumorigenic mechanisms in AA CRCs and better understand biological and clinical differences between CRCs that arise in AAs and NHWs, we analyzed the somatic mutational, copy number variation and methylation profiles in a group of AA patients from a series of newly diagnosed CRC cases.
Material and methods
Ascertainment, recruitment and biospecimen collection
The Chicago Colorectal Cancer Consortium (CCCC) ascertained incident CRC cases from surgery and endoscopy units and healthy controls undergoing routine screening colonoscopy from five large Chicago medical centers, including University of Illinois Hospital and Health Sciences System, Jesse Brown Veterans Administration, John H. Stroger Hospital of Cook County, University of Chicago Medicine and Rush University Medical Center, over a 2-year period (2011–2012). Biological specimens and patient information was collected as described previously (4). In this study, only prospectively ascertained AA patients with a diagnosis of adenocarcinoma of the colon or rectum were included (n = 54). Patients with CRC recurrence, inflammatory bowel disease, or non-adenocarcinoma tumors were excluded. All patients in the study provided written informed consent. The parent study received approval for human subjects research from the institutional review boards of each participating medical center, and the parent protocol was administered by the institutional review board at University of Illinois Hospital and Health Sciences System (2010-0168).
MSI was determined in paired DNA samples from tumor and uninvolved tissue as previously described (15). To estimate percentage of West African ancestry, a panel of 1000 ancestry informative markers was genotyped and analyzed as previously described (16).
DNA sequence analysis
Sequence analysis was performed as previously described (17). Briefly, genomic DNA (1 μg) from tumor and normal samples was sheared employing the Covaris system (Covaris, Woburn, MA) to target fragment sizes of 150–200 bp. Whole exome sequencing libraries were created using the TruSeq DNA Sample Prep Kit A and TruSeq Exome Kit (Illumina) using the manufacturer’s recommendations. Libraries were subsequently sequenced using Illumina TruSeq SBS v3 chemistry, to generate 83 cycles × 7 × 83 cycles run on the Illumina HiSeq 2000 system. All sequencing reads were converted to industry standard FASTQ files using the Bcl Conversion and Demultiplexing tool (Illumina). Population genotype principal component analysis was performed using SNP & Variation Suite v8.4.1 (Golden Helix, Bozeman, MT, www.goldenhelix.com) tool.
Sequencing reads were aligned to the GRCh37 reference genome using the MEM module of BWA v0.7.8 (18) and SAMtools v0.1.19 to produce BAM files. After alignment, the base quality scores were recalibrated and joint insertion/deletion realignment was performed on the BAM files using GATK v3.1-1 (19). Duplicate read pairs were marked using PICARD v1.111 (19). Final BAM files were then used to identify germline and somatic events. Germline SNP and insertion/deletions were identified using GATK HaplotypeCaller in the constitutional sample.
Single nucleotide variants (SNVs) and insertions/deletions were identified using Strelka somatic variant caller (20). To identify cancer drivers, analysis with MutSigCV (Mutation Significance) was performed (21) with set parameters of q < 0.1 and P < 0.05 to identify significant genes. Mutational Signature Analysis Tool (https://bitbucket.org/jtr4v/analysis-of-mutational-signatures) was used to determine mutation signatures.
Copy number analysis
Copy number analysis was conducted using the Affymetrix Cytoscan HD array carried out in the University of Illinois at Chicago Genomics Core according to the manufacturer’s instructions. CEL files were processed using Affymetrix Chromosome Analysis Suite (Version 3.0.0.42) using default quality control settings. TCGA CEL files were obtained from the Genomic Data Commons. Circular Binary Segmentation was run on samples using default settings (22). Cumulative copy number variation that exceeded a length threshold of 50% of a chromosome arm was recorded as an arm gain or loss, as this was the method adopted by the TCGA. Visualization of genome-wide copy number alterations was completed using Rawcopy (Version 1.0) with default settings. Chromosome-arm gains and losses were compared with frequencies reported by the TCGA by Fisher exact test and adjustment for multiple testing was made by the Bonferroni correction.
Methylation analysis
DNA methylation arrays were processed using the Infinium Human Methylation 450 BeadChip Kit (23,24) carried out in the Northwestern University Center for Genomic Medicine. Array data were processed using the ‘minfi’ package (25) and the ‘noob’ preprocessing scheme (26). We then employed the DMRcate procedure of Peters et al. (27) to identify statistically significant differences in regional DNA methylation. Differentially methylated regions (DMRs) were mapped to chromatin states using ChromHMM (28). Infinium Human Methylation 450 BeadChip DNA methylation data from The Cancer Genome Atlas (TCGA, n = 611 cases) were downloaded from the Genomic Data Commons and preprocessed as described earlier. APC mutation status was obtained from Mutect2 MAF files downloaded from Genomic Data Commons using TCGAbiolinks (Version 2.7.6). All NHW APC-mutation-negative samples (n = 15), a random subset of NHW APC-mutation-positive samples (n = 29) and all available NHW normal samples (n = 8) from the TCGA were used for subsequent analysis. Heatmaps were generated in R (version 3.4.3) using the ComplexHeatmap package (version 1.12.0).
Statistical analysis
Statistical tests of continuous variables were performed by the t-test and of categorical variables by the Fisher exact test or chi-square test, as appropriate. Wilcoxon non-parametric test was used for comparison of age at diagnosis between groups. To make comparisons with the TCGA exome sequence data, we used processed mutation data from 189 non-hypermutable CRCs downloaded from http://cbioportal.org (29,30), from which three were excluded based on failure to cluster with European populations in the principal component analysis. An additional 33 cases were available for the analysis of APC mutation and age. Statistical software packages such as R (https://www.r-project.org; Versions 3.1.1 & 3.3.2) and GraphPad Prism 7 (GraphPad Software, Inc.) were used to perform statistical analysis.
Results
To define the somatic mutational spectrum in AA CRCs, we conducted a whole exome analysis of selected tumors from the CCCC. Tumor samples collected fresh by the CCCC were subjected to MSI analysis (4). We excluded MSI tumors from the analysis and performed exome capture and DNA sequencing on 45 tumor-normal pairs. Sequence depth was greater than 100 genome equivalents on average in tumor DNA and 30 genome equivalents in normal DNA (Supplementary Table 1, available at Carcinogenesis Online). In the CCCC cohort, race was assigned by self-report. Mean West African ancestry was over 73% as assessed using genotype data from ancestry informative markers (Supplementary Table 2, available at Carcinogenesis Online). We further confirmed genetic ancestry using principal component analysis of the exome sequence data (31,32). All the CCCC AA CRC cases clustered with African-ancestry populations (Supplementary Figure 1, available at Carcinogenesis Online).
In CRC_46 and CRC_08, there were 4431 SNVs and 2923 SNVs per sample, respectively. CRC_46 and CRC_08 contained mutations in the proofreading domain of POLE, namely V411L in case CRC_46, which is in the active site of the exonuclease domain, and S459F in case CRC_08, which is in the RNAse H-like domain. Both mutations have been previously associated with hypermutation in CRC (8,33). The frequency of MSI-negative, POLE-associated hypermutable CRCs we detected in the CCCC series (2/45 = 4.4%) was not statistically different from the frequency reported by the TCGA (7/165 = 4.2%) and it was similar to the frequency reported for the Case Western series (2/41 = 4.9%) (14). For subsequent analyses, these two tumors were removed. In the remaining tumors, the average number of SNVs per tumor was 180.
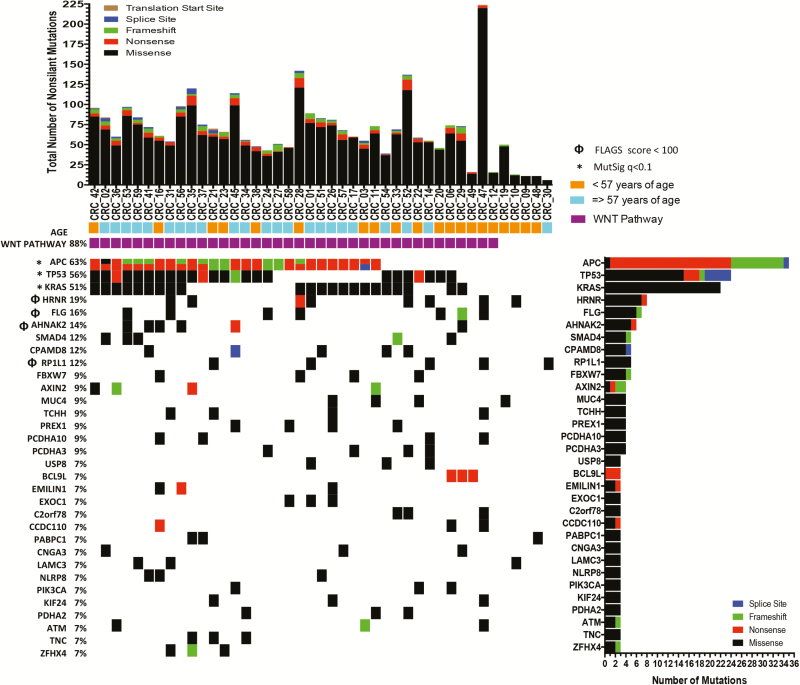
Using the list of somatic mutations generated by Strelka, we carried out mutation analysis using MutSig and ranked potential cancer driver genes based on the q value <0.1 (Figure 1). Each of the 32 genes in Figure 1 had at least three non-silent mutations in the CCCC AA CRC series. Seven of the 32 genes in Figure 1 are well established CRC driver genes, including APC, TP53, KRAS, SMAD4, FBXW7, PIK3CA and ATM. At least two novel genes represent reasonable driver gene candidates, including PREX1, which is a guanine nucleotide exchange factor for RAC that has been previously studied in breast and prostate cancer and in melanoma (34–36), and BCL9L (see later). We matched the MutSig output with their rank by frequently mutated genes (FLAGS) in public exomes (37) (Supplementary Table 3, available at Carcinogenesis Online) and observed that the mutations identified in FLG, HRNR, AHNAK2 and RP1L1 are very probably miscalled due to bioinformatics challenges with gene families. The mutations in CPAMD8, MUC4, TCHH and LAMC3 may also be questioned for the same reason even though these genes did not make the top 100 on the FLAGS list. We also found many of the specific mutations in these eight genes, which were called as somatic mutations, in the germline exomes of the ExAc database, suggesting a general problem with mutation calling for these genes. The complete list of somatic mutations detected in this sampling of tumors is provided in Supplementary Table 4, available at Carcinogenesis Online.
Figure 1.
Candidate driver genes in AA CRCs. The top panel shows the number of SNVs in each of the 43 tumors included in the analysis, coded by mutation type. The bottom panel shows the 32 genes in order of q value rank up to the threshold 0.1, matching the sample to the mutation, coded by mutation type.
APC-mutation-negative tumors are associated with early-onset CRC
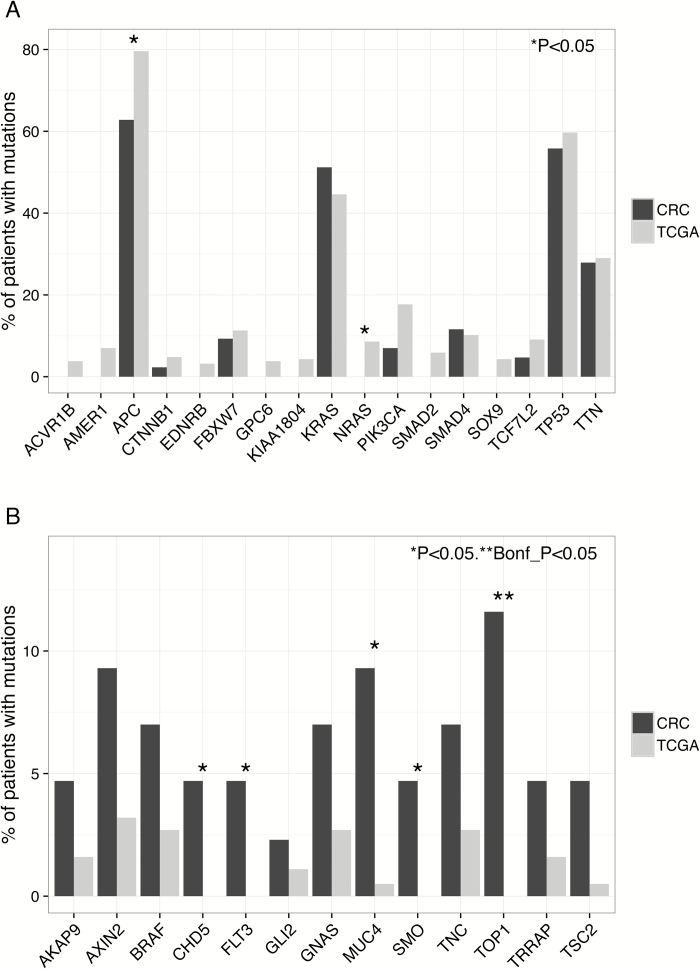
Mutations in the three most commonly mutated genes in CRC, namely APC, TP53 and KRAS were significantly associated with CRC in the MutSig analysis (P < 0.05). Although the frequencies of TP53 and KRAS mutations in AA CRCs were not statistically different from those in NHW CRCs, the frequency of APC mutations in AA CRCs was 63% (27 of 43), whereas in the TCGA NHWs it was 80% (151 of 186, P = 0.03; calculated from TCGA data based on 186 non-hypermutated CRCs in NHWs; Figure 2A). Because mutations in APC are less frequent in right-sided CRCs even taking into account MSI status (38–40) and AAs have a greater proportion of MSS right-sided CRCs (4), we considered the possibility that the difference in APC-mutation frequency could be explained by the proportion of right-sided CRCs in AAs. In the present series, there were 16 right-sided and 27 left-sided AA CRCs; 7 of 16 (0.56 of total) right-sided CRCs had at least one APC mutation whereas 18 of 27 (0.67 of total) of the left-sided CRC had at least one APC mutation (Table I). Thus, the frequency of APC mutations in the left-sided CRCs is still lower than what is reported in the TCGA.
Figure 2.
The contribution of known cancer-associated genes to AA CRCs. (A) Underrepresentation of mutations in the most frequently mutated CRC driver genes identified in TCGA. (B) Overrepresentation of cancer-associated genes.
Table 1.
Comparison of the clinical-pathological, epidemiological, and molecular features of African American CRC cases with and without an APC mutation
| Feature | All cases (n = 43) | APC mutation positive (n = 27) | APC mutation negative (n = 16) | P value |
|---|---|---|---|---|
| Clinical-pathological | ||||
| Gender, males/females | 26/17 | 19/8 | 7/9 | 0.11 |
| Age at diagnosis, median | 60 | 63 | 51.5 | 0.009 |
| Percent WAA2, mean (SD) | 77 | 80 | 73 | 0.019 |
| FDR3 with cancer <60 (%) | 16 (37) | 10 (37) | 6 (38) | 1 |
| Cases with a previous cancer (%) | 5 (12) | 1 (4) | 4 (25) | 0.06 |
| Tumor location (R/L) | 15/28 | 9/18 | 6/10 | 1 |
| TNM stage4 | 20/22 | 11/15 | 9/7 | 0.52 |
| Grade, low + moderate/high | 26/12 | 13/10 | 13/2 | 0.08 |
| Cytogenetic (stable/unstable)5 | 5/28 | 1/21 | 4/7 | 0.03 |
| Epidemiological | ||||
| Body mass index, median | 27 | 27 | 28 | 0.43 |
| Frequent exercise6 (%) | 8 (19) | 7 (26) | 1 (6) | 0.22 |
| Smoking, ever/never | 27/15 | 17/9 | 10/6 | 1 |
| Drinking, ever/never | 20/16 | 13/8 | 7/8 | 0.49 |
| High school education7 (%) | 26 (60) | 16 (59) | 10 (66) | 0.75 |
| Household income (<$25,000) (%) | 31 (72) | 19 (73) | 12 (80) | 0.72 |
| Molecular8 | ||||
| Total SNVs, median | 157 | 170 | 107 | 0.006 |
| TP53 mutation (%) | 24 (56) | 17 (63) | 7 (44) | 0.34 |
| KRAS mutation (%) | 22 (51) | 17 (63) | 5 (31) | 0.06 |
| BCL9L mutation (%) | 3 (7) | 0 (0) | 3 (19) | 0.04 |
| C2ORF78 mutation (%) | 3 (7) | 0 (0) | 3(19) | 0.04 |
1 P-values were calculated for APC-mutation-positive and APC-mutation-negative comparisons. Fisher exact test was performed for comparisons of categorical variables and Mann–Whitney for comparisons of continuous variables. Only non-hypermutable tumors were analyzed. P values less than 0.05 are shown in boldface.
2Percent West African ancestry (WAA) was estimated from ancestry informative markers using an algorithm implemented in STRUCTURE, and significance was tested by t-test. Ancestry informative markers were unavailable from 37 cases.
3FDR, first-degree relative with a cancer diagnosed before age 60.
4TNM stands for TNM Classification of Malignant Tumors, with T standing for tumor, N for nodes, and M for metastasis. Comparison of T0+TI+TII and TIII+TIV. Staging unavailable from two tumors.
5Chromosome stability was defined as no chromosome arms gained or lost in the tumor as determined by analysis of Affymetrix Cytoscan HD array data (n = 33).
6Frequent exercise was defined as three or more exercise sessions per week; does not take into account occupation-associated physical activity.
7Number of cases in which completion of high school was highest education level.
8Comparisons of mutation frequencies (APC mutation positive versus APC mutation negative) were performed on the 32 genes identified by q < 0.1 in the MutSig analysis. The P values were not adjusted for multiple testing; see text. The complete set of statistical comparisons is provided in Supplementary Table 1, available at Carcinogenesis Online.
The low frequency of APC mutations raised the question whether the APC-mutation-negative tumors are associated with a distinct tumor type (Table I). Gender, tumor location by side, tumor stage, histological grade, family history of cancer and epidemiological features were not different in the APC-mutation-positive and APC-mutation-negative tumors. On the other hand, the median mutation frequencies were significantly different (170 in APC mutation positive versus 107 in APC mutation-negative; P = 0.006, Mann–Whitney test). In addition, the APC-mutation-negative tumors were more likely to be classified chromosomally stable (see later). Importantly, APC-negative tumors were associated with younger age of diagnosis (P = 0.01). We also observed a trend for previous history of cancer in the APC-negative tumors (P = 0.06). Although the frequency of TP53 mutations was similar in the two groups (17 of 27 versus 7 of 16, respectively; P = 0.34), KRAS mutations trended toward lower frequency in the APC-mutation-negative group (17 of 27 versus 5 of 16; P = 0.06).
Because we found APC-negative tumors were associated with younger age of diagnosis, we investigated whether this association can be detected in TCGA NWH CRCs. From the cBioPortal server, we identified 219 NHW cases with a non-hypermutated tumor. Of these 219 cases, 181 tumors were APC mutation positive and 38 were APC mutation negative. The median age for the APC-mutation-positive group was 68 years, and for the APC-mutation-negative group it was 54.5. The association between APC-mutation-negative tumors and early-onset CRC was significant (P < 10−5).
BCL9L in the MutSig q <0.1 group was more frequently mutated than expected in the APC-mutation-negative tumors (P = 0.05; Figure 1). The cases CRC_06, CRC_29 and CRC_49 exhibited somatic mutations in BCL9L. These tumors contained the nonsense mutations Q654*, observed once, and R415*, observed twice. All three cases were female, with an average age at diagnosis of 46, and they had a striking family history of cancer. The mother and aunt of CRC_06 had breast cancer in their 40s. CRC_29 had a previous ovarian cancer and her aunt had a uterine cancer in her 40s. CRC_49 had a preceding breast cancer at 46 and her mother a breast cancer at 48. No germline mutations in breast-cancer-associated, homologous recombination genes (BRCA1, BRCA2, PALB2, etc.), nor in BCL9L itself were identified in these three cases; thus, the observation of family history of cancer and its possible connection to somatic mutations in BCL9L remains to be explained. Cases CRC_29 and CRC_49 had co-occurring mutations in BRAF—p.G469E and p.K601E, respectively—both of which activate the kinase domain. In the non-hypermutated, NHW TCGA series, BCL9L mutation was present in 2.6% of tumors, but it was not associated with APC-mutation-negative tumors.
Mutations in C2ORF78 were also more frequent than expected in the APC-mutation-negative group (P = 0.05). The non-silent mutation frequency in CRC in TCGA was 0.9%, which is not significantly associated with CRC. A possible role for C2ORF78 in colon carcinogenesis needs to be further investigated.
Underrepresentation of known driver genes in AA CRCs
We next considered the other most frequently mutated genes identified in the MutSig analysis by the TCGA (8), which, except for TTN, are considered to be driver genes for CRC (Figure 2A). In the AA CRCs, zero non-silent mutations were identified in ACVR1B, EDNRB, FAM123B, GPC6, KIAA1804, NRAS, SMAD2 and SOX9; only one mutation in CTNNB1; and two in TCF7L2. In contrast, in non-hypermutated, NHW CRCs from the TCGA, there were a total of 101 mutations in these genes. From the tumor-specific perspective, we found that only 3 AA CRCs of 43 tested (7%) carried a non-silent somatic mutation in at least 1 these 10 genes, whereas 79 NHW CRCs of 186 (41.8%) tested did. The underrepresentation of known driver genes predominantly affects the genes with lower frequencies of mutations, as the frequency of non-silent mutations at TP53, KRAS, FBWX7, SMAD4 and TTN was comparable in AA and NHW CRCs. To rule out inadequate sequence coverage as a trivial explanation for the observed difference, we manually checked the numbers of reads for each exon of each of these genes to confirm that there were at least 30 sequencing reads in each.
To determine whether other driver genes may compensate for the underrepresentation of TCGA driver genes, we identified all genes mutated in AA CRCs that were 2-fold or greater in frequency compared with non-hypermutated NHW CRCs in the TCGA, removing FLAGs genes that are frequently identified as mutated for technical reasons (37) and filtering the gene list using the merged lists of CRC-associated genes from the COSMIC and the Atlas of Genetics and Cytogenetics databases (Figure 2B). Five of the 13 genes that passed this screen are significantly associated with AA CRCs, including CHD5, FLT3, MUC4, SMO and TOP1. CHD5 was one of the genes of interest identified in the Case Western study (14) and it has been implicated in cancer development in methylation studies (41,42). None of the three oncogenic mutations in BRAF were V600E, but it is important to point out that these mutations are occurring in the context of MSS tumors. Mutations in AXIN2 are also overrepresented in AA CRCs. These data underscore the role of alternative driver genes operating in AA CRCs.
Copy number variation in AA CRCs
To examine the question whether differences in mutational mechanisms might underpin differences in APC-mutation-negative and mutation-positive tumors, we analyzed mutagenesis patterns and copy number variation. We compared the frequencies of mutant triplets in exome sequence data from CCCC and TCGA CRCs (Supplementary Figure 2, available at Carcinogenesis Online). As expected, hypermutable tumors with mutations of the proofreading exonuclease domain had a predominance of TCT to TAT and TCG to TTG mutations. MSS tumors had a predominance of C to T mutations in the CpG dinucleotide, reflecting a mutational process dominated by deamination of methylated cytosines. The frequencies of triplets in these two groups were indistinguishable in AA compared with NHW CRCs. Mutational signatures in APC-mutation-negative and mutation-positive tumors were also indistinguishable.
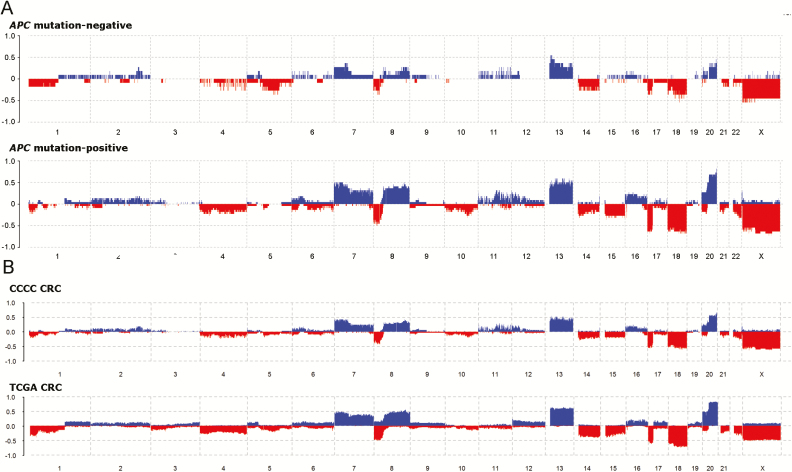
To address the question whether gross chromosomal abnormalities were different in APC-mutation-negative and mutation-positive tumors, we performed a copy number analysis in CCCC AA CRCs using Affymetrix Cytoscan HD array analysis on 33 that were co-analyzed by whole exome sequencing. We also compared results to TCGA data collected using the Affymetrix Genome-Wide Human SNP Array 6.0. In AA CRCs, the most frequently gained chromosome arms (those occurring in >20% of CRCs) were 7p and q, 8p and q, 12p, 13q, and 20p and q (Table 2). The most frequently lost chromosomes arms (those occurring in >20% of CRCs) were 4p and q, 8p, 14q, 15q, 17p, 18p and q, 20p, 21q, and 22q. As expected, loss of heterozygosity was frequently observed at APC (5q), TP53 (17p) and SMAD4 (18q) (Supplementary Table 5, available at Carcinogenesis Online). Using the R package Rawcopy, we observed substantially lower levels of chromosome-arm gains and losses in the APC-mutation-negative tumors compared with the APC-mutation-positive tumors (Figure 3A). Defining a chromosomally stable tumor as one that contains no chromosome arm gains or losses, we found that the APC-mutation-negative CRCs were associated with chromosome stability (Table 1; P = 0.03). The most frequent gains and losses in AA CRCs were the same as in NHWs (Figure 3B); however, the average frequencies of chromosome-arm gains was 8.1% lower in AA compared with NHW CRCs (P < 0.0001), and for chromosome-arm losses it was 4.4% lower (P < 0.0001). These lower frequencies are most probably explained by the lower levels of chromosome-arm gains and losses occurring in APC-mutation-negative cancers.
Table 2.
Frequency of chromosome arm gains and losses in CRCs from AA and NHW
| CCCC AA CRC | TCGA NWH CRC | |||||||
|---|---|---|---|---|---|---|---|---|
| Arm | Gain frequency | Loss frequency | Gain frequency | Loss frequency | Gain difference1 | Gain P value2 | Loss difference3 | Loss P value2 |
| 1p | 0 | 0.15 | 0.03 | 0.19 | 0.03 | 0.6 | 0.04 | 0.81 |
| 1q | 0.09 | 0.03 | 0.17 | 0.1 | 0.08 | 0.32 | 0.07 | 0.34 |
| 2p | 0.09 | 0 | 0.08 | 0.03 | −0.01 | 0.74 | 0.03 | 0.6 |
| 2q | 0.12 | 0 | 0.1 | 0.03 | −0.02 | 0.76 | 0.03 | 0.6 |
| 3p | 0.03 | 0 | 0.06 | 0.09 | 0.03 | 1 | 0.09 | 0.09 |
| 3q | 0 | 0 | 0.10 | 0.05 | 0.10 | 0.05 | 0.05 | 0.37 |
| 4p | 0 | 0.3 | 0.01 | 0.25 | 0.01 | 1 | −0.05 | 0.53 |
| 4q | 0 | 0.33 | 0.04 | 0.24 | 0.04 | 0.61 | −0.09 | 0.29 |
| 5p | 0.06 | 0.09 | 0.14 | 0.10 | 0.08 | 0.28 | 0.01 | 1 |
| 5q | 0 | 0.12 | 0.07 | 0.17 | 0.07 | 0.24 | 0.05 | 0.62 |
| 6p | 0.15 | 0.09 | 0.14 | 0.07 | −0.01 | 0.79 | −0.02 | 0.72 |
| 6q | 0.06 | 0.12 | 0.14 | 0.10 | 0.08 | 0.28 | −0.02 | 0.76 |
| 7p | 0.33 | 0 | 0.47 | 0.01 | 0.14 | 0.14 | 0.01 | 1 |
| 7q | 0.27 | 0.06 | 0.41 | 0.01 | 0.14 | 0.18 | 0.05 | 0.1 |
| 8p | 0 | 0.33 | 0.28 | 0.50 | 0.28 | 6 × 10 −5 | 0.17 | 0.09 |
| 8q | 0.12 | 0 | 0.46 | 0.12 | 0.34 | 1 × 10 −4 | 0.12 | 0.03 |
| 9p | 0.03 | 0.03 | 0.18 | 0.08 | 0.15 | 0.02 | 0.05 | 0.49 |
| 9q | 0 | 0.03 | 0.15 | 0.09 | 0.15 | 0.01 | 0.06 | 0.33 |
| 10p | 0 | 0.18 | 0.05 | 0.09 | 0.05 | 0.37 | −0.09 | 0.12 |
| 10q | 0 | 0.12 | 0.02 | 0.13 | 0.02 | 1 | 0.01 | 1 |
| 11p | 0.15 | 0.03 | 0.09 | 0.09 | −0.06 | 0.34 | 0.06 | 0.33 |
| 11q | 0.09 | 0.06 | 0.07 | 0.11 | −0.02 | 0.72 | 0.05 | 0.55 |
| 12p | 0.12 | 0.03 | 0.21 | 0.09 | 0.09 | 0.35 | 0.06 | 0.33 |
| 12q | 0.06 | 0.03 | 0.18 | 0.07 | 0.12 | 0.13 | 0.04 | 0.71 |
| 13q | 0.27 | 0 | 0.56 | 0.04 | 0.29 | 0.003 | 0.04 | 0.61 |
| 14q | 0.03 | 0.27 | 0.05 | 0.30 | 0.02 | 1 | 0.03 | 0.84 |
| 15q | 0 | 0.27 | 0.02 | 0.32 | 0.02 | 1 | 0.05 | 0.69 |
| 16p | 0.18 | 0 | 0.18 | 0.05 | <0.01 | 1 | 0.05 | 0.37 |
| 16q | 0.15 | 0.03 | 0.18 | 0.06 | 0.03 | 0.81 | 0.03 | 1 |
| 17p | 0.03 | 0.58 | 0.05 | 0.56 | 0.02 | 1 | −0.02 | 1 |
| 17q | 0.06 | 0.09 | 0.12 | 0.15 | 0.06 | 0.4 | 0.06 | 0.44 |
| 18p | 0.03 | 0.49 | 0.09 | 0.61 | 0.06 | 0.33 | 0.13 | 0.19 |
| 18q | 0 | 0.49 | 0.01 | 0.66 | 0.01 | 1 | 0.18 | 0.05 |
| 19p | 0.09 | 0.03 | 0.11 | 0.04 | 0.02 | 1 | 0.01 | 1 |
| 19q | 0.09 | 0 | 0.15 | 0.05 | 0.06 | 0.44 | 0.05 | 0.37 |
| 20p | 0.24 | 0.12 | 0.58 | 0.32 | 0.34 | 3 × 10 −4 | 0.20 | 0.02 |
| 20q | 0.36 | 0 | 0.72 | 0.15 | 0.36 | 1 × 10 −4 | 0.15 | 0.01 |
| 21q | 0.03 | 0.15 | 0.02 | 0.22 | −0.01 | 0.52 | 0.07 | 0.5 |
| 22q | 0 | 0.24 | 0.03 | 0.26 | 0.03 | 0.60 | 0.02 | 1 |
| Mean | 0.086 | 0.126 | 0.167 | 0.17 | 0.081 | <0.0001 | 0.044 | <0.0001 |
1The fractional difference in chromosome arm gains, subtracting the frequency of arm gains in CCCC AA CRC cases from the frequency in TCGA NHW CRC cases. Gains in excess of 15% are shaded bold.
2 P values were calculated by Fisher exact test. Values in bold are significant after Bonferroni correction.
3The fractional difference in chromosome arm losses, subtracting the frequency of arm losses in CCCC AA CRC cases from the frequency in TCGA NHW CRC cases. Losses in excess of 15% are shaded bold.
Figure 3.
APC-mutation-negative CRCs exhibit greater chromosome stability. (A) Comparison of copy number changes in APC-mutation-negative versus APC-mutation-positive CRCs. (B) Comparison of copy number changes in AA CCCC versus NHW TCGA CRCs.
Methylation patterns in APC-mutation-negative versus mutation-positive tumors
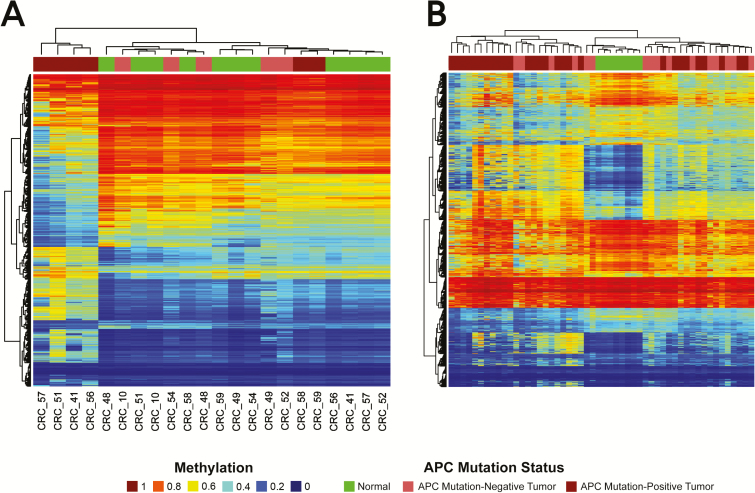
Because tumor mutation burden was lower in APC-mutation-negative tumors, we investigated whether distinct epigenetic mechanisms operate as drivers of carcinogenesis in these tumors. Illumina Infinium Human Methylation 450 BeadChip data from matched tumor and normal samples were available from a previous study of right-sided CRC cases. This previous study included 11 of the cases studied here—five APC-mutation-negative and six APC-mutation-positive tumors. Tumor-specific DNA methylation changes were identified, revealing 5923 DMRs (Figure 4A), of which 3596 (60%) were wider than one nucleosome footprint (200bp) and 4471 (75%) overlapped regions of tumor-specific hypermethylation. APC-mutation-positive tumors were broadly characterized by genome-wide hypomethylation with focal hypermethylation whereas APC-mutation-negative tumors were characterized by genome-wide hypermethylation, similar to levels in matched normal adjacent colon but easily distinguished from it.
Figure 4.
DMRs in APC-mutation-negative tumors cluster with adjacent normal tissue relative to APC-mutation-positive tumors. (A) Heatmap from unsupervised cluster analysis of the top 200 most variable DMRs in the six APC-mutation-positive and five APC-mutation-negative CRCs from the CCCC. (B) Unsupervised cluster analysis using the same 200 most variable DMRs in 15 APC-mutation-positive and 29 APC-mutation-negative CRCs from the TCGA.
Because APC-mutation-negative tumors in the CCCC series were associated with global hypermethylation, we tested whether the methylation signature identified in the Chicago series could be identified in TCGA NHW CRCs. Consequently, we performed cluster analysis of 44 samples from TCGA NHWs using the 200 most variable DMRs identified in the CCCC series (Figure 4B). Twelve of 15 (80%) of the APC-mutation-negative TCGA NHW CRCs clustered with normal tissue controls compared with 9 of 29 (31%; P = 0.004) of the APC-mutation-positive TCGA NHW CRCs. These results suggest that the collection of differentially methylation regions in APC-mutation-negative CRCs represents a novel epigenetic subtype of CRC.
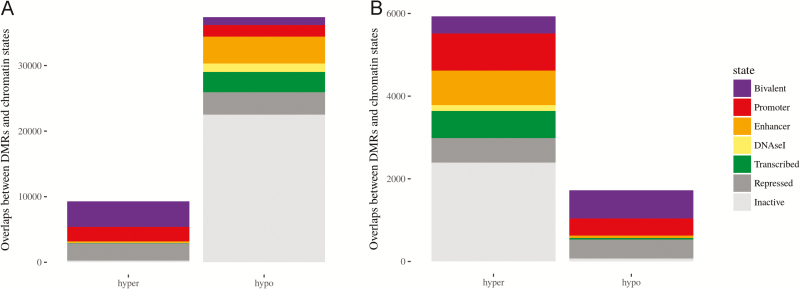
To gauge the impact of methylation changes on genomic regulatory features, we used the Reference Epigenome Mapping Consortium’s chromatin state map for normal colonic mucosa based on chromatin-immunoprecipitation-sequencing (ChIP-seq) data (43) to annotate DMRs. As noted earlier, DMRs in APC-mutation-positive tumors exhibited genome-wide hypomethylation with focal hypermethylation (Figure 5A). In particular, DNA hypermethylation strongly favored bivalent H3K4me3- and H3K27me3-associated promoters, which are critical regulatory switches in differentiation controlled by Polycomb repressor complex binding sites (44). In contrast, APC-mutation-negative tumors revealed substantially less dramatic gains and losses in DNA methylation at these sites relative to normal tissues (Figure 5B). There was proportionately greater methylation of CpG island regions in the APC-mutation-negative tumors; however, based on the five marker genes used to determine the CpG-island methylator phenotype (CIMP) (45), all five APC-mutation-negative CRCs are classified CIMP-0. There was an intriguing enrichment of DMRs in methylated enhancer regions (839) compared with enhancer regions in overall tumor vs normal DMRs (200). Finally, in the analysis of differentially methylated probes, three TCGA-identified cancer driver genes, including SOX9, GPC6 and KIAA1804, were hypermethylated in APC-mutation-negative tumors whereas the same sites were hypomethylated in APC-mutation-positive tumors. Differential hypermethylation between APC-mutation-negative and mutation-positive tumors was also observed in key WNT signaling pathway genes including GATA6, TET1, FAT1 and GLIS1.
Figure 5.
Co-localization of DMRs in APC-mutation-positive and mutation-negative tumors with seven chromatin states based on Reference Epigenome Mapping Consortium’s ChIP-seq data. (A) Comparison of 11 CRCs to adjacent normal tissue demonstrates global hypomethylation with focal hypermethylation. (B) Comparison of five APC-mutation-negative tumors with adjacent normal tissue demonstrates increased methylation in the absence of global hypomethylation.
Discussion
In our analysis of AA CRCs from the CCCC, we found that APC mutations were significantly less frequent in this series compared with the TCGA. The 16 APC-mutation-negative cases were younger and more often affected by previous cancer. The association between APC-mutation-negative CRC and earlier age of onset was also detected in the TCGA series, which is an independent validation of the result identified in the CCCC series. In addition, we found that APC-mutation-negative tumors exhibited lower numbers of somatic mutations, and they were more chromosome stable. In this context, known driver genes were strikingly underrepresented whereas other cancer-associated genes were overrepresented. Finally, APC-mutation-negative tumors exhibited a distinct methylation signature, characterized by hypermethylation of select regulatory regions, affecting in particular genes in the WNT signaling pathway. This methylation signature was also enriched in APC-mutation-negative CRCs in TCGA NHWs.
In three of the APC-mutation-negative cases, we found mutations in BCL9L, which is a negative regulator of the β-catenin. The BCL9L cases had a strong family history of early-onset cancer and was associated with somatic mutations in BRAF that activate the kinase domain, co-occurring in two of the three cases. BCL9 and BCL9L are homologs of the fruit fly legless gene, and they are essential components of WNT signaling, mediating interaction between beta-catenin and Pygopus homologs. There is a growing body of evidence that indicates that BCL9 and BCL9L are important in carcinogenesis (46–48); hypothetically, BCL9L acts as a tumor suppressor, with truncating mutations leading to oncogenic upregulation of β-catenin.
Our study is the second to focus on somatic mutations in AA CRC. The Case Western study performed exome sequencing of 31 AA CRC cases. They used a two-stage discovery–validation approach to identify novel cancer driver genes present in AA CRCs that had not been previously found in the white population, including somatic mutations in EPHA6 and FLCN. In the CCCC series, we found no mutations in EPHA6 and one missense allele in FLCN. We found two missense mutations in the chromatin remodeling gene CHD5, which was one of the genes overrepresented in the CCCC AAs compared with the TCGA NHWs (Figure 2B). Of the remaining 17 candidate cancer-driving genes reported in the Case Western series, we found non-silent mutations in four—GPR149, ZNF862, MGAT4C and WDR87—each in a single case. Clinical differences between the two series might explain some of the variance in results. For example, the Case Western series was entirely from the colon, whereas seven of the CCCC cases were rectal cancers and five were from the rectosigmoid junction. Other possibilities include the age distribution, which was not reported in that study, and the stage distribution. All but one of the Case Western cases were Stage IV, whereas only 10 of the CCCC cases were.
In our earlier characterization of CRC in Chicago AAs (4), we found the median age of diagnosis (n = 62) was younger compared with the median age in Chicago NHWs (n = 65), with over 15% of cases diagnosed before the age of 50 compared with 7% in NHWs. In the CCCC cases sequenced here, the median age (n = 60) was younger than in the CCCC AA series from as a whole, with 26% of the cases diagnosed before age 50. Thus, younger cases are oversampled in our exome sequence data, and the enrichment of APC-mutation-negative CRCs is very likely a consequence of this oversampling, reflecting differences in the process of carcinogenesis between early-onset CRC and late-onset CRC. Previous reports on sporadic early-onset CRCs have shown they are more frequently located in the distal colon or rectum than late-onset CRCs (49–51), they affect females and males nearly equally whereas patients with late-onset CRC are more frequently male (49) and they are relatively more frequent in Hispanics and AAs than in NHWs (4,5,52). Early-onset CRC more often presents with venous or lymphovascular invasion with aggressive features (signet ring or mucinous); it is more often poorly differentiated (53,54); and cases more often present with advanced disease (55). Previously reported molecular features that have been associated with early-onset CRC include microsatellite and chromosome stability (56,57), CIMP-low and CIMP-0 (58), and LINE1 ‘extreme hypomethylation’ (59). Expression profiles of early-onset CRCs are easily distinguishable from late-onset CRCs, characterized by upregulation of beta-catenin (60–62). Our analysis here is broadly consistent with this previous work. In addition, we have found an excess of APC-mutation-negative tumors, reduced tumor mutation burden and distinctive methylation changes.
Increased methylation in APC-mutation-negative tumors was observed at several genes that regulate the WNT signaling pathway, the dysregulation of which may help explain the absence of APC mutations. SOX9 is often overexpressed in CRCs, and its function has been linked to maintenance of stem cell phenotype through activation of WNT signaling (63,64). Similarly, GATA6 maintains stem cell phenotype by upregulating LGR5 expression and downregulating BMP2 (65), and GLIS1 is involved in the autocrine activation of WNT signaling (66). On the other hand, TET1 and FAT1 are tumor suppressors. TET1 binds to DKK gene inhibitors of WNTs to maintain their expression (67) and FAT1 binds directly to β-catenin to suppress its function (68). Future studies are needed to analyze the role of methylation in expression and function of these genes in APC-mutation-negative CRCs.
Limitations of the study include issues relating to comparisons between CCCC and TCGA datasets (differences in sequencing platforms and bioinformatics processing pipelines) and small sample size affecting the analysis of methylation data. Large-scale analysis is needed to validate the association between APC-mutation-negative cancer and early-onset CRC and the distinctive methylation profile observed in these tumors.
Together, the results on APC-mutation-negative CRCs point to a non-canonical carcinogenic process that is more dominated by epigenetic changes, less dominated by somatic mutational changes and occurring at earlier ages. We suggest that the development of early-onset CRC might often occur in an accelerated time frame spurred by environmental factors that impinge on epigenetic mechanisms, and recent analysis of SEER data on CRC incidence stratified by stage at diagnosis broadly supports this interpretation (69). What environmental factors associate with early-onset CRC is a matter of considerable interest. We have recently reported a strong association between AA CRC and sulfidogenic bacteria; in particular the taurine-respiring species Bilophila wadsworthia was 2.5 times more abundant in AAs compared with NHWs irrespective of disease status, and it was 1.9 times more abundant in AA CRC cases compared with AA controls (70). Moreover, we have found that serum 25-hydroxy-vitamin D levels were significantly lower in Chicago AAs compared with Chicago NHWs, and low vitamin D levels were associated with CRC in AAs (unpublished observations from the CCCC). How factors such as these might interact to impel higher rates of early-onset CRC in AAs is not understood. Further studies are needed to understand and prevent early-onset CRC in AAs and in other populations.
Funding
National Institutes of Health (U01 CA153060 and P30 CA023074 to N.A.E.); American Cancer Society Illinois Division (223187 to X.L.); Cancer Biology Training T32 from the National Institute of Health (CA009213 to G.J.A.); Training Program in Investigative Gastroenterology T32 (DK007017 to R.M.X.). Internal funds from the Office of the Vice Chancellor of the University of Illinois at Chicago (to N.A.E. and X.L.)
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the recruiters of the CCCC for their dedication and integrity, including Maggie Moran, Timothy Carroll, Katy Ceryes, Amy Disharoon, Archana Krishnan, Katie Morrissey, Maureen Regan and Katya Seligman. Zarema Arbieva and the University of Illinois at Chicago Genomics Core in the Research Resources Core performed the hybridization and initial analysis of Cytoscan HD arrays. Study sponsors had no role in the design of the study; in the collection, analysis, or interpretation of the data; in the writing of the manuscript and in the decision to submit the manuscript for publication.
Conflict of Interest Statement: None declared.
Abbreviations
- AAs
African Americans
- CCCC
Chicago Colorectal Cancer Consortium
- CRC
colorectal cancer
- MSI
microsatellite instability
- MSS
microsatellite stable
- NHWs
non-Hispanic whites
- TCGA
The Cancer Genome Atlas
References
- 1. Society A.C. (2016)Cancer Facts & Figures 2016. American Cancer Society Inc, Atlanta, GA: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures.html (31 December 2016, last date accessed). [Google Scholar]
- 2. Society A.C. (2014)Colorectal Cancer Facts & Figures 2014–16 American Cancer Society Inc, Atlanta, GA: https://www.cancer.org/research/cancer-facts-statistics/colorectal-cancer-facts-figures.html (31 December 2016, last date accessed). [Google Scholar]
- 3. Rex D.K., et al. ; American College of Gastroenterology (2009)American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected]. Am. J. Gastroenterol., 104, 739–750. [DOI] [PubMed] [Google Scholar]
- 4. Xicola R.M., et al. (2014)Excess of proximal microsatellite-stable colorectal cancer in African Americans from a multiethnic study. Clin. Cancer Res., 20, 4962–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DeSantis C.E., et al. (2016)Cancer statistics for African Americans, 2016: progress and opportunities in reducing racial disparities. CA. Cancer J. Clin., 66, 290–308. [DOI] [PubMed] [Google Scholar]
- 6. Irby K., et al. (2006)Emerging and widening colorectal carcinoma disparities between Blacks and Whites in the United States (1975–2002). Cancer Epidemiol. Biomarkers Prev., 15, 792–797. [DOI] [PubMed] [Google Scholar]
- 7. Thornton J.G., et al. (2007)Racial variation in colorectal polyp and tumor location. J. Natl. Med. Assoc., 99, 723–728. [PMC free article] [PubMed] [Google Scholar]
- 8. Network T.C.G.A. (2012)Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Augustus G.J., et al. (2018)Colorectal cancer disparity in African Americans: risk factors and carcinogenic mechanisms. Am. J. Pathol., 188, 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kupfer S.S., et al. (2010)Genetic heterogeneity in colorectal cancer associations between African and European Americans. Gastroenterology, 139, 1677–1685, 1685.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kupfer S.S., et al. (2014)Shared and independent colorectal cancer risk alleles in TGFβ-related genes in African and European Americans. Carcinogenesis, 35, 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang H., et al. (2013)Fine-mapping of genome-wide association study-identified risk loci for colorectal cancer in African Americans. Hum. Mol. Genet., 22, 5048–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seshagiri S., et al. (2012)Recurrent R-spondin fusions in colon cancer. Nature, 488, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guda K., et al. (2015)Novel recurrently mutated genes in African American colon cancers. Proc. Natl. Acad. Sci. U. S. A., 112, 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xicola R.M., et al. ; Gastrointestinal Oncology Group of the Spanish Gastroenterological Association (2007)Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J. Natl. Cancer Inst., 99, 244–252. [DOI] [PubMed] [Google Scholar]
- 16. Kupfer S.S., et al. (2009)Novel single nucleotide polymorphism associations with colorectal cancer on chromosome 8q24 in African and European Americans. Carcinogenesis, 30, 1353–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shanmugam V., et al. (2014)Whole genome sequencing reveals potential targets for therapy in patients with refractory KRAS mutated metastatic colorectal cancer. BMC Med. Genomics, 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H., et al. (2010)Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics, 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McKenna A., et al. (2010)The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saunders C.T., et al. (2012)Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics, 28, 1811–1817. [DOI] [PubMed] [Google Scholar]
- 21. Lawrence M.S., et al. (2013)Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature, 499, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olshen A.B., et al. (2004)Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics, 5, 557–572. [DOI] [PubMed] [Google Scholar]
- 23. Sandoval J., et al. (2011)Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics, 6, 692–702. [DOI] [PubMed] [Google Scholar]
- 24. Legendre C.R., et al. (2016)Pathway implications of aberrant global methylation in adrenocortical cancer. PLoS One, 11, e0150629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aryee M.J., et al. (2014)Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Triche T.J. Jr, et al. (2013)Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res., 41, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peters T.J., et al. (2015)De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin, 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ernst J., et al. (2012)ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods, 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao J., et al. (2013)Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cerami E., et al. (2012)The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang C., et al. ; FUSION Study (2014)Ancestry estimation and control of population stratification for sequence-based association studies. Nat. Genet., 46, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Auton A., et al. (2015)A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shinbrot E., et al. (2014)Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res., 24, 1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qin J., et al. (2009)Upregulation of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer metastasis. Oncogene, 28, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindsay C.R., et al. (2011)P-Rex1 is required for efficient melanoblast migration and melanoma metastasis. Nat. Commun., 2, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lucato C.M., et al. (2015)The phosphatidylinositol (3,4,5)-trisphosphate-dependent Rac Exchanger 1·Ras-related C3 botulinum toxin substrate 1 (P-Rex1·Rac1) complex reveals the basis of Rac1 activation in breast cancer cells. J. Biol. Chem., 290, 20827–20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shyr C., et al. (2014)FLAGS, frequently mutated genes in public exomes. BMC Med. Genomics, 7, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berger B.M., et al. (2003)Colon cancer-associated DNA mutations: marker selection for the detection of proximal colon cancer. Diagn. Mol. Pathol., 12, 187–192. [DOI] [PubMed] [Google Scholar]
- 39. Chang S.C., et al. (2016)Mutation spectra of common cancer-associated genes in different phenotypes of colorectal carcinoma without distant metastasis. Ann. Surg. Oncol., 23, 849–855. [DOI] [PubMed] [Google Scholar]
- 40. Lüchtenborg M., et al. (2004)APC mutations in sporadic colorectal carcinomas from The Netherlands Cohort Study. Carcinogenesis, 25, 1219–1226. [DOI] [PubMed] [Google Scholar]
- 41. Mokarram P., et al. (2009)Distinct high-profile methylated genes in colorectal cancer. PLoS One, 4, e7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fatemi M., et al. (2014)Epigenetic silencing of CHD5, a novel tumor-suppressor gene, occurs in early colorectal cancer stages. Cancer, 120, 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ernst J., et al. (2015)Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol., 33, 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hahn M.A., et al. (2014)Loss of the polycomb mark from bivalent promoters leads to activation of cancer-promoting genes in colorectal tumors. Cancer Res., 74, 3617–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weisenberger D.J., et al. (2006)CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet., 38, 787–793. [DOI] [PubMed] [Google Scholar]
- 46. de la Roche M., et al. (2008)The function of BCL9 in Wnt/beta-catenin signaling and colorectal cancer cells. BMC Cancer, 8, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deka J., et al. (2010)Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Res., 70, 6619–6628. [DOI] [PubMed] [Google Scholar]
- 48. Giannakis M., et al. (2016)Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep., 15, 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Siegel R., et al. (2014)Colorectal cancer statistics, 2014. CA. Cancer J. Clin., 64, 104–117. [DOI] [PubMed] [Google Scholar]
- 50. O’Connell J.B., et al. (2003)Rates of colon and rectal cancers are increasing in young adults. Am. Surg., 69, 866–872. [PubMed] [Google Scholar]
- 51. You Y.N., et al. (2012)Young-onset colorectal cancer: is it time to pay attention?Arch. Intern. Med., 172, 287–289. [DOI] [PubMed] [Google Scholar]
- 52. Siegel R.L., et al. (2015)Cancer statistics for Hispanics/Latinos, 2015. CA. Cancer J. Clin., 65, 457–480. [DOI] [PubMed] [Google Scholar]
- 53. Chang D.T., et al. (2012)Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod. Pathol., 25, 1128–1139. [DOI] [PubMed] [Google Scholar]
- 54. Yantiss R.K., et al. (2009)Clinical, pathologic, and molecular features of early-onset colorectal carcinoma. Am. J. Surg. Pathol., 33, 572–582. [DOI] [PubMed] [Google Scholar]
- 55. Stigliano V., et al. (2014)Early-onset colorectal cancer: a sporadic or inherited disease?World J. Gastroenterol., 20, 12420–12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chan T.L., et al. (2001)Early-onset colorectal cancer with stable microsatellite DNA and near-diploid chromosomes. Oncogene, 20, 4871–4876. [DOI] [PubMed] [Google Scholar]
- 57. Silla I.O., et al. (2014)Early-onset colorectal cancer: a separate subset of colorectal cancer. World J. Gastroenterol., 20, 17288–17296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Perea J., et al. (2014)Age at onset should be a major criterion for subclassification of colorectal cancer. J. Mol. Diagn., 16, 116–126. [DOI] [PubMed] [Google Scholar]
- 59. Baba Y., et al. (2010)Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol. Cancer, 9, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kirzin S., et al. (2014)Sporadic early-onset colorectal cancer is a specific sub-type of cancer: a morphological, molecular and genetics study. PLoS One, 9, e103159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Berg M., et al. ; INFAC-study group (2010)Distinct high resolution genome profiles of early onset and late onset colorectal cancer integrated with gene expression data identify candidate susceptibility loci. Mol. Cancer, 9, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jandova J., et al. (2016)Sporadic early-onset colon cancer expresses unique molecular features. J. Surg. Res., 204, 251–260. [DOI] [PubMed] [Google Scholar]
- 63. Matheu A., et al. (2012)Oncogenicity of the developmental transcription factor Sox9. Cancer Res., 72, 1301–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ma F., et al. (2016)SOX9 drives WNT pathway activation in prostate cancer. J. Clin. Invest., 126, 1745–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Whissell G., et al. (2014)The transcription factor GATA6 enables self-renewal of colon adenoma stem cells by repressing BMP gene expression. Nat. Cell Biol., 16, 695–707. [DOI] [PubMed] [Google Scholar]
- 66. Vadnais C., et al. (2014)Autocrine activation of the Wnt/β-Catenin Pathway by CUX1 and GLIS1 in Breast Cancers. Biol. Open, 3, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Neri F., et al. (2015)TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway. Oncogene, 34, 4168–4176. [DOI] [PubMed] [Google Scholar]
- 68. Morris L.G., et al. (2013)Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat. Genet., 45, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Augustus G.J., et al. (2018)Is increased colorectal screening effective in preventing distant disease?PLoS One, 13, e0200462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yazici C., et al. (2017)Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut, 66, 1983–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.