Abstract
The Gompertz equation describes survival in terms of initial mortality rate (parameter a), indicative of health, and age-dependent acceleration in mortality rate (parameter b), indicative of aging. Gompertz parameters were analyzed for several published studies. In Drosophila females, mating increases egg production and decreases median life span, consistent with a trade-off between reproduction and longevity. Mating increased parameter a, causing decreased median life span, whereas time parameter b was decreased. The inverse correlation between parameters indicates the Strehler–Mildvan (S-M) relationship, where loss of low-vitality individuals yields a cohort with slower age-dependent mortality acceleration. The steroid hormone antagonist mifepristone/RU486 reversed these effects. Mating and mifepristone showed robust S-M relationships across genotypes, and dietary restriction showed robust S-M relationship across diets. Because nutrient optima differed between females and males, the same manipulation caused opposite effects on mortality rates in females versus males across a range of nutrient concentrations. Similarly, p53 mutation in Drosophila and mTOR mutation in mice caused increased median life span associated with opposite direction changes in mortality rate parameters in females versus males. The data demonstrate that dietary and genetic interventions have sex-specific and sometimes sexually opposite effects on mortality rates consistent with sexual antagonistic pleiotropy.
Keywords: Aging, Drosophila, Trade-offs, Sexual antagonistic pleiotropy.
Sex-specific and sex-biased effects of aging interventions are common across species; however, the underlying mechanism(s) for sex-dimorphism in life-span regulation have not yet been fully elucidated (1–3). Sex-specific selective pressures are hypothesized to lead to gene functions that are suboptimal in one or both sexes, thereby potentially contributing to the aging phenotype (sexual antagonistic pleiotropy) (4,5). Life span is one of the most robust measures of aging and is typically reported in terms of median life span, sometimes including estimates of maximum life span. Fitting survival data to the Gompertz-Makeham (G-M) equation allows for more detailed analysis of the changes in animal mortality with age (6). In the G-M equation, the increase of mortality (μx) with age (x) is expressed as: μx = aebx+c. The G-M equation describes survival curves in terms of parameter a (initial mortality rate), parameter b (rate of exponential increase in mortality), and parameter c (age-independent mortality); a decrease in one or more of these parameters can produce an increase in median life span. The related Gompertz equation (μx = aebx) describes survival in terms of parameter a and parameter b without inclusion of parameter c.
Traditionally, the initial mortality rate (parameter a) has been thought to result from both the inherent failure rate of animal physiology and the mortality resulting from environmental challenges, including pathogens (7). When the life spans of model organisms are assayed in the laboratory, environmental challenges are minimized by providing constant food, temperature and humidity, and by limiting exposure to pathogens. For example, several of the Drosophila life-span data sets analyzed here were generated under conditions where the presence or absence of the commensal bacterial flora has been shown to have no detectable effect on the life span of males from long-lived starting strains (8). Similarly, the mouse mechanistic target of rapamycin (mTOR) mutant and control life spans were analyzed under specific pathogen-free conditions (9). Therefore, under these optimized conditions, the initial mortality rate should primarily indicate the inherent risk of mortality due to the health or vitality of the animal, without significant effect of environmental challenges, other than intentional manipulations. The age-associated acceleration in mortality rate (parameter b) is traditionally thought to reflect an internal aging process that might involve the accumulation of some as-yet unknown type of damage (7).
Over 50 years ago, Strehler and Mildvan analyzed cross-sectional survival data from model organisms and multiple human populations and found that when initial mortality rate (parameter a) was increased, the age-associated acceleration in mortality rate (parameter b) was decreased (10). This negative correlation between parameters a and b is since called the Strehler–Mildvan or S-M relationship. The S-M relationship is thought to result because an increased initial mortality rate (parameter a) preferentially causes the loss of the lowest-vitality individuals. This leaves a cohort comprised of more robust individuals and therefore a slower age-associated mortality acceleration (parameter b). Certain subsequent studies of cross-sectional human survival data suggested that the S-M relationship was less robust (11,12). However, more recent studies of human cohort survival data support the existence of the S-M relationship (13–16). The S-M relationship is also observed across male and female cohorts of wild-type Drosophila (17,18).
Dietary restriction (DR) can increase median life span in multiple species, and several mechanisms have been hypothesized, including increased investment of resources into somatic maintenance pathways important for longevity (19,20). The conclusions for mortality rate changes due to DR in rodents are mixed. Finch reported calculated values for parameter a and parameter b for a series of studies of DR in males of long-lived rat strains and found a consistent S-M relationship where parameter a was increased and parameter b was decreased (7); in that study, parameter b was reported as mortality rate doubling time, calculated as mortality rate doubling time = Ln 2/b. DR of both protein and sugar in a large cohort of male rats (21) caused decreased parameter b, resulting in increased median life span, and a coincident increase in parameter a (22), again consistent with a S-M relationship. In contrast, recent meta-analyses of DR interventions in male and female rodents (including rats and mice) have suggested that both parameters a and b are reduced (23) or that parameter b is decreased independent of alterations in parameter a (24). One possible explanation for the differing conclusions regarding mortality rate changes in rodent DR studies is possible differences between the responses of rats and mice, where mice are generally observed to have a smaller magnitude response (25). In addition, the response to DR in mice has been reported to differ in sign depending on the genotype (26). Another possibility, suggested by the data presented below, is that for certain analyses the S-M relationship may have been obscured by the pooling of male and female data with opposite-sign responses.
When DR was modeled in Drosophila by dilution of both yeast (the protein source) and sugar in the media, it produced increased median life span of both males and females through a reduction in initial mortality rate, with greater effect observed in females (27). Notably, the effects on initial mortality rate were largely reversible when flies were switched between diets (28,29). Subsequent studies revealed the importance of the yeast:sugar ratio and the concentration of specific nutrients in regulating life span and reproduction (30–32). For example, DR as produced by reduction of yeast with constant sugar in continuously mated animals yielded increased median life span in females but not in males (33). Possible S-M relationships in Drosophila DR responses remain largely unexplored.
In Drosophila females, mating and the transfer of male seminal proteins including sex peptide cause metabolic changes in the female that include increased reproduction and decreased median life span (34). These changes are generally interpreted as an induced trade-off between reproduction and life span, and might result from the diversion of metabolic resources to reproduction and away from somatic maintenance pathways required for optimal longevity. We have previously reported that feeding the drug mifepristone/RU486 to the mated females blocked the effects of mating, yielding decreased reproduction and median life span increases up to +68% (35). Assay of feeding behavior using the same nutrient-rich media and a dye-uptake assay indicated that mifepristone caused either no change or a slight increase in food intake, arguing against a DR mechanism for the life-span increase (35). The exact mechanism for mifepristone life-span increase is not yet clear; however, because mifepristone is a steroid hormone antagonist (36), the results implicate steroid hormone signaling in regulating the trade-off between reproduction and life span. Consistent with this idea, steroid hormone signaling has recently been shown to regulate adult Drosophila sexual differentiation and a female metabolic state that favors reproduction (37,38). The changes in Drosophila life span caused by mating and by mifepristone varied dramatically across genotypes, thereby facilitating analysis of mortality rates.
Methods
Survival data for multiple Drosophila genotypes, virgin versus mated and mated −/+ mifepristone are from Landis et al. (35). Flies were of the indicated homozygous genotypes or were the progeny of the indicated crosses, presented in the order: male genotype × female genotype. In crosses where either of the parental strains contained a balancer chromosome, progeny were selected that did not contain the balancer. Drosophila p53 null mutant and control data are for the “W” cohort, pooled data, reported in Waskar et al. (39). Drosophila control and DR (yeast and sugar) data are from table 2 reported by Magwere et al. (27). Drosophila control and DR (yeast) data are for individually cultured flies reported by Zajitschek et al. (33). Mouse mTOR wild-type and mTORdelta/delta mutant data are from Wu et al. (9). G-M modeling was conducted using WinModest software (version 1.0.0.1) as previously described (6,40). Briefly, the age-specific mortality rate (μx) was calculated using WinModest by binning the days over which deaths were counted (e.g., fly deaths were recorded every other day) such that μx = (−ln(Nx+ δx/Nx))/δx, where Nx is the number of flies alive at day x, δx is the bin size, and μx is the mortality rate at day x. Parameters (a, b, c) were calculated by WinModest based on a likelihood ratio test. Microsoft Excel was used to plot the full model (μx = aebx + c) as the trend line, along with data points indicating the calculated values for Ln (μx) at each age time point. Re-composed survival curves were generated by fitting the data to the Gompertz equation (μx= aebx), that is, omitting any age-independent mortality (G-M parameter c). The rebuilt survival curves were plotted using Microsoft Excel, using the reverse calculation of μx as follows: μx = aebx, Px = e-μx, where Px was the probability of surviving from age (x) to age (x+1). Percent survival at each day (Lx) was calculated as Lx = (Lx−1)(Px−1). For the re-composed survival curves, any value below 0.5% survival was considered to be the final data point. Linear regression and additional statistical analyses were conducted using R statistical environment (41).
Results
Here, survival data for several life-span interventions were fitted to the G-M equation separately for males and females to determine which parameters have been altered. Control and experimental groups were compared using WinModest software (6), which determines whether there was a statistically significant change for each parameter. In cases where parameter c was not significantly altered the analysis was repeated with c constrained and parameter a and parameter b were again compared to determine possible significance of changes.
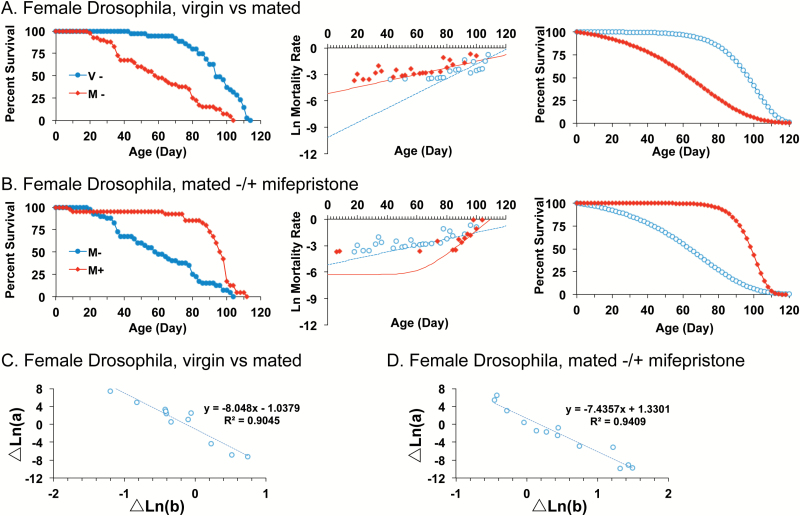
A long-lived hybrid genotype generated from two common laboratory strains of Drosophila was analyzed, where mating caused a −50% decrease in median life span of females (Figure 1A, left panel) (35). The WinModest software was used to calculate G-M parameter values, and the natural log mortality rate versus time was plotted (Figure 1A, center panel). The trend line represents the data fitted to the G-M equation and includes time points where finite mortality was measured (indicated by red and blue circles), as well as zero values for the time points where no mortality was observed (not shown), as well as the parameter c component representing age-independent mortality. A re-composed survival curve was then generated for the fitted data by excluding age-independent mortality (G-M parameter c) and fitting the data to the Gompertz equation (Figure 1A, right panel). The G-M modeling revealed that mating increased initial mortality rate (parameter a) and decreased age-dependent mortality (parameter b), indicating the S-M relationship (statistical summary in Table 1). The analysis was continued on a series of 10 additional homozygous and heterozygous genotypes generated from common wild-type and transgenic laboratory strains, and where the magnitude of the median life-span decrease caused by mating varied from −50% to no effect (35). Across the 11 genotypes, a robust S-M relationship was observed for the negative effect of mating on life span (Figure 1C, statistical summary in Supplementary Table S1). Feeding mifepristone to the mated females blocked the negative effect of mating on life span: mifepristone decreased initial mortality rate (parameter a), thereby yielding dramatically increased median life span of +64% (Figure 1B; Table 1). At the same time, mifepristone increased age-dependent mortality (parameter b), again indicating the S-M relationship. The analysis of mifepristone effects was conducted across the same series of 11 genotypes where the effect of mifepristone on median life span varied from +64% to no effect (35), and again a strict S-M relationship was observed (Figure 1D, Supplementary Table S1). In the longest-lived starting strains mifepristone also decreased parameter a and increased parameter b in virgin females (Supplementary Table S1), indicating that in long-lived genotypes, the trade-off is already partially active in virgin females prior to its activation by mating.
Figure 1.
Mating and mifepristone interventions in Drosophila reveal a strict S-M relationship across genotypes. (A and B) Left panel, raw survival data. Center panel, plot of natural log mortality rate versus time, and trend line for data fitted to the G-M model. Please note that the trend line represents the data fitted to the G-M equation and includes time points where finite mortality was measured (indicated by red and blue circles), as well as zero values for the time points where no mortality was observed (not shown), as well as the parameter c component representing age-independent mortality. Right panel, reconstructed survival curves for data fitted to the Gompertz model. (A) Female Drosophila, virgin versus mated. (B) Female Drosophila, mated −/+ mifepristone feeding. Statistical summary for (A and B) in Table 1. (C) Change in parameter a versus change in parameter b for female Drosophila, virgin versus mated, for each of 11 independent genotypes. Linear regression R[2] = 0.90, significance of predictor p = 6.93e-06. (D) Change in parameter a versus change in parameter b for mated female Drosophila, (−) mifepristone versus (+) mifepristone, for each of 11 independent genotypes, 13 independent experiments. Linear regression R[2] = 0.94, significance of predictor p = 4.22e-08. Statistical summary for (C and D) in Supplementary Table S1.
Table 1.
Statistical Summary
| Group | Parameter | Control | Experimental | χ2 | p | χ2 | p |
|---|---|---|---|---|---|---|---|
| Female Drosophila, virgin vs mated | c is constrained | ||||||
| A | 4.00×10–5 | 5.64×10–3 | 6.432 | 0.011 | 25.843 | <0.001 | |
| B | 1.66×10–1 | 7.31×10–2 | 5.556 | 0.018 | 14.894 | <0.001 | |
| C | 2.06×10–9 | 2.06×10–9 | 0 | 1 | |||
| Female Drosophila, mated, (−) drug vs (+) drug | c is contrained | ||||||
| A | 5.64×10–3 | 3.02×10–7 | 10.505 | 1.19E-03 | 35.929 | <0.001 | |
| B | 7.31×10–2 | 2.74×10–1 | 10.083 | 1.50E-03 | 33.68 | <0.001 | |
| C | 2.06×10–9 | 1.85×10–3 | 0.325 | 0.569 | |||
| Female Drosophila p53+/+ vs p53−/− | c is constrained | ||||||
| A | 4.0×10–5 | 3.85×10–7 | 18.835 | 1.43E-05 | 20.089 | <0.001 | |
| B | 2.88×10–1 | 4.04×10–1 | 11.431 | 7.22E-04 | 12.046 | <0.001 | |
| C | 2.06×10–9 | 9.0×10–5 | 0.064 | 0.801 | |||
| Male Drosophila p53+/+ vs p53−/− | c is constrained | ||||||
| A | 4.10×10–4 | 1.28×10–3 | 3.044 | 0.081 | 7.530 | 0.006 | |
| B | 2.05×10–1 | 1.23×10–1 | 15.660 | 7.58E-05 | 26.358 | <0.001 | |
| C | 2.06×10–9 | 1.16×10–3 | 0.445 | 0.505 | |||
| Female mice mTOR+/+ vs mTORdelta/delta | c is constrained | ||||||
| A | 2.23×10–3 | 1.00×10–4 | 3.497 | 0.061 | 5.900 | 0.015 | |
| B | 1.46×10–1 | 2.32×10–1 | 2.825 | 0.093 | 3.801 | 0.051 | |
| C | 2.06×10–9 | 2.06×10–9 | 0 | 1 | |||
| Male mice mTOR+/+ vs mTORdelta/delta | |||||||
| A | 2.30×10–4 | 4.20×10–4 | 0.099 | 0.753 | |||
| B | 2.76×10–1 | 2.08×10–1 | 0.767 | 0.381 | |||
| C | 2.06×10–9 | 2.06×10–9 | 0 | 1 | |||
| Female Drosophila DR (y), 1.0X vs 0.4X | |||||||
| A | 1.60×10–4 | 6.14×10–7 | 11.974 | <0.001 | 22.039 | <0.001 | |
| B | 9.75×10–2 | 1.76×10–1 | 11.073 | <0.001 | 19.597 | <0.001 | |
| C | 2.80×10–3 | 1.19×10–3 | 1.119 | 0.290 | |||
| Male Drosophila DR (y), 1.0X vs 0.4X | |||||||
| A | 1.60×10–4 | 2.40×10–4 | 0.300 | 0.584 | 0.300 | 0.584 | |
| B | 1.24×10–1 | 1.09×10–1 | 1.090 | 0.296 | 1.172 | 0.279 | |
| C | 2.06×10–9 | 2.06×10–9 | 0 | 1 | |||
Notes: Degrees of freedom equals 1 in each case. DR = Dietary restriction. Bold type indicates statistical significance at p < .05.
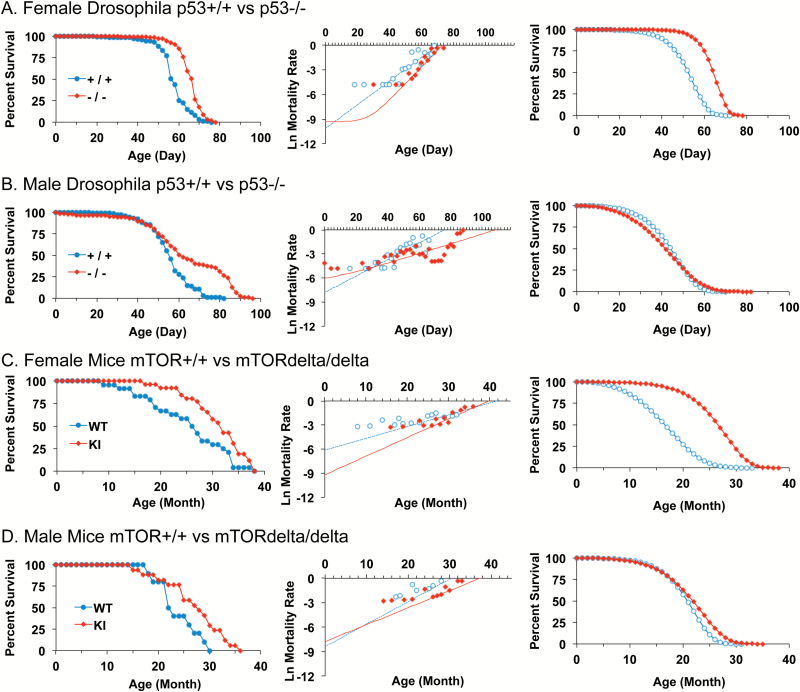
The p53 null mutation was previously found to increase median life span of mated female Drosophila, with smaller, more variable median life-span increases observed for males (39). G-M modeling revealed that in mated females the p53 null mutation decreased initial mortality rate (parameter a) and increased age-dependent mortality (parameter b) to yield increased median life span, similar to the effects of mifepristone, and indicating a S-M relationship (Figure 2A; Table 1). In contrast, in male Drosophila, the p53 null mutation had the opposite effect: in males the p53 null mutation increased parameter a and decreased parameter b (Figure 2B; Table 1), which yields a net smaller increase in median life span, than that was observed for females, and indicating a S-M relationship with opposite sign. Strikingly, a similar pattern of sex-specific parameter changes was observed for the life-span increase caused by a hypomorphic mTOR mutation in mice. A recent analysis of male and female mTORdelta/delta mutant mice revealed a statistically significant increase in median life span for the combined cohort, as well as for males and females analyzed separately (9). Here, the male and female mTOR mutant mouse survival data were analyzed separately by G-M modeling. In female mice, the mTORdelta/delta mutation decreased initial mortality rate (parameter a) to yield increased median life span (Figure 2C; Table 1). At the same time, age-dependent mortality (parameter b) showed a trend to increase that approached significance (p = .051), suggesting a S-M relationship. In contrast, in male mice the mTORdelta/delta mutation caused parameter changes in the opposite direction, but that did not reach statistical significance (Figure 2D, Table 1). In the future, it may be of interest to further analyze effects of mTORdelta/delta mutation in male mice using larger cohort sizes.
Figure 2.
Drosophila p53 null mutation and mouse mTOR hypomorphic mutation have sex-specific effects on mortality rates. (A–D) Left panel, raw survival data. Center panel, plot of natural log mortality rate versus time, and trend line for data fitted to the G-M model. Right panel, reconstructed survival curves for data fitted to the Gompertz model. Statistical summary in Table 1. (A) Female Drosophila p53+/+ versus p53−/−. (B) Male Drosophila p53+/+ versus p53−/−. (C) Female Mice mTOR+/+ (WT) versus mTORdelta/delta (KI). (D) Male Mice mTOR+/+ (WT) versus mTORdelta/delta (KI).
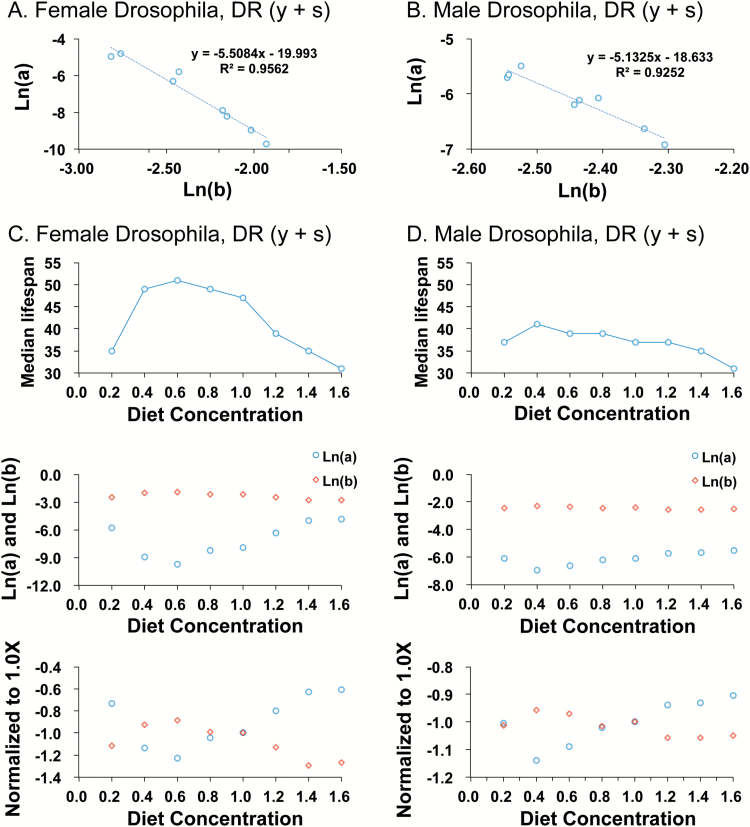
Finally, two different models for DR in Drosophila were analyzed. In the first DR model, in experiments conducted by Magwere et al. (27), both yeast (the dietary protein source) and sugar were varied together by dilution of the media, across a range from 0.2X to 1.6X relative to a standard 1.0X concentration. They found that the median life span of females was maximal at 0.6X, and this was interpreted as a life-span increase for females due to DR. Female life span was significantly reduced at 0.2X, and this was interpreted as a starvation response. In contrast, the median life span of males was maximal at 0.4X, and this was interpreted as a life span increase for males due to DR. In addition, those authors analyzed mortality rates by fitting the data to the Gompertz equation and found that for both males and females the increase in median life span was associated with a decrease in initial mortality rate (parameter a), with a greater decrease observed in females than in males. Regarding the age-associated acceleration in mortality rate (parameter b), they reported a significant increase for females at 0.6X concentration relative to the extremes (0.2X and 1.6X), and no significant changes for males. This lead them to conclude that ‘The rate at which mortality rate accelerates with age does not seem to play a significant role in the response of life span to DR or to starvation’. However, plotting their reported values for parameter a versus parameter b reveals a robust S-M relationship for both sexes across the various concentrations of media (Figure 3A and B). Comparison of median life span to mortality rates across the various diet concentrations illustrates the S-M inverse relationship between parameter a and parameter b for both females and males (Figure 3C and D; compare upper and middle panels). As diet concentration is varied, the S-M relationship changes in sign at the concentration coincident with the minimum value for parameter a for each sex: 0.6X for females and 0.4X for males. Because of this, a change from 0.6X to 0.4X diet has opposite effects on the mortality rate parameters in males versus females. Normalizing the values of parameter a and parameter b to the 1.0X diet concentration illustrates how both the direction of parameter change and the magnitude of parameter change can differ between males and females as diet is altered (Figure 3C and D; lower panels). This presentation also illustrates how the S-M relationship is altered in opposite direction as one adjusts the diet above or below the reference value of 1.0X.
Figure 3.
Effect of dietary restriction (DR) of both yeast and sugar on male and female mortality rates in Drosophila. (A and B) Change in parameter a versus change in parameter b across eight media concentrations: 0.2X, 0.4X, 0.6X, 0.8X, 1.0X, 1.2X, 1.4X, and 1.6X media. (A) Female Drosophila (mated). Linear regression R[2] = 0.95, significance of predictor p = 2.67e-05. (B) Male Drosophila (mated). Linear regression R[2] = 0.92, significance of predictor p = 1.35e-04. (C and D) Change in median life span compared to change in mortality rates across the eight media concentrations. Upper panels, median life span. Middle panels, natural log value for paramater a (blue circles) and natural log value for parameter b (red circles). Lower panels, natural log value for paramater a (blue circles) and natural log value for parameter b (red circles), with data normalized to the absolute value for the 1.0X media concentration. (C) Females. (D) Males.
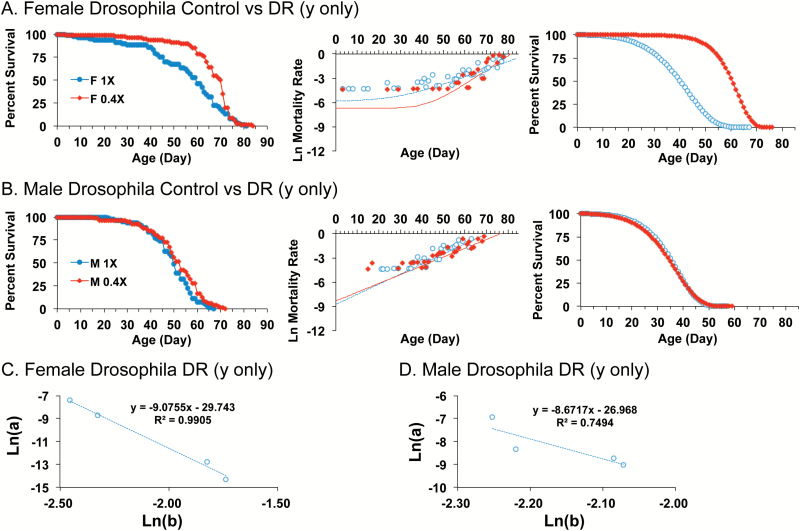
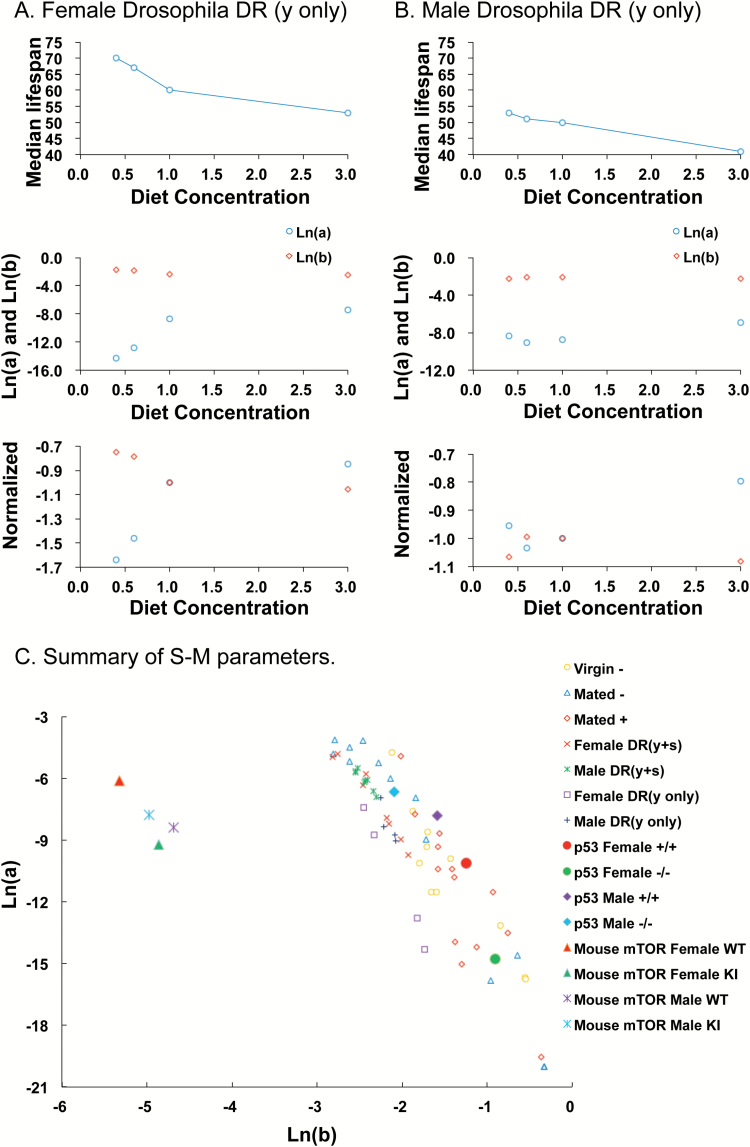
In the second model for DR, in experiments conducted by Zajitschek et al. (33), the concentration of yeast was varied with the concentration of sugar kept constant. Yeast concentrations were varied across concentrations 0.4X, 0.6X, 1.0X and 3.0X. In females, increased median life span was observed for 0.4X relative to 1.0X indicating a DR response, whereas no significant effect was observed for males (33). G-M equation modeling of the data revealed that in females, this DR caused decreased parameter a and increased parameter b (Figure 4A; Table 1), consistent with a S-M relationship. Indeed plotting parameter a versus parameter b confirmed a robust S-M relationship for females across all four yeast concentrations (Figure 4C). In contrast, in males, comparing 0.4X to 1.0X yeast concentration revealed G-M parameter changes in the opposite direction but that did not reach statistical significance (Figure 4B; Table 1). Comparison of median life span to mortality rates across the various dietary yeast concentrations illustrates the inverse relationship between parameter a and parameter b for both females and males (Figure 5A and B; compare upper and middle panels). In addition, it can be seen how a change from 0.6X to 0.4X has opposing effects on the mortality rate parameters in males versus females (Figure 5A and B; middle and lower panels). Finally, a summary of all the mortality rate parameters illustrates the S-M relationship for both mice and flies, and the much smaller values for parameter b for mice relative to flies (Figure 5C).
Figure 4.
Effect of DR of yeast with constant sugar on male and female mortality rates in Drosophila. (A and B) Comparison of 1.0X media to 0.4X media. Left panel, raw survival data. Center panel, plot of natural log mortality rate versus time, and trend line for data fitted to the G-M model. Right panel, reconstructed survival curves for data fitted to the Gompertz model. (A) Female Drosophila. (B) Male Drosophila. Statistical summary in Table 1. (C and D) Change in parameter a versus change in parameter b across four yeast concentrations: 0.4X, 0.6X, 1.0X, and 3.0X. (C) Female Drosophila. Linear regression R[2] = 0.99, significance of predictor p = 0.00474. (D) Male Drosophila. Linear regression R[2] = 0.75, significance of predictor p = 0.134. Statistical summary in Supplementary Table S1.
Figure 5.
Effect of DR of yeast with constant sugar on life span and mortality rates, and summary of S-M parameters. (A and B) Change in median life span compared to change in mortality rates across the four yeast concentrations: 0.4X, 0.6X, 1.0X, and 3.0X. Upper panels, median life span. Middle panels, natural log value for paramater a (blue circles) and natural log value for parameter b (red circles). Lower panels, natural log value for parameter a (blue circles) and natural log value for parameter b (red circles), with data normalized to the absolute value for the 1.0X media concentration. (A) Females. (B) Males. (C) Summary of S-M parameters. Values of Ln(a) and Ln(b) are plotted for each of the data sets analyzed. Here the values of parameter b for mice were converted from months (as in Figure 2 and Table 1) to days, to facilitate comparison to the Drosophila data.
Discussion
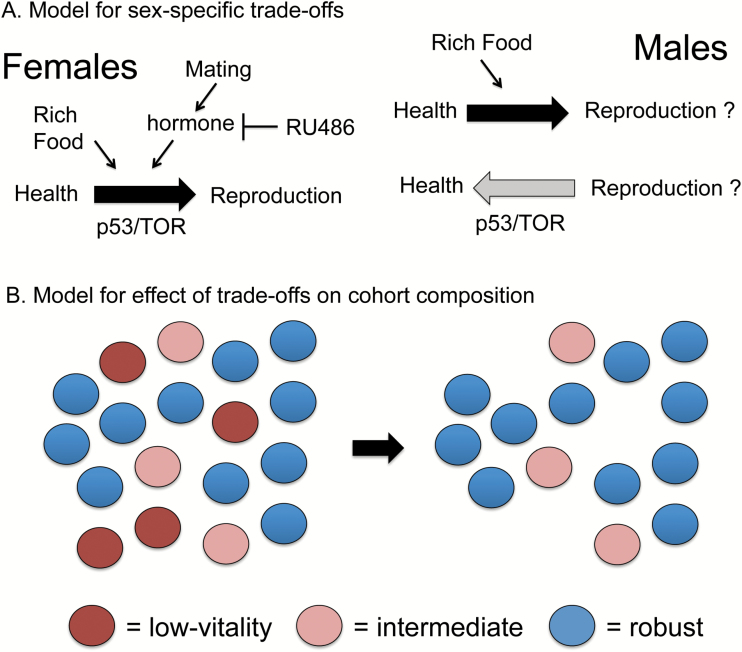
The analyses presented here demonstrate that several metazoan life-span interventions (mifepristone, p53 mutation, mTOR mutation, DR) can act in females to reduce initial mortality rate (parameter a), resulting in increased median life span. Because both mifepristone and DR can also reduce reproduction, these similarities suggest that these interventions may inhibit a trade-off between health and reproduction that is otherwise lethal to low-vitality individuals (model in Figure 6A). Coincidently, the age-dependent mortality rate (parameter b) was increased, leading to lesser or no increase in maximum life span, and indicating the S-M relationship. The S-M relationship indicates population heterogeneity where the same low-vitality individuals are preferentially susceptible to both increased initial mortality (as caused by trade-offs) and to mortality rate acceleration (Figure 6B).
Figure 6.
Model for sex-specific trade-offs and effects on cohort composition. (A) Model for sex-specific trade-offs. In female Drosophila, mating caused increased initial mortality rate and increased reproduction, consistent with an induced trade-off that causes lethality in the lowest-vitality individuals. The steroid hormone antagonist mifepristone/RU486 blocked the effects of mating, suggesting a role for steroid hormone signaling in the mating response. Increased concentration of yeast and sugar in the media also caused increased initial mortality rate and increased reproduction in female Drosophila, consistent with an induced trade-off. Finally, p53 mutation in female Drosophila and mTOR mutation in female mouse both decreased initial mortality rate, suggesting a normal role for these genes in diverting resources away from pathways important for health. In males, increased concentration of yeast and sugar in the media caused increased initial mortality rate, consistent with a diversion of resources away from pathways important for health. In contrast, p53 mutation in male Drosophila (and potentially mTOR mutation in male mouse) was associated with increased initial mortality rate, suggesting these genes normally regulate a diversion of resources towards health in the male. (B) Model for effect of trade-offs on cohort composition. Trade-offs between health and reproduction as shown in (A) preferentially cause lethality in the lowest-vitality individuals, thereby increasing initial mortality rate (parameter a). This leaves a cohort comprised of more robust individuals and therefore a slower age-dependent mortality rate acceleration (parameter b) (the S-M relationship). Similarly, during normal aging, the preferential loss of the lowest-vitality individuals can leave a cohort comprised of more robust individuals and a deceleration in mortality rates.
Evolutionary theory predicts trade-offs between longevity and reproductive fitness (42). The generality of this relationship has sometimes been questioned based on the findings that in certain instances increases in female life span can be produced without a detectable reduction in the number of progeny produced (43). However, the magnitude of life-span increases observed in the absence of reduced progeny number were generally modest, and reproductive fitness may include physiological changes that are costly for survival, but not necessarily represented by the number of offspring produced in the laboratory, for example, costly metabolic and behavioral changes and/or increased quality of offspring. When large-magnitude life-span increase is produced by DR in female Drosophila, it is typically associated with decreased number of progeny, consistent with a trade-off; however, the molecular mechanism(s) are not yet clear (31,44). Notably, life-span extension in female Drosophila due to DR (45) and life-span shortening due to mating (46) still occur in sterile females, indicating that the trade-off occurs upstream of the actual production of eggs.
The results obtained for males were more complex. For DR of both yeast and sugar in male Drosophila, the increase in median life span (27) was found to be associated with a robust S-M relationship, similar to the situation in females, but with smaller magnitude of effect. Notably, the diet concentration that produced the minimum value of parameter a was different for females (0.6X) compared to males (0.4X). Because of this, a change in diet concentration between these values produced opposite effects on mortality rate parameters in females versus males. A similar situation occurred with DR of yeast when the yeast was varied from 0.6X to 0.4X. These data indicate that the different nutrient optima of females versus males creates a range of nutrient concentrations where dietary interventions produce opposite effects on mortality rates in the sexes. Notably, a similar result was obtained for genetic manipulation of the p53/TOR pathway. The p53 null mutation in Drosophila (39) and the mTOR hypomorphic mutation in mice (9) can increase median life span of both males and females. However, the increased median life span caused by p53 null mutation in male Drosophila exhibited a S-M relationship with opposite sign relative to females, and for mouse mTOR mutation, the increase in median life span in males was associated with parameter changes that, while not reaching statistical significance, were also in the opposite direction relative to the changes in females. Notably, this pattern of S-M changes observed in male flies and mice (increased parameter a and decreased parameter b) is the same direction of S-M changes observed in several previous studies of DR in large cohorts of male rats (7,21,22). The negative correlation between parameters a and b in both males and females indicates that the S-M relationship is observed in both sexes. However, because the direction of these changes was sometimes opposite in males relative to females, it suggests that the p53/TOR pathway sometimes regulates a trade-off in opposite direction in males relative to females (model in Figure 6A), similar to the situation observed with DR. The p53 and TOR pathways exhibit extensive cross-talk in mammalian cells in regulating metabolism in response to diet, hormonal signals and stress (47), making this pathway a strong candidate for mediating trade-offs between reproduction and life span. Indeed in Drosophila, the TOR pathway modulates longevity in response to DR (48,49); however, it is not yet clear if TOR signaling is essential for the response (31). Previously, the male and female mTOR mutant mice were analyzed together for functional metrics of aging and displayed reduced tumor incidence and improved gait, memory and muscle strength relative to wild-type mice. In contrast, bone structure and immune function were reduced, suggestive of possible trade-offs (9). Notably, mating in female Drosophila causes reduced resistance to introduced pathogens and chronic inflammation (35,50,51). One possibility is that a diversion of resources away from normal immune function and towards reproduction renders the female less able to control the commensal flora, resulting in chronic inflammation associated with increased mortality.
The Drosophila interventions DR, mating and mifepristone are acting in adult animals, whereas Drosophila p53 mutation and mouse mTOR mutation might be acting during development and/or in adults. Previous conditional transgenic manipulations indicated that in Drosophila, p53 negatively regulates median life span in adult females and has smaller, positive effects on median life span in adult males, consistent with sexual antagonistic pleiotropy (39,52). The present data confirm and extend those observations and reveal that p53 has opposing effects on Drosophila mortality rates in males versus females. The results demonstrate that a robust S-M relationship is observed for multiple metazoan life-span interventions and that under appropriate conditions the sign of the parameter changes is opposite in males versus females. These results implicate the p53/TOR pathway, steroid hormone signaling and diet in sex-specific life-span regulation that may include opposing, sex-specific trade-offs between reproduction and life span.
Several previous studies have reported a deceleration in mortality rates at the latest ages for organisms including medfly (53), Drosophila (54), bean beetle (55) and Caenorhabditis elegans (56). Mortality rate deceleration at the latest ages has also been reported for certain human populations; however, the possible cause(s) for these observations in humans remain controversial (57–60). One hypothesis is that this phenomenon results from population heterogeneity, similar to the S-M relationship, such that early loss of the low-vitality individuals leaves a population comprised of more robust individuals and a slower mortality rate acceleration (61) (Figure 6B). This heterogeneity might be (epi)genetic, because even in inbred animals there may exist cryptic heterogeneity in epigenetic patterns and/or maternal contributions including mitochondrial genotypes. The related possibility is environmentally induced heterogeneity, resulting from interactions between the organism and micro-variation in the environment, as well as stochastic events. Finally, variation in microbial flora might be important, as trades-offs between reproduction and immune function could potentially cause a lethal loss of control over normally benign commensal species (8,62). Analysis of Caenorhabditis strains genetically selected for stress resistance (63) and of C. elegans and Drosophila containing reporters of stress response (64–66) have provided evidence in support of heterogeneity for aging. Experiments in Drosophila designed to modulate environmentally induced heterogeneity did not reduce mortality deceleration (67), arguing against environmentally induced heterogeneity and in favor of (epi)genetic heterogeneity. Our observation of robust S-M relationships in Drosophila in response to mating, mifepristone, p53 and DR argues in favor of the existence of heterogeneity in the starting population (Figure 6B). The S-M relationship indicates a subset of low-vitality individuals that is preferentially susceptible to both increased initial mortality (as caused by trade-offs) and to aging; however, the results do not distinguish whether this subset of low-vitality individuals results from (epi)genetic heterogeneity, environmentally induced heterogeneity, or some combination.
The present results may help explain certain patterns in human mortality. For example, the Bolivian Tsimane are forager-horticulturalists with a high pathogen burden: over half of documented deaths are from infectious and parasitic disease (68,69). Surprisingly, peripheral artery disease is absent and hypertension is reduced in the Tsimane relative to U.S. populations (70). One possibility is that loss of low-vitality individuals from the Tsimane population due to pathogens (increased initial mortality rate) yields a relatively robust cohort with a reduced rate of aging-associated cardiovascular disease (a S-M relationship). In humans, a subset of individuals exhibit a geriatric frailty syndrome characterized by sensitivity to stressors, chronic inflammation and increased mortality rate (71,72). Recent longitudinal studies of humans found heterogeneity in patterns of the epigenetic marker DNA methylation, which were predictive of earlier mortality (73,74). Taken together, the results caution that dietary and p53/TOR interventions might sometimes have opposite effect in men versus women, and suggest that identification of the underlying causes of heterogeneity may allow for interventions to be tailored to the individual.
Funding
This work was supported by the Department of Health and Human Services grant (AG011833 to J.T.), pilot project funds from the Southern California Environmental Health Sciences Center grant (5P30ES007048 to J.T.), a grant from National Natural Science Foundation of China (31500970 to J.S.), the Project-sponsored by SRF for ROCS, SEM (JS), and the Returned Overseas Chinese Scholars Research Merit Aid, Zhejiang [2014]115 (to J.S.).
Conflict of Interest
The authors declare that they have no conflict of interest.
Supplementary Material
Acknowledgments
We thank Caleb Finch and Felix Zajitschek for comments on the manuscript draft and thank Felix Zajitschek, Toren Finkel, J. Julie Wu, and Ilsa I. Rovira for generously providing the raw survival data for their previously published studies. Authors’ contributions: J.T. and J.S. designed the study. J.S., J.T., and G.N.L. conducted the data analyses. J.T. and J.S. wrote the manuscript. All authors read and approved the final manuscript.
References
- 1. Tower J. Mitochondrial maintenance failure in aging and role of sexual dimorphism. Arch Biochem Biophys. 2015;576:17–31. doi:10.1016/j.abb.2014.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morrow EH. The evolution of sex differences in disease. Biology of Sex Differences. 2015;6:5. doi:10.1186/s13293-015-0023-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burger JM, Promislow DE. Sex-specific effects of interventions that extend fly life span. Sci Aging Knowledge Environ. 2004;2004:pe30. doi:10.1126/sageke.2004.28.pe30 [DOI] [PubMed] [Google Scholar]
- 4. Tower J. Sex-specific regulation of aging and apoptosis. Mech Ageing Dev. 2006;127:705–718. doi:10.1016/j.mad.2006.05.001 [DOI] [PubMed] [Google Scholar]
- 5. Promislow D. Mate choice, sexual conflict, and evolution of senescence. Behav Genet. 2003;33:191–201. doi:10.1023/A:1022562103669 [DOI] [PubMed] [Google Scholar]
- 6. Pletcher SD. Model fitting and hypothesis testing for age-specific mortality data. J Evol Biol. 1999;12:430–439. doi:10.1046/j.1420-9101.1999.00058.x [Google Scholar]
- 7. Finch CE. Longevity, senescence and the genome. Chicago, IL: University of Chicago Press; 1990. [Google Scholar]
- 8. Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi:10.1016/j.cmet.2007.06.006 [DOI] [PubMed] [Google Scholar]
- 9. Wu JJ, Liu J, Chen EB, et al. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 2013;4:913–920. doi:10.1016/j.celrep.2013.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strehler BL, Mildvan AS. General theory of mortality and aging. Science. 1960;132:14–21. doi:10.1126/science.132.3418.14 [DOI] [PubMed] [Google Scholar]
- 11. Yashin AI, Begun AS, Boiko SI, Ukraintseva SV, Oeppen J. New age patterns of survival improvement in Sweden: do they characterize changes in individual aging? Mech Ageing Dev. 2002;123:637–647. doi:10.1016/S0047-6374(01)00410-9 [DOI] [PubMed] [Google Scholar]
- 12. Gavrilov LA, Gavrilova NS. The reliability theory of aging and longevity. J Theor Biol. 2001;213:527–545. doi:10.1006/jtbi.2001.2430 [DOI] [PubMed] [Google Scholar]
- 13. Hawkes K, Smith KR, Robson SL. Mortality and fertility rates in humans and chimpanzees: How within-species variation complicates cross-species comparisons. Am J Hum Biol. 2009;21:578–586. doi:10.1002/ajhb.20890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng H, Yang Y, Land KC. Heterogeneity in the Strehler-Mildvan general theory of mortality and aging. Demography. 2011;48:267–290. doi:10.1007/s13524-011-0013-8 [DOI] [PubMed] [Google Scholar]
- 15. Beltran-Sancheza H, Crimmins E, Finch C. Early cohort mortality predicts the rate of aging in the cohort: a historical analysis. J Dev Orig Health Dis. 2012;3:380–386. doi:10.1017/S2040174412000281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finch CE, Beltrán-Sánchez H, Crimmins EM. Uneven futures of human lifespans: reckonings from Gompertz mortality rates, climate change, and air pollution. Gerontology. 2014;60:183–188. doi:10.1159/000357672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krementsova AV, Roshina NV, Tsybul’ko EA, Rybina OY, Symonenko AV, Pasyukova EG. Reproducible effects of the mitochondria-targeted plastoquinone derivative SkQ1 on Drosophila melanogaster lifespan under different experimental scenarios. Biogerontology. 2012;13:595–607. doi:10.1007/s10522-012-9404-5 [DOI] [PubMed] [Google Scholar]
- 18. Zhikrevetskaya S, Peregudova D, Danilov A, et al. Effect of low doses (5-40 cGy) of gamma-irradiation on lifespan and stress-related genes expression profile in Drosophila melanogaster. PLoS One. 2015;10:e0133840. doi:10.1371/journal.pone.0133840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirkwood TB, Shanley DP. Food restriction, evolution and ageing. Mech Ageing Dev. 2005;126:1011–1016. doi:10.1016/j.mad.2005.03.021 [DOI] [PubMed] [Google Scholar]
- 20. Masoro EJ. Caloric restriction-induced life extension of rats and mice: a critique of proposed mechanisms. Biochim Biophys Acta. 2009;1790:1040–1048. doi:10.1016/j.bbagen.2009.02.01 [DOI] [PubMed] [Google Scholar]
- 21. Yu BP, Masoro EJ, Murata I, Bertrand HA, Lynd FT. Life span study of SPF Fischer 344 male rats fed ad libitum or restricted diets: longevity, growth, lean body mass and disease. J Gerontol. 1982;37:130–141. doi:10.1093/geronj/37.2.130 [DOI] [PubMed] [Google Scholar]
- 22. Pletcher SD, Khazaeli AA, Curtsinger JW. Why do life spans differ? Partitioning mean longevity differences in terms of age-specific mortality parameters. J Gerontol A Biol Sci Med Sci. 2000;55:B381–B389. doi:10.1093/gerona/55.8.B381 [DOI] [PubMed] [Google Scholar]
- 23. Nakagawa S, Lagisz M, Hector KL, Spencer HG. Comparative and meta-analytic insights into life extension via dietary restriction. Aging Cell. 2012;11:401–409. doi:10.1111/j.1474-9726.2012.00798.x [DOI] [PubMed] [Google Scholar]
- 24. Simons MJ, Koch W, Verhulst S. Dietary restriction of rodents decreases aging rate without affecting initial mortality rate—a meta-analysis. Aging Cell. 2013;12:410–414. doi:10.1111/acel.12061 [DOI] [PubMed] [Google Scholar]
- 25. Swindell WR. Dietary restriction in rats and mice: a meta-analysis and review of the evidence for genotype-dependent effects on lifespan. Ageing Res Rev. 2012;11:254–270. doi:10.1016/j.arr.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liao CY, Johnson TE, Nelson JF. Genetic variation in responses to dietary restriction–an unbiased tool for hypothesis testing. Exp Gerontol. 2013;48:1025–1029. doi:10.1016/j.exger.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Magwere T, Chapman T, Partridge L. Sex differences in the effect of dietary restriction on life span and mortality rates in female and male Drosophila melanogaster. J Gerontol A Biol Sci Med Sci. 2004;59:3–9. doi:10.1093/gerona/59.1.B3 [DOI] [PubMed] [Google Scholar]
- 28. Mair W, Goymer P, Pletcher SD, Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi:10.1126/science.1086016 [DOI] [PubMed] [Google Scholar]
- 29. Good TP, Tatar M. Age-specific mortality and reproduction respond to adult dietary restriction in Drosophila melanogaster. J Insect Physiol. 2001;47:1467–1473. doi:10.1016/S0022-1910(01)00138-X [DOI] [PubMed] [Google Scholar]
- 30. Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell. 2008;7:478–490. doi:10.1111/j.1474-9726.2008.00400.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tatar M, Post S, Yu K. Nutrient control of Drosophila longevity. Trends Endocrinol Metab. 2014;25:509–517. doi:10.1016/j.tem.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun X, Komatsu T, Lim J, et al. Nutrient-dependent requirement for SOD1 in lifespan extension by protein restriction in Drosophila melanogaster. Aging Cell. 2012;11:783–793. doi:10.1111/j.1474-9726.2012.00842.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zajitschek F, Zajitschek SR, Friberg U, Maklakov AA. Interactive effects of sex, social environment, dietary restriction, and methionine on survival and reproduction in fruit flies. Age (Dordr). 2013;35:1193–1204. doi:10.1007/s11357-012-9445-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wolfner MF. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity (Edinb). 2002;88:85–93. doi:10.1038/sj.hdy.6800017 [DOI] [PubMed] [Google Scholar]
- 35. Landis GN, Salomon MP, Keroles D, Brookes N, Sekimura T, Tower J. The progesterone antagonist mifepristone/RU486 blocks the negative effect on life span caused by mating in female Drosophila. Aging (Albany NY). 2015;7:53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baulieu EE. RU 486 (mifepristone). A short overview of its mechanisms of action and clinical uses at the end of 1996. Ann N Y Acad Sci. 1997;828:47–58. doi:10.1111/j.1749-6632.1997.tb48523.x [DOI] [PubMed] [Google Scholar]
- 37. Sieber MH, Spradling AC. Steroid signaling establishes a female metabolic state and regulates SREBP to control oocyte lipid accumulation. Curr Biol. 2015;25:993–1004. doi:10.1016/j.cub.2015.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fagegaltier D, König A, Gordon A, et al. A genome-wide survey of sexually dimorphic expression of Drosophila miRNAs identifies the steroid hormone-induced miRNA let-7 as a regulator of sexual identity. Genetics. 2014;198:647–668. doi:10.1534/genetics.114.169268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waskar M, Landis GN, Shen J, et al. Drosophila melanogaster p53 has developmental stage-specific and sex-specific effects on adult life span indicative of sexual antagonistic pleiotropy. Aging (Albany NY). 2009;1:903–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen J, Ford D, Landis GN, Tower J. Identifying sexual differentiation genes that affect Drosophila life span. BMC Geriatr. 2009;9:56. doi:10.1186/1471-2318-9-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 42. Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi:10.1016/j.cell.2005.01.027 [DOI] [PubMed] [Google Scholar]
- 43. Flatt T. Survival costs of reproduction in Drosophila. Exp Gerontol. 2011;46:369–375. doi:10.1016/j.exger.2010.10.008 [DOI] [PubMed] [Google Scholar]
- 44. Tatar M. The plate half-full: status of research on the mechanisms of dietary restriction in Drosophila melanogaster. Exp Gerontol. 2011;46:363–368. doi:10.1016/j.exger.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mair W, Sgro CM, Johnson AP, Chapman T, Partridge L. Lifespan extension by dietary restriction in female Drosophila melanogaster is not caused by a reduction in vitellogenesis or ovarian activity. Exp Gerontol. 2004;39:1011–1019. doi:10.1016/j.exger.2004.03.018 [DOI] [PubMed] [Google Scholar]
- 46. Ueyama M, Fuyama Y. Enhanced cost of mating in female sterile mutants of Drosophila melanogaster. Genes Genet Syst. 2003;78:29–36. doi:10.1266/ggs.78.29 [DOI] [PubMed] [Google Scholar]
- 47. Feng Z, Lin M, Wu R. The regulation of aging and longevity: a new and complex role of p53. Genes Cancer. 2011;2:443–452. doi:10.1177/1947601911410223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi:10.1016/j.cub.2004.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou S, Mackay T, Anholt RR. Transcriptional and epigenetic responses to mating and aging in Drosophila melanogaster. BMC Genomics. 2014;15:927. doi:10.1186/1471-2164-15-927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Short SM, Wolfner MF, Lazzaro BP. Female Drosophila melanogaster suffer reduced defense against infection due to seminal fluid components. J Insect Physiol. 2012;58:1192–1201. doi:10.1016/j.jinsphys.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fedorka KM, Linder JE, Winterhalter W, Promislow D. Post-mating disparity between potential and realized immune response in Drosophila melanogaster. Proc Biol Sci. 2007;274:1211–1217. doi:10.1098/rspb.2006.0394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shen J, Tower J. Drosophila foxo acts in males to cause sexual-dimorphism in tissue-specific p53 life span effects. Exp Gerontol. 2010;45:97–105. doi:10.1016/j.exger.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carey JR, Liedo P, Orozco D, Vaupel JW. Slowing of mortality rates at older ages in large medfly cohorts. Science. 1992;258:457–461. doi:10.1126/science.1411540 [DOI] [PubMed] [Google Scholar]
- 54. Curtsinger JW, Fukui HH, Townsend DR, Vaupel JW. Demography of genotypes: failure of the limited life-span paradigm in Drosophila melanogaster. Science. 1992;258:461–463. doi:10.1126/science.1411541 [DOI] [PubMed] [Google Scholar]
- 55. Tatar M, Carey JR, Vaupel JW. Long-term cost of reproduction with and without accelerated senescence in Callosobruchus maculatus: analysis of age-specific mortality. Evolution. 1993;47:1302–1312. doi:10.2307/2410149 [DOI] [PubMed] [Google Scholar]
- 56. Vaupel JW, Johnson TE, Lithgow GJ. Rates of mortality in populations of Caenorhabditis elegans. Science. 1994;266:826. doi:10.1126/science.7973640 [DOI] [PubMed] [Google Scholar]
- 57. Vaupel JW, Carey JR, Christensen K, et al. Biodemographic trajectories of longevity. Science. 1998;280:855–860. doi:10.1126/science.280.5365.855 [DOI] [PubMed] [Google Scholar]
- 58. Koopman JJ, Rozing MP, Kramer A, et al. Calculating the Rate of Senescence From Mortality Data: An Analysis of Data From the ERA-EDTA Registry. J Gerontol A Biol Sci Med Sci. 2015. doi: 10.1093/gerona/glv042 [DOI] [PubMed] [Google Scholar]
- 59. Gavrilov LA, Gavrilova NS. Mortality Measurement at Advanced Ages: A Study of the Social Security Administration Death Master File. North American actuarial journal: NAAJ. 2011;15:432–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Missov TI, Vaupel JW. Mortality Implications of Mortality Plateaus. SIAM Review. 2015;57:61–70. doi:10.1137/130912992 [Google Scholar]
- 61. Kirkwood TB. Deciphering death: a commentary on Gompertz (1825) ‘On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies’. Philos Trans R Soc Lond B Biol Sci. 2015;370. doi:10.1098/rstb.2014.0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Clark RI, Salazar A, Yamada R, et al. Distinct Shifts in Microbiota Composition during Drosophila Aging Impair Intestinal Function and Drive Mortality. Cell Rep. 2015;12:1656–1667. doi:10.1016/j.celrep.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen HY, Zajitschek F, Maklakov AA. Why ageing stops: heterogeneity explains late-life mortality deceleration in nematodes. Biol Lett. 2013;9:20130217. doi:10.1098/rsbl.2013.0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu D, Rea SL, Yashin AI, Johnson TE. Visualizing hidden heterogeneity in isogenic populations of C. elegans. Exp Gerontol. 2006;41:261–270. doi:10.1016/j.exger.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 65. Yang J, Tower J. Expression of hsp22 and hsp70 transgenes is partially predictive of drosophila survival under normal and stress conditions. J Gerontol A Biol Sci Med Sci. 2009;64:828–838. doi:10.1093/gerona/glp054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Landis GN, Abdueva D, Skvortsov D, et al. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:7663–7668. doi:10.1073/pnas.0307605101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Khazaeli AA, Pletcher SD, Curtsinger JW. The fractionation experiment: reducing heterogeneity to investigate age-specific mortality in Drosophila. Mech Ageing Dev. 1998;105:301–317. doi:10.1073/pnas.0307605101 [DOI] [PubMed] [Google Scholar]
- 68. Gurven M, Kaplan H, Supa AZ. Mortality experience of Tsimane Amerindians of Bolivia: regional variation and temporal trends. Am J Hum Biol. 2007;19:376–398. doi:10.1002/ajhb.20600 [DOI] [PubMed] [Google Scholar]
- 69. Vasunilashorn S, Crimmins EM, Kim JK, et al. Blood lipids, infection, and inflammatory markers in the Tsimane of Bolivia. Am J Hum Biol. 2010;22:731–740. doi:10.1002/ajhb.21074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gurven M, Kaplan H, Winking J, et al. Inflammation and infection do not promote arterial aging and cardiovascular disease risk factors among lean horticulturalists. PLoS One. 2009;4:e6590. doi:10.1371/journal.pone.0006590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chang SF, Lin PL. Frail phenotype and mortality prediction: a systematic review and meta-analysis of prospective cohort studies. Int J Nurs Stud. 2015;52:1362–1374. doi:10.1016/j.ijnurstu.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 72. Chen X, Mao G, Leng SX. Frailty syndrome: an overview. Clin Interv Aging. 2014;9:433–441. doi:10.2147/CIA.S45300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Christiansen L, Lenart A, Tan Q, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2015. doi: 10.1111/acel.12421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Moore AZ, Hernandez DG, Tanaka T, et al. Change in epigenome-wide DNA methylation over 9 years and subsequent mortality: results from the InCHIANTI Study. J Gerontol A Biol Sci Med Sci. 2015. doi: 10.1093/gerona/glv118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.