Abstract
Here, we report the biochemical characterization of the mono-ADP-ribosyltransferase 2,3,7,8-tetrachlorodibenzo-p-dioxin poly-ADP-ribose polymerase (TIPARP/ARTD14/PARP7), which is known to repress aryl hydrocarbon receptor (AHR)-dependent transcription. We found that the nuclear localization of TIPARP was dependent on a short N-terminal sequence and its zinc finger domain. Deletion and in vitro ADP-ribosylation studies identified amino acids 400–657 as the minimum catalytically active region, which retained its ability to mono-ADP-ribosylate AHR. However, the ability of TIPARP to ADP-ribosylate and repress AHR in cells was dependent on both its catalytic activity and zinc finger domain. The catalytic activity of TIPARP was resistant to meta-iodobenzylguanidine but sensitive to iodoacetamide and hydroxylamine, implicating cysteines and acidic side chains as ADP-ribosylated target residues. Mass spectrometry identified multiple ADP-ribosylated peptides in TIPARP and AHR. Electron transfer dissociation analysis of the TIPARP peptide 33ITPLKTCFK41 revealed cysteine 39 as a site for mono-ADP-ribosylation. Mutation of cysteine 39 to alanine resulted in a small, but significant, reduction in TIPARP autoribosylation activity, suggesting that additional amino acid residues are modified, but loss of cysteine 39 did not prevent its ability to repress AHR. Our findings characterize the subcellular localization and mono-ADP-ribosyltransferase activity of TIPARP, identify cysteine as a mono-ADP-ribosylated residue targeted by this enzyme, and confirm the TIPARP-dependent mono-ADP-ribosylation of other protein targets, such as AHR.
Keywords: aryl hydrocarbon receptor, mass spectrometry, mono-ADP-ribose, poly-ADP-ribose polymerase, TCDD-inducible poly-ADP-ribose polymerase
Introduction
The poly-ADP-ribose polymerase (PARP) family, also referred to as ADP-ribosyltransferase diphtheria toxin-like (ARTD) family, consists of 17 members that share a conserved catalytic PARP domain [1,2]. PARPs use nicotinamide adenine dinucleotide (NAD+) as a substrate to transfer an ADP-ribose (ADPr) monomer onto specific amino acid acceptor residues in target proteins or multiple ADPr monomers onto the same amino acid resulting in an ADPr polymer, known as poly-ADP-ribose (PAR) [2]. ADP-ribosylation of proteins alters their enzymatic activity, stability, and ability to interaction with other proteins [3]. PARP family members have diverse cellular distribution and expression patterns, indicating wide and distinct biological functions. The majority of PARPs have mono-ADP-ribosylation (MARylation) and not poly-ADP-ribosylation (PARylation) activities [4]. Because of this, a new nomenclature has been proposed to more accurately describe their ADP-ribosyltransferase activity [2]. ADP-ribosylation by PARPs can be reversed by enzymes that remove ADP-ribose, including poly(ADP-ribose) glycohydrolase (PARG), ADP-ribosyl hydrolases, terminal ADP-ribose protein glycohydrolase TARG1/C6orf130, and macrodomain-containing proteins [5,6].
PARP1 (ARTD1) is the founding and best characterized member of the PARP family. PARP1 catalyses the transfer of PAR to acceptor proteins and has important roles in cell proliferation, protein degradation, transcription, RNA biology, and cellular stress responses, such as DNA damage [7]. PARP inhibitors represent a promising strategy for cancer therapy [8,9]. Our current understanding of which amino acid residues are modified by PARPs is for the most part based on data from PARP1 [10]. Mass spectrometry studies reveal that PARP1 modifies glutamates, asparates, lysines, and arginines [11]. Although no consensus peptide sequence has been identified, a recent study identified several potential consensus motifs [12]. Less is known about MARylation and mono-ADP-ribosylating PARPs [4,13]; however, mass spectrometry studies confirmed that both acidic and lysine residues as well as cysteine residues are targets of these enzymes [4]. Recent proteome-wide studies have also identified PARP-specific targets for both poly and mono-ADP-ribosylating PARP family members, providing insights into their distinct cellular roles [14,15].
The TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin)-inducible poly-ADP-ribose polymerase TIPARP (also known as ARTD14/PARP7) is a mono-ADP-ribosylating PARP that modifies itself, core histones and transcription factors, such as aryl hydrocarbon receptor (AHR) and liver X receptors (LXRs) [16–18]. TIPARP contains an uncharacterized N-terminus, followed by a CCCH-zinc finger domain, a WWE (tryptophan–tryptophan–glutamate) domain, and a conserved PARP catalytic domain [1,16]. TIPARP is predominantly localized in the nucleus; however, no nuclear localization signal (NLS) has been reported [16,19]. Interestingly, TIPARP translocates to the cytosol in response to viral infection, reactive oxygen species (ROS), and mitochondrial damage [20]. TIPARP's zinc finger domain binds to Sindbis virus RNA to induce its degradation; however, the role of TIPARP's zinc finger domain in the absence of viral infection is unclear [20]. In addition, TIPARP influences the pluripotency of embryonic stem cells and more recently, it has been shown to regulate astrocyte autophagy and activation [21].
AHR is a ligand-activated transcription factor and a member of the basic helix-loop-helix Period-AHR nuclear translocator (ARNT)-singled minded (bHLH-PAS) family. Ligand-bound AHR regulates its target genes as a heterodimer complex with ARNT. This complex binds to specific DNA sequences located in the upstream regulatory regions of AHR target genes, such as cytochrome P450 1A1 (CYP1A1) and TIPARP [22]. AHR is best known for its ability to mediate the toxic responses of the environmental contaminant TCDD [23,24]. AHR also regulates immune function, inflammation, and stem differentiation, and plays a role in cancer [25]. TIPARP functions as part of a negative feedback loop regulating AHR activity [16]. TIPARP mono-ADP-ribosylates AHR in vitro, and the ability of TIPARP to repress AHR activity is dependent on its zinc finger domain and its catalytic activity. In vivo loss of Tiparp expression in mice increases their sensitivity to TCDD-induced toxicities and lethality [17]. TIPARP has been reported to regulate stem cell pluripotency, viral replication, innate immunity, and platelet-derived growth factor (PDGF)-dependent responses [26–29]. Although multiple protein targets of TIPARP have been identified, characterization of its enzymatic activity and the identification of the amino acids targeted by TIPARP have not been reported.
Herein, we report the biochemical characterization of TIPARP's MARylating activity. Using truncations and site-directed mutations, we identified a nuclear localization sequence in TIPARP and mapped its minimum auto-MARylation region. We identified several modified peptides in TIPARP and in its target protein AHR, and used electron transfer dissociation mass spectrometry to map a modified cysteine residue in the TIPARP N-terminus. Our findings characterize the cellular localization and MARylation activity of TIPARP, provide further insights into how TIPARP acts as a transcriptional repressor of AHR, and confirm cysteine as a target of mono-ADP-ribosylating PARPs.
Experimental procedures
Chemicals
3-Aminobenzoamide (3-ABA), (N-(6-oxo-5,6-dihydrophenanthridin-2-yl)(N,N-dimethylamino) acetamide) (PJ34), meta-iodobenzylguanidine (MIBG), iodoacetamide (IAM), and NH2OH (hydroxylamine) were purchased from Sigma-Aldrich (Oakville, Ontario). 4-[(3-[(4-Cyclopropylcarbonyl)piperazin-4-yl]carbonyl)-4-fluoropheny]methyl(2H)phthalazin-1-one (Olaparib) and 4-[[4-fluoro-3-[(hexahydro-1H-1,4-diazepin-1-yl)carbonyl]phenyl]methyl]-1(2H)-phthalazinone (Ku-0058948) were purchased from Toronto Research Chemicals, Inc. (Toronto, Canada). All other chemicals and biological reagents were purchased from other vendors and were of the highest quality.
Plasmids
The plasmids pGEX-TIPARP and pGEX-AHR 430–848 have been described elsewhere [16,17]. TIPARP N-terminal truncated cDNAs were PCR-amplified using pcDNA-TIPARP as a template. pGEX-TIPARP C39A and pGEX-TIPARP H532A were generated by PCR-mediated site-directed mutagenesis. The pGEX and pcDNA3.1 N-terminal deletions of TIPARP used for the 32P-NAD+-ribosylation assays or 35S protein shift assays, respectively, were generated by PCR using a common reverse primer and amino acid-specific forward primers. The PCR products were digested with EcoRI and SalI and cloned into the appropriately digested pGEX-4T1 or pcDNA3.1 plasmids. The GFP-TIPARP, GFP-TIPARPΔ1-32, GFP-TIPARPΔ1-102, GFP-TIPARP C243A, GFP-TIPARP H532A, GFP-TIPARPΔ1-244, pRC-AHR and pCMV-ARNT plasmids have been described elsewhere [16]. GFP-TIPARPΔ1-52 was generated by PCR. The PCR product was digested with EcoRI and SalI and cloned into the appropriately digested pGFP-C2 plasmid. GFP-TIPARP K41A and GFP-TIPARP K41A;C243A were generated by PCR-mediated site-directed mutagenesis using GFP-TIPARP and GFP-TIPARPC243A as templates, respectively. All cloning and site-directed mutagenesis primers are listed in Supplementary Table S1.
Immunofluorescence
Human hepatoma HuH7 cells were cultured in high glucose (4.5 g/l) DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (PEST). All cell lines were maintained at 37°C and 5% CO2 and subcultured every 2–3 days or when cells reached 80% confluency. HuH7 cells were seeded onto glass cover slips in a six-well plate (3.5 × 105 cells/well). HuH7 cells were transfected with 500 ng of GFP-TIPARP, GFP-TIPARP K41A, GFP-TIPARPΔ1-32, GFP-TIPARPΔ1-52, GFP-TIPARPΔ1-102, GFP-TIPARP C243A, GFP-TIPARP K41A;C243A, or GFP-TIPARPΔ1-244 using Lipofectamine 2000. After 24 h, cells were fixed with 4% paraformaldehyde, permeabilized with 0.4% Triton X-100, and mounted onto slides with Vectashield mounting medium containing DAPI (4,6-diamidino-2-phenylindole) to stain DNA (Vector Laboratories). GFP-expressing cells were visualized using a Zeiss AxioImager M2 epifluorescence microscope and images were captured with a Zeiss AxioCam MRm camera and Zeiss AxioVision software. For these studies, 10 cells were evaluated per condition. The 10 cells were randomly chosen under low magnification based on a moderate level of GFP expression. Cells with very high and blotchy protein expression patterns were excluded, and cells with very low expressing proteins where the signal was not bright enough to withstand multiple z-focus imaging were excluded.
Generation of stable and inducible cell line
We generated a doxycycline (dox)-inducible TIPARP hemagglutinin (HA)-overexpressing MCF-7 cell line using a Tet-ON 3G system. To achieve this, we transfected MCF-7 cells with CMV-Hygro plasmid and used hygromycin to select for MCF-7/Tet 3G cells. MCF-7/Tet 3G cells were then transfected using lentivirus carrying a TRE3G-TIPARP-HA-Puro construct. After the transfection, both hygromycin and puromycin were used for the selection of stable dox-inducible TIPARP-HA-overexpressing MCF-7 cells.
Reporter gene assays
HuH7 cells were seeded in 12-well plates (1.0 × 105 cells/well) and transfected with 400 ng of the various GFP-TIPARP deletion and mutation variants, and 200 ng of pCYP1B1-luc using Lipofectamine 2000. All reactions included 50 ng of pCH110-β-Gal to normalize for transfection efficiency. After 24 h, cells were treated with TCDD or DMSO (dimethyl sulfoxide) for 24 h before completing luciferase and β-galactosidase assays.
Western blots
HuH7 cells were seeded in six-well plates and transfected with 1.5 µg of pEGFP-TIPARP deletion and mutation variants. Forty-eight hours after transfection, cell pellets were collected and lysed in cell lysis buffer (20 mM Tris–HCl, pH 7.4, 1% NP40, and 150 mM NaCl). After centrifugation, the protein concentration of the clarified cell extract was determined by the BCA assay according to the manufacturer's instructions (Bio-Rad). Ten microgram of total protein was separated by SDS–PAGE and transferred to a PVDF membrane. Membranes were incubated with anti-GFP antibody (TaKaRa; JL8) and stripped and then incubated with anti-β-actin antibody (Sigma; A-2228).
In vitro ADP-ribosylation assays
Glutathione S-transferase (GST)-tagged recombinant proteins were expressed and purified as previously described [16]. Auto-ribosylation assays were carried out in 30 µl reaction volumes at 30°C for 20 min in 1× reaction buffer (50 mM Tris-base, pH 8.0, 4 mM MgCl2, and 0.2 mM DTT) and 2 µCi 32P-NAD+ (Perkin-Elmer) and/or 500 µM β-NAD+ (Sigma). Approximately 1 µg of purified GST-tagged proteins were used. For hetero-ribosylation experiments, 4 µg of target proteins were added to the reactions. Reactions were stopped by the addition of 1× sample buffer. Samples were separated by SDS–PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. PVDF membranes were visualized by GelCode Blue (GCB) Stain Reagent (Thermo Scientific) to ensure equal loading before ADP-ribosylated proteins were visualized by autoradiography.
Co-immunoprecipitation assays
COS-1 cells were seeded in six-well plates (2.0 × 10−5 cells/well) in DMEM containing 10% FBS and 1% PEST. The next morning, cells were transfected with plasmid GFP-TIPARP, GFP-TIPARPC243A, or GFP-TIPARPH532A and pRC-AHR, pCMV-ARNT, GFP-TIPARP1–234, or GFP-TIPARP1–448 using Lipofectamine 2000. Twenty-four hours later, cells were washed with ice-cold PBS and lysed in cell lysis buffer (200 mM NaCl, 1% NP40 and 20 mM HEPES at pH 7.4, and supplemented with protease inhibitor cocktail [Sigma]). Lysates were clarified by centrifugation and incubated with anti-GFP (3E6; Life technologies), anti-AHR (H211; Santa Cruz), or anti-FLAG (M2; Sigma). Protein complexes were captured with protein A or G Dynal beads (Life Technologies). Beads were washed four times with cell lysis buffer and eluted in 1× sample buffer. Samples were separated by SDS–PAGE and transferred to PVDF membranes. Membranes were probed with anti-PAN-ADP-ribose (Millipore), anti-GFP (JL8; Clontech), or anti-AHR (H211) followed by incubation with the appropriate secondary antibodies. TIPARP-HA-overexpressing MCF7 cells were plated at a density of 2 million cells per 15 cm dish in DMEM containing 10% FBS and 1% PEST. The next morning, water or 1.5 µg/ml dox was added to the dish. Six hours later, the cells were treated with DMSO or 10 nM TCDD. Twenty-four hours later, cells were washed with ice-cold PBS and lysed in cell lysis buffer as described above. Lysates were clarified by centrifugation and incubated with anti-AHR (H211; Santa Cruz). Protein complexes were captured with protein A Dynal beads (Life Technologies). Samples were separated by SDS–PAGE and transferred to PVDF membranes. Membranes were probed with anti-mono-ADP-ribose-binding reagent (Millipore; MABE1076), anti-AHR (H-211), or anti-HA (Roche 3F10) followed by incubation with the appropriate secondary antibodies. For GFP-TIPARP immunoprecipitations analyzed by mass spectrometry, cell extracts were prepared from eight 15 cm dishes transfected with 10 µg of GFP-TIPARP. The pooled extracts were immunoprecipitated with anti-GFP (3E6) and the entire reaction was separated by SDS–PAGE. The gel was stained with Coomassie blue and the immunoprecipitated band corresponding to GFP-TIPARP was excised.
Inhibition of auto-ribosylation of TIPARP by different PARP inhibitors
Approximately 2 µg of GST-TIPARP protein was pre-incubated with PARP inhibitors: 3-aminobenzamide (3-ABA; 0.01–1.5 mM), PJ34 (0.1–250 µM), olaparib (0.05–1.25 µM), and Ku-0058948 (0.05–1.25 µM) or MIBG (0.5–500 µM) for 5 min at room temperature in 1× reaction buffer with varying concentrations. Reactions were initiated with the addition of 2 µCi 32P-NAD+ and incubated for 20 min at 30°C and stopped by the addition of 1× sample buffer. Samples were separated by SDS–PAGE and transferred to PVDF membranes as described above. IC50 values for the PARP inhibitors for PARP1 and TIPARP were calculated in GraphPad Prism 5 after densitometric analysis (ImageJ) of the autoradiography films.
Methylation by IAM
Approximately 2 µg of purified GST-TIPARP was pre-incubated with IAM (0.02–2 mM) for 30 min at 37°C. Control reactions were pre-incubated with 2 mM NaCl. Reactions were initiated with addition of 2 µCi 32P-NAD+ and incubated for 20 min at 30°C and stopped by the addition of 1× sample buffer. Samples were separated by SDS–PAGE and transferred to PVDF membranes as described above.
Hydrolysis with hydroxylamine
Approximately 2 µg of purified GST-TIPARP was incubated with 2 µCi 32P-NAD+ in 1× reaction buffer for 20 min at 30°C. Reactions were then incubated with neutral 1 M hydroxylamine for 1 or 2 h at 30°C. Control reactions were incubated with 1 M NaCl. Reactions were stopped with 1× sample buffer. Samples were separated by SDS–PAGE and transferred to PVDF membranes as described above.
35S Protein shift assays
Full-length pcDNA-TIPARP [30] and various TIPARP truncation proteins were generated using 35S-methionine (Perkin-Elmer) and the TNT Coupled Reticulocyte Lysate System according to the manufacturer's instructions (Promega). Five percent of 35S-labeled TIPARP proteins were incubated in 1× reaction buffer with or without 500 µM β-NAD+ (Sigma) for 30 min at 30°C. Reactions were stopped by the addition of 1× SDS sample buffer and proteins were separated by SDS–PAGE and visualized by autoradiography.
Peptide synthesis and overlay
The complete sequence of human TIPARP protein (amino acids 1–657; NP_001171646) was synthesized as 20-mer peptides four amino acid offsets on cellulose membranes using a Multipep automated peptide synthesizer (INTAVIS Bioanalytical Instruments, Koeln, Germany) as described previously [31]. Peptide arrays were blocked in 1% casein in Tris-buffered saline + Tween 20 (TBST) for 1 h at room temperature. The overlays were done using 1 µg/ml GST-TIPARP fusion protein or GST protein in 1% casein TBST overnight at 4°C. Membranes were then washed three times for 10 min in TBST at room temperature. Bound proteins were detected by immunoblotting with 1 : 5000 dilution of anti-GST-HRP (ab58626, Abcam).
In-gel protein digestion
Ten micrograms of purified GST-TIPARP was incubated with 500 µM NAD+ (Sigma) in 1× reaction buffer for 30 min at 30°C. Reactions were stopped by the addition of 1× SDS sample buffer and proteins were separated by SDS–PAGE and Coomassie blue stained. The stained gel slices containing ADP-ribosylated TIPARP were destained, reduced with dithiothreitol (Sigma), alkylated with IAM (Sigma), and the extraction of protease-generated peptides was performed as previously described [32]. Proteolytical digestions used 4 µg trypsin (Sigma) in 50 mM NH4HCO3 at 37°C or 0.5 µg chymotrypsin (Sigma) in 100 mM Tris, 10 mM CaCl2·2H2O, pH 7.8 at 25°C, each for 20 h.
Reverse-phase nano LC-MS2 analysis of proteolytic peptides
The analysis of in-gel digested TIPARP peptides was done by reverse-phase nanoflow liquid chromatography (nLC) coupled to a nanoelectrospray LTQ Orbitrap XL mass spectrometer utilizing higher energy C-cell dissociation (HCD) and electron transfer dissociation (ETD) fragmentation (RP nLC-ESI MS2). The RP nLC was performed as recently described [33], but only a Zorbax 300 SB-C18 5 µm pre-column (Agilent) was used. nESI was achieved by applying a 1.2 kV between the 8 µm diameter emitter (PicoTip emitter, New objectives, Woburn, MA) and the capillary entrance of the Orbitrap. Mass spectra was acquired in the positive ion mode with one survey scan (m/z 300–2000, target value 1 000 000 ions, with a resolution of 60 000 at m/z 400) followed by HCD MS2 of the three most intense ions in the Orbitrap. The following parameters were used: target value of 5000 ions, ion selection threshold 500 counts, and dynamic exclusion of selected ions for 90 s. In addition, the samples were analyzed on a QExactive Orbitrap mass spectrometer as recently described [34].
ETD parameters
The two most intense ions on the parent mass list, consisting of both the 12C and one 13C precursor masses of identified ADP-ribosylated peptides, were selected for ETD. The minimal selection threshold was set at 500 with a 1 Da isolation window. The intensity of the reagent gas (at m/z 202) was at a minimum of 1 × 106 before analysis and the ETD reaction time was set at 150 ms. No supplement activation was used.
Bioinformatics analysis
nLC-MS2 spectra of TIPARP and AHR peptides were searched with the MaxQuant software [35] with default settings against an in-house generated protein database containing the GST-TIPARP amino acid sequence for ADP-ribosylation and allowing for up to three dynamic ADP-ribosylation modifications of 541.0611 Da per peptide [36]. Positively flagged peptides were manually inspected using Thermo Xcalibur (v. 3.0.63) for the presence of reporter ions specific for ADP-ribosylation at m/z 250.0932 (adenosine-H2O; composition C10H11N5O3), at m/z 348.0701 (adenosine monophosphate; composition C10H14N5O7P), and at m/z 428.0364 (adenosine diphosphate; composition C10H15N5O7P2) [36,37]. Moreover, all MS2 spectra were manually investigated for the presence of ADP-ribose reporter ions by flagging all ADP-ribose reporter ions in the nLC-MS2 chromatogram with a 10 ppm accuracy using Thermo Xcalibur and subsequently manually identified using the PeptideMass (ExPASy) and ProteinProspector (UCSF) tools. Quantitation of relative amounts of ADP-ribosylated peptides was done by extracting the precursor mass (MS1) elution peak of all the ADP-ribose-modified and -unmodified peptides. The area under the curve for modified and unmodified peptides was determined by the add peak feature in Proteome Discoverer (v. 3.0.63), setting the curve limits as close to the beginning and end of the elution peak as possible (excluding tailing). The relative amount (in %) of each modified peptide compared to the unmodified version was calculated by the value for the area under curve.
Statistical analyses
All data were presented as means and standard error of the mean (SEM). One-way analysis of variance (ANOVA) followed by Tukey's multiple comparison tests or two-tailed Student's t-tests was used to assess statistical significance (P < 0.05).
Results
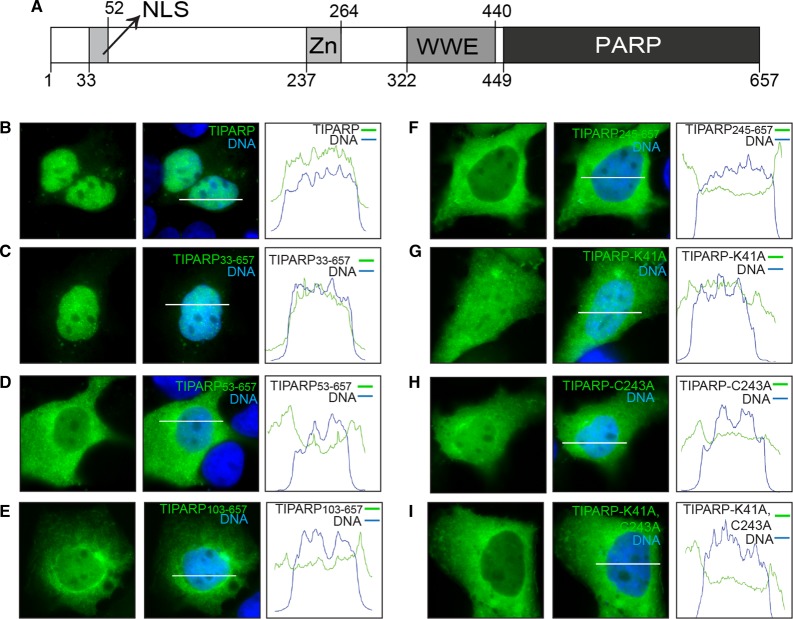
The N-terminus and an intact zinc finger domain are required for the nuclear localization of TIPARP
TIPARP is a CCCH-zinc finger-type member of the PARP family that contains a WWE domain and a C-terminal PARP catalytic region. However, no known functional domains have been identified within the first 236 amino acids of TIPARP. TIPARP localizes primarily as small nuclear foci in HuH7 cells [19], but it has also been reported to exhibit weak cytosolic staining in Hela and MCF7 cells [19,38]. To identify possible functional domains in the N-terminus of TIPARP, we did a sequence analysis and found a putative NLS that included amino acids 41-KKKDQKR-47 (Figure 1A). We then made various site-directed mutants and N-terminal deletion mutants of TIPARP in which the putative NLS was removed and studied their localization by direct fluorescence in HuH7 cells. Representative cells were imaged and protein localization was analyzed across a bisection of each cell (white bars, Figure 1B–I). The adjacent histograms illustrate pixel intensity (y-axis) for the DNA (blue) and respective GFP-tagged protein (green) across this line (x-axis). The amplitude of the green histogram was compared across the area under the blue histogram (nucleus) to outside of this area (cytoplasm) to measure the relative localization and pattern of the GFP-tagged proteins. GFP-TIPARP and GFP-TIPARP 33–657 localized primarily to the nucleus as visible foci and the corresponding histograms showed moderate changes in amplitude of the green histogram across the DNA region, confirming nuclear localization with a pattern of many bright foci (Figure 1B,C). GFP-TIPARP 53–657 and GFP-TIPARP 103–657 showed significant localization to the cytoplasm, but the protein that remained nuclear maintained a speckled pattern (Figure 1D,E). GFP-TIPARP 245–657 localized almost exclusively to the cytoplasm with only faint and diffuse nuclear staining, which corresponded to very small changes in the amplitude of the green histogram across the DNA region (Figure 1F).
Figure 1. Subcellular localization of TIPARP in HuH7 cells.
Transient expression of GFP-tagged wild-type TIPARP and TIPARP mutants in HuH7 cells. (A) Diagram of TIPARP protein showing its functional domains: NLS, nuclear localization sequence; Zn, CCCH-type Zn finger domain; WWE, tryptophan–tryptophan–glutamate domain; PARP, poly(ADP-ribose) polymerase catalytic domain. (B–I) Representative images of GFP-tagged TIPARP proteins that were transiently transfected into HuH7 cells grown on coverslips (63× magnification): (B) TIPARP, (C) TIPARP 33–657, (D) TIPARP 53–657, (E) TIPARP 103–657, (F) TIPARP 245–657, (G) TIPARP K41A, (H) TIPARP C243A, and (I) TIPARP K41A;C243A. DNA was stained with DAPI to show the nucleus. The white bars (B–I) correspond to the x-axis of the adjacent histograms. All bars are 25 µm. The histograms illustrate relative pixel intensity (y-axis) for the DNA (blue) and respective GFP-tagged protein (green) across the line measured. The area under the blue histogram represented DNA, and the amplitude of the green histogram was compared across this area (nucleus) to outside of this area (cytoplasm) to measure the relative localization of the GFP-tagged protein. The data are representative of 10 randomly chosen cells per condition.
Since residues 41–47 contained a characteristic lysine-rich NLS sequence, we mutated lysine 41 to alanine to determine its impact on the cellular localization of TIPARP. GFP-TIPARP K41A showed significant localization to the cytoplasm, but the remaining nuclear portion of protein maintained a speckled pattern. This also corresponded with moderate changes in amplitude of the green histogram across the DNA region (Figure 1G). Because the TIPARP 245–657 removed the first cysteine of the CCCH-type zinc finger domain (spanning amino acids 237–264), we created a GFP-TIPARP C243A to determine the impact of the zinc finger domain on TIPARP localization. GFP-TIPARP C243A localized to both the cytoplasm and the nucleus. However, unlike the other mutants that exhibited staining in both cellular compartments, GFP-TIPARP C243A exhibited a diffuse nuclear staining pattern, which was reflected by small changes in amplitude of the green histogram across the DNA region (Figure 1H). GFP-TIPARP K41A;C243A exhibited an almost exclusively cytoplasmic staining pattern with little nuclear staining supported by relatively low pixel intensity and minor changes in amplitude of the green histogram across the DNA region (Figure 1I). This pattern was similar to that observed for GFP-TIPARPΔ1-244 (Figure 1F), suggesting that nuclear localization of TIPARP requires an N-terminal NLS region and an intact zinc finger domain. Collectively, these findings suggested that an NLS was present within amino acids 33-ITPLKTCFKKKDQKRLGTGT-52 and that a CCCH-type zinc finger domain also influences the cellular localization of TIPARP.
Auto-ADP-ribosylation activity of TIPARP
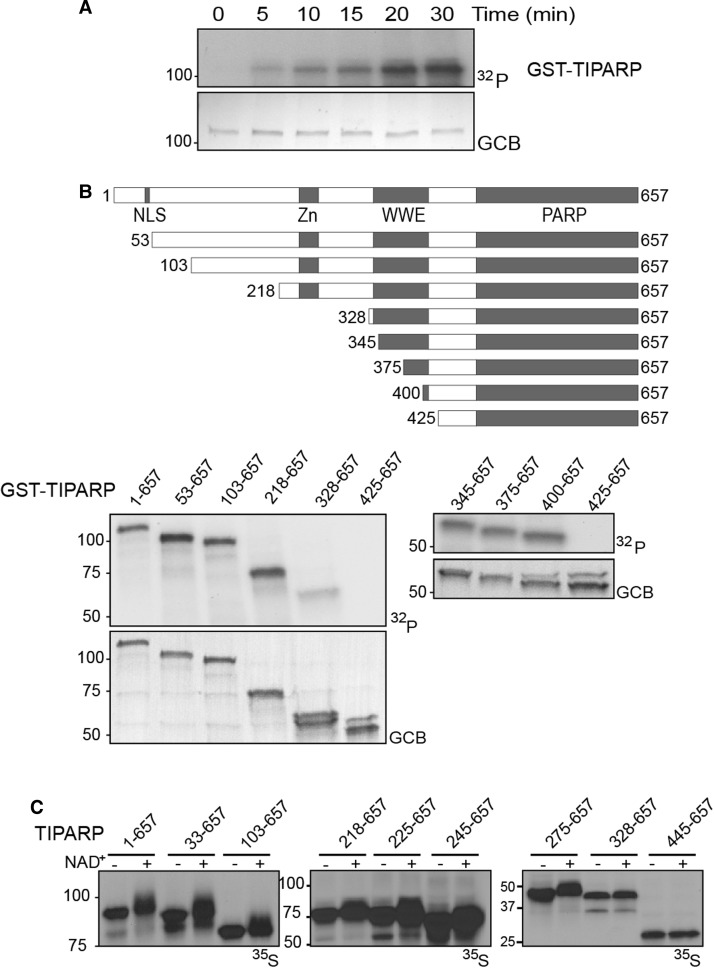
Our group and others have reported that TIPARP mono-ADP-ribosylates itself, core histones, LXRα/β and AHR [4,16–18]. However, the identification of the mono-ADP-ribosylated sequences within TIPARP and the minimal catalytically active truncation of TIPARP have not been described. To this end, we performed kinetic ADP-ribosylation assays using recombinant GST-TIPARP (full length 1–657) protein that was incubated with 32P-NAD+. We used a GST-tag to purify TIPARP since purification of 6x-histidine tagged TIPARP proteins was unsuccessful due to insolubility (data not shown). In agreement with previous findings, GST-TIPARP exhibited mono-ADP-ribosyltransferase rather than ADP-polymerase activity [4,16], as evidenced by the lack of shift in its mobility during SDS–PAGE. Mono-ADP-ribosylation was detected as early as 5 min after incubation with 32P-NAD+ and reached a peak after 20 min (Figure 2A).
Figure 2. Characterization of the auto-mono-ADP-ribosyltransferase activity of TIPARP.
(A) Time course studies of TIPARP auto-ADP-ribosylation using 32P-NAD+. The auto-ADP-ribosylation of TIPARP was done by incubating 1 µg of GST-tagged TIPARP with 2 µCi 32P-NAD+ in a PARP buffer for the times indicated. (B) Mapping of the TIPARP's auto-ADP-ribosylation region using GST-tagged N-terminal TIPARP deletion proteins. One microgram of GST-tagged truncation TIPARP protein was incubated with 2 µCi 32P-NAD+ for 20 min at room temperature and ADP-ribosylation was detected by autoradiography after SDS–PAGE. Domains in the TIPARP protein are shown: Zn, CCCH-type Zn finger domain; WWE, tryptophan–tryptophan–glutamate module; PARP, poly-ADP-ribose polymerase catalytic domain. GCB staining shows levels of proteins loaded into SDS–PAGE gel. (C) Auto-ADP-ribosylation of in vitro translated TIPARP full-length and truncated proteins. The indicated TIPARP deletion proteins were in vitro translated using the TNT Coupled Reticulocyte Lysate System in the presence of 35S-methionine. Five percent of the reaction was incubated with or without NAD+ as described in the Materials and Methods. The data are representative of three independent experiments.
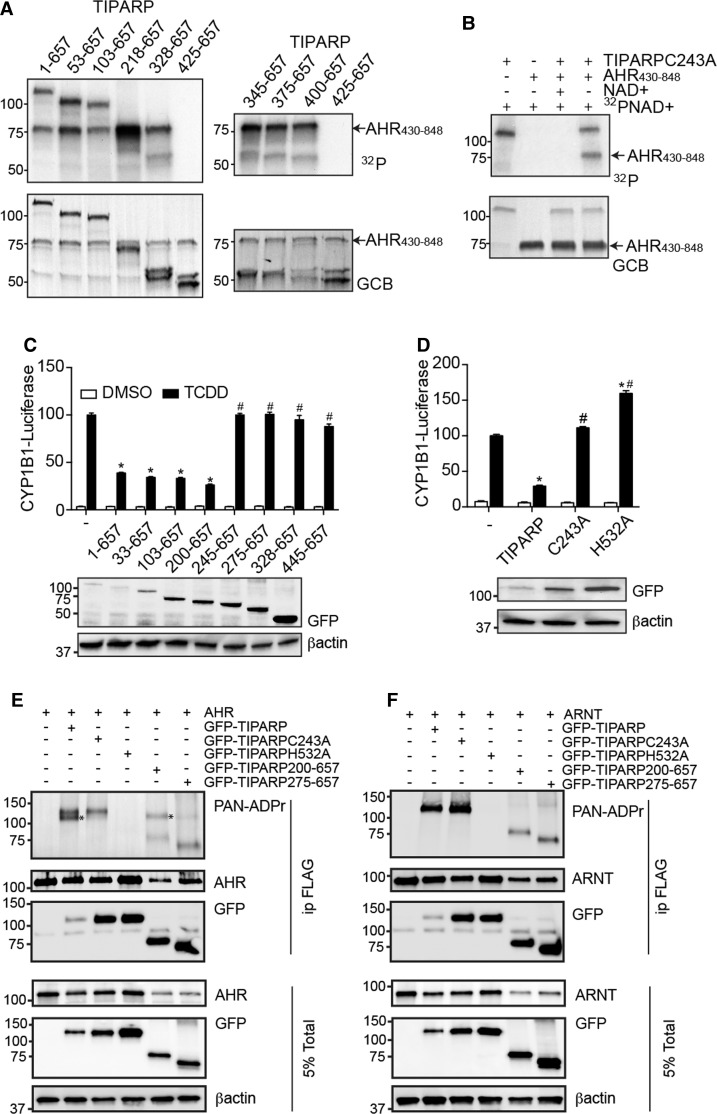
To identify the ADP-ribosylated region of TIPARP, we tested the auto-ADP-ribosylation activity of various truncated GST-TIPARP proteins. The auto-MARylation activity was similar for full length and 53–657, 103–657 and 218–657 N-terminal-deleted TIPARP proteins, but markedly reduced for TIPARP 328–657 (Figure 2B). No auto-MARylation activity was detected for TIPARP 425–657. To more finely map the minimum auto-MARylation region of TIPARP, we tested the auto-MARylation activity of TIPARP 345–657, 375–657, and 400–657. The results from these studies mapped the minimum catalytically active region of TIPARP to amino acids 400–657 (Figure 2B). To confirm these findings, we used a protein shift assay in which TIPARP was in vitro translated with 35S-methionine in the presence or absence on NAD+. Auto-MARylation activity of TIPARP or that of the different truncations was evident by a small shift or smear in the migration of the proteins during SDS separation (Figure 2C). This is in contrast to the PARylation activity of PARP1, which resulted in a large increase in protein mass evidenced by the large shift in migration of 35S-labeled PARP1 after incubation with NAD+ (Supplementary Figure S1A). No shift or smear was observed with the catalytic mutant of TIPARP H532A (Supplementary Figure S1B). In support of the results from the ADP-ribosylation assays using 32P-NAD+, a small shift or smear in the migration of the proteins was evident for full length and all truncated versions of TIPARP that were tested with the exception of TIPARP 445–657.
TIPARP interacts with and mono-ADP-ribosylates other TIPARP molecules
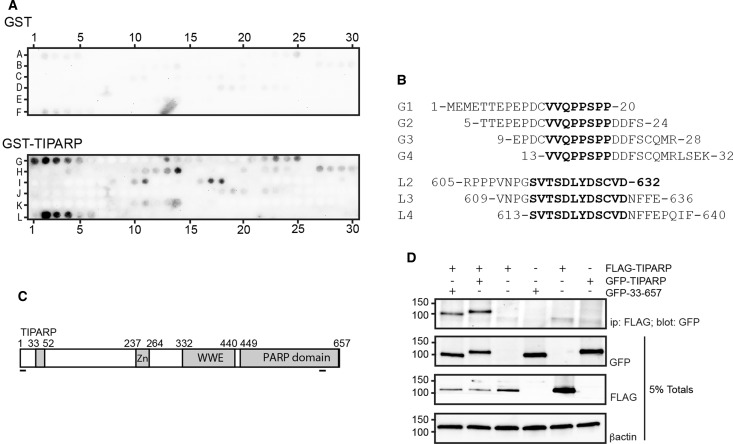
Since we mapped the auto-ribosylation domain to amino acids 1–400 of TIPARP, we examined specific interaction sites between bacterially expressed GST-TIPARP and TIPARP using peptide arrays covering the entire TIPARP protein sequence. Binding events that included at least three consecutive peptides or spots were considered positive interactions. No binding of GST to spotted TIPARP was observed. However, GST-TIPARP bound to two sequences of spotted TIPARP (Figure 3A,B). The first interaction site mapped to sequences within the N-terminus (amino acids 1–32) with an overlap sequence of 13VVQPPSPP20. The other interaction site mapped to a sequence within the catalytic domain and had the overlap sequence of 613SVTSDLYDSCVD632 (Figure 3C). These findings suggested that these sequences may play a role in the self-association of two TIPARP molecules. To test this, we determined whether TIPARP (1–657) interacted with itself or TIPARP 33–657. However, the loss of amino acids 1–32 in TIPARP did not affect its ability to interact with a full-length TIPARP molecule (Figure 3D). Deletion of the sequence of 613SVTSDLYDSCVD632 was not tested, because it would disrupt the catalytic domain and potentially affect TIPARP catalytic activity.
Figure 3. TIPARP interacts with specific sequences in its N-terminus and catalytic domains.
(A) TIPARP interacts with sequences in its N-terminal region and its catalytic domain. 20-mer peptides with four amino acid offset covering the entire sequence of TIPARP were spotted on a membrane that was incubated with GST or GST-TIPARP that were detected by immunoblotting for GST. (B) The peptide sequences that specifically bound GST-TIPARP are shown in bold font. (C) Schematic showing the relative position of the interacting peptide sequences to known functional in TIPARP. (D) COS1 cells were transfected with FLAG-TIPARP1-657 and GFP-TIPARP (full-length; 1–657) or GFP-TIPARP 33–657. Cell extracts were prepared 24 h later and immunoprecipitated with anti-FLAG followed by western blotting with the indicated antibodies. The data are representative of three independent experiments.
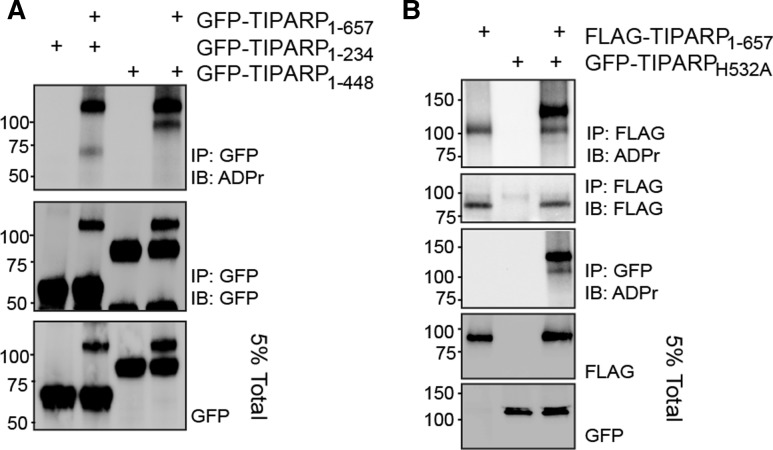
We next determined whether TIPARP auto-MARylation activity occurred in cis (modification of the same TIPARP protein) and/or trans (one TIPARP molecule modifies a different TIPARP molecule). For these assays, we transfected GFP-TIPARP with either GFP-TIPARP 1–234 or GFP-TIPARP 1–448, C-terminal deletion mutants that do not contain a catalytic domain. Both TIPARP truncation mutants were MARylated in the presence of full-length GFP-TIPARP (Figure 4A). No ADP-ribosylation was detected in the absence of GFP-TIPARP (Figure 4A). To further verify these findings, we tested whether FLAG-TIPARP could modify the catalytically inactive GFP-TIPARP H532A mutant. The addition of the GFP tag was necessary to distinguish WT TIPARP from TIPARP H532A. The findings reveal that GFP-TIPARP H532A is efficiently MARylated in the presence of WT TIPARP, which was observed in immunoprecipitations for FLAG or GFP (Figure 4B). These data also show that TIPARP is able to modify other TIPARP molecules.
Figure 4. TIPARP mono-ADP-ribosylates itself and other TIPARP molecules.
(A) COS1 cells were transfected with GFP-TIPARP1-657 and GFP-TIPARP1-234 or GFP-TIPARP1-448, or with FLAG-TIPARP1-657 and (B) GFP-TIPARP H532A. Cell extracts were prepared 24 h later and immunoprecipitated with anti-GFP followed by western blotting with anti-ADPr reagent, which binds to mono-ADP-ribosylated proteins. The data are representative of three independent experiments.
Inhibition of TIPARP by various PARP inhibitors
We next examined the ability of a panel of known PARP inhibitors to block TIPARP catalytic activity using concentration ranges previously reported to inhibit PARP1 auto-PARylation activity [39]. A comparison of the inhibition of PARP1 and TIPARP by known PARP inhibitors revealed that KU0058948 and PJ34 were more selective and potent inhibitors of PARP1 than of TIPARP. KU0058948 treatment resulted in no more than 50% inhibition of TIPARP activity at the highest concentration, and thus, an IC50 value could not be determined. The calculated IC50 value for PJ34 was approximately 500-fold higher for TIPARP compared with PARP1. However, the IC50 values for olaparib and 3-ABA were only about fivefold higher for TIPARP than for PARP1 (Table 1 and Supplementary Figure S2A–D).
Table 1. Comparison of the inhibition activity of various PARP inhibitors between PARP1 and bacterially expressed GST-TIPARP.
| Inhibitor | IC50 (M) | |
|---|---|---|
| PARP1 | TIPARP | |
| 3ABA | 1.7 ± 0.4 × 10−4* | 9.8 ± 4.0 × 10−4 |
| PJ34 | 5.5 ± 1.4 × 10−8* | 2.6 ± 1.1 × 10−5 |
| Olaparib | 1.4 ± 0.04 × 10−7* | 8.0 ± 2.0 × 10−7 |
| KU0058948 | 7.6 ± 0.4 × 10−8 | n.d. |
Abbreviations: n.d., not determined because greater than 50% inhibition was not achieved.
Significantly different compared with treatment matched TIPARP.
Cysteines and/or acidic amino acids are putative ADP-ribosylated targets in TIPARP
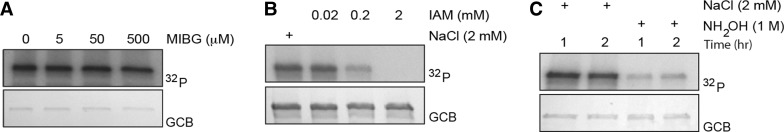
Since the chemical bonds between ADP-ribose and various amino acids are differentially sensitive to acidic, alkaline, and/or other treatments, we investigated the chemical stability of auto-modified TIPARP. The arginine-specific mono-ADP-ribosyltransferase inhibitor, MIBG [40], did not affect the enzymatic activity of TIPARP (Figure 5A), indicating that arginine residues are most likely not modified in TIPARP. The catalytic activity of TIPARP was sensitive to IAM, suggesting that cysteine residues are modified or are important for enzymatic activity (Figure 5B). We tested the ability of HgCl2 treatment, which cleaves the linkage between cysteine and ADP-ribose, to affect TIPARP activity. However, HgCl2 treatment reduced TIPARP stability and altered its migration in SDS–PAGE, making it difficult to draw conclusions about the impact of HgCl2 on cysteine ADP-ribosylation under these conditions (data not shown). The catalytic activity of TIPARP was sensitive to hydroxylamine treatment, which hydrolyses ester bonds between ADP-ribose and acidic amino acids (Figure 5C), indicating that ADP-ribosylation occurs on glutamate and/or aspartate residues in TIPARP under our experimental conditions.
Figure 5. Sensitivity of auto-ADP-ribosylation of TIPARP to different treatment conditions.
(A) GST-TIPARP was incubated with different concentrations of MIBG for 5 min and auto-ribosylation was initiated with the addition of 32P-NAD+ as described in the Materials and Methods. (B) GST-TIPARP was incubated with 32P-NAD+ for 20 min at room temperature and then treated with 1 M neutral hydroxylamine (NH2OH) or NaCl for 1 or 2 h. (C) GST-TIPARP was incubated with different concentrations of IAM or 2 mM NaCl for 30 min at 37°C and the reaction was initiated with the addition of 32P-NAD+ as described in Materials and Methods. ADP-ribosylation was detected by autoradiography after SDS–PAGE followed by transfer onto a PVDF membrane. GCB staining shows levels of proteins loaded into SDS–PAGE gel. 3-ABA, 3-aminobemzamide. The data are representative of at least two independent experiments.
TIPARP mono-ADP-ribosylates and represses AHR
We have reported that TIPARP functions as part of a negative feedback loop to repress AHR activity through a mechanism that depends on TIPARP's catalytic activity. TIPARP has been reported to mono-ADP-ribosylate AHR, but this has not been fully characterized [17]. To this end, we determined if the TIPARP deletion mutants retained their ability to modify AHR by incubating them with GST-AHR 430–848. All of the different TIPARP deletion mutants, with the exception of the auto-ribosylation deficient variant, TIPARP 425–657, mono-ADP-ribosylated AHR (Figure 6A). Consistent with these data, a point mutation in the Zn finger domain (C243A) did not prevent its ability to mono-ADP-ribosylate AHR in vitro (Figure 6B). Transfection of a series of N-terminal deletion mutants of TIPARP revealed that deletion of 1–199 did not reduce, while removal of 1–244 abolished the ability of TIPARP to repress AHR-regulated CYP1B1 luciferase activity (Figure 6C). This region overlapped with TIPARP's Zn finger domain. However, point mutants C243A and a catalytically inactivating H532A mutation abolished the ability of TIPARP to repress AHR-regulated CYP1B1 reporter gene activity [16]. Although TIPARP catalytic activity was required to repress AHR, the in vitro ribosylation assays showed that many of the truncations and the C243A TIPARP mutant modified AHR, but did not repress AHR activity (Figure 6A–D). To reconcile the discrepancies between the ability of TIPARP to mono-ADP-ribosylate AHR and TIPARP's ability to repress AHR, we tested whether TIPARP, TIPARP C243A, or TIPARP H532A mono-ADP-ribosylated AHR in cells. Immunoprecipitated AHR was probed with the anti-PAN-ADPr reagent that detects ADP-ribosylated proteins. Mono-ADP-ribosylation was detected on both ectopically expressed AHR and TIPARP. TIPARP C243A retained its auto-ADP-ribosylation activity, but failed mono-ADP-ribosylate AHR. No ADP-ribosylation of AHR or TIPARP was detected in cells transfected with the catalytic mutant TIPARP H532A (Figure 6E). In agreement with previous in vitro ribosylation assay studies [17], TIPARP failed to ADP-ribosylate ARNT when tested under similar conditions (Figure 6F). Combined with the cell localization data (Figure 1), these data suggest that TIPARP C243A fails to mono-ADP-ribosylate AHR in cells due to altered cellular localization rather than reduced activity.
Figure 6. TIPARP mono-ADP-ribosylates AHR in vitro.
(A,B) One microgram of GST-tagged TIPARP protein was incubated with 2 µg of GST-AHR 430–848 and 2 µCi 32P-NAD+. Mono-ADP-ribosylation of proteins was detected by autoradiography after SDS–PAGE and levels of proteins loaded were visualized by GCB staining. Repression of TCDD-induced CYP1B1-luc reporter gene activity by (C) TIPARP N-terminal truncations, (D) TIPARP zinc-finger (C243A) and catalytic mutant (H532A) point mutants. Full-length TIPARP (1–657), TIPARP truncations, or point mutant constructs were co-overexpressed with pCYP1B1-luc in HuH-7 cells. Twenty-four hours after transfection, cells were treated with DMSO or 10 nM TCDD for 24 h. Corresponding western blots of transfected HuH7 cells of GFP-TIPARP proteins and βactin are shown below reporter gene data. Data were presented as means ± SEM from three independent experiments. *P < 0.05 compared with non-TIPARP-transfected cells treated with TCDD. #P < 0.05 compared with TIPARP-transfected cells treated with TCDD. (E) In cell mono-ADP-ribosylation of AHR by TIPARP, and different TIPARP point mutants and truncation variants. Mono-ADP-ribosylated AHR and TIPARP was detected by anti-PAN-ADPr reagent. (F) TIPARP does not mono-ADP-ribosylate ARNT in transient transfected COS1 cells. The data (E,F) are representative of three independent experiments.
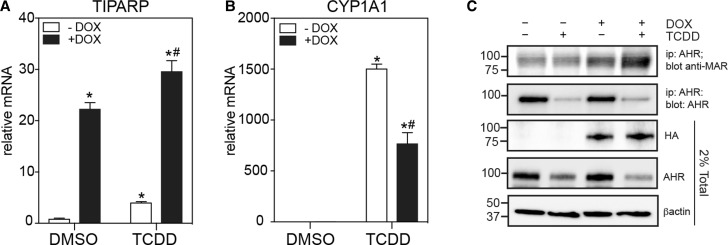
To determine whether increased TIPARP expression correlates with increased AHR mono-ADP-ribosylation and reduced TCDD-induced and AHR-regulated CYP1A1 transcription, we created a Tet-ON TIPARP overexpression cell line, in which the expression of HA-tagged TIPARP is regulated by doxycycline. Treatment with 1.5 µg/ml dox resulted in a 20-fold increase in TIPARP mRNA levels (Figure 7A). TCDD-induced CYP1A1 mRNA levels were reduced in the presence of dox compared with TCDD alone (Figure 7B). We next tested the ability of TIPARP to mono-ADP-ribosylate endogenous AHR in the presence and absence of dox and after treatment with DMSO of TCDD. Immunoprecipitated AHR was probed with the anti-mono-ADP-binding reagent (anti-MAR). TCDD treatment resulted in proteolytic degradation of AHR, which was reflected by the reduced AHR protein in the total cell extracts and after immunoprecipitation. Despite this reduction, increased mono-ADP-ribosylation was detected in the presence of dox for both the DMSO and TCDD treatments compared with no dox (Figure 7C). Multiple bands were detected as a smear, which could represent AHR degradation products and/or other MARylated proteins, including TIPARP since it interacts with AHR. These data show that TIPARP's ability to repress AHR activity is correlated with its ability to mono-ADP-ribosylate AHR in cells.
Figure 7. Increased TIPARP expression increases mono-ADP-ribosylation of AHR and represses AHR signalling.
MCF7 cells expressing a Tet-ON regulated HA-tagged TIPARP were cultured in the presence or absence of dox and treated with DMSO or 10 nM of TCDD for 20 h. The mRNA expression levels of (A) TIPARP and (B) CYP1A1 were determined as described in the Materials and Methods. (C) The increased mono-ADP-ribosylation of AHR in the presence of increased expression of TIPARP. Mono-ADP-ribosylation of immunoprecipitated AHR by TIPARP was detected by anti-MAR reagent. The data are representative of three independent experiments. *P < 0.05 compared with no dox DMSO-treated cells. #P < 0.05 compared with TIPARP-transfected cells treated with TCDD.
Identification of mono-ADP-ribosylated peptides in TIPARP and AHR
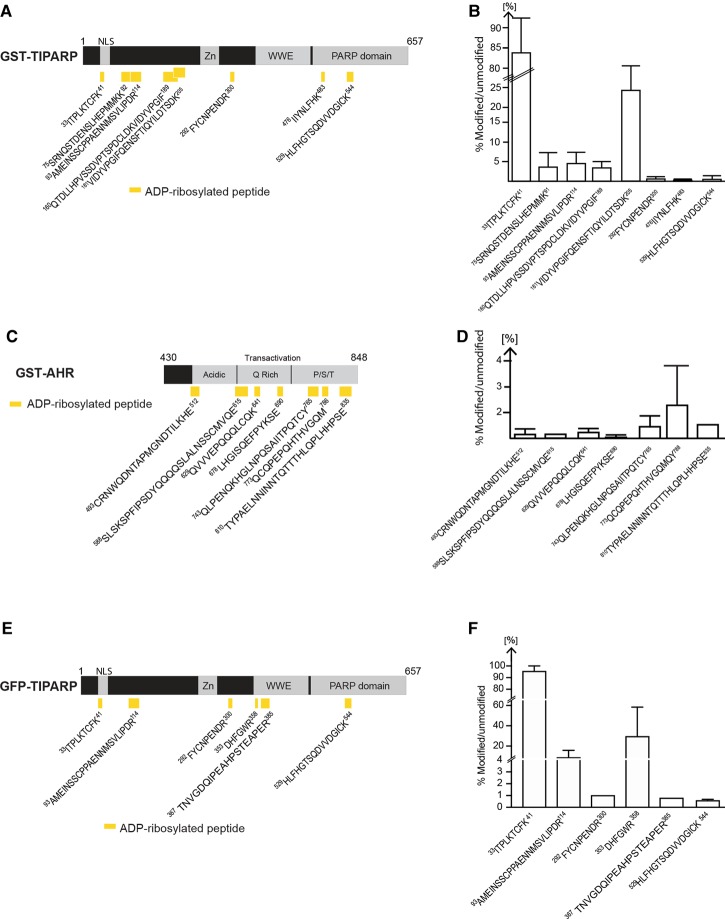
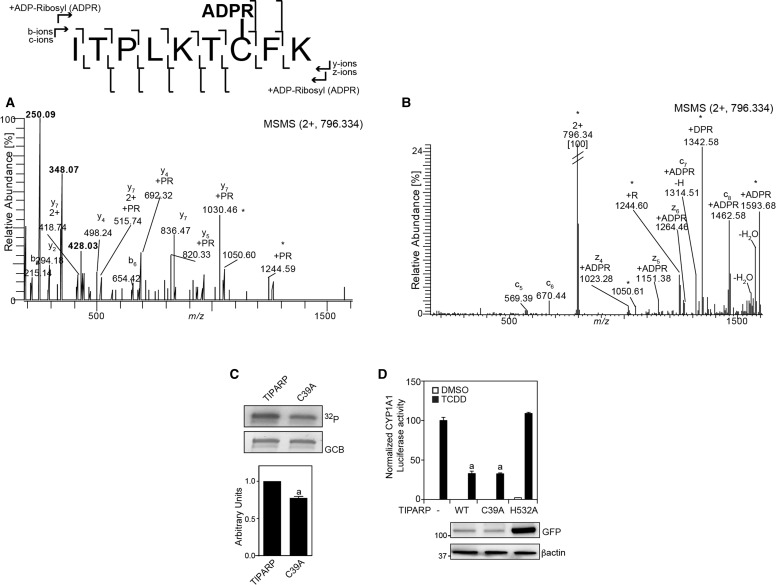
To determine the location of the ADP-ribosylated sites in TIPARP, we analyzed trypsin or chymotrypsin-digested ADP-ribosylated TIPARP peptides on an Orbitrap XL by mass spectrometry using HCD fragmentation and investigated the generated MS2 peptide spectra for the presence of ADP-ribosylation. Using this approach, we identified eight peptides in TIPARP that were modified by a single ADP-ribose moiety on one of their residues (Figure 8A and Supplementary Figure S3A–H). Six of the peptides were located within the first 300 amino acids of TIPARP and two peptides were located within the catalytic domain. The percentage of modified versus unmodified peptide was calculated from the total peptide population that represented all peptides formed, including peptide forms with sequence overlap due to missed trypsin cleavage sites, sodium adducts, and oxidized forms (Figure 8B). The MARylated peptides showed varying levels of ADP-ribosylation with the smallest peptide that corresponded to 33ITPLKTCFK41 showing the highest relative level of modification.
Figure 8. ADP-ribosylated peptides in bacterially expressed GST-TIPARP.
(A) Schematic representation of the domain structure of TIPARP and the location of the ADP-ribosylated peptides (yellow rectangles) identified by MS in GST-TIPARP. Peptide sequences are numbered according to the unmodified full-length protein. (B) Mono-ADP-ribosylated peptides in GST-TIPARP show varying levels of ADP-ribosylation. (C) Schematic representation of the domain structure of AHR and the location of the ADP-ribosylated peptides (yellow rectangles) identified by MS in GST-AHR. (D) Mono-ADP-ribosylated peptides in AHR show varying levels of ADP-ribosylation. (E) Schematic representation of the domain structure of TIPARP and the location of the mono-ADP-ribosylated peptides (yellow rectangles) identified by MS in immunoprecipitated GFP-TIPARP. (F) ADP-ribosylated peptides in GFP-TIPARP show varying levels of ADP-ribosylation. The bar graphs (B,D,F) show the relative abundance of the ADP-ribosylated peptides. The percentage of modified versus unmodified peptide was calculated from the total peptide population that represents all peptide forms including peptide forms with sequence overlap due to missed trypsin cleavage sites, sodium adducts, and oxidized forms. Data are expressed as means ± SEM from at least two independent experiments.
We then used the same strategy to identify the location of ADP-ribosylated sites in AHR amino acids 430–848. We identified seven MARylated peptides in AHR that were spread across the acidic, Q-rich, and P/S/T regions in its transactivation domain (Figure 8C and Supplementary Figure S4A–G). The percentage of modified versus unmodified peptide was calculated as above and is shown in Figure 8D. The mono-ADP-ribosylated peptides in AHR had varying levels of modification. However, the overall level of AHR MARylation was lower than that observed for TIPARP. A recent study demonstrated that mono-PARPs auto-MARylated glutamate, aspartate, lysine, and cysteine residues [4]. All of the MARylated peptides identified in TIPARP and AHR contained at least one of these residues.
Since it is possible that the ADP-ribosylated peptides identified using bacterially expressed TIPARP and in vitro ADP-ribosylation assays might not represent those present in TIPARP isolated from mammalian cells, we immunoprecipated overexpressed GFP-TIPARP from COS1 cells and analyzed trypsin- or chymotrypsin-digested ADP-ribosylated GFP-TIPARP peptides by mass spectrometry. We identified six peptides, four of which overlapped with those identified in GST-TIPARP (Figure 8E,F and Supplementary Figure S5A–F). This suggests that TIPARP auto-ADP-ribosylation exhibits a certain level of plasticity and/or that the ADP-ribosylation of some peptides is influenced by the assay conditions and source of the enzyme. However, in agreement with our observations for GST-TIPARP, a peptide containing C39 (33-ITPLKYCFK-41) was also the most highly modified one in GFP-TIPARP, with greater than 90% modified compared with unmodified.
Identification of mono-ADP-ribosylated cysteine 39 in GST-TIPARP
To identify the modified amino acid residues, we used ETD on targeted ADP-ribosylated trypsin generated peptides. For the ADP-ribosylated peptide 33ITPLKTCFK41, the ETD spectrum provided the necessary peptide backbone fragmentation to assign Cys39 as the site for ADP-ribosylation (Figure 9A,B). The modification was located by the detection of fragment ions at m/z 1314.51 and at m/z 1462.58 corresponding to the c7 (ITPLKTC) and c8 (ITPLKTCF) ions, respectively. These ions were the only ions of the c-series from c5 to c8 with mass increases corresponding to ADP-ribose (541.061 Da). No ion corresponding to the peptide with attached ADP-ribose could be detected N-terminally of c7 (i.e. within the peptide 33-ITPLKT-38). Moreover, from the analysis of the z-ion series, we detected an ion at m/z 1023.26 corresponding to z4 (TCFK) with an ADP-ribosylation. The TIPARP peptide 33ITPLKTCFK41 was therefore ADP-ribosylated at Cys39. Moreover, nLC-MS2 of a C39A mutant showed only unmodified 33ITPLKTAFK41 peptide (data not shown). An approximate 20% decrease in auto-ADP-ribosylation of GST-TIPARP after incubation with 32P-NAD+ was observed in the C39A mutant compared with wild-type protein (Figure 9C), confirming that this residue was modified in TIPARP.
Figure 9. TIPARP auto-ADP-ribosylates cysteine 39.
(A) HCD MS2 spectrum of the trypsin generated [M+2H+] precursor ion at m/z 796.334 corresponding to the ADP-ribosylated peptide 32ITPLKTCFK41. The ADP-ribose specific reporter ions can be seen in the low mass area at m/z 428.03, m/z 348.07, and m/z 250.09. (B) ETD MS2 spectrum of the trypsin generated [M+2]2+ precursor ion at m/z 796.334 corresponding to the ADP-ribosylated peptide 32ITPLKTCFK41shows the site of modification by detection of fragment ions c7 and c8, the only c-ions with a mass addition of 541.061 Da corresponding to an ADP-ribosylation. (C) Mutation of cysteine 39 to alanine reduced auto-ADP-ribosyltransferase activity of TIPARP. One µg of GST-tagged TIPARP or TIPARPC39A was incubated with 2 µCi 32P-NAD+ for 20 min at room temperature and auto-ADP-ribosylation was detected by autoradiography after SDS–PAGE followed by transfer to a PVDF membrane. A representative experiment is shown and densitometric (ImageJ) analysis was used to quantify changes in TIPARP autoribosylation activity compared with wild-type after normalization to GCB protein levels. (D) HuH7 cells were transfected with CYP1A1-Luc reporter and pcDNA-TIPARP (wild type), C39A or H532A as described in Materials and Methods. Twenty-four hours after transfection, cells were treated with DMSO or 10 nM TCDD for 24 h. Data are expressed as means ± SEM from three independent experiments. aP < 0.0001 compared with TCDD-treated no TIPARP (−).
Because we previously reported that TIPARP represses AHR activity [16], we determined the ability of TIPARP-C39A to repress TCDD-induced AHR-regulated CYP1A1 luciferase activity. No significant differences were observed in the ability of TIPARP C39A to repress CYP1A1 luciferase activity compared with wild-type TIPARP (Figure 9D). The introduction of the C39A mutation did not affect the cellular localization of TIPARP (data not shown). The catalytic mutant, TIPARP H532A, failed to reduce CYP1A1 luciferase activity (Figure 9D). Taken together, our data suggest that C39 is mono-ADP-ribosylated in TIPARP, but this modification is independent of its ability to repress AHR.
Discussion
In this study, we characterized the cellular localization and biochemical properties of TIPARP. TIPARP is a MARylating PARP that exhibits auto- and hetero-MARylation activity [16,17,38]. We identified an NLS sequence in TIPARP's N-terminus that works together with its Zn finger domain to maintain TIPARP in a predominantly nuclear location, consistent with its role as a regulator of transcription [16]. We showed that TIPARP's ability to repress AHR activity depends on its ability to MARylate AHR. We also identified multiple ADP-ribosylated peptides in TIPARP and AHR and mapped one of the mono-ADP-ribosylation sites to C39 of TIPARP.
Similar to previous studies investigating the catalytic activity of PARPs, we used recombinant proteins overexpressed as GFP or GST fusions. Although the GFP tag does not influence the ability of TIPARP to function as a transcriptional repressor or affect its catalytic activity [19,38], we cannot completely rule out that the catalytic activity of bacterially expressed GST-TIPARP differs from that of overexpressed and endogenous TIPARP. Bacterially expressed TIPARP failed to ADP-ribosylate C-terminal deletion mutants in vitro; however, both TIPARP deletion mutants were ADP-ribosylated by full-length TIPARP when transiently transfected into mammalian cells. Whether these differences were due to reduced or altered catalytic activity of bacterially expressed TIPARP or the result of altered folding of the truncated TIPARP proteins was not determined and cannot be ruled out.
TIPARP, together with PARP12 (ARTD12) and PARP13 (ARTD13), belongs to a subfamily of PARPs that contain a CCCH-type zinc finger that has been shown to bind viral and cytoplasmic RNAs [41,42]. PARP13 is catalytically inactive but regulates viral RNA proliferation by binding and degrading retroviral RNA via its four zinc finger domains [43]. PARP12 localizes to stress granules where it was found to block mRNA translation [44]. Both processes require PARP12's five zinc finger domains and its catalytic domain [44]. Similar to PARP13, TIPARP also regulates viral replication by binding to Sindbis virus (SINV) RNA via its zinc finger domain [20]. Deletion of the catalytic domain did not prevent TIPARP–SINV RNA interaction, showing that this interaction was independent of TIPARP's catalytic activity. TIPARP localizes in the nucleus, but exhibits weak cytosolic staining in a cell-type dependent manner [19,38]. TIPARP's cytosolic staining is also increased during viral infection and during ROS-induced mitochondrial damage [20,29]. Our data show that TIPARP's lone zinc finger domain regulated its subcellular localization and ability to repress AHR activity. Whether this is through specific interactions with nuclear RNAs or through other mechanism remains to be determined.
TIPARP was less sensitive to inhibition by some PARP inhibitors, which is consistent with PARP inhibitor profiling where the majority of PARP inhibitors interacts with several PARP family members but show a greater preference for PARP1–4 compared with mono-ADP-ribosylating PARPs [45]. However, small molecule inhibitors that show preference for PARP15 (ARTD7), PARP16 (ARTD8), or PARP10 (ARTD10) have been described [46]. Caution should be used when interpreting data generated using current PARP inhibitors to probe for TIPARP function. Identifying TIPARP-selective inhibitors are needed to determine its cellular protein targets and the importance of its catalytic activity for its biological and cellular action. Molecular modeling approaches using crystal structures of several MARylating PARP members will be important in identifying such inhibitors. Alternatively, using targeted gene editing to create catalytically mutant TIPARP variants followed by proteomic studies will help to determine the importance of TIPARP catalytic cell function and physiology, as well as to identify the TIPARP-dependent mono-ADP-ribosylome using immunoprecipitation methods [15,47,48]. These approaches could be complemented by chemical genetic proteomics strategies to determine TIPARP-specific targets to better understand its cellular function.
Consistent with PARP1 and other MARylating PARPs, acidic, lysine, and cysteine residues were modified in TIPARP. The most strongly auto-modified region mapped to the N-terminal half of TIPARP, which agreed well with the location of the modified peptides identified by mass spectrometry. Our findings suggest that TIPARP is able to modify other TIPARP molecules in trans (TIPARP molecule modifies another TIPARP molecule); however, we cannot exclude the possibility that TIPARP also modifies itself in cis (TIPARP molecule modifies itself). TIPARP and PARP10 (ARTD10) auto-modify similar residues including cysteines [4], but modify different cellular proteins. TIPARP ADP-ribosylates AHR, LXR, and TBK1 [17,18,29], while PARP10 modifies many cellular targets including NF-κ-B essential modulator (NEMO) and glycogen synthase kinase 3 beta (GSK3β) [14,49].
The identification of C39 as being auto-modified in TIPARP confirms that cysteine is targeted by mono-ADP-ribosylating PARPs [4]. Mutation of cysteine 39 to alanine caused a slight but significant reduction in auto-ribosylation activity, though it did not affect the ability of TIPARP to repress AHR transcriptional activity. This is not unexpected since the N-terminal deletion mutant TIPARP 200–657 is as effective as full-length TIPARP at repressing AHR activity [16]. A catalytically active TIPARP and an intact zinc finger are required to repress transcription, but whether it needs to be auto-modified remains an open question. We were unable to identify the specific modified residues in the remaining five peptides, which prevented us from trying to create a catalytically active TIPARP in terms of hetero-ribosylation activity but one that lacks auto-modification activity. Mutation of C39A does not affect TCDD-induced reporter gene activity, but whether it affects increased sensitivity to TCDD-induced toxicity observed in Tiparp null mice remains to be determined [17,50]. We cannot exclude the possibility that the ADP-ribosylation of C39 is important for other properties of TIPARP activity that were not tested in our study.
It is also possible that the ADP-ribosylated residues we identified using bacterially expressed protein may differ from modified residues in endogenous TIPARP. However, a recent whole-cell ADP-ribosylome analysis of HeLa cells also identified mono-ADP-ribosylation of C39 in endogenous TIPARP [47]. Moreover, we also identified a highly mono-ADP-ribosylated peptide containing C39 from immunopreciptated GFP-TIPARP overexpressed in mammalian cells. Taken together, this suggests a role for mono-ADP-ribosylation of C39 in regulating TIPARP's function. Creation of a mouse model harboring a TIPARP C39A mutation will be needed to determine the physiological important of mono-ADP-ribosylation of this residue. Moreover, advances in mass spectrometric approaches and enrichment strategies will be needed to improve the identification and mapping of modified residues in endogenous TIPARP and its target proteins, as well as be used to characterize the TIPARP-dependent ADP-ribosylome.
There is increasing interest in understanding the importance of MARylating PARPs in regulating diverse cell functions such as membrane organelle regulation, signal transduction, the unfolded protein response, stress responses, transcription, and DNA damage [51]. The consequences of auto- and hetero-MARylation on protein function are still not well understood. Our findings characterize the cellular localization and mono-ADP-ribosyltransferase activity of TIPARP and identify cysteine as an ADP-ribosylated residue targeted by this enzyme. Identifying TIPARP-specific recognition sequences, selective inhibitors, and intracellular protein targets as well as using genetically modified mouse models will be important future steps needed to fully characterize the importance of this enzyme and to determine how TIPARP regulates the activity of transcription factors, such as AHR.
Acknowledgements
The authors thank all members of the Matthews and Grant laboratories for their help with the preparation of the manuscript. The authors thank Prof. Michael Denison (University of California Davis, CA) for providing the pCYP1A1-luciferease (pGudluc) plasmid.
Abbreviations
- 3-ABA
3-aminobenzoamide
- ADPr
ADP-ribose
- AHR
aryl hydrocarbon receptor
- ARNT
aryl hydrocarbon nuclear translocator
- ARTD
ADP-ribosyltransferase diptheria toxin-like
- CYP1A1
cytochrome P450 1A1
- DAPI
4,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- Dox
doxycycline
- ETD
electron transfer dissociation
- FBS
fetal bovine serum
- GCB
Gelcode blue
- GST
glutathione S-transferase
- HA
hemagglutinin
- HCD
higher energy C-cell dissociation
- IAM
iodoacetamide
- Ku-0058948
4-[[4-fluoro-3-[(hexahydro-1H-1,4-diazepin-1-yl)carbonyl]phenyl]methyl]-1(2H)-phthalazinone
- LXRs
liver X receptors
- MARylation
mono-ADP-ribosylation
- MIBG
meta-iodobenzylguanidine
- MS
mass spectrometry
- NAD+
nicotinamide adenine dinucleotide
- nLC
nanoflow liquid chromatography
- NLS
nuclear localization signal
- Olaparib
4-[(3-[(4-cyclopropylcarbonyl)piperazin-4-yl]carbonyl)-4-fluoropheny]methyl(2H) phthalazin-1-one
- PAR
poly-ADP-ribose
- PARylation
poly-ADP-ribosylation
- PARP
poly-ADP-polymerase
- PEST
penicillin–streptomycin
- PJ34
N-(6-oxo-5,6-dihydrophenanthridin-2-yl)(N,N-dimethylamino) acetamide
- PVDF
polyvinylidene difluoride membrane
- TBST
Tris-buffered saline + Tween 20
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TIPARP
TCDD-inducible poly-ADP-ribose polymerase
- WWE
tryptophan–tryptophan–glutamate
Author Contribution
A.G., C.B., S.V.S., G.G., D.H., L.M., S.A., T.C., L.T., and J.M. carried out most of the experiments and analyzed the data. A.M. and J.H.A. performed the mass spectrometry experiments. D.M.G., H.I.N. and J.M. designed the study. J.M. supervised the project and wrote the manuscript with critical input from all co-authors.
Funding
This work was supported by Canadian Institutes of Health Research (CIHR) operating grants [MOP-494265 and MOP-125919]; CIHR New Investigator Award; an Early Researcher Award from the Ontario Ministry of Innovation [ER10-07-028]; the Johan Throne Holst Foundation; Novo Nordic Foundation; and the Norwegian Cancer Society to J.M. This work was also funded by grants from the Johan Throne Holst Foundation; and the Novo Nordic Foundation to H.I.N.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Supplementary Material
References
- 1.Hassa P.O. and Hottiger M.O. (2008) The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13, 3046–3082 10.2741/2909 [DOI] [PubMed] [Google Scholar]
- 2.Hottiger M.O., Hassa P.O., Lüscher B., Schüler H. and Koch-Nolte F. (2010) Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 35, 208–219 10.1016/j.tibs.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 3.Cohen M.S. and Chang P. (2018) Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat. Chem. Biol. 14, 236–243 10.1038/nchembio.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vyas S., Matic I., Uchima L., Rood J., Zaja R., Hay R.T. et al. (2014) Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 5, 4426 10.1038/ncomms5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer-Ficca M.L., Meyer R.G., Coyle D.L., Jacobson E.L. and Jacobson M.K. (2004) Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 297, 521–532 10.1016/j.yexcr.2004.03.050 [DOI] [PubMed] [Google Scholar]
- 6.Barkauskaite E., Jankevicius G., Ladurner A.G., Ahel I. and Timinszky G. (2013) The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 280, 3491–3507 10.1111/febs.12358 [DOI] [PubMed] [Google Scholar]
- 7.Ryu K.W., Kim D.S. and Kraus W.L. (2015) New facets in the regulation of gene expression by ADP-ribosylation and poly(ADP-ribose) polymerases. Chem. Rev. 115, 2453–2481 10.1021/cr5004248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drew Y. and Plummer R. (2009) PARP inhibitors in cancer therapy: two modes of attack on the cancer cell widening the clinical applications. Drug Resist. Updat. 12, 153–156 10.1016/j.drup.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 9.Dizdar O., Arslan C. and Altundag K. (2015) Advances in PARP inhibitors for the treatment of breast cancer. Expert Opin. Pharmacother. 16, 2751–2758 10.1517/14656566.2015.1100168 [DOI] [PubMed] [Google Scholar]
- 10.Chapman J.D., Gagné J.P., Poirier G.G. and Goodlett D.R. (2013) Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 12, 1868–1880 10.1021/pr301219h [DOI] [PubMed] [Google Scholar]
- 11.Hottiger M.O. (2015) Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu. Rev. Biochem. 84, 227–263 10.1146/annurev-biochem-060614-034506 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Wang J., Ding M. and Yu Y. (2013) Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods 10, 981–984 10.1038/nmeth.2603 [DOI] [PubMed] [Google Scholar]
- 13.Verheugd P., Forst A.H., Milke L., Herzog N., Feijs K.L., Kremmer E. et al. (2013) Regulation of NF-kappaB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat. Commun. 4, 1683 10.1038/ncomms2672 [DOI] [PubMed] [Google Scholar]
- 14.Carter-O'Connell I., Jin H., Morgan R.K., Zaja R., David L.L., Ahel I. et al. (2016) Identifying family-member-specific targets of mono-ARTDs by using a chemical genetics approach. Cell Rep. 14, 621–631 10.1016/j.celrep.2015.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels C.M., Ong S.E. and Leung A.K. (2014) Phosphoproteomic approach to characterize protein mono- and poly(ADP-ribosyl)ation sites from cells. J. Proteome Res. 13, 3510–3522 10.1021/pr401032q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacPherson L., Tamblyn L., Rajendra S., Bralha F., McPherson J.P. and Matthews J. (2013) 2,3,7,8-tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 41, 1604–1621 10.1093/nar/gks1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed S., Bott D., Gomez A., Tamblyn L., Rasheed A., MacPherson L. et al. (2015) Loss of the mono-ADP-ribosyltransferase, TIPARP, increases sensitivity to dioxin-induced Steatohepatitis and lethality. J. Biol. Chem. 290, 16824–16840 10.1074/jbc.M115.660100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bindesboll C., Tan S., Bott D., Cho T., Tamblyn L., MacPherson L. et al. (2016) TCDD-inducible poly-ADP-ribose polymerase (TIPARP/PARP7) mono-ADP-ribosylates and coactivates liver X receptors. Biochem. J. 473, 899–910 10.1042/BJ20151077 [DOI] [PubMed] [Google Scholar]
- 19.MacPherson L., Ahmed S., Tamblyn L., Krutmann J., Förster I., Weighardt H. et al. (2014) Aryl hydrocarbon receptor repressor and TiPARP (ARTD14) use similar, but also distinct mechanisms to repress aryl hydrocarbon receptor signaling. Int. J. Mol. Sci. 15, 7939–7957 10.3390/ijms15057939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozaki T., Komano J., Kanbayashi D., Takahama M., Misawa T., Satoh T. et al. (2017) Mitochondrial damage elicits a TCDD-inducible poly(ADP-ribose) polymerase-mediated antiviral response. Proc. Natl Acad. Sci. U.S.A. 114, 2681–2686 10.1073/pnas.1621508114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han B., Zhang Y., Zhang Y., Bai Y., Chen X., Huang R. et al. (2018) Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy 14, 1164–1184 10.1080/15548627.2018.1458173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitlock J.P., Jr (1999) Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 39, 103–125 10.1146/annurev.pharmtox.39.1.103 [DOI] [PubMed] [Google Scholar]
- 23.Poland A. and Knutson J.C. (1982) 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 22, 517–554 10.1146/annurev.pa.22.040182.002505 [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Salguero P.M., Hilbert D.M., Rudikoff S., Ward J.M. and Gonzalez F.J. (1996) Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 140, 173–179 10.1006/taap.1996.0210 [DOI] [PubMed] [Google Scholar]
- 25.Stockinger B., Di Meglio P., Gialitakis M. and Duarte J.H. (2014) The aryl hydrocarbon receptor: multitasking in the immune system. Annu. Rev. Immunol. 32, 403–432 10.1146/annurev-immunol-032713-120245 [DOI] [PubMed] [Google Scholar]
- 26.Roper S.J., Chrysanthou S., Senner C.E., Sienerth A., Gnan S., Murray A. et al. (2014) ADP-ribosyltransferases Parp1 and Parp7 safeguard pluripotency of ES cells. Nucleic Acids Res. 42, 8914–8927 10.1093/nar/gku591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmahl J., Raymond C.S. and Soriano P. (2007) PDGF signaling specificity is mediated through multiple immediate early genes. Nat. Genet. 39, 52–60 10.1038/ng1922 [DOI] [PubMed] [Google Scholar]
- 28.Atasheva S., Frolova E.I. and Frolov I. (2014) Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication. J. Virol. 88, 2116–2130 10.1128/JVI.03443-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada T., Horimoto H., Kameyama T., Hayakawa S., Yamato H., Dazai M. et al. (2016) Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat. Immunol. 17, 687–694 10.1038/ni.3422 [DOI] [PubMed] [Google Scholar]
- 30.MacPherson L., Lo R., Ahmed S., Pansoy A. and Matthews J. (2009) Activation function 2 mediates dioxin-induced recruitment of estrogen receptor alpha to CYP1A1 and CYP1B1. Biochem. Biophys. Res. Commun. 385, 263–268 10.1016/j.bbrc.2009.05.060 [DOI] [PubMed] [Google Scholar]
- 31.Knævelsrud H., Søreng K., Raiborg C., Håberg K., Rasmuson F., Brech A. et al. (2013) Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J. Cell Biol. 202, 331–349 10.1083/jcb.201205129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anonsen J.H., Vik A., Egge-Jacobsen W. and Koomey M. (2012) An extended spectrum of target proteins and modification sites in the general O-linked protein glycosylation system in Neisseria gonorrhoeae. J. Proteome Res. 11, 5781–5793 10.1021/pr300584x [DOI] [PubMed] [Google Scholar]
- 33.Fæste C.K., Moen A., Schniedewind B., Haug Anonsen J., Klawitter J. and Christians U. (2016) Development of liquid chromatography-tandem mass spectrometry methods for the quantitation of Anisakis simplex proteins in fish. J. Chromatogr. A. 1432, 58–72 10.1016/j.chroma.2016.01.002 [DOI] [PubMed] [Google Scholar]
- 34.Małecki J., Aileni V.K., Ho A.Y.Y., Schwarz J., Moen A., Sørensen V. et al. (2017) The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A). Nucleic Acids Res. 45, 4370–4389 10.1093/nar/gkx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox J. and Mann M. (2008) Maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 36.Matic I., Ahel I. and Hay R.T. (2012) Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nat. Methods 9, 771–772 10.1038/nmeth.2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao Z., Gao P. and Liu H.W. (2009) Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 131, 14258–14260 10.1021/ja906135d [DOI] [PubMed] [Google Scholar]
- 38.Vyas S., Chesarone-Cataldo M., Todorova T., Huang Y.H. and Chang P. (2013) A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 4, 2240 10.1038/ncomms3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loseva O., Jemth A.S., Bryant H.E., Schüler H., Lehtiö L., Karlberg T. et al. (2010) PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 285, 8054–8060 10.1074/jbc.M109.077834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loesberg C., van Rooij H. and Smets L.A. (1990) Meta-iodobenzylguanidine (MIBG), a novel high-affinity substrate for cholera toxin that interferes with cellular mono(ADP-ribosylation). Biochim. Biophys. Acta 1037, 92–99 10.1016/0167-4838(90)90106-P [DOI] [PubMed] [Google Scholar]
- 41.Guo X., Carroll J.W., Macdonald M.R., Goff S.P. and Gao G. (2004) The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 78, 12781–12787 10.1128/JVI.78.23.12781-12787.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X., Ma J., Sun J. and Gao G. (2007) The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl Acad. Sci. U.S.A. 104, 151–156 10.1073/pnas.0607063104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todorova T., Bock F.J. and Chang P. (2014) PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat. Commun. 5, 5362 10.1038/ncomms6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welsby I., Hutin D., Gueydan C., Kruys V., Rongvaux A. and Leo O. (2014) PARP12, an interferon-stimulated gene involved in the control of protein translation and inflammation. J. Biol. Chem. 289, 26642–26657 10.1074/jbc.M114.589515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahlberg E., Karlberg T., Kouznetsova E., Markova N., Macchiarulo A., Thorsell A.G. et al. (2012) Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 30, 283–288 10.1038/nbt.2121 [DOI] [PubMed] [Google Scholar]
- 46.Ekblad T., Lindgren A.E., Andersson C.D., Caraballo R., Thorsell A.G., Karlberg T. et al. (2015) Towards small molecule inhibitors of mono-ADP-ribosyltransferases. Eur. J. Med. Chem. 95, 546–551 10.1016/j.ejmech.2015.03.067 [DOI] [PubMed] [Google Scholar]
- 47.Larsen S.C., Hendriks I.A., Lyon D., Jensen L.J. and Nielsen M.L. (2018) Systems-wide analysis of serine ADP-ribosylation reveals widespread occurrence and site-specific overlap with phosphorylation. Cell Rep. 24, 2493–2505.e2494 10.1016/j.celrep.2018.07.083 [DOI] [PubMed] [Google Scholar]
- 48.Gibson B.A., Zhang Y., Jiang H., Hussey K.M., Shrimp J.H., Lin H. et al. (2016) Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353, 45–50 10.1126/science.aaf7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feijs K.L., Kleine H., Braczynski A., Forst A.H., Herzog N., Verheugd P. et al. (2013) ARTD10 substrate identification on protein microarrays: regulation of GSK3beta by mono-ADP-ribosylation. Cell Commun. Signal. 11, 5 10.1186/1478-811X-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutin D., Tamblyn L., Gomez A., Grimaldi G., Soedling H., Cho T. et al. (2018) Hepatocyte-specific deletion of TIPARP, a negative regulator of the aryl hydrocarbon receptor, is sufficient to increase sensitivity to dioxin-induced wasting syndrome. Toxicol. Sci. 165, 347–360 10.1093/toxsci/kfy136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bütepage M., Eckei L., Verheugd P. and Lüscher B. (2015) Intracellular mono-ADP-ribosylation in signaling and disease. Cells 4, 569–595 10.3390/cells4040569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.